Pih1d3 promotes the stability and assembly of axonemal outer and inner dynein arms to allow for cytoplasmic preassembly of dynein complexes in spermatogenic cells.
Abstract
Axonemal dynein complexes are preassembled in the cytoplasm before their transport to cilia, but the mechanism of this process remains unclear. We now show that mice lacking Pih1d3, a PIH1 domain–containing protein, develop normally but manifest male sterility. Pih1d3−/− sperm were immotile and fragile, with the axoneme of the flagellum lacking outer dynein arms (ODAs) and inner dynein arms (IDAs) and showing a disturbed 9+2 microtubule organization. Pih1d3 was expressed specifically in spermatogenic cells, with the mRNA being most abundant in pachytene spermatocytes. Pih1d3 localized to the cytoplasm of spermatogenic cells but was not detected in spermatids or mature sperm. The levels of ODA and IDA proteins were reduced in the mutant testis and sperm, and Pih1d3 was found to interact with an intermediate chain of ODA as well as with Hsp70 and Hsp90. Our results suggest that Pih1d3 contributes to cytoplasmic preassembly of dynein complexes in spermatogenic cells by stabilizing and promoting complex formation by ODA and IDA proteins.
Introduction
Cilia are long appendages that extend from the cell body. There are two types of cilia: motile cilia and immotile cilia (also known as primary cilia). In mammals, motile cilia are present in the respiratory epithelium, reproductive system (for example, the oviduct), and central nervous system (for example, the ependyma). Motile cilia that are present singly or in small numbers per cell are referred to as flagella, as in single-cell protozoa such as Chlamydomonas and in mammalian sperm. Primary cilia are found in most mammalian cell types, including those of the skin, kidney (renal tubular epithelial cells), and blood vessels (endothelial cells; Wheatley et al., 1996).
Cilia contain a microtubule-based axoneme covered by a specialized ciliary membrane that is continuous with the plasma membrane of the cell. The axoneme is composed of nine peripheral microtubule doublets surrounding a central core that may or may not contain two central microtubules (9+2 for motile cilia and 9+0 for primary cilia, respectively). The axoneme of motile cilia specifically contains other associated structures such as outer dynein arms (ODAs) and inner dynein arms (IDAs), radial spokes, and nexin links.
Given that cilia do not contain DNA or any machinery necessary for protein synthesis, all ciliary proteins are synthesized in the cytoplasm and then transported to the site of cilium assembly by a process known as intraflagellar transport (Rosenbaum and Witman, 2002). The history of ciliary proteins before such transport, including their state in the cytoplasm and how they are sorted, has remained unclear, however.
In Chlamydomonas, ciliary proteins are stocked in a cytoplasmic pool before they are sorted (Rosenbaum and Child, 1967; Rosenbaum et al., 1969). In the cytoplasm, components of multi-subunit structures such as ODAs, IDAs, and radial spokes have been detected in preassembled complexes, rather than individually (Fok et al., 1994; Piperno and Mead, 1997; Fowkes and Mitchell, 1998; Qin et al., 2004), as the result of a process known as cytoplasmic preassembly (Kobayashi and Takeda, 2012).
The molecular components of the axoneme and mechanism of cytoplasmic preassembly have also been investigated in Chlamydomonas. ODAs have thus been found to be composed of three heavy chains (HCs; α, β, and γ, each of ∼500 kD), two intermediate chains (IC1, 78 kD; IC2, 69 kD), 10 light chains (LC1 to LC10, 8 to 22 kD), and a 7S factor comprising three proteins that form an ODA attachment site or docking complex (DC105, DC62.5, and DC25). IDAs are structurally more diverse. Immunoprecipitation analysis has revealed that the three HCs and two ICs of ODAs exist in the cytoplasm as a complex (Fowkes and Mitchell, 1998).
Analysis of dynein-deficient mutants of Chlamydomonas has identified four proteins that are required for cytoplasmic preassembly of dynein complexes: PF13 (also known as Ktu or DNAAF2; Omran et al., 2008), ODA7 (also known as LRRC50 or DNAAF1; Duquesnoy et al., 2009; Loges et al., 2009), MOT48 (Yamamoto et al., 2010), and PF22 (also known as DNAAF3; Mitchison et al., 2012). PF13 and ODA7 are required for stability of the three ODA HCs in the cytoplasm, PF22 for preassembly of ODAs and IDAs, and MOT48 for preassembly of IDAs. Two of these four proteins, DNAAF1 and MOT48, contain a PIH1 domain, a motif that was first identified in the Saccharomyces cerevisiae protein PIH1 (also known as Nop17p; Gonzales et al., 2005). This yeast protein functions to maintain or promote the assembly of the BOX C/D small nucleolar RNP, a prerRNA-processing complex, by interacting with the molecular chaperone HSP90 (Zhao et al., 2008).
Twister is another protein that contains a PIH1 domain. The Twister gene was first identified by genetic screening in zebrafish as one of the genes whose mutation resulted in the formation of kidney cysts, a phenotype reminiscent of polycystic kidney disease (PKD) in humans (Sun et al., 2004). The precise function of Twister has remained unknown, however. We have now generated a mutant mouse that lacks the Twister-like gene Pih1d3. Analysis of the mutant animals suggests that Pih1d3 is required for the motility of sperm and is a dynein axonemal assembly factor (DNAAF) that promotes the stability and assembly of HCs and ICs of axonemal ODAs and IDAs in the cytoplasm of mouse sperm. Pih1d3 is the first Dnaaf found to be required for the cytoplasmic preassembly of ICs and for the 9+2 organization of microtubules.
Results
Pih1d3−/− male mice are sterile
Two genes in the mouse genome show homology to Twister of zebrafish: RIKEN 4930521A18 (referred to as Pih1d3 in this study; its human homologue is called PIH1D3) and E230019M04 (Twister2; Fig. 1 A). Pih1d3 and Twister2 possess highly similar coding regions, but they differ in their 3′ untranslated regions. We focused on Pih1d3 in the present study. To analyze the function of Pih1d3 in mouse, we generated a mutant allele (Pih1d3neo) with a standard gene-targeting approach (Fig. S1). Pih1d3– and Pih1d3flox alleles were subsequently generated from Pih1d3neo with the use of Cre and Flp recombinases, respectively. An Ires-lacZ cassette present in the 3′ untranslated region of the Pih1d3flox allele would allow monitoring of Pih1d3 expression by staining of embryos with the LacZ substrate X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
Figure 1.
