Abstract
BACKGROUND:
Recurrent pregnancy loss is a common occurrence and a matter of concern for couples planning the pregnancy. Chromosomal abnormalities, mainly balanced rearrangements, are common in couples with repeated miscarriages.
PURPOSE:
The purpose of this study is to evaluate the contribution of chromosomal anomalies causing repeated spontaneous miscarriages and provide detailed characterization of a few structurally altered chromosomes.
MATERIALS AND METHODS:
A retrospective cytogenetic study was carried out on 4859 individuals having a history of recurrent miscarriages. The cases were analyzed using G-banding and fluorescence in situ hybridization wherever necessary.
RESULTS:
Chromosomal rearrangements were found in 170 individuals (3.5%). Translocations were seen in 72 (42.35%) cases. Of these, reciprocal translocations constituted 42 (24.70%) cases while Robertsonian translocations were detected in 30 (17.64%) cases. 7 (4.11%) cases were mosaic, 8 (4.70%) had small supernumerary marker chromosomes and 1 (0.6%) had an interstitial microdeletion. Nearly, 78 (1.61%) cases with heteromorphic variants were seen of which inversion of Y chromosome (57.70%) and chromosome 9 pericentromeric variants (32.05%) were predominantly involved.
CONCLUSIONS:
Chromosomal analysis is an important etiological investigation in couples with repeated miscarriages. Characterization of variants/marker chromosome enable calculation of a more precise recurrent risk in a subsequent pregnancy thereby facilitating genetic counseling and deciding further reproductive options.
Keywords: Break points, chromosomal abnormalities, recurrent loss of pregnancy, small supernumerary marker chromosome, translocations
Introduction
Recurrent pregnancy loss is classically defined as the occurrence of three or more consecutive abortions; however, the American Society of Reproductive Medicine has recently redefined recurrent pregnancy loss as two or more abortions.[1] Pregnancy loss is a clinically recognized pregnancy, involuntarily ending before 20 weeks of gestation.[1] About 10 and 15% of the clinically recognizable pregnancies result in spontaneous miscarriages, with an additional preclinical loss of 22%.[2,3] Nearly, 50% of the cases, the etiology remains unknown, although numerous different factors have been suggested to be involved including parental genetic makeup, uterine abnormalities, hormonal imbalance, hematological and immunological disorders as well as a variety of other phenomena such as infections, environmental exposures and more.[4]
The frequency of chromosomal abnormalities among couples with recurrent miscarriage varies from 2% to 8%.[3,5,6,7,8,9,10] The most commonly observed rearrangements are either reciprocal or Robertsonian translocations,[9] which lead to an unequal dispersion of chromosome content during meiosis that predispose to genetically unbalanced gametes thus resulting in a non-viable fetus. Studies of couples who are balanced translocation carriers indicate that as many as 80% of the pregnancies may end-up in spontaneous miscarriage. Furthermore, two-thirds of balanced autosomal translocation carriers are observed in couples experiencing two or more pregnancy losses. This is 30 times more than the rate being detected in the general population as shown by Fryns and Van Buggenhout.[9] Therefore, parental chromosomal assessment is important to understand the etiology for recurrent miscarriage.[3]
In this study, we aim to identify the types of microscopically visible structural abnormalities and their frequencies in the parents with recurrent miscarriages and compare the findings with that present in the literature. Moreover, comprehensive characterization of variants and marker chromosome was carried out, which could help in providing precise recurrent risk for the subsequent pregnancy.
