Abstract
The path from gene (DNA) to gene product (RNA or protein) is the foundation of genotype giving rise to phenotype. Comparison of genomic analyses (DNA) with paired transcriptomic studies (mRNA) is critical to evaluating the pathogenic processes that give rise to human disease. The ability to analyze both DNA and mRNA from the same sample is not only important for biologic interrogation but also to minimize variance (e.g. sample loss) unrelated to the biology. Existing methods for RNA and DNA purification from a single sample are typically time consuming and labor intensive or require large sample sizes to split for separate RNA and DNA extraction procedures. Thus, there is a need for more efficient and cost effective methods to purify both RNA and DNA from a single sample. To address this need, we have developed a technique, termed SNARE (Selective Nucleic Acid Removal via Exclusion), that uses pinned oil interfaces to simultaneous purify mRNA and DNA from a single sample. A unique advantage of SNARE is the elimination of dilutive wash and centrifugation processes that are fundamental to conventional methods where sample is typically discarded. This minimizes loss and maximizes recovery by allowing non-dilutive re-interrogation of the sample. We demonstrate that SNARE is more sensitive than commercially available kits; robustly and repeatably achieving mRNA and DNA purification from extremely low numbers of cells for downstream analyses. In addition to sensitivity, SNARE is fast, easy to use, cost-effective and requires no laboratory infrastructure or hazardous chemicals. We demonstrate the clinical utility of the SNARE with prostate cancer circulating tumor cells to demonstrate its ability to perform both genomic and transcriptomic interrogation on rare cell populations that would be difficult to achieve with any current method.
INTRODUCTION
Biological complexity emerges from different organizational levels in highly regulated and coordinated processes, involving the path from gene (DNA) to gene product (RNA). Full understanding of these links is beginning to unlock the secrets of cell differentiation, development, aging and pathological conditions. But for a more complete picture, techniques that allow for integrated RNA and DNA extraction within the same biological sample will be essential1. For example, recent studies using paired genomic and transcriptomic analysis in cancer have begun to identify driver genes that could possibly serve as potential therapeutic intervention candidates or be involved in the mechanisms of disease progression2. These methods are also increasingly important as many biological samples are difficult to obtain, valuable and of limited size, leading to the need to extract as much information as possible from a small amount of material3. In addition, simultaneous RNA and DNA extraction helps reduce potential errors and variation in data due to experimental differences and sample loss. While techniques exist for the extraction of RNA and DNA from the same sample, they are often not capable of rare cell analysis due to sample damage and loss during processing. To overcome these obstacles, we present a simple and rapid method for the extraction and purification of mRNA and DNA from a single sample.
Until recently, the traditional approach to analyzing RNA and DNA from the same sample was to split the sample. But even when sample size was not limited, researchers feared losing data or introducing error, especially when trying to correlate genomic changes with gene expression changes1. These traditional techniques for simultaneous nucleic acid (NA) purification include cesium chloride step-gradient ultracentrifugation4 or a phase separation guanidinium thiocyanate-phenol-chloroform procedure5. While several variations of the guanudinium-based technique now exist (Trizol, TriFAST or Tri-Reagent) they are time consuming, labor intensive, require use of hazardous reagents and require relatively large sample sizes (>1000 cells)6. Alternatives to the phase separation guanidinium-based methods include spin column technologies6,7, which are faster and avoid the use of toxic reagents. These are now commercially manufactured by Qiagen, GE Healthcare, Macherey-Nagel, Norgen Biotek and Serva3. Of these manufacturers, only Qiagen’s DNA/RNA Allprep Micro kit is recommended for small sample sizes. However, the Qiagen kit requires an increased number of processing steps, such as centrifugation and must use carrier RNA if fewer than 100 cells are used. Klein and colleagues also used olgio(dt)25 Dynabeads® to purify mRNA and collected all the wash buffers to precipitate out the DNA8. While the technique showed single cell sensitivity, it was laborious and took over 24 hours to complete. Additionally, several RNA and DNA microfluidic purification devices have been developed that promise to reduce laboratory time, human interaction, and reagent and equipment costs9. While a few microfluidic devices have been developed to purify RNA or DNA interchangeably10, to our knowledge none have been developed to purify RNA and DNA simultaneously from a single sample in a cost-effective and time-efficient way.
Another area of emerging interest for simultaneous mRNA and DNA purification is rare cells, however current methods for lower sample sizes are limited due to sample loss, poor reproducibility and incompatibility with whole genome and transcriptomic amplification methods11. These rare cells are important in a range of clinical and biological spheres, including the characterization of circulating tumor cells (CTCs) for disease prognosis and personalized treatment12,13; circulating fetal cells for prenatal diagnosis14; T-cells for immune monitoring15; and stem cells for analysis of biochemical and developmental processes16. High importance is placed on these rare cells as they can be captured from blood replacing painful and expensive biopsies and permitting more frequent testing. While numerous publications have described methods for rare cell capture, the main end point has been enumeration with little focus on molecular interrogation of these cells due to lack of tools. Such analysis could allow us to predict therapeutic benefit and select optimal treatment strategies on a per-patient basis.