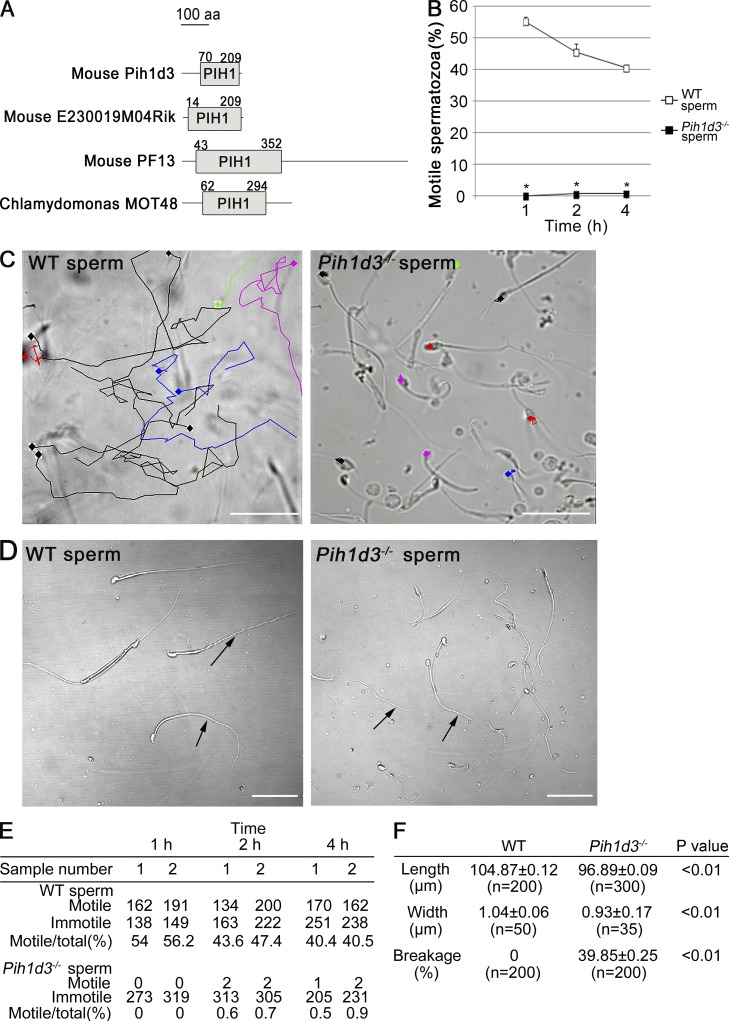
Sperm of Pih1d3−/− mice are immotile and morphologically abnormal. (A) Structural representation of four proteins that contain a PIH1 domain. The numbers indicate the amino acid residues that correspond to predicted PIH1 domain regions. (B) Motility of sperm from adult wild-type (WT) and Pih1d3−/− mice as evaluated by determination of the percentage of motile sperm at 1, 2, and 4 h after release from the cauda epididymis. Two batches of at least 100 sperm were examined for each mouse. Data are means ± SEM (n = 2 batches from one mice). *, P < 0.01 versus the corresponding value for WT sperm (Student’s t test). (C) Movement of 10 WT or Pih1d3−/− sperm was traced with TEMA software over 5 s. The movement of individual sperm cells is indicated by the different colored lines. Bars, 50 µm. (D) Light microscopy of WT and Pih1d3−/− spermatozoa collected from the cauda epididymis. Whereas the head of sperm from Pih1d3−/− males appeared normal, the tail was thinner, shorter, and more susceptible to breakage (arrows) compared with that of WT sperm. Images were obtained with a 40× objective. Bars, 50 µm. (E) Original data for B. (F) Sperm length, the width of the midpiece region, and the frequency of broken sperm are shown for WT and Pih1d3−/− sperm. The number of samples examined is shown as n.
Intercrossing of Pih1d3+/− mice yielded Pih1d3−/− offspring at the expected Mendelian frequency at birth, and the mutant animals showed no obvious phenotypic abnormalities. The knockout mice thus did not manifest kidney cysts or other abnormalities related to ciliary defects (such as situs inversus and respiratory infection), with the exception that Pih1d3−/− males proved to be infertile. Mating of Pih1d3−/− males with Pih1d3+/− or Pih1d3+/+ females thus yielded no offspring, whereas Pih1d3−/− females showed normal fertility.
Sperm of Pih1d3−/− mice are immotile and morphologically abnormal
To investigate the cause of the sterility of Pih1d3−/− males, we examined the mature spermatozoa stocked in the cauda epididymis of adult mice. Light microscopy revealed that sperm from Pih1d3−/− males failed to move any distance (Fig. 1, B–E; and Table S1). Although a small proportion of the sperm cells shook their tails weakly, they were not able to move forward (Videos 1 and 2). Tracking of sperm movement with software confirmed the immotility of the Pih1d3-deficient sperm (Fig. 1 C). Light microscopy also showed that, whereas the head of sperm from Pih1d3−/− males appeared normal, the tail was frequently broken (Fig. 1, D and F). The mutant sperm was slightly shorter and thinner than wild-type (WT) sperm (Fig. 1 F).
We also examined other motile cilia of the Pih1d3−/− mouse. Cilia of the respiratory tract were normal in their motility (Videos 3 and 4) and ultrastructure (Fig. S2 A). Adult Pih1d3−/− mouse did not show hydrocephalus (Fig. S2 B), suggesting that cilia of ependymal cells are also normal.
Structural defects of Pih1d3-deficient sperm
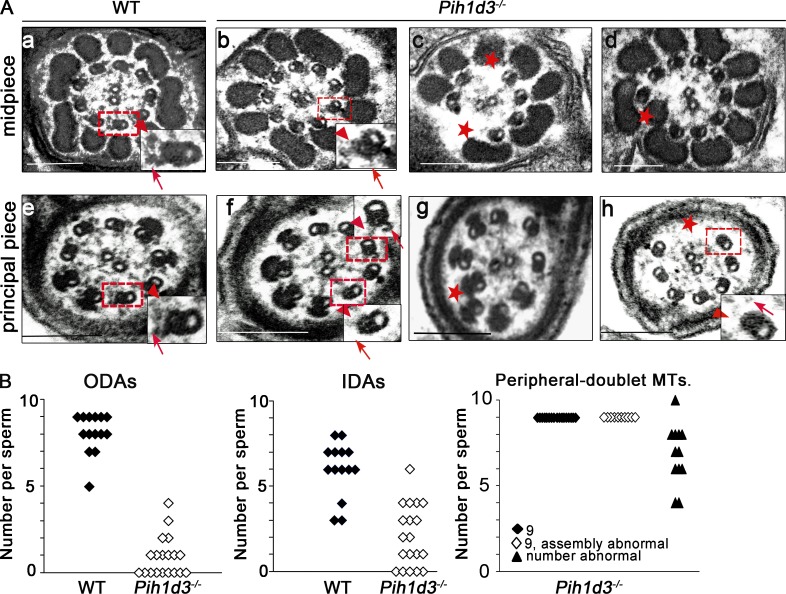
Sperm motility is generated by the sliding of microtubules forced by ODAs and IDAs. Given the loss of motility and fragile tail of Pih1d3-deficient sperm, we examined the ultrastructure of the mutant cells by transmission electron microscopy (TEM). Although the 9+2 organization of microtubules was apparent in WT sperm, this structural pattern was disrupted in the axoneme of the mutant sperm (Fig. 2, A–J). ODAs were thus almost entirely missing from the 19 Pih1d3−/− sperm examined, whereas IDAs were lost in some but not all of the mutant sperm. We counted 0.89 ODAs and 2.05 IDAs per mutant sperm, compared with values of 8.07 and 6.00 per WT sperm (Fig. 2 K). Furthermore, in 22 of 39 Pih1d3-null sperm examined, peripheral doublet microtubules were either reduced in number or all present but disorganized (Fig. 2).
Figure 2.
Disruption of axoneme integrity and dynein arm defects in sperm of Pih1d3−/− mice. (A) TEM of the midpiece (a–d) and principal piece (e–h) of sperm collected from the cauda epididymis of adult WT (a and e) or Pih1d32/2 (b–d and f–h) mice. Boxed areas are shown at higher magnification in the insets. Red arrows and arrowheads indicate ODAs and IDAs, respectively, in WT sperm or their loss in Pih1d3−/− sperm. Red asterisks indicate loss of the 9+2 structural organization of microtubules in Pih1d3-deficient sperm. Bars, 200 nm. (B) Quantitation of ODAs, IDAs, and peripheral doublet microtubules in sperm of WT and Pih1d3−/− mice. The numbers of ODAs and IDAs in TEM sections of 14 WT sperm and 19 Pih1d3−/− sperm were counted, and the average number of each per sperm was calculated: 0.89 ODAs and 2.05 IDAs per mutant sperm, compared with 8.07 ODAs and 6.00 IDAs per WT sperm. Peripheral-doublet microtubules in 39 Pih1d3−/− sperm were also counted, with 17 sperm showing nine intact doublets (closed diamonds), 10 sperm showing nine abnormal doublets (open diamonds), and 12 sperm showing a reduced number of doublets (filled triangles).
