Abstract
Assigning molecular functions and revealing dynamic connections between large numbers of partially characterized proteins in regulatory networks are challenges in systems biology. We showed that functions of signaling proteins can be discovered with a differential equations model of the underlying signaling process to extract specific molecular parameter values from single-cell, time-course measurements. By analyzing the effects of 250 small interfering RNAs on Ca2+ signals in single cells over time, we identified parameters that were specifically altered in the Ca2+ regulatory system. Analysis of the screen confirmed known functions of the Ca2+ sensors STIM1 (stromal interaction molecule 1) and calmodulin and of Ca2+ channels and pumps localized in the endoplasmic reticulum (ER) or plasma membrane. Furthermore, we showed that the Alzheimer’s disease–linked protein presenilin-2 and the channel protein ORAI2 prevented overload of ER Ca2+ and that feedback from Ca2+ to phosphatidylinositol 4-kinase and PLCδ (phospholipase Cδ) may regulate the abundance of the plasma membrane lipid PI(4,5)P2 (phosphatidylinositol 4,5-bisphosphate) to control Ca2+ extrusion. Thus, functions of signaling proteins and dynamic regulatory connections can be identified by extracting molecular parameter values from single-cell, time-course data.
INTRODUCTION
Mammalian signal transduction relies on feedback-connected regulatory networks that integrate cues from both the inside and outside of cells to control specific cell functions. The quest to assign molecular functions and to reveal dynamic connections between large numbers of putative or partially characterized proteins in such a network is formidable because only a limited number of molecular parameters can be monitored in live cells. Here, we exploited the fact that a measured time course of a cell’s response is differentially shaped by multiple underlying molecular parameters, such that the response trajectory reflects changes in internal states and activities that are otherwise difficult to measure. Extraction of hidden parameters from time-resolved measurements using ordinary differential equations has been applied in engineering. In biology, this parameter extraction approach has been used to interpret population dynamics, such as the kinetics of viral load (1) or bacterial growth (2), and to analyze phosphorylation kinetics from bulk cell measurements (3). However, the comprehensive use of parameter estimation in an entire signaling system is challenged by potentially incomplete model structures and the need to identify experimental protocols that sufficiently constrain model parameters (4). Moreover, the reaction rates of a signaling system often depend nonlinearly on the concentrations of signaling molecules that vary considerably from cell to cell. Therefore, population-averaged time courses not only blunt characteristic features of single-cell responses, such as the steepness, curvature, or peak time of a signaling trajectory, but the combination of nonlinearity with in-herent variability also precludes the accurate representation of a population-averaged signal trajectory by a molecular kinetic model. Therefore, we asked whether a kinetic model can instead be constrained by analyzing large numbers of variable single-cell trajectories. Such a strategy is becoming broadly applicable with the increased availability of automated live-cell imaging and with the availability of massively parallel computing of numerically stable algorithms for parameter estimation, such as the multiple-shooting method (5), which we used here. Combined with systematic small interfering RNA (siRNA) perturbations, this model-based analysis of single-cell dynamics allowed us to identify molecular roles of putative regulatory proteins and to reconstruct control principles of a complex signaling system using a single scalable assay.
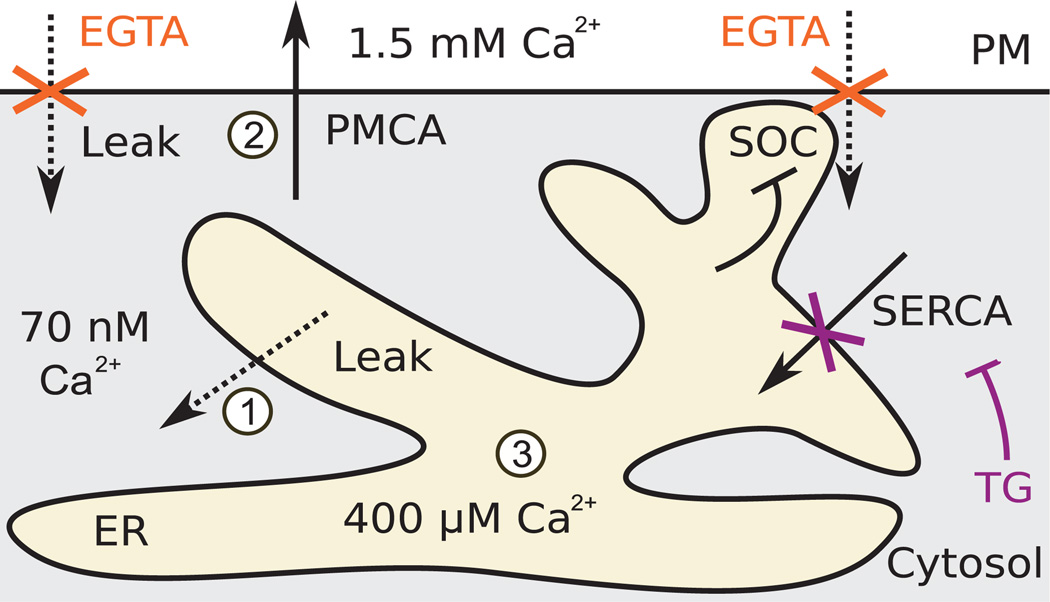
We applied this approach to mammalian Ca2+ signaling because of the documented complexity of regulation that governs a core system of pumps and channels (Fig. 1). This core part of the cellular Ca2+ system can be engaged by numerous types of receptors that open Ca2+ channels in the plasma membrane (PM) or endoplasmic reticulum (ER) (6). PM Ca2+ pumps (PMCA1, PMCA2, PMCA3, PMCA4) extrude Ca2+ to the outside, maintaining a basal cytosolic concentration of ~70 nM against an extracellular concentration of 1.5mM, and ER Ca2+ pumps (SERCA1, SERCA3,SERCA3) load Ca2+ into the ER to ~400 µM, which is available for receptor-triggered release through inositol trisphosphate–sensitive Ca2+ channels [IP3Rs (inositol 1,4,5-trisphosphate receptors)]. Both types of pumps counter the effect of leak through constitutively active Ca2+ leak channels, as well as Ca2+ signals arising from activation of channels in the ER and the PM. The ER and PM are connected by feedback through a family of Ca2+-binding STIM (stromal interaction molecule) proteins that sense decreased ER Ca2+ concentrations to induce PM Ca2+ influx through activation of ORAI Ca2+ channels [store-operated Ca2+ influx (SOC)] (7).
Fig. 1. Schematic of the core Ca2+ regulatory system.