Here we present a microfluidic device termed Selective Nucleic Acid Removal via Exclusion (SNARE), which has been designed to overcome limitations of current technology. SNARE builds on previous work that exploited the dominance of surface tension over gravity at the microscale to establish “virtual walls” between immiscible and aqueous phases. These virtual walls were used to separate the complex upstream from the downstream solution for purification of mRNA, DNA or cells17–19. Specifically in the previous work, we described the physical principles of using immiscible phase for mRNA extraction18. Additionally, we demonstrated purification of specific cell populations19,20 and DNA extraction for detection of botulism neurotoxin from complex food matrices17. To operate, any analyte bound to paramagnetic particles (PMPs) is translocated across the immiscible phases using a simple handheld magnet. One unique advantage conferred by this system is the ability to resample the original input material as it is never lost by aspiration, transfer, dilutive or centrifugation based processes, an important advantage when dealing with rare biological samples. This non-destructive sampling method allows repeated interrogation of the original input material by the sequential addition of paramagnetic particles (PMPs) of varying chemistries and different lysis/binding buffers to the input well to isolate mRNA and DNA from the same sample. Overall, SNARE requires less time, labor, resources and laboratory equipment than current methods with the potential for high throughput automation and robotic processing.
In this manuscript, we demonstrate that SNARE technology is able to extract and purify mRNA and DNA from a single sample. To benchmark SNARE, we utilize the only commercially available spin column technology recommended for DNA and RNA extraction from low cell numbers. We demonstrate the sensitivity of SNARE to perform low cell number (<10) extraction of both mRNA and DNA by qPCR. We further show that purified mRNA and DNA is suitable for Sanger sequencing from the same cell population. Finally, we use SNARE to isolate mRNA and DNA from CTCs, a rare cell population. The ease of use and sensitivity of SNARE make it a unique technique for purification of mRNA and DNA from a single, rare sample.
EXPERIMENTAL SECTION
SNARE Fabrication
SNARE was manufactured from 2 mm thick polystyrene (PS, Goodfellow, UK) using a CNC mill (PCNC770, Tormach, USA). The complete device was 19 × 27 mm, in order to increase ergonomic handling. The input and middle well consisted of two through holes, 3 mm in width and 5 mm in height. The mRNA and DNA output well has the same dimensions as the input well with a 1.5 mm depth. Each well was connected by a trapezoid with a height ranging from 2 mm down to 0.8 mm and was milled to a depth of 0.3 mm (See Figure 1). The back was mirrored based on the front piece. The front and back were solvent bonded using acetonitrile so that the input and middle well had an approximate volume of 40–60 µL and the output well 15–20 µL. Pressure sensitive adhesive film (MicroAmp, Applied Biosystems, USA) was then applied to the front and back of the device as walls to contain the fluids.
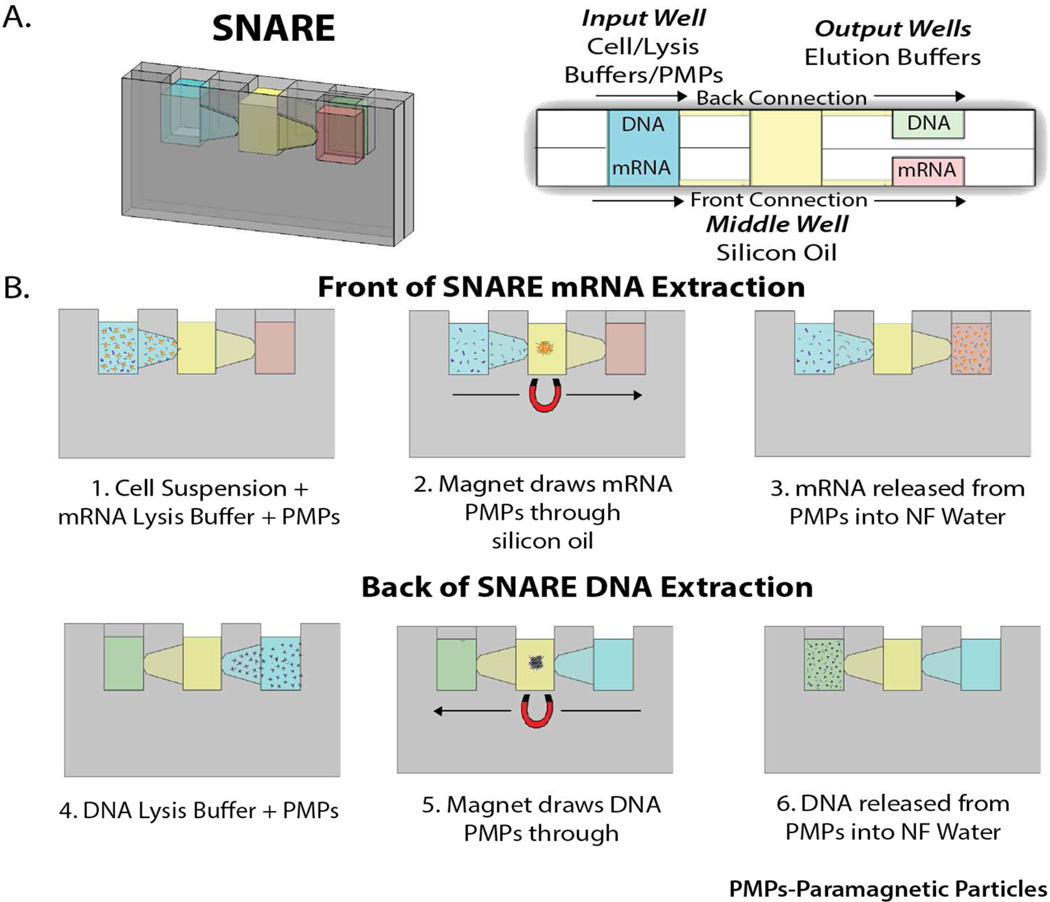
Figure 1.