Pih1d3 is expressed in spermatogenic cells and encodes a cytoplasmic protein
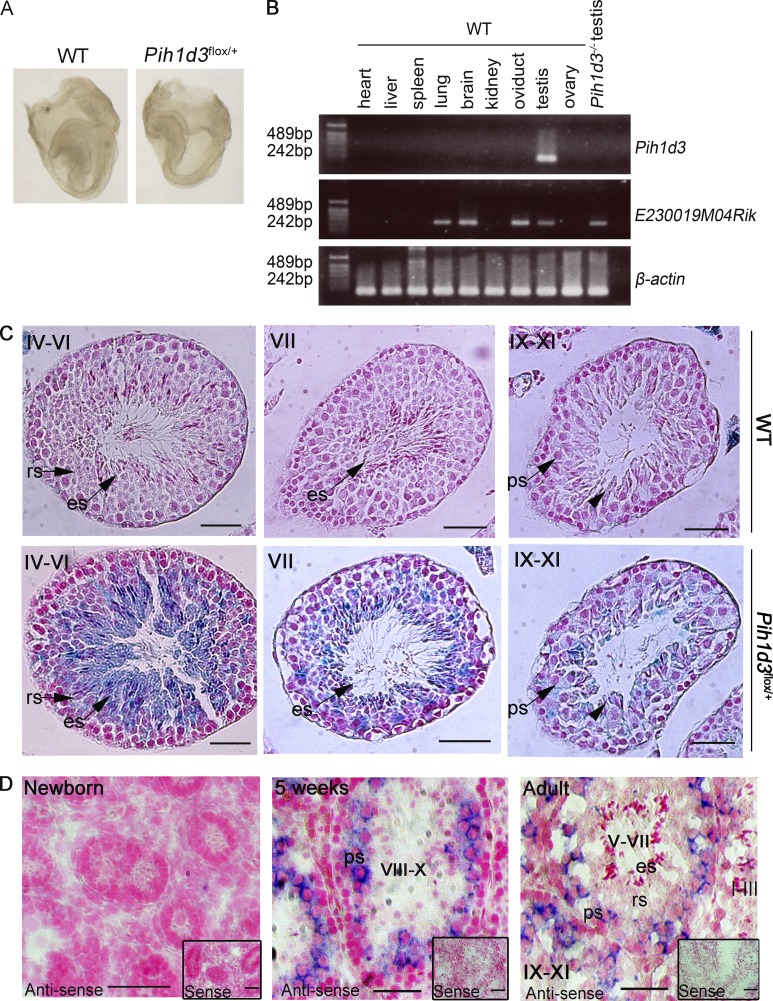
We next examined the expression of Pih1d3 at various stages of development. Staining of Pih1d3flox/+ embryos with X-gal at embryonic day (E) 8.0, when breaking of left-right symmetry takes place in the node, did not reveal expression of the Ires-lacZ cassette present in the 3′ untranslated region of the Pih1d3flox allele, suggesting that Pih1d3 is not expressed at this time (Fig. 3 A). In expression databases (such as UniGene), Pih1d3 transcripts were found only in the testis, both in mouse and human. In RT-PCR analysis, Pih1d3 mRNA was detected only in the testis among various organs examined, whereas E230019M04 (Twister2) mRNA was detected in organs with motile cilia such as the lung, brain, oviduct, and testis (Fig. 3 B).
Figure 3.
Pih1d3 expression during sperm development. (A) WT (control) and Pih1d3flox/+ embryos at E8.0 were stained with X-gal. Pih1d3 is not expressed in mouse embryos at E8.0. Bars, 0.5 mm. (B) Investigation of Pih1d3 and E230019M04Rik expression in the indicated visceral organs from adult WT mouse and adult Pih1d3−/− testis (negative control) by RT-PCR. β-Actin serves as a loading control. (C) X-gal staining of the testis of adult Pih1d3flox/+ or WT (control) mice. The sections were counterstained with nuclear fast red. Roman numerals indicate stages of the cycle of the seminiferous epithelium. ps, pachytene spermatocyte; rs, round spermatids; es, elongated spermatids. The arrowheads indicate elongating spermatids. Bars, 50 µm. (D) In situ hybridization analysis of Pih1d3 mRNA in the testis of newborn, 5-wk-old, and adult WT mice with an antisense probe. Insets show staining with the corresponding sense probe as a control. All sections were counterstained with nuclear fast red. Stages of the cycle of the seminiferous epithelium are indicated with roman numerals. Bars, 50 µm.
Staining of the testis from adult Pih1d3flox/+ mice with X-gal revealed that spermatogenic cells ranging from pachytene spermatocytes to elongated spermatids were positive for Pih1d3 expression (Fig. 3 C). We also examined Pih1d3 expression in the testis of newborn, 5-wk-old, and adult WT mice by in situ hybridization (Fig. 3 D). Although Pih1d3 mRNA was not detected in the newborn testis, it was apparent in pachytene spermatocytes at 5 wk of age as well as in pachytene and diplotene spermatocytes of adult animals. The fact that Pih1d3 expression was detected in elongating and elongated spermatids of Pih1d3flox/+ mice by X-gal staining but not in those of WT mice by in situ hybridization suggested that Pih1d3 mRNA may undergo posttranscriptional regulation in the testis.