Materials and Methods
A 19 years retrospective study was carried out in couples with a clinical diagnosis of recurrent miscarriages from the period of January 1994 to December 2012. Institutional ethics approval as well as patient informed consent was obtained. A total of 4859 individuals (2428 couples and three single female parents) were investigated for chromosomal abnormalities. In all the cases, detailed reproductive case histories were taken and karyotypes were generated. Metaphase chromosome preparations from the peripheral blood cultures were made according to the standard cytogenetic protocols. Cytogenetic analysis was performed by GTG-banding at approximately 550-band level. Molecular cytogenetic techniques such as fluorescence in situ hybridization (FISH) and/or array-comparative genomic hybridization (CGH) were performed to comprehensively characterize small supernumerary marker chromosomes (sSMCs). FISH was performed using commercially available whole chromosome paint (WCP) probes for chromosomes 13, 14, 15, 20 and 22. Microdeletion was confirmed using probe set obtained from Kreatech. Further characterization of structurally altered chromosomes was carried out using a probe for Yq12, centromeric probes for chromosome 13/21, 14/22 and 15, as well as a homemade probes set called subcentromeric multicolor FISH (subcenM-FISH).[11] Besides bacterial artificial chromosome-probes RP11-446P9 in 15q12, RP11-408F10 in 15q13.1 as well as a WCP15 probes were applied in one case. A total of 30 metaphases were analyzed for all the patients, but in cases of abnormalities and mosaicism, the number was extended to 100 metaphases. Chromosomal abnormalities were reported according to the current International System for Human Cytogenetic Nomenclature.[12] Along with the structural rearrangements and aneuploidies, common chromosomal variants were also studied.
Results
This study comprises of 4859 parents with the history of repeated miscarriages. Constitutional chromosomal abnormalities were detected in 170 (3.5%) whereas variants were observed in 78 (1.6%) individuals. Among the 170 individuals with abnormal karyotype, 121 (71.2%) were females while 49 (28.8%) were males, producing female to male ratio of 2.1:1 as shown in Table 1. Ten patients out of total studied cases had one or two deliveries of congenitally abnormal child or fetal/child death preceding the episodes of repeated miscarriages at the time of this study.
Table 1.
Summary of cytogenetically detected chromosomal rearrangements
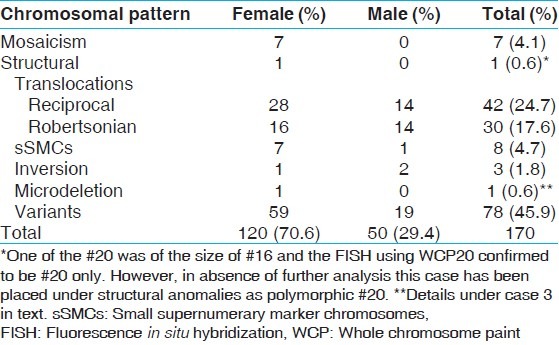
The mean age and range of parents with recurrent miscarriages were 30.6 years and 19-46 years, respectively. The mean age for females was 28.7 years and 32.9 years for males. Cytogenetically detected chromosomal rearrangements are shown in Table 1. Of the 170 abnormal cases, the predominant rearrangements detected were translocations (n = 72; 42.35%) with reciprocal (n = 42; 24.70%) translocations being the majority. This is followed by sSMCs (n = 8; 4.70%), mosaicism (n = 7; 4.11%), inversions (n = 3; 1.76%) and one case each (0.58%) of interstitial microdeletion and polymorphism of chromosome 20 centromeric region. Tables 2 and 3 show the detected chromosomal breakpoints involved in the reciprocal and Robertsonian translocations, respectively. Reciprocal translocations involving three chromosomes were also observed in two cases. Female to male ratio was 2:1 for being a reciprocal translocation carrier when compared with 1.6:1 in Robertsonian translocation carriers. In cases of mosaicism, involvement of only X chromosome was observed. Out of eight cases with sSMC, four were comprehensively characterized using FISH and/or array-CGH of which details of three sSMCs are described elsewhere.[13] The remaining four patients declined further analysis [Table 4]. Chromosome 13 and 14 were the most frequently observed acrocentric chromosomes, whereas chromosomes 3, 4, 8 and 11 predominated among the autosomes with non-involvement of X chromosome amongst reciprocal translocations, as shown in Table 4 and Figure 1.
Table 2.