A) (left) Picture of SNARE device with dimensions labeled and (right) top down schematic of SNARE device with wells labeled. Note the two fluid paths. One on the front of the device and one on the back. mRNA extraction occurs along the front and DNA extraction occurs along the back. B) Operation of SNARE for mRNA and DNA extraction and purification from a single sample. Steps 1–3 show front side of SNARE for mRNA isolation. Steps 4–6 show backside of SNARE for DNA isolation. (PMPs: Paramagnetic Particles)
Lysis/Binding Buffer Optimization
Three separate lysis/mRNA binding buffers were evaluated to determine which resulted in the highest relative GAPDH signal, signifying better nucleic acid capture. The SNARE protocol was performed as described below except different lysis/mRNA binding buffers were used including; 1× RIPA buffer (Milipore), LIDS (Life Technologies, USA) and a less stringent Modified LIDS buffer. While the Modified LIDS buffer is described in detail in the SNARE Operation section, the only difference from the commercially available LIDS buffer is the replacement of the ionic detergent lithium dodecyl sulfate (LDS) with a nonionic detergent Igepal® CA-630. GAPDH detection by qPCR was performed on both mRNA and DNA. Relative GAPDH signals were determined and a Student’s two-tailed t-test performed for comparison of each lysis/mRNA binding buffer with p<0.05 considered significant.
SNARE Operation
Operation of SNARE is outlined in Figure 1B. To operate SNARE, 15 µL of nuclease free water was added to both output wells. Next, 10 µL of cells suspended in 1×PBS was added to the input well, followed by 15 µL lysis and mRNA binding buffer, referred to as Modified LIDS, (10 mM Tris-HCL, 500 mM lithium chloride, 1 % Igepal® CA-630 (Sigma-Aldrich, USA), 5 mM ethylenediaminetetraacetic acid (EDTA), 5 mM dithiothreitol, pH 7.5) containing 30 Lg olgio(dt)25 Dynabeads® (Life Technologies, USA). To complete filling of the device, 40 µL silicon oil (Fisher, USA) was added to the middle well. After 5 minutes a permanent magnet (B333-N52 K&J Magnetics) was introduced to the front side of the input well to gather the olgio(dt) PMPs. Next, the magnet was manually pulled across the front until the olgio(dt)25 PMPs reached the RNA output well. Next, 25 µL of DNA binding buffer (10 mM Tris-HCL, 6 M GTC, 0.1 % Igepal® CA-630, pH 7.5) containing 1 µL MagneSil® PMPs (Promega, Madison) was added to the input well. After 5 minutes, the MagneSil® PMPs were transferred across the back side of the device to the DNA output well using the permanent magnet. The elution buffers along with PMPs were collected for further downstream analysis.
Quantitative PCR
The mRNA elution sample containing PMPs was reverse transcribed using a High Capacity cDNA Reverse Transcript kit (ABI, Foster City, CA) according to manufacturer’s directions. For GAPDH assays, 4 µL of cDNA template was mixed with 10 µL LightCycler 480® probes master mix (Roche, USA), 1 µL TaqMan® Gene Expression Assay (Specified in Table S1, Life Technologies, USA) and 5 µL nuclease free (NF) water. Each reaction was amplified for 50 cycles (denatured at 95 °C for 15 seconds followed by annealing at 60°C for 1 minute) using a LightCycler® 480 Real Time PCR System (Roche, USA). Relative GAPDH signal levels were quantified and normalized using, (2−(45−Cp)).
Cell Culture
Prostate cancer epithelial cells (LNCaPs) were cultured at 37 °C and maintained under 5 % CO2 in polystyrene flasks in Cornig Cellgro® RPMI 1640 Medium (VWR) containing 10 % fetal bovine serum (Gibco®), 1 % Penicillin Streptomycin (Gibco®), 1 % MEM-nonessential amino acids (Gibco®) and 1 % NaPyruvate (Cornig Cellgro®) until confluent. Cells were released using a 0.05% trypsin/EDTA solution and neutralized using media for collection via centrifugation.
SNARE Comparison to Qiagen AllPrep DNA/RNA Microextraction Kit
A 1:10 serial dilution of 100,000 to 100 LNCaP cells/mL in 1× PBS was performed for three seperate experiments. Ten µL of each serial dilution (n=2) was processed using SNARE to equal 1000, 100, 10 and 1 LNCaP per device. Ten µL of the same serial dilutions were added to 65 µL RLT buffer and processed according to Qiagen AllPrep DNA/RNA Micro Kit manufacturer’s directions. For all samples containing 100 cells or fewer, carrier RNA was added to the Qiagen samples as recommended in the manufacturer’s protocol. A control sample containing no cells was performed with each methodology. GAPDH detection by qPCR was performed for direct comparison of both methods.
Sequencing of the Androgen Receptor from mRNA and genomic DNA
For mRNA purified from 10 LNCaP cells using the SNARE procedure, exon 5 and 6 of the androgen receptor (primers shown in Supplementary Table 1) were amplified by qPCR according to directions above (primers shown in Supplementary Table 1). After amplification PCR products were purified using the MinElute PCR Purification Kit (Qiagen, USA). The product was cloned using the pGEM®-T Easy Vector System (Promega, Madison).
For DNA purified from 10 LNCaP cells using the SNARE procedure, exon 8 of the androgen receptor (primers shown in Supplementary Table 1) was amplified using Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Scientific, USA) according to manufacturer’s directions. The reaction was completed using Bio-Rad C1000 Thermo Cycler (Bio-Rad, USA) with initial denaturation at 98°C for 30 sec, denature at 98° C for 10 seconds, anneal and extend at 72° C for 20 seconds, which was repeated for 35 cycles with final extension at 72° C for 10 minutes. Samples were sent to the Wisconsin Biotechnology Center where a Big Dye (Life Technologies, USA) reaction was performed and PCR products directly sequenced (ABI 3730xl). Samples were analyzed using ABI Sequence Scanner Version 1 and nucleotide NCBI blast.