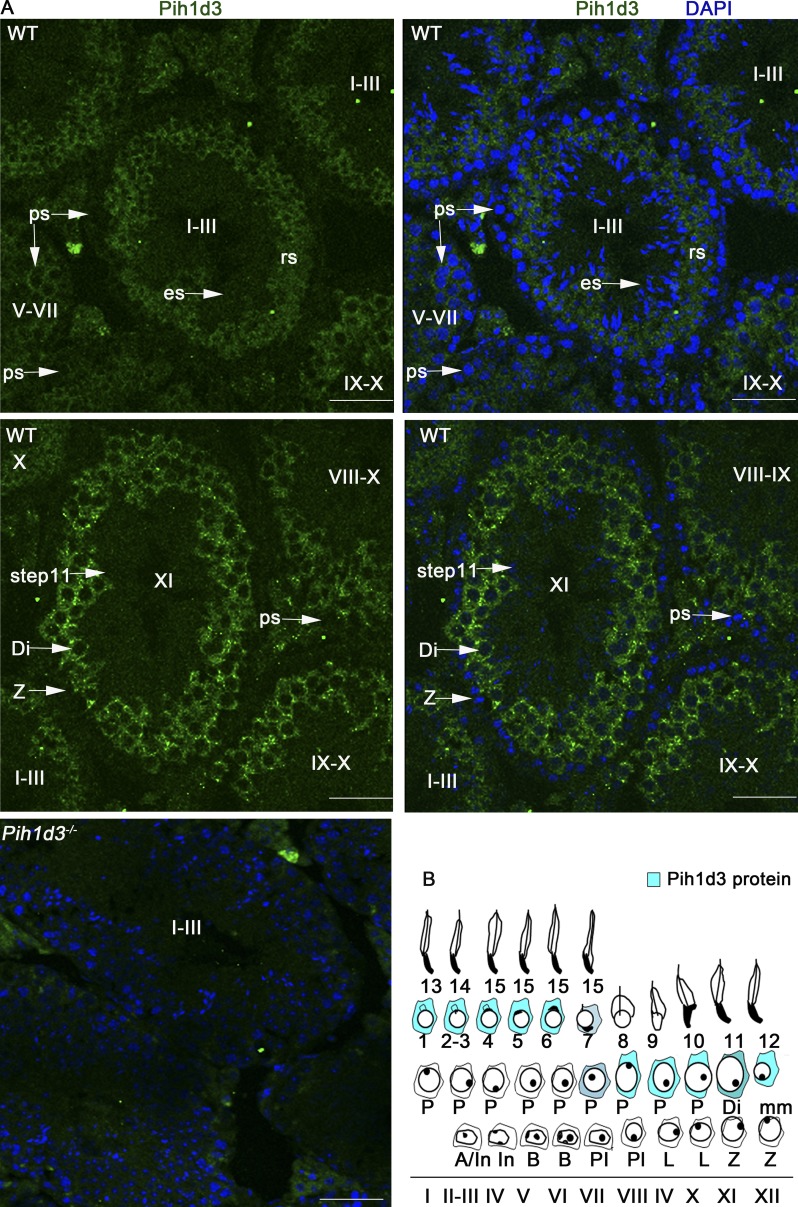
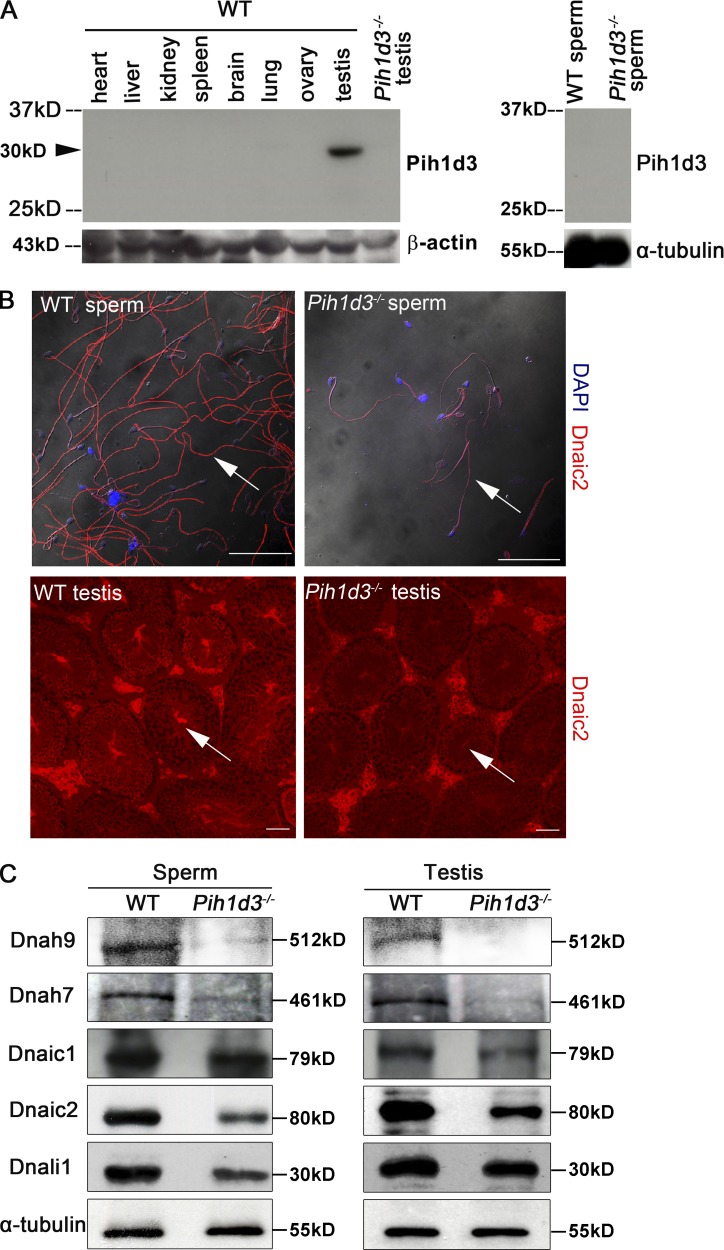
Immunofluorescence staining of the testis of adult WT mice revealed that Pih1d3 is localized to the cytoplasm of spermatogenic cells, ranging from late pachytene spermatocytes to round spermatids, with the abundance of the protein being highest in pachytene and diplotene spermatocytes (Fig. 4, A and B). Such staining was not detected in the testis of Pih1d3−/− males (Fig. 4 A), indicating the high specificity of the antibodies. Like Pih1d3 mRNA (Fig. 3 D), Pih1d3 protein was not detected in elongating or elongated spermatids, suggesting that Pih1d3 is not involved in intramanchette transport. Immunoblot analysis also showed that Pih1d3 is expressed in the testis of adult WT mice, but it was not detected in mature sperm (Fig. 5 A).
Figure 4.
Subcellular localization of Pih1d3 in adult mouse testis. (A) Immunofluorescence staining of the testis of adult WT or Pih1d3−/− mice with antibodies to Pih1d3. Pih1d3 (green fluorescence) was detected in the cytoplasm of WT sperm cells from late pachytene spermatocytes to round spermatids (rs), with its abundance being highest at the diplotene (Di) and pachytene stages. Nuclei were stained with DAPI (blue fluorescence). Stages of the cycle of the seminiferous epithelium are indicated with roman numerals. Images were obtained with a 40× objective lens. Bars, 50 µm. (B) Staging diagram for Pih1d3 protein expression in the adult testis. Circles around the nucleus indicate cytoplasm, with the intensity of the blue color representing the expression level of Pih1d3.
Figure 5.
Down-regulation of dynein subunits in the testis of Pih1d3−/− mice. (A) Immunoblot analysis of visceral organs from adult WT mouse and the sperm and testis of adult WT and Pih1d3−/− mice with antibodies to Pih1d3. The Pih1d3 protein (∼30 kD) is indicated by the arrowhead. (B) Immunofluorescence staining of sperm and the testis of adult WT and Pih1d3−/− mice with antibodies to Dnaic2. The abundance of Dnaic2 (red fluorescence) was reduced in the sperm and testis of Pih1d3−/− mice compared with those of WT mice (white arrows). Nuclei were stained with DAPI (blue fluorescence). Images were taken with a 40× objective lens (sperm) and 20× objective lens (testis). Bars, 50 µm. (C) Immunoblot analysis of dynein subunits and α-tubulin (loading control) in the sperm and testis of adult WT and Pih1d3−/− mice.
Cytoplasmic preassembly of dynein arms requires Pih1d3
Sperm are produced in the seminiferous tubules of the testis. Given that Pih1d3 protein was detected in the cytoplasm of spermatogenic cells and that Pih1d3-deficient sperm have structural defects, we next examined whether spermatogenesis proceeds normally in the testis of Pih1d3−/− mice. Hematoxylin-eosin (H&E) staining revealed that most seminiferous tubules and the cauda epididymis of adult Pih1d3−/− males were normal, although a small number of seminiferous tubules showed the absence of spermatogenic cells, with the accumulation of elongated spermatids in the lumen (Fig. S3, A–C; and Table S2). Immunochemical staining failed to detect the cleaved form of caspase-3 in the Pih1d3−/− testis (Fig. S3 E), suggesting that the frequency of apoptosis is not increased in the mutant.
SMART and Pfam programs predicted that mouse Pih1d3 contains a PIH1 domain spanning amino acid residues 70 to 209 (Fig. 1 A; Fig. S4). PF13 and MOT48 both contain a PIH1 domain (Fig. 1 A) and are required for cytoplasmic preassembly of dynein arm components in Chlamydomonas. PF13 is thus responsible together with ODA7 for the folding and stability of ODA HCs as well as for HC–IC complex assembly (Omran et al., 2008; Mitchison et al., 2012), whereas MOT48 contributes to IDA assembly (Yamamoto et al., 2010). These observations together with our present results suggested that Pih1d3 may play a similar role in dynein preassembly.