PM Ca2+ pump PMCA maintains a large gradient of Ca2+ across the PM. Ca2+ pump SERCA in the ER membrane fills an ER Ca2+ store that is used in rapid signaling events. Both pump proteins maintain these gradients against the effects of leak through regulated and constitutively active Ca2+ channels. Depletion of the ER Ca2+ store triggers influx of Ca2+ from the outside in a process termed SOC, which is inhibited by ER Ca2+. Our experimental approach combined thapsigargin (TG) to block SERCA with EGTA to chelate external Ca2+, thus blocking entry of Ca2+ from the outside. Dashed arrows indicate passive, but possibly regulated, leak. Solid arrows indicate ATP (adenosine 5′-triphosphate)–driven pump activities. Numbers in white circles reference model terms in Fig. 2D.
RESULTS
Quantitative model to probe the Ca2+ regulatory system
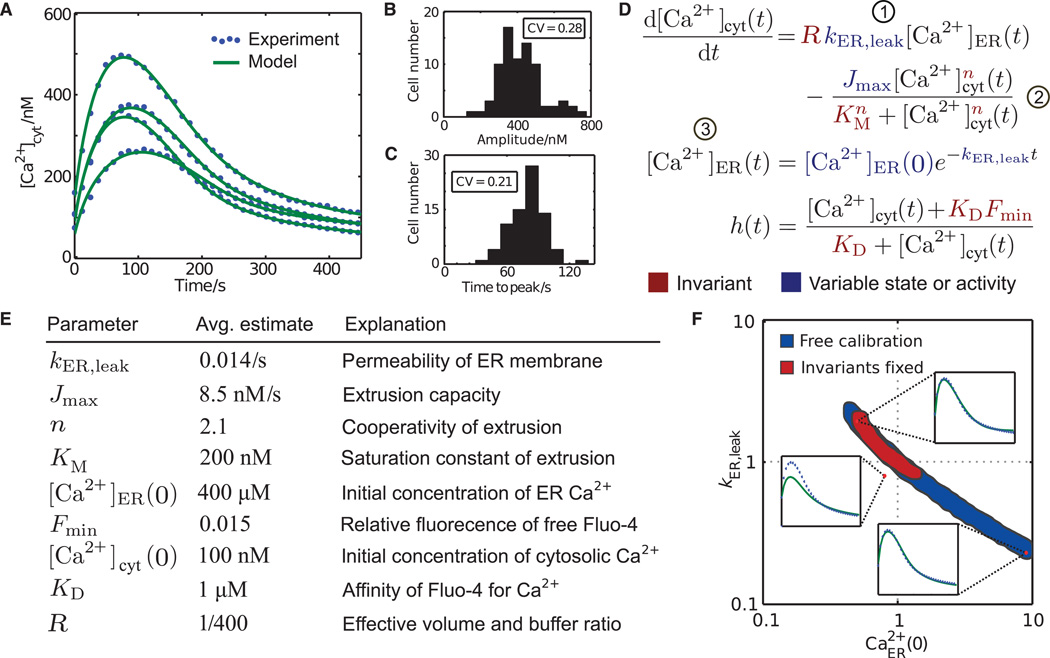
We rapidly chelated extracellular Ca2+ with EGTA to prevent Ca2+ influx and added thapsigargin to inhibit ER SERCA pumps (8), and then using the cytosolic Ca2+ indicator Fluo-4, we monitored the resulting release of Ca2+ from ER Ca2+ stores into the cytoplasm and its clearance out of the cell (Fig. 2A). For each cell, we also measured SOC (kSOC) by addition of external Ca2+ (7) at the end of the experiment. The response of individual cells after EGTA and thapsigargin treatment was highly variable, as shown by substantial coefficients of variation for amplitudes and peak times (Fig. 2, B and C).
Fig. 2. Extraction of molecular parameter values from time-resolved, single-cell Ca2+ measurements.
(A) Single-cell Ca2+ responses upon addition of EGTA and thapsigargin. (B and C) Cell-to-cell variability of amplitude (B) and peak timing (C). CV, coefficient of variation. (D) Differential equations model that was used to infer parameter values. Numbers in circles denote terms illustrated in Fig. 1. (E) Population averages of single-cell parameter estimates. KD and R were linear with [Ca2+]cyt or [Ca2+]ER(0), respectively, and thus were always fixed (see Materials and Methods). (F) Exemplary slice through the seven-dimensional parameter space in which parameter sets were projected onto the plane spanned by the axes of ER leak (kER,leak) and the initial concentration of ER Ca2+ [Ca2+ER(0)]. Blue shading indicates the range of fold changes away from a simulated “true” parameter set in the center for which nearly identical fits could be obtained when all parameters were variable. Red shading shows the reduced range of parameters when the invariant parameters n, KM, and Fmin were constrained. The upper right and lower left insets show examples of parameter combinations that fit the same data. The upper left inset shows an example where this is not the case despite much smaller deviation from the true parameter set than the other two example parameter combinations.
We developed a quantitative model that describes the resulting single-cell responses (change in the concentration of cytosolic Ca2+ [Ca2+]cyt) as the result of a leak of Ca2+ out of the ER into the cytosol at a rate kER,leak and the pumping of Ca2+ from the cytoplasm to the outside with a capacity Jmax, an affinity KM, and cooperativity n (9). We used the measured Fluo-4 intensity to derive [Ca2+]cyt, assuming saturable Ca2+ binding and a residual fluorescence Fmin in the absence of Ca2+ (Fig. 2D).
Model calibration using a multiple-shooting Gauss-Newton–type algorithm (5)with all seven parameters left open resulted in close agreement between model output and individual single-cell trajectories (Fig. 2A). Moreover, the average parameter estimates obtained from large numbers of individual single-cell traces agreed well with previously reported values (Fig. 2E; see Materials and Methods). However, the parameters of individual cells were poorly defined and co-dependent (Fig. 2F and fig. S1, A and B). For example, two of the insets in Fig. 2F (upper right and lower left) show examples in which parameter combinations within the blue-shaded region correspond to virtually identical fits despite more than 10-fold differences in parameter values. Additionally, even small deviations from this region generated highly discrepant predictions (Fig. 2F, upper left insert). We considered that the regulated parameters would be better constrained (that is, the region in parameter space that matched the response trajectory would be more limited) if the invariant parameters n, KM, and Fmin were constrained independently on the basis of previous knowledge (Fig. 2F, red-shaded region). However, the challenge of such an approach is that even small errors in fixing the invariant parameters would preclude any possible fit, rendering all other parameters inaccessible for analysis [fig. S1B; see also (4)].