Patient Data
Prostate circulating tumor cells defined as EpCAM positive, intact nuclei based on Hoescht, and CD45 negative were collected under a University of Wisconsin IRB-approved protocol and isolated in a method previously described20. mRNA and DNA were extracted from the prostate CTCs using the SNARE method. Cycle threshold (Ct) values for AR and GAPDH were determined by qPCR according to directions above (See Supplementary Table 1 & Table 2 for primers & probes).
RESULTS and DISCUSSION
SNARE Operation
SNARE uses exclusion-based immiscible filtration assisted by surface tension (IFAST)18 to extract and purify mRNA and DNA from the same sample (Figure 1A). Immiscible phase filtration takes advantage of the ability of aqueous and oil phases to be loaded side-by-side, without stratification, to form virtual walls21. This principle is based on the dominance of surface tension over gravity at the microscale, as defined by the dimensionless Bond Number (Bo<1)18. To operate SNARE, PMPs functionalized with oligo(dt)25 and a lysis/binding buffer optimized to bind mRNA are added to the input well (Figure 1B, step 1). Following mRNA binding, an external magnet draws the mRNA –bound PMPs through the middle well containing silicon oil (Figure 1B, step 2) to the front output well (Figure 1B, step 3). Next, silica PMPs and a lysis/binding buffer optimized to bind DNA (Figure 1B, step 4) are added to the input well. Following DNA binding, the DNA- bound silica PMPs are moved through the middle well along the backside of the device (Figure 1B, step 5) to the back output well (Figure 1B, step 6). Samples can then be collected and used for downstream mRNA or DNA assays. It should be noted, in applications were only mRNA or DNA is required one could choose to collect either or.
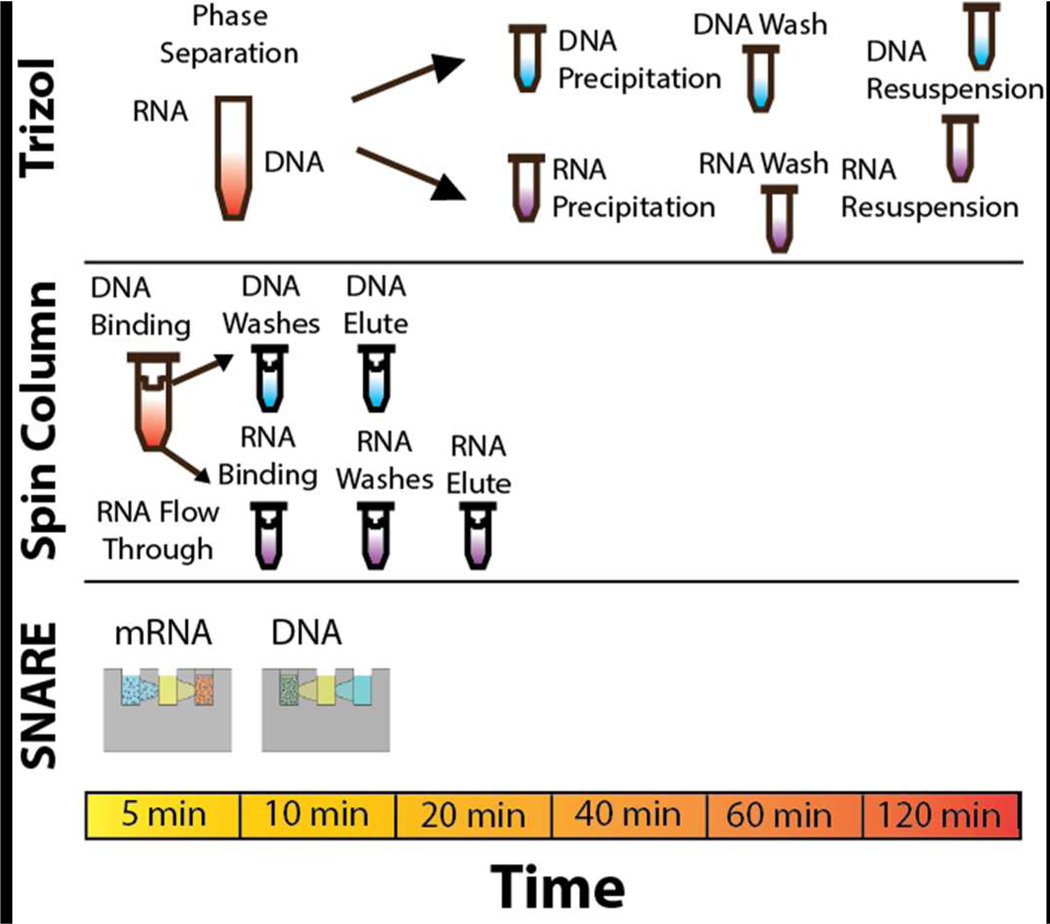
SNARE was designed to simplify purification of mRNA and DNA from a single sample by minimizing work flow and preparation time while maximizing sample recovery for downstream analyses. Figure 2 illustrates a comparison of SNARE to the current methods to isolate RNA and DNA from a single sample. The traditional method is guanidinium thiocyanate-phenol-chloroform commericially known as Trizol, which uses phase separation to extract RNA and DNA. However, Trizol requires several processing steps that are time consuming, laborious and require high reagent and material consumption. Trizol also uses toxic chemicals and is not recommended for small sample sizes. While spin columns are faster than Trizol they still require high reagent and material consumption. In addition, the multiple centrifugation and pipetting steps are still time consuming and can result in sample loss due to dilution and transfer steps. In contrast, the SNARE process takes only 10 minutes to complete reducing time and labor needed. As SNARE only requires the use of a pipette and a handheld magnet to operate, the cost of use is greatly reduced. However, if a large number of samples need to be processed, SNARE has the potential to be easily automated22. Lastly, SNARE’s simplistic design also helps to lower the cost as it could be manufactured using standard methods.