Cases with reciprocal translocation
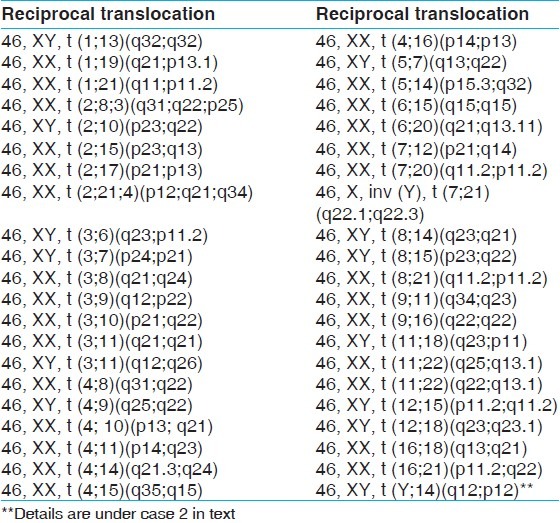
Table 3.
Cases with robertsonian translocation
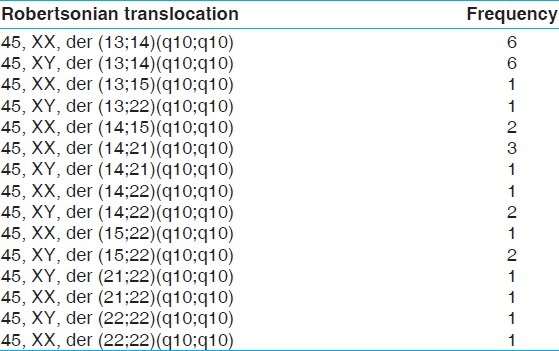
Table 4.
Summary of cases having mosaicism and marker chromosomes
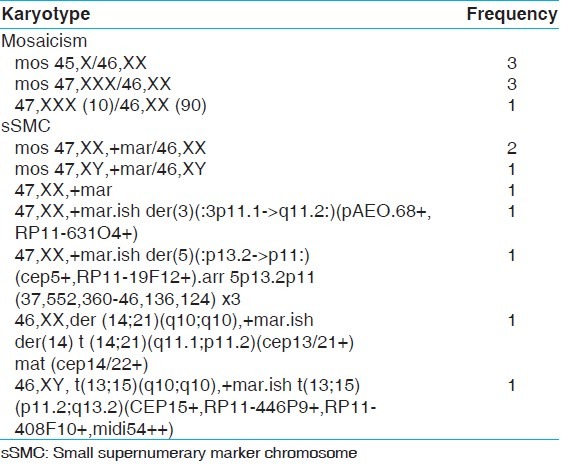
Figure 1.
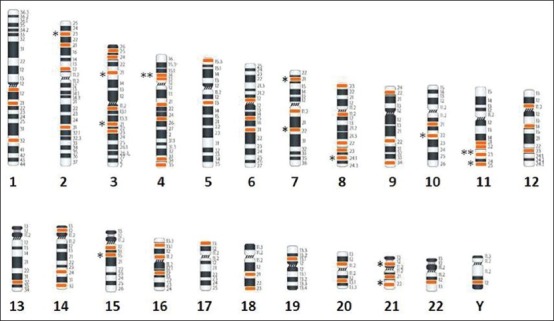
Breakpoint distribution is based on cytogenetic data shown in Table 3. Chromosomes are arranged according to the number except X chromosome as no break point was detected. Below each chromosome the black color numerical indicates the chromosome number and the break points are colored according to the number of break observed in the study. Each star represent on the left of chromosome indicates if a breakpoint was observed in more than one case
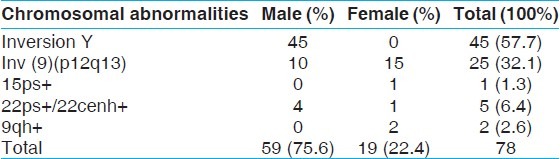
Among variants, inversion Y was the most commonly seen polymorphism (n = 45; 57.70%) while chromosome 9 heterochromatin inversion was observed in 32% (n = 25) of the cases. Remaining cases showed variations in the satellite regions of acrocentric chromosomes and in the pericentric heterochromatic region of chromosome 9 [Table 5].