We therefore examined the abundance of ODA and IDA subunits in sperm and testis of adult Pih1d3−/− mice. Immunoblot analysis revealed that the amounts of Dnah9 (mouse orthologue of Chlamydomonas ODA HCβ), Dnaic1 (ODA IC1), Dnaic2 (ODA IC2), and Dnah7 (mouse orthologue of Chlamydomonas IDA HC) were all decreased in Pih1d3−/− sperm and testis compared with those of WT mice (Fig. 5 B), suggesting that the cytoplasmic preassembly of ODA and IDA subunits is impaired in spermatogenic cells of the mutant. The reduced abundance of Dnaic2 in Pih1d3-deficient sperm and testis was also confirmed by immunofluorescence analysis (Fig. 5 C). Pih1d3 may similarly promote the stability of ODA and IDA HCs. The abundance of DNALI1 (mouse orthologue of Chlamydomonas IDA LC p28) was decreased in sperm but not in the testis of Pih1d3−/− mice (Fig. 5 B), suggesting that the stability of IDA LCs may not be greatly affected by the absence of Pih1d3.
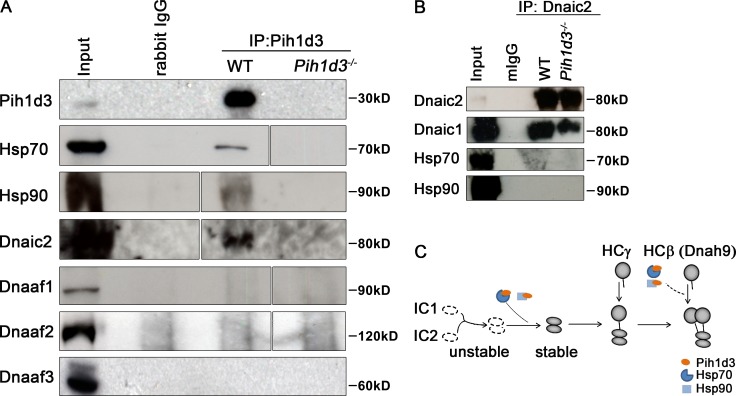
The yeast protein PIH1 interacts with HSP90 (Zhao et al., 2008), whereas the PIH1 domain–containing protein PF13 interacts with HSP70 (Omran et al., 2008). We therefore next tested whether Pih1d3 also interacts with Hsp70 or Hsp90. Co-immunoprecipitation analysis revealed that Hsp70 and Hsp90 were indeed detected in immunoprecipitates prepared from the testis of WT mice with antibodies to Pih1d3 (Fig. 6 A), indicating that Pih1d3 is associated with these two molecular chaperones. Furthermore, Pih1d3 was found to form a complex with Dnaic2 (Fig. 6 A), suggesting that it may function as a co-chaperone to help maintain the stability of dynein HCs and ICs. However, Pih1d3 was not found to be associated with Dnaaf1, Dnaaf2, and Dnaaf3 (Fig. 6 A), suggesting that Pih1d3 functions at a different step during the assembly of dynein complex.
Figure 6.
Pih1d3 interacts with Hsp70, Hsp90, and Dnaic2 but not with other Dnaafs. (A) A testis extract prepared from adult WT mice and Pih1d3−/− mice was subjected to immunoprecipitation (IP) with antibodies to Pih1d3 or control rabbit immunoglobulin G (rIgG), and the resulting precipitates as well as the original extract (input) were subjected to immunoblot analysis with antibodies to Hsp70, Hsp90, Dnaic2, Dnaaf1, Dnaaf2, and Dnaaf3. An equivalent amount of lysate (containing 2 or 5 µg protein) from the wild-type and the mutant testes was tested. White lines indicate the removal of intervening lanes for presentation purposes. (B) Testis extract prepared from adult WT mice and Twister−/− mice were subjected to immunoprecipitation (IP) with antibodies to Dnaic2 or control mouse immunoglobulin G (mIgG), and the resulting precipitates as well as the original extract (input) were subjected to immunoblot analysis with antibodies to Dnaic2, Dnaic1, Hsp70, and Hsp90. (C) Model for Pih1d3 function in cytoplasmic preassembly of ODA dyneins. A complex of Pih1d3 with the chaperones Hsp70 or Hsp90 may promote the stability of IC1 and IC2 by stabilizing newly formed complex. Pih1d3 acting in concert with the molecular chaperones may also promote HC stability by facilitating the proper folding of HCs or their assembly with ICs.
Pih1d3 may be required for assembly of the Dnaic1–Dnaic2 complex, given that formation of this complex is the first step of ODA assembly and that the levels of IC1 and IC2 are reduced in Chlamydomonas IC2 and IC1 mutants, respectively, indicating that IC1 and IC2 molecules unable to form the IC1–IC2 complex are unstable (Fowkes and Mitchell, 1998). However, Dnaic1 was found to coprecipitate with Dnaic2 in the testis of WT and Pih1d3−/− mice (Fig. 6 B), suggesting that the IC1–IC2 complex can be formed in the absence of Pih1d3. While Pih1d3 formed a complex with Dnaic2, Hsp70, and Hsp90 (Fig. 6, A and B), Hsp70/90 was not coprecipitated with Dnaic2 (Fig. 6 B). These results suggest that Pih1d3 may be stabilized by interaction with Hsp70/90, which in turn promotes efficient formation of IC–HC complex (Fig. 6 C).
Discussion
Absence of Pih1d3 protein in mature sperm, its localization in the cytoplasm of spermatogenic cells, and the phenotype of Pih1d3 mutant mice collectively suggest that Pih1d3 is an essential factor for cytoplasmic preassembly of axonemal dyneins. Cytoplasmic preassembly of axonemal dyneins has been most well characterized in Chlamydomonas, with four DNAAFs having been identified to date: DNAAF1, DNAAF2, DNAAF3, and MOT48 (Omran et al., 2008; Yamamoto et al., 2010; Mitchison et al., 2012). Our data now suggest that Pih1d3 is a novel Dnaaf with unique features. Cytoplasmic preassembly of dynein HCs has been found to require at least two steps (Yamamoto et al., 2010). The first step involves the folding of the globular dynein head domain, which is required for HC stability. DNAAF1 and DNAAF2 contribute to this step together with HSP70. The second step involves formation of the HC–IC complex, which requires DNAAF3. Pih1d3 may be involved in a different step during axonemal dynein assembly because it does not interact with the known Dnaafs (Fig. 6 A).
Unlike other DNAAFs, however, we have now shown that Pih1d3 interacts with Dnaic2 (ODA IC2) as well as with Hsp70 and Hsp90, and the lack of Pih1d3 resulted in down-regulation of Dnaic1 and Dnaic2 in mouse testis. The formation of IC1–IC2 complex is required for maintenance of the stability of these two factors in the cytoplasm. Mutation of Chlamydomonas IC70 (IC1) thus renders IC78 (IC2) unstable, whereas mutation of IC78 results in a reduction in the abundance of IC70 in the cytoplasm (Fowkes and Mitchell, 1998). However, the IC1–IC2 complex was formed normally in the absence of Pih1d3 (Fig. 6 B), whereas the complex was rendered unstable in the mutant testis. Pih1d3 may be required to maintain the stability of the IC1–IC2 complex by directly interacting with the complex. Alternatively, Pih1d3 may promote the formation of the IC–HC complex, and the absence of Pih1d3 may indirectly reduce the level of Dnaic and Dnahc proteins in the testis. Because Hsp70/90 was not coimmunoprecipitated with Dnaic2, the role of Hsp70/90 may be to maintain the stability and integrity of Pih1d3.