Figure 2.
RNA and DNA extraction methods from a single sample, using the traditional Trizol (guanidinium thiocyanate-phenol-chloroform) or Spin Column methods as compared to SNARE
Lysis/Binding Buffer Optimization
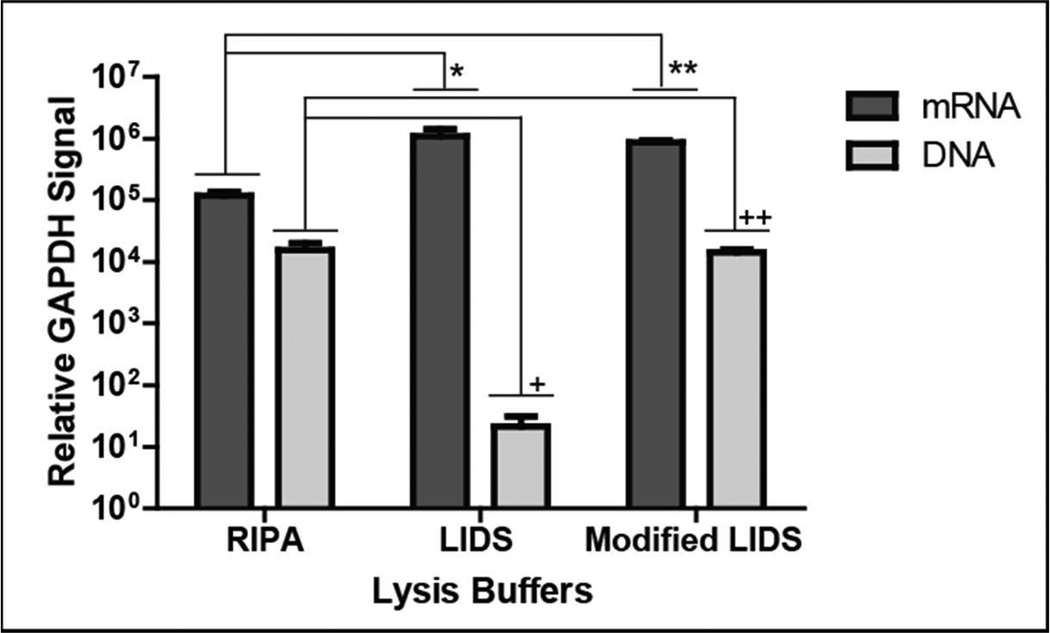
For successful NA isolation using SNARE, selection of PMPs with optimized binding chemistries and binding buffers is critical to affect cell lysis and facilitate NA-PMP binding interactions. To achieve maximum RNA and DNA extraction efficiency, three different lysis mRNA binding buffers (RIPA, LIDS, Modified LIDS) were evaluated using SNARE and the relative GAPDH signal was calculated for both mRNA and DNA from 1000 LNCaP cells. LNCaPs were chosen for this study and subsequent analysis as a representative model system for rare prostate cancer CTCs. Relative GAPDH signal was detected by qPCR because traditional methods to determine purity and amount (Agilent Bioanlayzer, absorbance at 260 nm & flourimeter) were not applicable for the limited amount of material isolated from low numbers of cells. GAPDH was also used as it is a commonly used reference gene and is expressed in LNCaPs. There was no difference in mRNA isolation as measured by relative GAPDH signal between LIDS or Modified LIDS (p>0.5) (Figure 3). However, use of either LIDS or Modified LIDS resulted in a higher relative mRNA GAPDH signal as compared to RIPA (p<0.03 and p<0.001, respectively). The relative increase in GAPDH mRNA signal could be due to the differences in the concentration of salts used in the RIPA (150 mM NaCl) as compared to LIDS and Modified LIDS (500 mM LiCl). Especially as binding is dependent on the poly(A)+ tail of mRNA forming stable hybrids with the functionalized oglio(dT) PMPs under high-salt conditions23. The ability to efficiently extract DNA from samples after using these lysis/binding buffers was also tested. We observed higher relative GAPDH DNA signal for RIPA (p<0.04) and Modified LIDS (p<0.001) compared to LIDS, meaning a greater sensitivity was observed. No statistical difference was seen between RIPA and Modified LIDS (p>0.8).
Figure 3.
Relative mRNA and DNA GAPDH signal isolated using SNARE for the comparison of different lysis/mRNA binding buffers. Based on this data, Modified LIDS was recommended for use in SNARE *p<0.03, ** p<0.001, + p<0.001, ++ p<0.04 Sample size per a group n=6.