The uncertainty of parameter estimates can also be described by a local approximation of the covariance matrix based on the sensitivity of measurement predictions to parameter values (10). Such calculations showed that a predominant direction, or eigenvector, in parameter space exists, along which parameter values are uncertain (fig. S1C). This direction spans several of the parameter axes and constitutes, for example, the negative correlation of kER,leak and [Ca2+]ER(0) seen in Fig. 2F (indicated in fig. S1C by a difference in sign of the corresponding components) and correlations of these parameters with invariant parameters, such as Fmin. This interdependence explains why previous knowledge of one invariant parameter can constrain estimates of other parameters. In particular, such calculations predicted that if n, KM, and Fmin were known, then the regulated parameters Jmax, Ca2+ER(0), and kER,leak would be well determined even if PMCA or, to a lesser degree, SERCA was knocked down (fig. S1, D and E).
Predicting protein function from single-cell, time-resolved siRNA data using a kinetic model
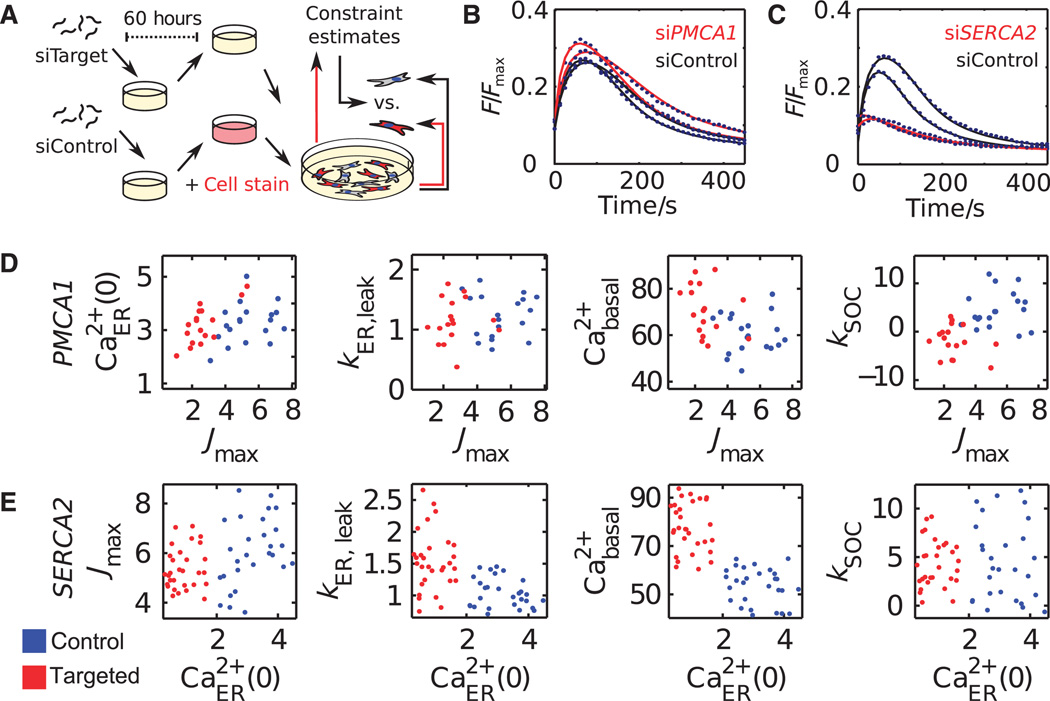
We established cocultures of human embryonic kidney (HEK) 293T cells transfected with siRNAs to knock down proteins of interest and fluorescently marked control cells and used these cocultured cells to obtain model-derived estimates for n, KM, and Fmin. We analyzed large numbers of single-cell responses of control cells cultured in conditions identical to those of the siRNA-targeted cells. Although poorly defined when estimated from individual cells, we used the average estimates of these invariant parameters obtained from hundreds of individual control cells per well to constrain a second round of single-cell parameter estimation in both the control and the siRNA-targeted cells (Fig. 3A, see Materials and Methods). Overlaying the model trajectories on the experimental single-cell responses after PMCA1 or SERCA2 knockdown (Fig. 3, B and C) showed that the constrained model retained its ability to recapitulate variable single-cell responses and that it could do so in different siRNA perturbation backgrounds.
Fig. 3. Mapping of siRNA effects into a molecular parameter space of protein function.
(A) Coculture assay used to constrain the parameter estimation problem. (B and C) Single-cell calcium traces of control cells and cells expressing siRNA targeting PMCA1 (B) or SERCA2 (C). (D and E) Single-cell parameter estimates from cells in which PMCA1 (D) or SERCA2 (E) was knocked down (red) or from control cells (blue). Center 80% values from each condition are shown.
Moreover, decomposing the single-cell time courses into distinct molecular parameters correctly assigned the known cellular roles to the PMCA and SERCA Ca2+ pumps. Only Jmax was reduced in cells in which PMCA1 was knocked down (Fig. 3D). The PMCA1 knockdown cells also had increased basal [Ca2+]cyt (Fig. 3D, third graph) and showed adaptive down-regulation of SOC (Fig. 3D, fourth graph) as previously observed in insect cells (11). Likewise, knockdown of SERCA2 predominantly reduced the ER load [Ca2+]ER(0), but not Jmax, and also resulted in increased basal [Ca2+]cyt due to the feedback activation of SOC (Fig. 3E). Together, this showed that parameter shifts induced by siRNAs targeting proteins that function in the regulatory network can directly predict protein function and provide insight into mechanisms of adaptation.