The level of Dnah9 and Dnah7 was severely reduced in the testis and sperm from the Pih1d3−/− mouse (Fig. 5 B). Given that formation of the IC1–IC2 complex precedes formation of the IC–HC complex, the reduced levels of Dnaic1 and Dnaic2 in sperm cells of Pih1d3−/− mice may be responsible for the associated down-regulation of dynein HCs. Alternatively, Pih1d3 may also directly regulate the stability of ODA and IDA HCs as well as that of ODA ICs in the cytoplasm by serving as a co-chaperone of Hsp70 and Hsp90 (Fig. 6 C).
The phenotype of mouse Dnaaf mutants or humans deficient in DNAAFs shows substantial variation. Humans that lack DNAAF1 or DNAAF2 show a typical primary ciliary dyskinesia (PCD) phenotype characterized by recurrent pneumonia, situs inversus, and immotile sperm (Omran et al., 2008; Loges et al., 2009), whereas those lacking DNAAF3 manifest a spectrum of PCD phenotypes (Mitchison et al., 2012). On the other hand, mutation of Pih1d3 appears to affect only the motility of sperm, with other motile cilia such as those in the trachea remaining unaffected. This relatively mild phenotype might be due to the presence of Twister2. Twister2 transcripts (RIKEN E230019M04) were indeed detected in various organs with motile cilia including the lung, brain, and oviduct (Fig. 3 B). Interestingly, Twister2 mRNA was also found in the testis of WT and Pih1d3−/− mice. These observations suggest that Pih1d3 and Twister2 may have distinct roles in the testis although their amino acid sequences are similar, or that Twister2 transcripts may be translationally arrested in the testis. Alternatively, there may be functional redundancy between Pih1d3 and other DNAAFs such as PF13/Dnaaf2. However, this possibility is also unlikely given that humans that lack PF13 and Pih1d3−/− mice show distinct ciliary defects. A more likely explanation may be that Pih1d3 contributes to a mechanism of dynein arm assembly that is required specifically for formation of the motile flagellum of sperm, but not for that of other motile cilia. In this regard, the structure of the sperm flagellum shows differences from that of motile cilia in the trachea. For example, DNAH5 is localized only to the proximal region of sperm flagella but is present throughout the axoneme of respiratory cilia in humans (Fliegauf et al., 2005). The precise mechanism of ODA and IDA preassembly, as well as the composition of ODAs and IDAs, may thus differ between sperm flagella and motile cilia of the trachea.
Unlike Twister-null zebrafish, Pih1d3−/− mice do not manifest polycystic kidney disease. This difference may be attributable to structural differences in kidney cilia between these two species. Whereas kidney cilia in mammals are 9+0 immotile cilia, those in zebrafish are 9+2 motile cilia with ODAs and IDAs (Kramer-Zucker et al., 2005). Alternatively, Twister 2 may compensate the absence of Pih1d3 in the kidney.
Some DNAAF mutants show a loss of dynein arms but maintain an intact 9+2 microtubular organization. However, Pih1d3−/− sperm exhibit a disorganized pattern of axonemal microtubules, with some of the pairs in the 9+2 arrangement being absent. Given that Pih1d3 was found to interact with both Hsp70 and Hsp90, it is possible that it may perform other functions in addition to serving as a Dnaaf. Hsp90 also regulates the stability of axonemal β-tubulin in multiciliated cells of the airway (Takaki et al., 2007). Pih1d3, together with Hsp90, may therefore also play a role in other aspects of axonemal assembly, such as in the cytoplasmic preassembly of intramanchette transport factors or their association with axonemal components.
Materials and methods
Generation of Pih1d3 mutant mice
A targeting vector for Pih1d3 was constructed with a strategy based on flanking LoxP sites (Fig. S1 A). A DNA sequence for the Myc epitope tag, an internal ribosome entry site (Ires) and lacZ expression cassette, and an FRT-flanked phosphoglycerate kinase gene (Pgk) promoter–driven neomycin resistance gene (neo) cassette were positioned downstream of the Pih1d3 exon, and the entire region was flanked by LoxP sites. The targeting vector was introduced into embryonic stem (ES) cells, and clones that had undergone homologous recombination were identified by Southern blot analysis and introduced into ICR morulas at the 4- to 16-cell stage. Chimeric mice were distinguished on the basis of their coat color, with skin cells derived from the ES clones and ICR cells being black and white, respectively. Chimeras with a coat that was 80–100% black were crossed with ICR mice, and those showing germline transmission of the modified DNA were mated with CAG-Flp mice to generate Pih1d3+/flox offspring or with CAG-Cre mice to generate Pih1d3+/– offspring. Pih1d3−/− mice were generated by intercrossing of Pih1d3+/− heterozygotes.
Southern blot and PCR analyses
Genomic DNA of ES clones or mice was digested with KpnI and EcoRV, and the resulting fragments were subjected to Southern blot analysis with a 5′ external probe (Fig. S1, A and B). The WT allele generated a hybridizing fragment of 9.4 kb, whereas the neo, flox, and null alleles yielded a hybridizing fragment of 8.7 kb. PCR analysis was also performed to detect the WT, flox, and null alleles (Fig. S1, A and C). The forward primer P1 (5′-CTGAGGACGTATACCTAGGA-3′) and reverse primer P2 (5′-GACGACACACTAACTGGTAC-3′) were used to detect the WT allele, the forward primer P3 (5′-GCTACCATTACCAGTTGGTC-3′) and reverse primer P2 were used to detect the flox allele, and the forward primer P4 (5′-TGATGGATCCCACAGCCTAA-3′) and reverse primer P2 were used to detect the null allele.
Histological analysis
The testis and cauda epididymis of adult WT or Pih1d3−/− mice were dissected, fixed overnight in Bouin’s solution at 4°C, dehydrated, and embedded in paraffin wax. Serial sections (7-µm thickness) were then prepared from the tissue and stained with H&E according to standard procedures. To examine the cerebrum, the cerebrum of adult WT or Pih1d3−/− mice were fixed in 4% paraformaldehyde in PBS at 4°C for 48 h, washed in PBS for 30 min three times, exposed consecutively to glucose solutions of 10, 15, 20, and 30%, and embedded in OCT compound for preparation of frozen sections (7-mm thickness). Sections were then stained with H&E according to standard procedures. Histological photos were taken using a microscope (Diaphot; Nikon) and camera (model DP21; Olympus).
Assessment of sperm motility and sperm count
Cauda epididymis of adult WT or Pih1d3−/− mice were recovered, immersed in prewarmed HTF medium (2.0 mM CaCl2-2H2O, 2.5 mM glucose, 5.0 mM KCl, 0.4 mM KH2PO4, 0.2 mM MgSO4-7H2O, 100 mM NaCl, 25 mM NaCHO3, 18.5 mM sodium lactate, 0.3 mM sodium pyruvate, 0.2 mM penicillin G sodium salt, 0.3 mM streptomycine sulfate, 4.00 g/liter BSA, and 2 mg/l phenol red [Jin et al., 2007]), and cut into several pieces to let sperm swim out. To assure that sperm stored in the cauda epididymis were all released, tubules of cauda epididymis were squeezed thoroughly with tweezers under microscopy. Sperm were then capacitated by placing the suspension in an incubator (37°C, 5% CO2 in air). The motility of sperm was evaluated by determination of the percentage of motile sperm with the use of a hemocytometer at 1, 2, and 4 h after their release from the cauda epididymis. Two lots of at least 100 sperm were examined for each mouse. Movement of sperm was also traced with the use of TEMA software (Photron), with 10 WT or Pih1d3−/− sperm being monitored over a period of 5 s. The number of sperm in each sample was counted with a hemocytometer. For adult WT and Pih1d3−/− mice, three mice were examined for sperm number, with cauda epididymis on both sides examined separately.