Table 5.
Cases with other forms of chromosomal alterations (variants)
Three selected cases of this study, in which further molecular cytogenetic characterization was carried out, are described below.
Case 1
An elderly couple was referred for chromosomal analysis to rule out inheritance of Robertsonian translocation [t(13;15)] detected in one of the son who expired at the age of 15 years due to uncontrolled weight gain. In addition, the couple had three first trimester fetal losses and one child expired at the age of 9 months due to uncontrolled seizures. Mother was chromosomally normal, whereas Robertsonian translocation was detected in the father along with a small marker chromosome presented in all cells (46,XY,der (13;15)(q10;q10), +mar]. Further characterization of sSMC was carried out using subcenM-FISH for chromosome 13 and probes for chromosome 15 as mentioned above. FISH confirmed the sSMC to be a “by-product” of a Robertsonian translocation involving chromosome 13 and 15. Breakpoints were located in 13p11.2 and 15q13.2 region and the karyotype redefined as 46,XY,t(13;15)(p11.2;q13.2). Although the second child was not investigated for chromosomal analysis, the uncontrolled seizure could be due to the proximal imbalance on #15 [15pter→15q13.2] as seen in 38% of the cases with Angelman syndrome.[14] Hence, rearranged chromosome t(13;15) and sSMC when found together in the fetus, no phenotypic features are likely to be seen as observed in the father. This further confirms the utility of various FISH probes in the comprehensive characterization of sSMCs.
Case 2
A young couple was studied to rule out chromosomal anomalies as one child expired soon after birth due to gastroesophageal reflux. Although the female was chromosomally normal, the karyotype of the male showed additional genetic material on one of the acrocentric short arms, i.e., a karyotype 46,XY,der(14)add(14)(p11.2). Various FISH probes such as subcenM-FISH Mix for chromosome 14 and cep 15 with a probe for Yq12 were used to confirm the derivative chromosome 14 as a reciprocal translocation involving #14p and #Yq (46,XY,t(Y;14)(q12;p12)]. It is a known heteromorphism without any clinical significance. This possibly point toward a different underlying pathology resulting in neonatal death rather than the involvement of altered chromosomes.
Case 3
A young couple had a history of two stillbirths at 25 weeks of gestation due to cardiac anomalies. Both parent karyotypes were apparently normal at 550-band resolution. After analyzing the family history, a FISH study using the probe for DiGeorge syndrome critical region was carried out and was found to be positive in the mother. This couple carries 50% risk of having same anomalies during the subsequent pregnancy.
In 10 cases, parents had a child born with free trisomy 21 in addition to one structural rearrangement of chromosome other than 21. In these cases, one parent was carrying a structural rearrangement, which the child partly inherited. Thus, these children were bearing two types of chromosomal aberrations: One was free trisomy 21 and the other type was inherited from either parent carrying that particular aberration.