Identifying function from dynamic siRNA perturbation data
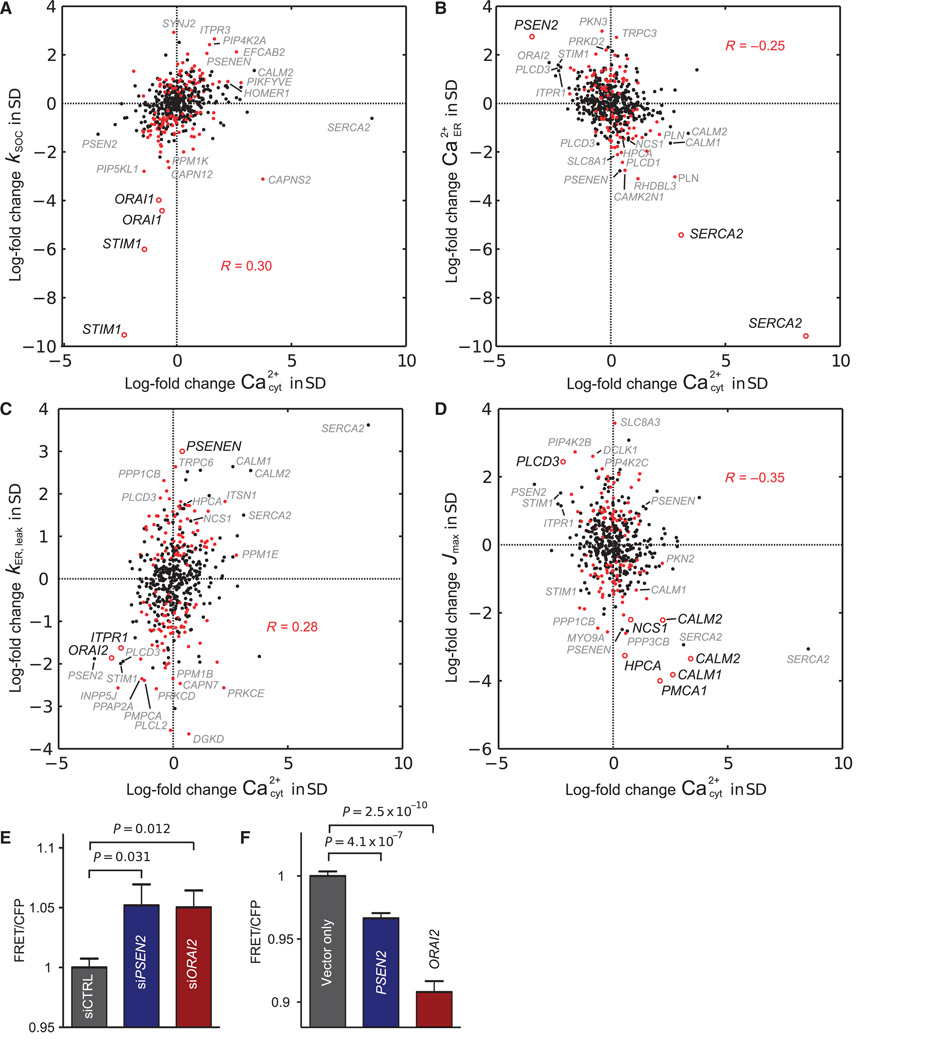
We extended this analysis to an siRNA set targeting 250 known or putative regulators of Ca2+ (table S1).We plotted the effects of the siRNAs on kSOC (Fig. 4A), [Ca2+]ER(0) (Fig. 4B), kER,leak (Fig. 4C), or Jmax (Fig. 4D) against the siRNA-evoked changes of basal [Ca2+]cyt that we measured before adding thapsigargin and EGTA. We calculated all changes as log-fold changes relative to the control cells that we cultured in each well (table S2), median-centered and normalized by the SD along each dimension per independent replicate screen. When analyzing the effect of the perturbation set on kSOC, we observed a significant correlation with basal [Ca2+]cyt (Fig. 4A). The two canonical SOC molecules STIM1 and ORAI1 were at the lower left of the distribution, indicating that knockdown of either of these proteins reduced basal [Ca2+]cyt and reduced SOC compared to control cells. In contrast, we observed an anticorrelation between the ER Ca2+ load and [Ca2+]cyt (Fig. 4B). Our finding that not only reductions in [Ca2+]ER but also increases led to opposing changes in [Ca2+]cyt indicated that SOC also operates in cells at rest (12).
Fig. 4. Five-dimensional parameter analysis of an siRNA screen and validation of model predictions for the roles of PSEN2 and ORAI2.
(A to D) Projections of the five-dimensional parameter readout onto the planes of basal cytosolic Ca2+ and (A) store-operated Ca2+ influx kSOC, (B) initial ER load Ca2+ER(0), (C) ER Ca2+ leak kER,leak, and (D) extrusion capacity Jmax are shown as log-fold changes relative to control cells, median-centered and normalized per replicate by the SD σ along each dimension. Red data points denote siRNA effects that changed the parameters predominantly along the respective dimension shown in (A), (B), (C), or (D). Parameter changes labeled with the name of the targeted gene were outliers with |σ| > 2.1 or were predicted to have functional changes of |σ| > 1 due to secondary effects or because they were one of a pair of replicates that was below the threshold. Parameter changes further analyzed and discussed in the text are indicated by open red circles and are labeled with emphasis in the projection where the primary change in function was observed. (E and F) FRET ratios showing differences in resting ER Ca2+ concentrations in cells when PSEN2 or ORAI2 was knocked down (E; n = 12) or overexpressed (F; n ≥ 22). Shown are Bonferroni-corrected P values from two-tailed Student’s t test; error bars show SEM.
Although knockdown of the ER Ca2+ pump SERCA2 produced the largest decrease in ER Ca2+, one of the presenilin-2 (encoded by PSEN2) knockdowns produced a large increase in ER Ca2+ load. Presenilins are the catalytically active constituent of the γ-secretase complex (13), and mutations in PSEN2 are associated with familial cases of Alzheimer’s disease (14). Loss of presenilin function has been proposed to either increase or decrease [Ca2+]ER (15–17), or not to change [Ca2+]ER (18). Given the current controversy, we tested the predicted physiological role of PSEN2 in regulating ER Ca2+ load using a fluorescence resonance energy transfer (FRET) probe based on the design of D1ER to measure changes in ER Ca2+ directly (11). Knockdown of PSEN2 significantly increased ER Ca2+ load when compared to control cells (Fig. 4E and fig. S2), and overexpression of PSEN2 resulted in the opposite effect (Fig. 4F).
Knockdown of presenilin enhancer 2 homolog (encoded by PSENEN), which is required for the proteolytic processing of presenilins (19), yielded a large increase in kER,leak, whereas knockdown of PSEN2 decreased kER,leak (Fig. 4C). Such a role of PSENEN in reducing kER,leak may be due to its function in reducing the amount of unprocessed PSEN2 (19), because the unprocessed form of presenilins contributes to ER Ca2+ leak (15, 17). However, knock-down of the ER Ca2+ release channel IP3R1 (encoded by ITPR1) or ORAI2 also had strong effects on kER,leak (Fig. 4C). Although the identification of IP3R1 is consistent with previous observations of a basal activity of IP3Rs (16), a function for ORAI2 in ER Ca2+ leak has previously not been described. We used the ER Ca2+ FRET probe to measure changes of ER Ca2+ in cells in which ORAI2 was knocked down or overexpressed. Knockdown of ORAI2 significantly increased [Ca2+]ER (Fig. 4E), whereas overexpression had the opposite effect (Fig. 4F). Because ORAI2 fails to rescue SOC in ORAI1 knockout cells (20),we confirmed further that the observed decrease in [Ca2+]ER was not due to a dominant-negative effect of the overexpressed ORAI2 on SOC entry by measuring basal [Ca2+]cyt. In cells overexpressing ORAI2, basal [Ca2+]cyt was increased as expected for an ER Ca2+ leak channel that triggers SOC as a result of lowering of [Ca2+]ER under basal conditions (fig. S3). Thus, our findings suggested that the ER Ca2+ leak may result from the cumulative conductance of many proteins, including unprocessed presenilins, basally active IP3Rs, and Ca2+ channels, such as ORAI2, that conduct Ca2+ as they progress through the ER en route to the PM.