Originally only RIPA and LIDS buffers were tested for mRNA extraction but upon addition of lysis/DNA binding buffer, physical examination revealed clumping between the DNA PMPs using LIDS. We hypothesized this difference was due to the ionic detergent lithium dodecyl sulfate (LDS) used in LIDS binding to the PMPs resulting in competitive binding with DNA. To circumvent this issue, the ionic LDS detergent was replaced with the non-ionic detergent Igepal CA-630 in Modified LIDS to achieve efficiency comparable to RIPA. Therefore, the Modified LIDS allowed us to maintain GAPDH signals that were not statistically different from LIDS without compromising DNA GAPDH signal. For DNA lysis/binding buffers, two different buffers containing either 6 M or 8 M guanidinium thiocyanate (GTC) were tested. While the DNA PMPs use silica for DNA binding they also have the ability to bind RNA under the right conditions (i.e. salt, pH), however the buffer was chosen to limit RNA binding and contamination. No differences were seen between RIPA buffers containing 6 M or 8 M GTC. However, the 8 M GTC buffer was poorly soluble, making operation difficult due to salt precipitation when the devices were kept on ice. Therefore, the Modified LIDS was selected with 6 M GTC for lysis/DNA binding buffer.
SNARE Comparison to Qiagen AllPrep DNA/RNA Micro Kit
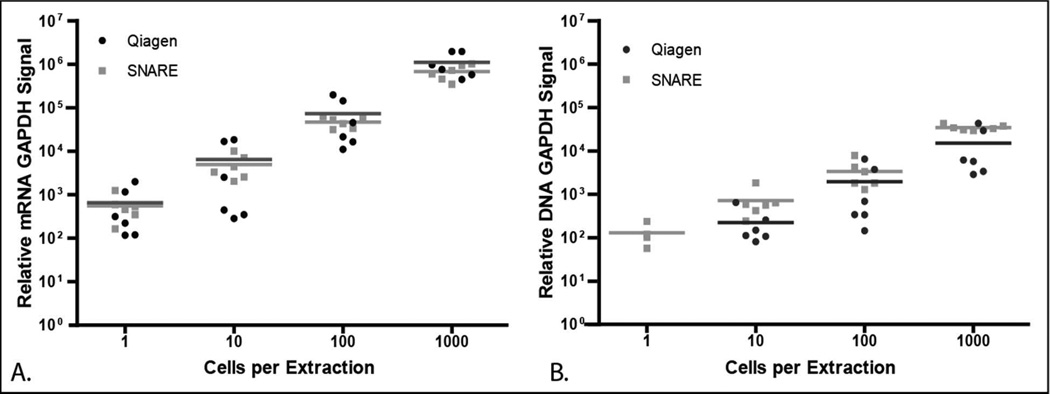
We used a Qiagen Allprep DNA/RNA Micro kit as a benchmark to SNARE as it is the most widely used sensitive commercially available technique. SNARE achieved higher relative mRNA and DNA signal compared to the Qiagen kit, which used carrier RNA since the kit does not purify using a polyadenylated mRNA tail (Figure 4). Using either SNARE or the Qiagen kit for mRNA extraction we were able to detect GAPDH in all of the samples, including sample dilutions containing a single cell (Figure 4A). To assess for possible NA contamination from the device or buffers, a control sample containing no cells was processed and no amplification was seen. Higher variability in mRNA isolation was observed for the Qiagen technique as cell number decreased, with the average coefficient of variance across all cell dilutions being 48.7±15.1 % for Qiagen and 28.9±7.0 % for SNARE. The differences in relative signal could be due to decreased yield through additional fluid transfer steps, fluid shear stresses and partial elution in wash buffers. In Figure 4B, GAPDH DNA signal was detected in 50 % of the single cell dilutions using SNARE. In contrast, no GAPDH DNA signal was observed for the same dilution using the Qiagen kit. While SNARE showed higher DNA sensitivity, the signal was not always positive at a single cell level likely due to stochasticity. We also confirmed the efficiency of the relative mRNA and DNA GAPDH signal using a standard curve (Figure S-1). In addition to GAPDH, we were also able to detect by qPCR androgen receptor (AR) and prostate serum antigen (PSA) with greater sensitivity as compared to Qiagen (Figure S-2). Finally, we used SNARE to isolate mRNA and DNA from the same sample using two other cell lines (THP-1: Human acute monocytic leukemia cell line, HMF: Human myocardial fibroblasts) to demonstrate its broad utility (Figure S-3).
Figure 4.
Comparison of A) relative GAPDH mRNA, and B) GAPDH DNA signal purified from 1000, 100, 10 or 1 LNCaPs using SNARE (grey dots) or Qiagen (black dots). Each dot represents a nucleic acid purification procedure with horizontal lines representing the mean of the individual experiments. Sample size per a group n=6.
SNARE Enables Sequencing of Clinically Relevant Mutations
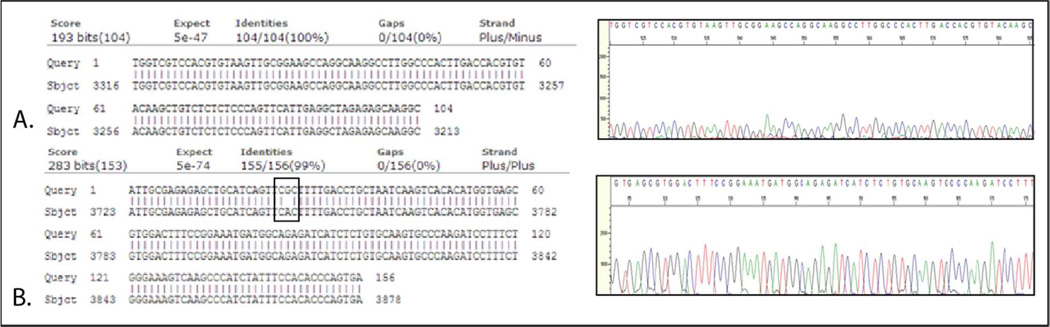
Deciphering nucleic acid sequences is essential for virtually all branches of biological research especially cancer pathogenesis, which is driven by inherited genetic variation and acquired somatic mutations. Therefore, we demonstrate mRNA and DNA extracted from 10 LNCaPs using SNARE could be used in Sanger sequencing. We specifically sequenced amplified regions of the AR, as it is a major driver of prostate cancer24 from which the LNCaP cell line was derived. Figure 5A shows that the amplicon of the AR from SNARE-isolated mRNA was correctly amplified. Figure 5B shows that exon 8 of the AR was also correctly amplified from SNARE isolated DNA. A known mutation found in LNCaPs at T887A was also identified, as expected25. These data demonstrate the utility of SNARE for Sanger sequencing applications.