Discussion
The prevalence of 3.5% of chromosomal abnormality detected in the present study is in the lower limit of the range of 2-8% reported in most studies [Figure 2]. Such variability among different studies is likely to be related to the number of subjects analyzed and the different composition of the populations examined as well as referral bias. The low frequency obtained in this study may be due to the large number of subjects involved with recurrent pregnancy wastages. It could also be an indication that more subtle or submicroscopic molecular causes that can’t be detected by cytogenetic analysis could be responsible for such losses among the subjects. However, the finding of this study is in agreement with the study of Dubey et al.[5] on 742 couples, which showed 2% risk of chromosomal abnormalities in subjects with recurrent spontaneous abortions, of which structural abnormalities formed the largest group. Approximately, twice the number of females was seen with chromosomal rearrangements compared with males. Translocations constituted about 70.5% of all the chromosomal abnormalities, which makes it the most common chromosomal abnormality. The study showed that females and males were involved in translocation with frequencies of 44 (61.11%) and 28 (38.89%) cases respectively. In the study of Al-Hussain et al.,[15] a frequency of 86% was obtained with female to male ratio of 1.5:1. The study of Shafeghati et al.[16] is also in agreement with our findings. Reciprocal translocations were seen in twice the number of females compared with males, whereas no such sex predilection for Robertsonian translocation was observed. This sex predilection may be due to the fact that chromosomal abnormalities that are compatible with fertility in females may be associated with sterility in males. Supporting this hypothesis is the study, which showed a high frequency of structural chromosomal abnormalities among sperm cells with poor motility.[17] It is well-established that carriers of balanced translocations are subjected to an increased risk of abnormal chromosomal segregation during meiosis. The consequences include infertility, multiple pregnancy losses and live born of abnormal offspring with multiple congenital malformations. The recurrent risk in the subsequent pregnancy depends upon the chromosomes involved, exact breakpoint and sex of the carrier. This study did not show a definite pattern of translocation with regards to the type of the involved chromosome, however, chromosome 3, 4, 8 and 11 were most commonly involved in reciprocal translocations while chromosome 13 and 14 ranked first amongst the chromosomes involved in Robertsonian translocations [Figure 1 and Table 4]. Two cases of reciprocal translocations in which three chromosomes were involved in a particular translocation event was observed in our present study. Similar translocations were also observed before in the study of Manvelyan et al.[18] We were unable to find any correlation between specific break point and increased risk of miscarriages in the present study.
Figure 2.
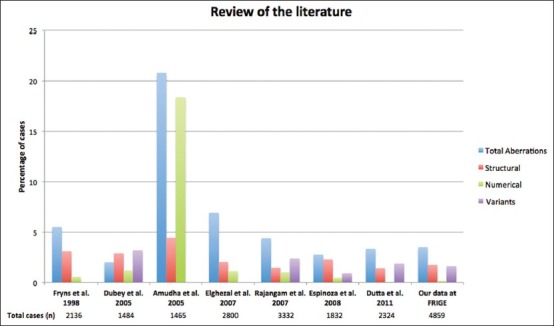
Various chromosomal aberrations (structural, numerical and variants) observed in the present study and its comparison with the literature
Inversions were noted in three of our cases involving chromosome 2, 3 and 8 among, which males predominated. Two were paracentric (46,XY,inv (3)(q21q25) and 46,XY,inv(2)(q13q20)] and a pericentric inversion was observed in one case (46,XX,inv (8)(p21q21.2)]. Though pericentric inversions have been detected in all chromosomes with varying frequencies, chromosomes 2, 5, 7, 9 and 10 are statistically more prone to rearrangements. As many as 1:100 people exhibit such inversions in the heterochromatic region of chromosome 9, which is considered to be a population variant.[19] Similarly, but less frequently, inversions are also found in chromosome 1 and Y.[19,20,21] This study showed that inversion Y chromosome is the most common variant. It was seen in 1.87% of all male partners in this study and its relative high prevalence suggested that inversion was the most common chromosomal alteration associated with repeated pregnancy loss.[22,23,24] The incidence observed was as high as 1 in 4 males having primary infertility or repeated miscarriages.[25] According to the report of Sheth et al.,[26] the incidence of the trait in Gujarati subjects with inversion Y was 2.2% while in China, Zhou et al.[27] showed that inversion Y is seen in 22% males with recurrent miscarriages. In general, inversion of chromosome 9 has been considered to be a normal polymorphism, as it mainly consists of centromeric heterochromatin with no known coding regions. However, the association of chromosome 9 inversion with subfertility has been documented and it has been hypothesized that during an inversion event, there might be loss or suppression of the euchromatin chromosome region due to the position variegation effect, leading to abnormalities in children.[28,29] In the study of subfertility by García-Peiró et al.,[30] it was discovered that there were significantly high meiotic alterations and aneuploidy rates, high-sperm deoxyribonucleic acid fragmentation and altered seminogram parameters among males with inversion Y chromosome.