The effect of the perturbation set on the inferred extrusion capacity Jmax was anti-correlated with basal [Ca2+]cyt (Fig. 4D), with multiple targeted proteins affecting Ca2+ extrusion. Large parameter shifts were caused by knockdown of the PMCA isoform encoded by PMCA1 or knockdown of calmodulin isoforms encoded by CALM1 or CALM2, consistent with a critical role of calmodulin in PMCA activation (6). We identified two additional Ca2+-binding proteins, hippocalcin (encoded by HPCA) and neuronal calcium sensor-1 (NCS1; a homolog of the fly and yeast Ca2+-binding protein frequenin), as putative regulators of Ca2+ extrusion. Although a role for hippocalcin in Ca2+ extrusion from hippocampal neurons has been suggested previously (21), Saccharomyces cerevisiae frequenin and NCS1 have been shown to function as Ca2+-activated positive regulators of phosphatidylinositol (PI) 4-kinase, which converts PI to PI(4)P (phosphatidylinositol 4-phosphate) (22, 23).
Identifying a role for PI(4,5)P2 in regulation of Ca2+ extrusion
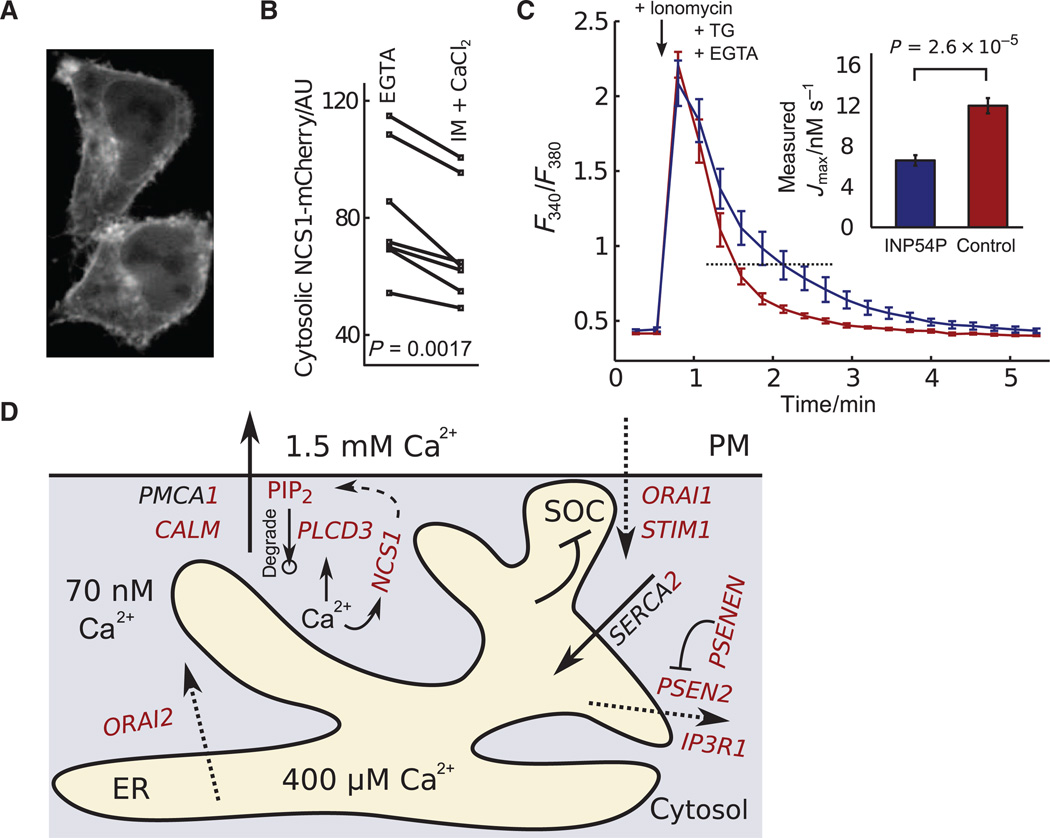
Consistent with a regulatory role for NCS1 at the PM, we observed a small but significant increase in the PM localization of an NCS1-mCherry fusion protein after cytosolic Ca2+ was increased by addition of ionomycin and extracellular Ca2+ (Fig. 5, A and B, and movie S1). Because PI(4)P is the main precursor of PI(4,5)P2 (phosphatidylinositol 4,5-bisphosphate), it was intriguing to also identify phospholipase Cδ3 (PLCδ3; encoded by PLCD3), which hydrolyzes PI(4,5) lipids, as a strong regulator opposing Ca2+ extrusion (Fig. 4D). PI(4,5)P2 is required in reconstituted membranes for purified PMCA to be active (24); thus, the effects of knocking down NCS1 or PLCδ3 may result from altering PMPI(4,5)P2 and thereby affecting PMCA activity.
Fig. 5. PI(4,5)P2 regulates Ca2+ extrusion and a more detailed model of the Ca2+ regulatory system.
(A) PM localization of NCS1-mCherry increased when cytosolic Ca2+ was raised (see also movie S1). (B) Quantification comparing NCS1 localization in low and high cytosolic Ca2+ (P value from two-tailed paired Student’s t test, n = 7). AU, arbitrary unit. (C) Fura-2 measurements of Ca2+ extrusion after rapid release from the ER by ionomycin. Inset shows quantification of the effect of reducing PI(4,5)P2 abundance by INP54P overexpression on Ca2+ extrusion (P value from two-tailed Student’s t test, n≥6; error bars show SEM). (D) More detailed molecular model of the Ca2+ regulatory systeminHEK293T cells. Specific isoforms and regulators are shown with their predicted functions derived from our computational analysis of time-course differences in the siRNA perturbation experiments. The circle below PIP2 represents a sink resulting from the degradation of this molecule. PIP2, PI(4,5)P2.
To test whether PI(4,5)P2 enhanced PMCA activity in a cellular context, we lowered the concentration of PI(4,5)P2 by expressing a fusion protein containing the catalytic domain of the PI(4,5)P2 5-phosphatase (INP54P) from S. cerevisiae in HeLa cells. Indeed, Ca2+ that was rapidly released from the ER by addition of ionomycin and thapsigargin in the presence of extracellular EGTA was eliminated at a twofold slower rate from cells expressing INP54P than from control cells (Fig. 5C). Thus, the regulation of PMCA activity by PI(4,5)P2 revealed a network with two feedbacks by which Ca2+-mediated activation of NCS1 increases and activation of PLCδ3 reduces the concentration of PI(4,5)P2.