Figure 5.
A) Sequencing chromatogram and alignment of exon 5 and 6 of the AR from mRNA purified from 10 LNCaP cells using the SNARE method. B) Sequencing chromatogram and alignment of exon 8 of the AR from DNA purified from 10 LNCaPs using the SNARE method. The T887A LNCaP mutation was identified (black box).
Patient Data
SNARE was shown to be efficient for extracting mRNA and DNA from LNCaPs serving as a model for rare prostate cancer CTCs. To demonstrate that SNARE can extract both mRNA and DNA from clinical samples for molecular interrogation, we processed CTCs from three patients with prostate cancer and examined relative GAPDH and AR signal by qPCR. This is a critical step forward as we move from the end point of CTC enumeration to the focus of molecular interrogation11. Within these patient samples, we were able to detect GAPDH and AR for both mRNA and DNA (Table 1). When CTCs were present we were able to amplify AR, a CTC specific gene which PBMCs do not express. While future molecular characterization will be dependent on the purity and efficiency of upstream rare cell capture methods, SNARE represents a method for sequential extraction of mRNA and DNA that maximizes the amount of information received from a single rare cell population. Importantly, SNARE is not limited to CTC mRNA and DNA extraction but can be expanded to use with other samples of interest.
Table 1.
GAPDH and AR relative mRNA and DNA Threshold Cycle (CT) from nucleic acids purified using SNARE from CTCs in three different patients diagnosed with prostate cancer.
| Patient # | CTC # | mRNA Threshold Cycle | DNA Threshold Cycle | ||
|---|---|---|---|---|---|
| GAPDH | AR | GAPDH | AR | ||
| 1 | 47 | 30.46 | 27.06 | 30.24 | 35.03 |
| 2 | 7 | 33.31 | 38.07 | 30.35 | 38.44 |
| 3 | 0 | 35.52 | 28.17 | 34.5 | |
CONCLUSION
We have shown SNARE can sequentially isolate both mRNA and DNA from a single sample by using immiscible phase exclusion. This method is advantageous when working with rare cell populations as it eliminates dilutive and centrifugation processes that result in sample loss due to increased fluid manipulation and purification time. In addition, SNARE enhances yield and reduces inter-experimental variability as no splitting of the original sample is needed. And given the increase in paired genomic and transcriptomic studies2 the advantages and need of SNARE are becoming more apparent, especially when analyzing rare cell populations, such as CTCs. In the future, whole genome and transcriptome amplification may be incorporated into the analysis to further expand the range of molecular assays that can be performed, including microarray analysis and whole genome/transcriptome sequencing applications. SNARE can be further expanded to integrate with other, previously developed microfluidic devices for rare cell isolation and analysis13. SNARE’s reduction in time, cost and equipment needed make it amenable to widespread adoption for low cell number nucleic acid isolation in both the research lab and for clinical use.
In summary, SNARE was shown to isolate as much or more mRNA and DNA from 1–10 cells as compared to the Qiagen Allprep DNA/RNA micro kit as demonstrated by qPCR. We also demonstrated the mRNA and DNA extracted from a low number of cells could be used as template for Sanger sequencing. Finally, the utility of SNARE to isolate mRNA and DNA from rare cell populations was shown using CTCs as a model. Detection of both relative GAPDH and AR signal was achieved from collected prostate cancer CTCs. While CTCs are just one example of a real world sample, the mRNA and DNA isolated using SNARE could allow for expansion into early disease detection, monitoring of treatment response, selection of targeted therapies and understanding of disease development.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Ms. Stephanie Thiede for her efforts cloning for sequencing. This work was supported by a grant from the Bill & Melinda Gates Foundation through the Grand Challenges in Global Health initiative, NIH Grant #5R33CA137673, NSF GRFP DGE-0718123 awarded to Lindsay Strotman, DOD grant W81XWH-12-1-0052 and the Prostate Cancer Foundation Young Investigator awarded to Dr. Lang. Scott Berry hold equity in Salus Discovery, LLC. David J. Beebe holds equity in Bellbrook Labs, LLC, Ratio, Inc and Salus Discovery, LLC.
Footnotes
Supporting Information.