In the present study, the numerical chromosome aberrations were observed in seven cases of sex chromosome mosaicism altogether producing a frequency of 4.11%.
Mosaicism was rare and it has been postulated by some studies that low level sex chromosome mosaicism is more common among phenotypically normal individuals being karyotyped for a history of recurrent abortion and infertility.[31,32] If sex chromosome mosaicism is shown to be the predictor of early menopause, then its association with increased miscarriage could be explained.[33] Characterization of sSMC in case 1 revealed it to be a product of Robertsonian translocation of chromosome 13 and 15 each. Thus, the pregnancy loss and the early death of two children could be explained by the loss of the proximal part of chromosome 13 and 15 as one of the children had inherited the translocated chromosome and not the marker. Absence of this would have led to uncontrolled weight gain and early death. In case 2, it was chromosome Y that was presenting with an additional material on #14. This shows different underlying etiology for the fetal death. These cases represent the utility of comprehensive characterization of structurally altered chromosome and detailed family history in concluding the cause of fetal death, as seen in case 3.
Carriers of balanced chromosome rearrangements, although phenotypically normal, experience recurrent adverse pregnancy outcomes as a consequence of formation of unbalanced gametes. In most cases, this chromosomal imbalance in zygotes is incompatible with viability expressed in the form of arrested embryos, failed implantation or spontaneous abortion. By selecting chromosomally balanced oocytes or embryos, in vitro fertilization success rates would be expected to improve for carriers of structurally altered chromosomes. Further characterization of such chromosomes would benefit both couples as well as genetic counselors in deciding the reproductive options and estimating the recurrent risk in subsequent pregnancies, respectively.
We conclude that chromosomal disorders contribute to the underlying basis of reproductive failure in a varying proportion of cases. The identification of chromosomal abnormality as the etiology of recurrent miscarriage will facilitate counseling and appropriate patient management. Cytogenetic analysis of both males and females with decreased reproductive fitness is essential for predicting the success of assisted reproductive procedures. From the data, it can be hypothesized that inversion Y and 9 might be associated with high frequency and occurrence of repeated pregnancy wastages; their significance in the present study needs to be proved by further studies on patients with recurrent miscarriages. Furthermore, comprehensive characterization of variant/marker chromosome could enable to calculate precise recurrent risk in the subsequent pregnancy.
Acknowledgments
The authors would like to express their thanks to Manisha Desai and Bhumika Patel for processing of samples and Dr. Stuti Tewari and Mr. Victor Olawale for part entry of the data, referring clinicians for their support and cooperation and patients for being a part of this study. Authors are grateful to FRIGE for partly supporting this study.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Practice Committee of the American Society for Reproductive Medicine. Evaluation and treatment of recurrent pregnancy loss: A committee opinion. Fertil Steril. 2012;98:1103–11. doi: 10.1016/j.fertnstert.