DISCUSSION
Our understanding of signal transduction systems is challenged by a high degree of variation of signaling responses within a cell population and by the limited number of reporters available to track specific signaling events over time. Our study showed that a single signaling reporter, combined with rapid perturbation and a mechanistically explicit differential equations model, can be used to determine multiple molecular parameters from single-cell data even for a complex signaling system.
The value of single-cell traces over population-averaged traces for our analysis method is exemplified by considering the extrusion term in our model. We found in our analysis of variability (Fig. 2, A to C) that cells have peak cytosolic Ca2+ concentrations as different as ~600 and ~200 nM shortly after addition of thapsigargin and EGTA. The instantaneous flux rate of extrusion for these cells calculates to 7.73 and 4.25 nM/s, respectively. On average, cytosolic Ca2+ will be extruded out of these cells at a rate of 5.99 nM/s. However, extrusion calculated on the basis of the average concentration of cytosolic Ca2+ at 400 nM calculates to 6.89 nM/s. This discrepancy is due to cooperativity and saturation (Fig. 2D, term labeled “2”) and would result in a substantial error in inferring other values in the same equation, such as the cooperativity of the PM Ca2+ pump or the ER leak rate. Thus, for a nonlinear process with high cell-to-cell variability, only single-cell data can accurately constrain the parameters of a kinetic model.
We combined this approach with systematic siRNA perturbations to reconstruct the regulatory control principles of the Ca2+ homeostatic system and assign molecular roles to putative regulatory proteins. Most notably, our analysis revealed a role of PI(4,5)P2 in controlling the rate of Ca2+ extrusion, confirmed a controversial hypothesis that PSEN2 has a role in preventing ER Ca2+ overload, and provided evidence that ORAI2 functions as a leak channel in the ER (Fig. 5D). The same single cell–based deconvolution approach that we introduce here can likely be used to identify molecular targets of drugs and their effect on dynamic regulatory mechanisms or to distinguish molecular causes of a disease that may differ within a patient population.
MATERIALS AND METHODS
Live-cell imaging
Extracellular buffer (ECB) contained 5 mM KCl, 125 mM NaCl, 1.5 mM CaCl2, 1.5 mM MgCl2, 10 mM d-glucose, and 20 mM Hepes (pH 7.4) and was supplemented freshly with probenecid (0.385 mg/ml) when Ca2+ dyes were used. Cells were loaded with 75 µl of Fluo-4 AM(1 µg/ml) or Fura-2 AM in the presence of F-127 (0.625 µl/ml) [20% solution in dimethyl sulfoxide (DMSO)] for 30 to 45 min, and then replaced with 50 µl of ECB. Hoechst 33342 (1 µg/ml) was in the loading buffer of Fluo-4 to provide a Fluo-4–independent mask for image analysis. Store-depletion traces were evoked by adding 50 µl of 8 mM EGTA and 4 µM thapsigargin in ECB. SOC was assayed 7.5 min later by adding 100 µl of 16 mM CaCl2 and 2 µM thapsigargin in ECB. For the purpose of calibration, Fluo-4 was saturated by adding 200 µl of 10 µMionomycin, 2 µM thapsigargin, and 5 mM CaCl2 at the end of every experiment. Hoechst 33342, Fluo-4 AM, Fura-2 AM, probenecid, F-127, and thapsigargin were from Invitrogen; ionomycin was from EMD Biosciences; and Hepes, DMSO, inorganic salts, and glucose were from Sigma. All Ca2+ imaging was done on an IX5000 automated epi-fluorescence microscope (Molecular Devices) equipped with a 4× S Fluor objective. Filters for Hoechst 33342 were D360/40× to D460/50m; Fluo-4, S470/30× to S510/30; and Fura-2, D340/12×, D360/10×, or D380/12× to D510/80m. For ER Ca2+ imaging by FRET, filters for CFP (cyan fluorescent protein) were S430/25 to 480/40; YFP (yellow fluorescent protein), S500/20 to S535/30; and FRET, S430/25 to S535/30. NCS1-mCherry was imaged on a custom-built spinning disc confocal microscope with a 100-mW, 593.5-nm solid-state laser (Changchun New Industries) with 594/10 excitation and 630/60 emission filters and an Apochromat 63× 1.4 numerical aperture oil objective (Carl Zeiss) mounted in an Axiovert 200M frame (Carl Zeiss) equipped with an Ultra VIEW spinning disk confocal scanner (PerkinElmer) and a CoolSNAP HQ CCD (charge-coupled device) camera (Photometrics).
Cell culture and transfection
NCS1 translocation experiments with HeLa cells were done in four-well Lab-Tek chambered coverslips (Nunc) coated with collagen (Advanced BioMatrix). All other experiments were done in 96-well plates (Corning) also coated with collagen. HEK293T cells were transfected with 75-ng siRNA using 0.2 µl of DharmaFECT I (Dharmacon) per well ~60 hours before imaging. Control cells were stained with 2.5 µM CMTPX (CellTracker Red, Invitrogen) in Opti-MEM I for 45 min, and then washed and quenched with Dulbecco’s modified Eagle’s medium + 10% fetal bovine serum for 45 min before all cells were replated on collagen a few hours before imaging. Cells that were transfected with targeting siRNA were treated identically, except that no CMTPX was mixed into Opti-MEM I. CMTPX had no effect on parameter estimates (fig. S4). Plasmids were transfected into HEK293T and HeLa cells with 3 µl of FuGENE 6 (Roche) per 1 µg of cDNA (complementary DNA) according to the manufacturer’s instructions. For expression of PSEN2 in HEK293T cells or YFP–FKBP (FK505 binding protein)–INP54P in HeLa cells, cells were transfected 24 hours before imaging. For expression of NCS1-mCherry in HeLa cells, cells were transfected 10 hours before imaging. The T1ER construct was co-transfected with siRNA using DharmaFECT Duo (Dharmacon).
cDNA constructs and siRNA
PSEN2 was amplified from the human ORFeome collection by polymerase chain reaction with primers 5′-ACTGCTCGAGATGCTCACATTCATGGCCTCTGAC-3′ (forward) and 5′-CAGTGAATTCTCAGATGTAGAGCTGATGGGAGG-3′ (reverse), cut with Xho I and Eco RI, and ligated into pIRES2-DsRed2-Express (Clontech). ORAI2 was amplified with primers 5′-ACTGCTCGAGATGAGTGCTGAGCTTAACGTGCCT-3′ (forward) and 5′-CAGTGAATTCTCACAAGACCTGCAGGCTGCGC-3′ (reverse), cut with Xho I and Eco RI, and ligated into pIRES2-DsRed2-Express. INP54P was expressed as a fusion construct YFP-FKBP-INP54P, as described in (25). YFP-FKBP served as the control. NCS1 was recombined from the human ORFeome collection into pmCherry-N-DEST/TO. T1ER was described in (11). The sequence of each construct was verified. Diced siRNA was synthesized according to a previously published protocol (26), with bacterial ampR serving as control. Primer sequences are listed in table S1. Synthetic siRNAs targeting PSEN2 with individual sequences 5′-GAGCGAAGCACGUGAUCAU-3′ (siPSEN2–4), 5′-GGAGGACCCUGACCGCUAU-3′ (siPSEN2-3), 5′-GAGCGGACGUCCCUAAUGU-3′ (siPSEN2-2), and 5′-CAUAUUCCCUGCCCUGAUA-3′ (PSEN2-1) and lamin A/C control siRNA(D-001050)were purchased from Dharmacon.