TEM
The cauda epididymis of adult WT and Pih1d3−/− mice were dissected, fixed with 2% paraformaldehyde and 1% glutaraldehyde in PBS for 16 h at 4°C, cut into small pieces, exposed to 2% osmium tetroxide, dehydrated, and processed for TEM according to standard procedures. The tracheas of adult WT and Pih1d3−/− mice were treated with 0.01% Triton X-100 for 5 min, fixed with 2% paraformaldehyde and 2.5% glutaraldehyde in cacodylate buffer and tannic acid (wt/vol; 1:1,000) for 16 h at 4°C, and processed similarly for TEM. All samples were observed with an electron microscope (JEM-1200EX; JEOL).
X-gal staining
X-gal staining of WT or Pih1d3flox/+ embryos was performed as described previously (Saijoh et al., 1999). E8.0 embryos were dissected, fixed in 1% PFA, 0.2% glutaraldehyde, and 0.02% NP40 in PBS for 15 min on ice, and rinsed in PBS/0.02% NP-40 for 5 min, three times. Then embryos were incubated overnight in X-gal staining solution, including 2 mM MgCl2, 4 mM K3Fe(CN)6, 4 mM K4Fe(CN)6 in PBS, and 1 mg/ml X-gal. Mouse testis were stained according to the same procedure, with the exception that the tissue was initially fixed for 2 h on ice and that paraffin sections were prepared after staining with X-gal and were then counterstained with nuclear fast red. Stages of seminiferous tubules were determined according to the position and morphology of spermatogenetic cells (Ahmed and de Rooij, 2009): at stage I–III, early pachytene spermatocytes localized close to the basal membrane; at stage IV–VI, pachytene spermatocytes gradually move toward the lumen; at stage VII, elongated spermatids lined up at the lumen side of the tubule; at stage IX–XI, shape of elongating spermatids began to change. All photos were taken using a microscope (MZ APO [Leica] or Diaphot [Nikon]) and camera (model DP21; Olympus).
RT-PCR
Visceral organs of adult WT mice or Pih1d3−/− mice were dissected and homogenized in Trizol reagent (Ambion). RNAs were recovered according to procedures of Trizol, then purified using the RNeasy Mini kit (QIAGEN). After concentration of yielded RNA was determined, 1 mg of RNA was used for synthesis of first strand of cDNA (PrimeScript RT reagent kit with gDNA Eraser; Takara Bio Inc.). PCR primers for Pih1d3 are 5′-AGCAAGCACTCTACCCTTGA-3′ (forward) and 5′-CTTGAAGGGACCAGGCAGAA-3′ (reverse); primers for E230019M04Rik, 5′-ATTTTCGTCCCCTAGGCTGTG-3′ (forward) and 5′-AGGCGGTCCAATATTCCCAG-3′ (reverse); and primers for β-actin, 5′-TGTGACGTTGACATCCGTAA-3′ (forward) and 5′-TGCTAGGAGCCAGAGCAGTA-3′ (reverse).
In situ hybridization
Testis were fixed overnight at 4°C with 4% paraformaldehyde, exposed consecutively to glucose solutions of 10, 15, 20, and 30%, and embedded in OCT compound for preparation of frozen sections. A plasmid containing a partial fragment of mouse Pih1d3 cDNA including the 3′ untranslated region was used to produce digoxigenin-labeled sense and antisense cRNA probes with the use of T3 and T7 RNA polymerases in separate reactions. Hybridization was performed at 55°C for 16 h, after which the slides were washed, incubated overnight at 4°C with alkaline phosphatase–conjugated antibodies to digoxigenin (Roche), and exposed to nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate for detection of the immune complexes. The sections were counterstained with nuclear fast red and mounted under glass coverslips with 2.5% 1,4-diazabicyclo[2.2.2]octane, 0.02% NaN3, and 50% glycerol in PBS. All photos were taken using a microscope (Diaphot; Nikon) and camera (model DP21; Olympus).
Generation of antibodies to Pih1d3 and immunostaining
Antibodies to Pih1d3 were prepared by injection of rabbits with a peptide corresponding to amino acids 201–218 (ETLEVRMTVORDLDFNIS) of the mouse protein. For immunofluorescence staining of the testis for Pih1d3 and Dnaic2, frozen sections were prepared as described above for in situ hybridization. Antigen retrieval was performed by boiling sections for 5 min in 10 mM citrate buffer (pH 6.0) for staining of Dnaic2. The sections were washed three times in PBS containing 0.1% Triton X-100, exposed to 20% sheep serum for 1 h at room temperature, incubated overnight at 4°C with mouse antibody to DNAI2 (1:100 dilution; M01, clone IC8; Abnova) or rabbit antibody to Pih1d3 (1:100 dilution), and washed again before incubation for 1 h at room temperature with Alexa Fluor 568–conjugated goat antibody to mouse IgG (Molecular Probes) or Alexa Fluor 488–conjugated goat antibody to rabbit IgG (Molecular Probes) counterstaining with DAPI. For immunofluorescence staining of sperm, cells collected from the cauda epididymis were fixed with 4% paraformaldehyde for 30 min at room temperature, transferred to slides, dried, and stored at −80°C until further analysis. The sperm were then treated with 0.1% Triton X-100 for 10 min at room temperature, exposed to 0.5% dried skim milk for 1 h at room temperature, and incubated consecutively overnight at 4°C with antibodies to Dnaic2 (1:100 dilution) and for 1 h at room temperature with Alexa Fluor 568–conjugated goat antibodies to rabbit IgG. All images were acquired with a confocal microscope (Fluoview FV 1000; Olympus). For immunohistochemical analysis of cleaved caspase-3, paraffin-embedded sections of the testis were prepared, subjected to antigen retrieval by boiling in 10 mM citrate buffer (pH 6.0) for 5 min, and treated consecutively at room temperature with 0.1% Triton X-100 for 10 min, 3% H2O2 for 30 min, and 5% BSA for 1 h. The sections were then incubated overnight at 4°C with rabbit antibodies to the cleaved form of caspase-3 (1:100 dilution; Cell Signaling Technology), after which immune complexes were detected with HRP-conjugated goat antibodies to rabbit IgG (ImmPRESS reagent kit; Vector Laboratories) and DAB (Dojindo).
Immunoblot analysis and immunoprecipitation
For immunoblot analysis, protein extracts were prepared from sperm or visceral organs with radioimmunoprecipitation (RIPA) buffer and subjected to SDS-PAGE. The separated proteins were transferred to a polyvinylidene difluoride membrane (GE Healthcare) and exposed to primary antibodies, and immune complexes were detected with the use of HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.; or R&D Systems) and ECL reagents (GE Healthcare). The primary antibodies included those to Pih1d3, DNAI2 (M01, clone IC8; Abnova), DNAI1 (SAB4501181; Sigma-Aldrich), Dnali1/DNALI1 (sc-160296; Santa Cruz Biotechnology, Inc.), Dnah7/DNAH7 (sc-167657; Santa Cruz Biotechnology, Inc.), Dnah9/DNAH9 (Omran et al., 2008; Matsuo et al., 2013), and α-tubulin (DM1A; Sigma-Aldrich) and β-actin (sc-130656; Santa Cruz Biotechnology, Inc.). The antibody to Dnah9 was raised in rabbits against a peptide corresponding to amino acid residues 1004–1352 of the mouse Dnah9 protein. For coimmunoprecipitation analysis, protein extracts of testis were prepared in a solution containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.8% NP-40, 1 mM PMSF, and a protease inhibitor cocktail (Roche). The extracts were incubated overnight with rotation at 4°C with antibodies to Pih1d3 or control rabbit IgG, after which Dynabeads G (Invitrogen) were added and the mixtures were incubated for an additional 4 h. The resulting immunoprecipitates were separated by SDS-PAGE and subjected to immunoblot analysis with antibodies to Pih1d3, HSP70-(610607; BD), HSP90 (610418; BD), DNAI2/Dnaic2 (M01, clone IC8; Abnova), DNAAF1/Dnaaf1(sc-133762; Santa Cruz Biotechnology, Inc.), Dnaaf2 (rabbit anti–mouse Ktu antibody raised against a full-length mouse Ktu protein; Omran et al., 2008; Matsuo et al., 2013), and DNAAF3-(AP11356c; Abgent). Co-immunoprecipitation of Dnaic2 with Dnaic1, Hsp70, and Hsp90 was performed using the same method and same antibodies except that the anti-DNAIC1 antibody (12756–1-AP; Proteintech) was used.