Additional information is available as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Xu C, Houck JR, Fan W, Wang P, Chen Y, Upton M, Futran ND, Schwartz SM, Zhao LP, Chen C, Mendez E. The Journal of molecular diagnostics : JMD. 2008;10:129–134. doi: 10.2353/jmoldx.2008.070131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirilo PD, Marchi FA, Barros Filho Mde C, Rocha RM, Domingues MA, Jurisica I, Pontes A, Rogatto SR. PloS one. 2013;8:e57901. doi: 10.1371/journal.pone.0057901. [DOI] [PMC free article] [PubMed] [Google Scholar]; Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Group M, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu C, Wyatt AW, Lapuk AV, McPherson A, McConeghy BJ, Bell RH, Anderson S, Haegert A, Brahmbhatt S, Shukin R, Mo F, Li E, Fazli L, Hurtado-Coll A, Jones EC, Butterfield YS, Hach F, Hormozdiari F, Hajirasouliha I, Boutros PC, Bristow RG, Jones SJ, Hirst M, Marra MA, Maher CA, Chinnaiyan AM, Sahinalp SC, Gleave ME, Volik SV, Collins CC. The Journal of pathology. 2012;227:53–61. doi: 10.1002/path.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathieson W, Thomas GA. Analytical biochemistry. 2013;433:10–18. doi: 10.1016/j.ab.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Grzendowski M, Riemenschneider MJ, Hawranke E, Stefanski A, Meyer HE, Reifenberger G, Stuhler K. Proteomics. 2009;9:4985–4990. doi: 10.1002/pmic.200800902. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P. BioTechniques. 1993;15:532–534. 536–537. [PubMed] [Google Scholar]
- 6.Tolosa JM, Schjenken JE, Civiti TD, Clifton VL, Smith R. BioTechniques. 2007;43:799–804. doi: 10.2144/000112594. [DOI] [PubMed] [Google Scholar]
- 7.Hummon AB, Lim SR, Difilippantonio MJ, Ried T. BioTechniques. 2007;42:467–470. 472. doi: 10.2144/000112401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein CA, Seidl S, Petat-Dutter K, Offner S, Geigl JB, Schmidt-Kittler O, Wendler N, Passlick B, Huber RM, Schlimok G, Baeuerle PA, Riethmuller G. Nature biotechnology. 2002;20:387–392. doi: 10.1038/nbt0402-387. [DOI] [PubMed] [Google Scholar]
- 9.Kim JJM, Hill P, Gale BK. Integrative Biology. 2009;1:574–586. doi: 10.1039/b905844c. [DOI] [PubMed] [Google Scholar]
- 10.Kim JJM, Hill P, Sonkul RS, Kim J, Gale BK. JOURNAL OF MICROMECHANICS AND MICROENGINEERING. 2011;22 [Google Scholar]; Witek MA, Hupert ML, Park DS, Fears K, Murphy MC, Soper SA. Analytical chemistry. 2008;80:3483–3491. doi: 10.1021/ac8002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zborowski M, Chalmers JJ. Analytical chemistry. 2011;83:8050–8056. doi: 10.1021/ac200550d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lianidou ES, Markou A. Clinical chemistry. 2011;57:1242–1255. doi: 10.1373/clinchem.2011.165068. [DOI] [PubMed] [Google Scholar]; Lowes LE, Goodale D, Keeney M, Allan AL. Methods in cell biology. 2011;102:261–290. doi: 10.1016/B978-0-12-374912-3.00010-9. [DOI] [PubMed] [Google Scholar]
- 13.Dharmasiri U, Witek MA, Adams AA, Soper SA. Annual review of analytical chemistry. 2010;3:409–431. doi: 10.1146/annurev.anchem.111808.073610. [DOI] [PubMed] [Google Scholar]
- 14.Steele CD, Wapner RJ, Smith JB, Haynes MK, Jackson LG. Clinical obstetrics and gynecology. 1996;39:801–813. doi: 10.1097/00003081-199612000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Daniele N, Scerpa MC, Caniglia M, Bernardo ME, Rossi C, Ciammetti C, Palumbo G, Locatelli F, Isacchi G, Zinno F. Pathology, research and practice. 2012;208:67–73. doi: 10.1016/j.prp.2011.10.006. [DOI] [PubMed] [Google Scholar]; Cheng X, Liu YS, Irimia D, Demirci U, Yang L, Zamir L, Rodriguez WR, Toner M, Bashir R. Lab on a chip. 2007;7:746–755. doi: 10.1039/b705082h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangopadhyay NN, Shen H, Landreneau R, Luketich JD, Schuchert MJ. Journal of immunological methods. 2004;292:73–81. doi: 10.1016/j.jim.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Strotman LN, Lin G, Berry SM, Johnson EA, Beebe DJ. The Analyst. 2012;137:4023–4028. doi: 10.1039/c2an35506j. [DOI] [PubMed] [Google Scholar]
- 18.Berry SM, Alarid ET, Beebe DJ. Lab on a chip. 2011;11:1747–1753. doi: 10.1039/c1lc00004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berry SM, Strotman LN, Kueck JD, Alarid ET, Beebe DJ. Biomedical microdevices. 2011;13:1033–1042. doi: 10.1007/s10544-011-9573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casavant BP, Guckenberger DJ, Berry SM, Tokar JT, Lang JM, Beebe DJ. Lab on a chip. 2013;13:391–396. doi: 10.1039/c2lc41136a. [DOI] [PubMed] [Google Scholar]
- 21.Atencia J, Beebe DJ. Nature. 2005;437:648–655. doi: 10.1038/nature04163. [DOI] [PubMed] [Google Scholar]
- 22.Berry SM, Regehr KJ, Casavant BP, Beebe DJ. Journal of laboratory automation. 2013;18:206–211. doi: 10.1177/2211068212462023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rio DC, Ares M, Jr., Hannon GJ, Nilsen TW. Cold Spring Harbor protocols. 2010;2010 doi: 10.1101/pdb.prot5439. pdb prot5454. [DOI] [PubMed] [Google Scholar]
- 24.Friedlander TW, Ryan CJ. The Urologic clinics of North America. 2012;39:453–464. doi: 10.1016/j.ucl.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Veldscholte J, Berrevoets CA, Ris-Stalpers C, Kuiper GG, Jenster G, Trapman J, Brinkmann AO, Mulder E. The Journal of steroid biochemistry and molecular biology. 1992;41:665–669. doi: 10.1016/0960-0760(92)90401-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.