Parameter estimation
To identify the molecular parameters of the model (Fig. 2D) from single-cell traces, we formulated a prediction h(t) for the ionomycin-calibrated Fluo-4 measurements as a function of cytosolic [Ca2+]cyt.
| (1) |
KD is the affinity of Fluo-4 for Ca2+, and Fmin is the fluorescence of Fluo-4 in the absence of Ca2+. KD and R were undetermined and scaled linearly with [Ca2+]cyt or [Ca2+]ER, respectively. Because KD and R had strictly no influence on the model other than scaling other parameters, those variables were always fixed. KD = 1 µM is a widely used estimate for the affinity of Fluo-4 for intracellular Ca2+ (Fig. 2E) (27).
All other parameters p were determined by minimizing the discrepancy between model prediction h(ti; p) and actual Fluo-4 data ηi in the form of a least-squares functional with measurement errors σi:
| (2) |
σi was assumed to be constant at 1% of maximal Fluo-4 fluorescence Fmax. Equation 2 was minimized for each single-cell trace with a multiple-shooting Gauss-Newton–type algorithm implemented by PARFIT (5). The multiple-shooting approach discretizes the model trajectory, allowing it to initialize the model at each node close to the measurements and only then to consider boundary constraints. As a consequence, fast convergence, relative independence of initial guesses, and increased resistance to poor local optima can be observed. The calculations were performed on a computer cluster assembled from 552 Intel E5345 processors with 2208 cores in total.
Representative population estimates shown in Fig. 2F are consistent with previous knowledge: Fmin is often assumed to be Fmin = 1/40 = 0.025. The value of [Ca2+]ER(0) = 400 µM is obtained for a ratio of volume and buffering between ER and cytosol of R = 1/400, consistent with much larger cytoplasmic volume and buffering of Ca2+ than in the ER (28). Without correction for differences in volume and buffering, an effective concentration of 1 µM in the ER is consistent with the typical height of a cytosolic Ca2+ peak induced by rapid release from the ER with ionomycin. KM = 200 nM and n = 2.1 are consistent with previously reported measurements for PMCA (9).
We calculated turnover at the ER membrane as JER = kER,leak [Ca2+]ER(0) = 14 nM/s.
We calculated turnover at the PM by substituting a basal cytosolic calcium concentration of 70 nM for [Ca2+]cyt(t) in:
| (3) |
This model prediction that turnover at the ER is many times faster than at the PM is consistent with the interpretation that intracellular Ca2+ signaling can in the short term be described by rapid uptake and release from Ca2+ stores (29).
To measure JPM, we recorded cytosolic calcium traces after Ca2+ was released from the ER by 1.25 µM ionomycin together with 1 µM thapsigargin to inactivate SERCA, and 5 mM extracellular EGTA. The slope of [Ca2+]cyt was measured where [Ca2+]cyt crossed KM = 200 nM downwards, corresponding to the half-maximal extrusion capacity Jmax/2. This measured Jmax in the range of 10 nM/s was consistent with the parameter estimate from the model (Fig. 2E). kSOC was not a model parameter and was instead measured for each cell as the initial slope of cytosolic Ca2+ when external Ca2+ was added back after store depletion.
Parameter uncertainty
We used the HYPERSPACE package (30) to sample the parameter space over two orders of magnitude around a representative parameter set (Fig. 2F and fig. S1, A and B). In addition, we characterized parameter uncertainty by a local measure based on the sensitivity of the measurement predictions h to the parameter values p. It can be shown that
| (4) |
and is a linear approximation of the parameter covariance matrix with
| (5) |
where Σ are the diagonalized measurement errors σi, and h are the vectorized model predictions at time points ti (10). We used the VPLAN package (10) to compute C in fig. S1C or for different knockdown efficiencies in fig. S1, D and E. Figure S1C shows the eigenvalue spectrum λ of C in unperturbed cells and the orientation of the largest eigenvector in parameter space, suggesting that our model has similar properties with respect to parameter uncertainty compared to all those characterized in (4).
For siRNA experiments, population estimates of Fmin, KM, and n were determined from control cells in coculture with the goal to minimize the influence of experimental variability and systematic errors. For the screen in Fig. 4, we considered that such effects could vary between cells (for example, due to variable abundance of PMCA isoforms or effects of cell shape) and considered imperfect calibration of Fluo-4. To this end, we added regularization terms to the least-squares functional Eq. 2, yielding
| (6) |
and reintroduced Fmax = 1 into Eq. 1, yielding
| (7) |
pk was the parameter Fmin, KM, n, or Fmax regularized to pk,0 (determined each from the control cells of the entire screen, or Fmax,0 = 1, respectively), each with an error estimate σk of 20%.
Supplementary Material
Acknowledgments
T7 RNA polymerase and Giardia Dicer were gifts from B.-O. Park. pmCherry-N-DEST/TO was contributed by A. Hayer. An ORAI2 template was provided by the R. Dolmetsch Lab. We thank J. Schlöder, S. Körkel, and H. G. Bock for advice on the use of PARFIT and VPLAN. J. Ferrell, M. Covert, R. Dolmetsch, and many members of T.M.’s laboratory were helpful with comments. We appreciate practical help by A. Seki and K. Han. Funding: The Bio-X2 cluster was funded by NSF award CNS-0619926 for computational resources. This work was supported by NIH grant GM030179 to T.M. and by the Swedish Society for Medical Research Postdoctoral Fellowship to S.M.
Footnotes
Author contributions: S.B. and T.M. conceived the study; S.B. and S.M. performed the experiments and analyzed the data; and S.B., S.M., and T.M. wrote the manuscript.