Online supplemental material
Fig. S1 shows strategy for generation of Pih1d3−/− mice. Fig. S2 shows motile cilia in the trachea and brain of Pih1d3−/− mouse. Fig. S3 shows spermatogenesis in seminiferous tubules of Pih1d3−/− mice Fig. S4 shows amino acid sequence alignment of four PIH1 proteins. Video 1 shows movies of WT sperm. Video 2 shows movies of Pih1d3−/− sperm. Video 3 shows movies of WT tracheas. Video 4 shows movies of Pih1d3−/− tracheas. Table S1 lists testis weight and sperm count of WT and Pih1d3−/− mice. Table S2 lists frequency of abnormal seminiferous tubules in Pih1d3−/− mice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201304076/DC1.
Supplementary Material
Acknowledgments
We thank Y. Ikawa and H. Nishimura for technical assistance.
This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan; by Core Research for Evolutional Science and Technology (CREST) of the Japan Science and Technology Corporation (JST); and by “Nanotechnology Platform” (project no. 12024046) of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Abbreviations used in this paper:
- DNAAF
- dynein axonemal assembly factor
- H&E
- hematoxylin-eosin
- HC
- heavy chain
- IC
- intermediate chain
- IDA
- inner dynein arm
- LC
- light chain
- ODA
- outer dynein arm
- TEM
- transmission electron microscopy
- WT
- wild type
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
References
- Ahmed E.A., de Rooij D.G. 2009. Staging of mouse seminiferous tubule cross-sections. Methods Mol. Biol. 558:263–277 10.1007/978-1-60761-103-5_16 [DOI] [PubMed] [Google Scholar]
- Duquesnoy P., Escudier E., Vincensini L., Freshour J., Bridoux A.M., Coste A., Deschildre A., de Blic J., Legendre M., Montantin G., et al. 2009. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 85:890–896 10.1016/j.ajhg.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M., Olbrich H., Horvath J., Wildhaber J.H., Zariwala M.A., Kennedy M., Knowles M.R., Omran H. 2005. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 171:1343–1349 10.1164/rccm.200411-1583OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok A.K., Wang H., Katayama A., Aihara M.S., Allen R.D. 1994. 22S axonemal dynein is preassembled and functional prior to being transported to and attached on the axonemes. Cell Motil. Cytoskeleton. 29:215–224 10.1002/cm.970290304 [DOI] [PubMed] [Google Scholar]
- Fowkes M.E., Mitchell D.R. 1998. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell. 9:2337–2347 10.1091/mbc.9.9.2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales F.A., Zanchin N.I., Luz J.S., Oliveira C.C. 2005. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J. Mol. Biol. 346:437–455 10.1016/j.jmb.2004.11.071 [DOI] [PubMed] [Google Scholar]
- Jin J., Jin N., Zheng H., Ro S., Tafolla D., Sanders K.M., Yan W. 2007. Catsper3 and Catsper4 are essential for sperm hyperactivated motility and male fertility in the mouse. Biol. Reprod. 77:37–44 10.1095/biolreprod.107.060186 [DOI] [PubMed] [Google Scholar]
- Kobayashi D., Takeda H. 2012. Ciliary motility: the components and cytoplasmic preassembly mechanisms of the axonemal dyneins. Differentiation. 83:S23–S29 10.1016/j.diff.2011.11.009 [DOI] [PubMed] [Google Scholar]
- Kramer-Zucker A.G., Olale F., Haycraft C.J., Yoder B.K., Schier A.F., Drummond I.A. 2005. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development. 132:1907–1921 10.1242/dev.01772 [DOI] [PubMed] [Google Scholar]
- Loges N.T., Olbrich H., Becker-Heck A., Häffner K., Heer A., Reinhard C., Schmidts M., Kispert A., Zariwala M.A., Leigh M.W., et al. 2009. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 85:883–889 10.1016/j.ajhg.2009.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M., Shimada A., Koshida S., Saga Y., Takeda H. 2013. The establishment of rotational polarity in the airway and ependymal cilia: analysis with a novel cilium motility mutant mouse. Am. J. Physiol. Lung Cell. Mol. Physiol. 304:L736–L745 10.1152/ajplung.00425.2012 [DOI] [PubMed] [Google Scholar]
- Mitchison H.M., Schmidts M., Loges N.T., Freshour J., Dritsoula A., Hirst R.A., O’Callaghan C., Blau H., Al Dabbagh M., Olbrich H., et al. 2012. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 44:381–389: S1–S2 10.1038/ng.1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omran H., Kobayashi D., Olbrich H., Tsukahara T., Loges N.T., Hagiwara H., Zhang Q., Leblond G., O’Toole E., Hara C., et al. 2008. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 456:611–616 10.1038/nature07471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Mead K. 1997. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA. 94:4457–4462 10.1073/pnas.94.9.4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Diener D.R., Geimer S., Cole D.G., Rosenbaum J.L. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255–266 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J.L., Child F.M. 1967. Flagellar regeneration in protozoan flagellates. J. Cell Biol. 34:345–364 10.1083/jcb.34.1.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J.L., Witman G.B. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813–825 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Rosenbaum J.L., Moulder J.E., Ringo D.L. 1969. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell Biol. 41:600–619 10.1083/jcb.41.2.600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijoh Y., Adachi H., Mochida K., Ohishi S., Hirao A., Hamada H. 1999. Distinct transcriptional regulatory mechanisms underlie left-right asymmetric expression of lefty-1 and lefty-2. Genes Dev. 13:259–269 10.1101/gad.13.3.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Amsterdam A., Pazour G.J., Cole D.G., Miller M.S., Hopkins N. 2004. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 131:4085–4093 10.1242/dev.01240 [DOI] [PubMed] [Google Scholar]
- Takaki E., Fujimoto M., Nakahari T., Yonemura S., Miyata Y., Hayashida N., Yamamoto K., Vallee R.B., Mikuriya T., Sugahara K., et al. 2007. Heat shock transcription factor 1 is required for maintenance of ciliary beating in mice. J. Biol. Chem. 282:37285–37292 10.1074/jbc.M704562200 [DOI] [PubMed] [Google Scholar]
- Wheatley D.N., Wang A.M., Strugnell G.E. 1996. Expression of primary cilia in mammalian cells. Cell Biol. Int. 20:73–81 10.1006/cbir.1996.0011 [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Hirono M., Kamiya R. 2010. Discrete PIH proteins function in the cytoplasmic preassembly of different subsets of axonemal dyneins. J. Cell Biol. 190:65–71 10.1083/jcb.201002081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R.L., Boone C., Hughes T.R., et al. 2008. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180:563–578 10.1083/jcb.200709061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.