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 2.Rull K, Nagirnaja L, Laan M. Genetics of recurrent miscarriage: Challenges, current knowledge, future directions. Front Genet. 2012;3:34. doi: 10.3389/fgene.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutta UR, Rajitha P, Pidugu VK, Dalal AB. Cytogenetic abnormalities in 1162 couples with recurrent miscarriages in southern region of India: Report and review. J Assist Reprod Genet. 2011;28:145–9. doi: 10.1007/s10815-010-9492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jauniaux E, Farquharson RG, Christiansen OB, Exalto N. Evidence-based guidelines for the investigation and medical treatment of recurrent miscarriage. Hum Reprod. 2006;21:2216–22. doi: 10.1093/humrep/del150. [DOI] [PubMed] [Google Scholar]
- 5.Dubey S, Chowdary MR, Prahlad B, Kumar V, Mathur R, Hamilton S, et al. Cytogenetic causes for recurrent spontaneous abortions-An experience of 742 couples (1484 cases) Indian J Hum Genet. 2005;11:94–8. [Google Scholar]
- 6.Rajangam S, Tilak P, Aruna N, Devi R. Karyotyping and counseling in bad obstetric history and infertility. Iran J Reprod Med. 2007;5:7–12. [Google Scholar]
- 7.Amudha S, Aruna N, Rajangam S. Consanguinity and chromosomal abnormality. Indian J Hum Genet. 2005;11:108–10. [Google Scholar]
- 8.Elghezal H, Hidar S, Mougou S, Khairi H, Saâd A. Prevalence of chromosomal abnormalities in couples with recurrent miscarriage. Fertil Steril. 2007;88:721–3. doi: 10.1016/j.fertnstert.2006.11.160. [DOI] [PubMed] [Google Scholar]
- 9.Fryns JP, Van Buggenhout G. Structural chromosome rearrangements in couples with recurrent fetal wastage. Eur J Obstet Gynecol Reprod Biol. 1998;81:171–6. doi: 10.1016/s0301-2115(98)00185-7. [DOI] [PubMed] [Google Scholar]
- 10.Meza-Espinoza JP, Anguiano LO, Rivera H. Chromosomal abnormalities in couples with reproductive disorders. Gynecol Obstet Invest. 2008;66:237–40. doi: 10.1159/000147170. [DOI] [PubMed] [Google Scholar]
- 11.Liehr T, Mrasek K, Weise A, Dufke A, Rodríguez L, Martínez Guardia N, et al. Small supernumerary marker chromosomes - Progress towards a genotype-phenotype correlation. Cytogenet Genome Res. 2006;112:23–34. doi: 10.1159/000087510. [DOI] [PubMed] [Google Scholar]
- 12.Karger S, Basel JC, Shaffer LG, McGowan-Jordan J, Schmid M, editors. An International System for Human Cytogenetic Nomenclature. 2013. [Google Scholar]
- 13.Sheth F, Andrieux J, Ewers E, Kosyakova N, Weise A, Sheth H, et al. Characterization of sSMC by FISH and molecular techniques. Eur J Med Genet. 2011;54:247–55. doi: 10.1016/j.ejmg.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Mignon-Ravix C, Depetris D, Luciani JJ, Cuoco C, Krajewska-Walasek M, Missirian C, et al. Recurrent rearrangements in the proximal 15q11-q14 region: A new breakpoint cluster specific to unbalanced translocations. Eur J Hum Genet. 2007;15:432–40. doi: 10.1038/sj.ejhg.5201775. [DOI] [PubMed] [Google Scholar]
- 15.Al-Hussain M, Al-Nuaim L, Abu Talib Z, Zaki OK. Cytogenetic study in cases with recurrent abortion in Saudi Arabia. Ann Saudi Med. 2000;20:233–6. doi: 10.5144/0256-4947.2000.233. [DOI] [PubMed] [Google Scholar]
- 16.Shafeghati Y, Nejad MK, Zangeneh M, Nejad RK, Mohammadi QB, Almadani N, et al. Abstract in European Human Genetics Conference. Strasbourg, France: European Society of Human Genetics; 2002. Results of chromosome studies in 664 Iranian couples with the history of recurrent early pregnancy loss; p. 5. [Google Scholar]