Competing interests: T.M. is a member of the scientific advisory board of Molecular Devices, Sunnyvale, CA. All other authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 2.Cairns BJ, Timms AR, Jansen VAA, Connerton IF, Payne RJH. Quantitative models of in vitro bacteriophage-host dynamics and their application to phage therapy. PLoS Pathog. 2009;5:e1000253. doi: 10.1371/journal.ppat.1000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleiman LB, Maiwald T, Conzelmann H, Lauffenburger DA, Sorger PK. Rapid phospho-turnover by receptor tyrosine kinases impacts downstream signaling and drug binding. Mol. Cell. 2011;43:723–737. doi: 10.1016/j.molcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 2007;3:1871–1878. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bock HG, Kostina E, Schlöder JP. Numerical methods for parameter estimation in nonlinear differential algebraic equations. GAMM-Mitteilungen. 2007;30:376–408. [Google Scholar]
- 6.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 7.Carrasco S, Meyer T. STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu. Rev. Biochem. 2011;80:973–1000. doi: 10.1146/annurev-biochem-061609-165311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thastrup O, Cullen PJ, Drøbak BK, Hanley MR, Dawson AP. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2+-ATPase. Proc. Natl. Acad. Sci. U.S.A. 1990;87:2466–2470. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bautista DM, Hoth M, Lewis RS. Enhancement of calcium signalling dynamics and stability by delayed modulation of the plasma-membrane calcium-ATPase in human T cells. J. Physiol. 2002;541:877–894. doi: 10.1113/jphysiol.2001.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer I, Bock HG, Körkel S, Schlöder JP. Numerical methods for optimum experimental design in DAE systems. J. Comput. Appl. Math. 2000;120:1–25. [Google Scholar]
- 11.Abell E, Ahrends R, Bandara S, Park BO, Teruel MN. Parallel adaptive feedback enhances reliability of the Ca2+ signaling system. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14485–14490. doi: 10.1073/pnas.1018266108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li YM, Lai MT, Xu M, Huang Q, DiMuzio-Mower J, Sardana MK, Shi XP, Yin KC, Shafer JA, Gardell SJ. Presenilin 1 is linked with γ-secretase activity in the detergent solubilized state. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6138–6143. doi: 10.1073/pnas.110126897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruts M, Van Broeckhoven C. Presenilin mutations in Alzheimer’s disease. Hum. Mutat. 1998;11:183–190. doi: 10.1002/(SICI)1098-1004(1998)11:3<183::AID-HUMU1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 15.Tu H, Nelson O, Bezprozvanny A, Wang Z, Lee SF, Hao YH, Serneels L, De Strooper B, Yu G, Bezprozvanny I. Presenilins form ER Ca2+ leak channels, a function disrupted by familial Alzheimer’s disease-linked mutations. Cell. 2006;126:981–993. doi: 10.1016/j.cell.2006.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasri NN, Kocks SL, Verbert L, Hébert SS, Callewaert G, Parys JB, Missiaen L, De Smedt H. Up-regulation of inositol 1,4,5-trisphosphate receptor type 1 is responsible for a decreased endoplasmic-reticulum Ca2+ content in presenilin double knock-out cells. Cell Calcium. 2006;40:41–51. doi: 10.1016/j.ceca.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Brunello L, Zampese E, Florean C, Pozzan T, Pizzo P, Fasolato C. Presenilin-2 dampens intracellular Ca2+ stores by increasing Ca2+ leakage and reducing Ca2+ uptake. J. Cell. Mol. Med. 2009;13:3358–3369. doi: 10.1111/j.1582-4934.2009.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shilling D, Mak DO, Kang DE, Foskett JK. Lack of evidence for presenilins as endoplasmic reticulum Ca2+ leak channels. J. Biol. Chem. 2012;287:10933–10944. doi: 10.1074/jbc.M111.300491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T. The role of presenilin cofactors in the γ-secretase complex. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 20.Gwack Y, Srikanth S, Oh-Hora M, Hogan PG, Lamperti ED, Yamashita M, Gelinas C, Neems DS, Sasaki Y, Feske S, Prakriya M, Rajewsky K, Rao A. Hair loss and defective T- and B-cell function in mice lacking ORAI1. Mol. Cell. Biol. 2008;28:5209–5222. doi: 10.1128/MCB.00360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuo Y, Ogura A, Kobayashi M, Masaki T, Furuta Y, Ono T, Takamatsu K. Hippocalcin protects hippocampal neurons against excitotoxin damage by enhancing calcium extrusion. Neuroscience. 2007;145:495–504. doi: 10.1016/j.neuroscience.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Hendricks KB, Wang BQ, Schnieders EA, Thorner J. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat. Cell Biol. 1999;1:234–241. doi: 10.1038/12058. [DOI] [PubMed] [Google Scholar]
- 23.De Barry J, Janoshazi A, Dupont JL, Procksch O, Chasserot-Golaz S, Jeromin A, Vitale N. Functional implication of neuronal calcium sensor-1 and phosphoinositol 4-kinase-b interaction in regulated exocytosis of PC12 cells. J. Biol. Chem. 2006;281:18098–18111. doi: 10.1074/jbc.M509842200. [DOI] [PubMed] [Google Scholar]
- 24.Missiaen L, Raeymaekers L, Wuytack F, Vrolix M, de Smedt H, Casteels R. Phospholipid- protein interactions of the plasma-membrane Ca2+-transporting ATPase. Biochem. J. 1989;263:687–694. doi: 10.1042/bj2630687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abe N, Inoue T, Galvez T, Klein L, Meyer T. Dissecting the role of PtdIns(4,5)P2 in endocytosis and recycling of the transferrin receptor. J. Cell Sci. 2008;121:1488–1494. doi: 10.1242/jcs.020792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers JW, Jones JT, Meyer T, Ferrell JE., Jr Recombinant dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- 27.Thomas D, Tovey SC, Collins TJ, Bootman MD, Berridge MJ, Lipp P. A comparison of fluorescent Ca2+ indicator properties and their use in measuring elementary and global Ca2+ signals. Cell Calcium. 2000;28:213–223. doi: 10.1054/ceca.2000.0152. [DOI] [PubMed] [Google Scholar]
- 28.Mogami H, Gardner J, Gerasimenko OV, Camello P, Petersen OH, Tepikin AV. Calcium binding capacity of the cytosol and endoplasmic reticulum of mouse pancreatic acinar cells. J. Physiol. 1999;518:463–467. doi: 10.1111/j.1469-7793.1999.0463p.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer T, Stryer L. Molecular model for receptor-stimulated calcium spiking. Proc. Natl. Acad. Sci. U.S.A. 1988;85:5051–5055. doi: 10.1073/pnas.85.14.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zamora-Sillero E, Hafner M, Ibig A, Stelling J, Wagner A. Efficient characterization of high-dimensional parameter spaces for systems biology. BMC Syst. Biol. 2011;5:142. doi: 10.1186/1752-0509-5-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.