- 17.Rybouchkin A, Benijts J, De Sutter P, Dhont M. Disintegration of chromosomes in dead sperm cells as revealed by injection into mouse oocytes. Hum Reprod. 1997;12:1693–8. doi: 10.1093/humrep/12.8.1693. [DOI] [PubMed] [Google Scholar]
- 18.Manvelyan M, Schreyer I, Höls-Herpertz I, Köhler S, Niemann R, Hehr U, et al. Forty-eight new cases with infertility due to balanced chromosomal rearrangements: Detailed molecular cytogenetic analysis of the 90 involved breakpoints. Int J Mol Med. 2007;19:855–64. doi: 10.3892/ijmm.19.6.855. [DOI] [PubMed] [Google Scholar]
- 19.Kumar M, Thatai A, Chapadgaonkar SS. Homozygosity and heterozygosity of the pericentric inversion of chromosome 9 and its clinical impact. J Clin Diagn Res. 2012;6:816–20. [Google Scholar]
- 20.De Braekeleer M, Dao TN. Cytogenetic studies in couples experiencing repeated pregnancy losses. Hum Reprod. 1990;5:519–28. doi: 10.1093/oxfordjournals.humrep.a137135. [DOI] [PubMed] [Google Scholar]
- 21.Wolf GC, Mao J, Izquierdo L, Joffe G. Paternal pericentric inversion of chromosome 4 as a cause of recurrent pregnancy loss. J Med Genet. 1994;31:153–5. doi: 10.1136/jmg.31.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishna DS, AL-Awadi SA, Farag TI. Pericentric inversion and recombinant aneusomy and other associated chromosomal aberrations: Random or non-random. Am J Hum Genet. 1992;51:A291. [Google Scholar]
- 23.Madon PF, Athalye AS, Parikh FR. Polymorphic variants on chromosomes probably play a significant role in infertility. Reprod Biomed Online. 2005;11:726–32. doi: 10.1016/s1472-6483(10)61691-4. [DOI] [PubMed] [Google Scholar]
- 24.Motos Guirao MA. Pericentric inversion of the human Y chromosome. An Esp Pediatr. 1989;31:583–7. [PubMed] [Google Scholar]
- 25.Verma RS, Rodriguez J, Dosik H. The clinical significance of pericentric inversion of the human Y chromosome: A rare “third” type of heteromorphism. J Hered. 1982;73:236–8. doi: 10.1093/oxfordjournals.jhered.a109627. [DOI] [PubMed] [Google Scholar]
- 26.Sheth FJ, Shah UJ, Desai M, Sheth JJ. Clinical profile of inversion Y in people of Gujarat, West India. Int J Hum Genet. 2011;11:245–8. [Google Scholar]
- 27.Zhou B, Tang YP, Liu YZ. Mechanism of the formation of a special inv (Y): A case study. Yi Chuan. 2006;28:148–52. [PubMed] [Google Scholar]
- 28.Thomas IM. Cytogenetic basis of recurrent abortions. Perinatology. 1999;1:181–7. [Google Scholar]
- 29.Dávalos IP, Rivas F, Ramos AL, Galaviz C, Sandoval L, Rivera H. inv (9)(p24q13) in three sterile brothers. Ann Genet. 2000;43:51–4. doi: 10.1016/s0003-3995(00)00013-7. [DOI] [PubMed] [Google Scholar]
- 30.García-Peiró A, Oliver-Bonet M, Navarro J, Abad C, Guitart M, Amengual MJ, et al. Sperm DNA integrity and meiotic behavior assessment in an infertile male carrier of a 9qh+++ polymorphism. J Biomed Biotechnol. 2011;2011:730847. doi: 10.1155/2011/730847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nazmy NA. Cytogenetic studies of couples with reproductive failure in alexandria, egypt. J Egypt Public Health Assoc. 2008;83:255–71. [PubMed] [Google Scholar]
- 32.Mau UA, Bäckert IT, Kaiser P, Kiesel L. Chromosomal findings in 150 couples referred for genetic counselling prior to intracytoplasmic sperm injection. Hum Reprod. 1997;12:930–7. doi: 10.1093/humrep/12.5.930. [DOI] [PubMed] [Google Scholar]
- 33.Kline J, Kinney A, Levin B, Warburton D. Trisomic pregnancy and earlier age at menopause. Am J Hum Genet. 2000;67:395–404. doi: 10.1086/303009. [DOI] [PMC free article] [PubMed] [Google Scholar]


