SUMMARY
Growth factors activate Ras, PI3K and other signaling pathways. It is not well understood how these signals are translated by individual cells into a decision to proliferate or differentiate. Here, using single-cell image analysis of nerve growth factor (NGF)-stimulated PC12 cells, we identified a two-dimensional phospho-ERK (pERK)-phospho-AKT (pAKT) response map with a curved boundary that separates differentiating from proliferating cells. The boundary position remained invariant when different stimuli were used or upstream signaling components perturbed. We further identified Rasa2 as a negative feedback regulator that links PI3K to Ras, placing the stochastically distributed pERK-pAKT signals close to the decision boundary. This allows for uniform NGF stimuli to create a subpopulation of cells that differentiates with each cycle of proliferation. Thus, by linking a complex signaling system to a simpler intermediate response map, cells gain unique integration and control capabilities to balance cell number expansion with differentiation.
INTRODUCTION
Growth factor stimuli can induce different cell fates by activating Ras, PI3K, Src, PLCγ and other signaling pathways (Lemmon and Schlessinger, 2010). It is not well understood how cells integrate such complex signaling responses to make all-or-none cell fate decisions. One hypothesis is that cells use multiple pathways to better monitor the presence of neighboring cells, growth factors, hormones, nutrient availability and intracellular stress. These pathways may then get integrated at specific signaling steps that function as “bottlenecks” or “hubs” (Albert, 2005; Barabasi and Oltvai, 2004). In turn, multiple downstream targets may link such an integration point to a cell fate. It is often implicitly assumed in pharmacological or genetic studies that signaling or transcriptional networks have such an hourglass or hub organization with a single intermediate integration point where a key decision is made (Friedman and Perrimon, 2007).
We investigated if and how such signaling hubs contribute to cell fate decisions by focusing on the PI3K and Ras pathways. These pathways are likely particularly important given their ubiquitous roles in regulating proliferation and differentiation and their dominant role in promoting cancer progression (Crespo and Leon, 2000; Katso et al., 2001; Okkenhaug and Vanhaesebroeck, 2003). We chose PC12 cells as a model system since nerve growth factor (NGF) activates both pathways and triggers a decision between proliferation and differentiation into sympathetic-like neuronal cells (Greene and Tischler, 1976). We also selected this cell model since it had suitable cell uniformity, speed of differentiation and transfectability that was difficult to match using differentiation-proliferation models in an in vivo setting. This offered the opportunity to ask systematic and quantitative questions about signaling processes at the single-cell level.
We used automated imaging and single-cell image analysis to compare the NGF-induced cell fate to the activation of the multifunctional protein kinases ERK and AKT, important downstream targets of Ras and PI3K signaling (Chambard et al., 2007; Manning and Cantley, 2007). This led to the unexpected finding that a two dimensional pERK-pAKT response map with a curved boundary separates regions with proliferation and differentiation cell fates. The same NGF stimulus caused significant cell-to-cell variation of pERK and pAKT signals, placing cells on both sides of the boundary, producing proliferating and differentiating subpopulations. Furthermore, the boundary position remained invariant when we used EGF, NGF or serum to stimulate cells or when we used small molecule inhibitors or siRNA knockdown to perturb upstream regulators. Finally, using a targeted siRNA screen, we identified Rasa2 as a regulator that places the distributed pERK-pAKT signals close to the boundary. We show that Rasa2 is a late NGF-induced PI3K-regulated RasGAP that connects PI3K to Ras signaling by negative feedback. Together, our study shows that cell fate decisions can be encoded by signaling response maps that function as intermediate integration and decision points. Such a response map provides mechanistic insights how identical populations of cells are split into subpopulations with different cell fates and how the number of differentiating cells can be regulated within a uniform population.
RESULTS
A two dimensional pERK-pAKT response map for proliferation
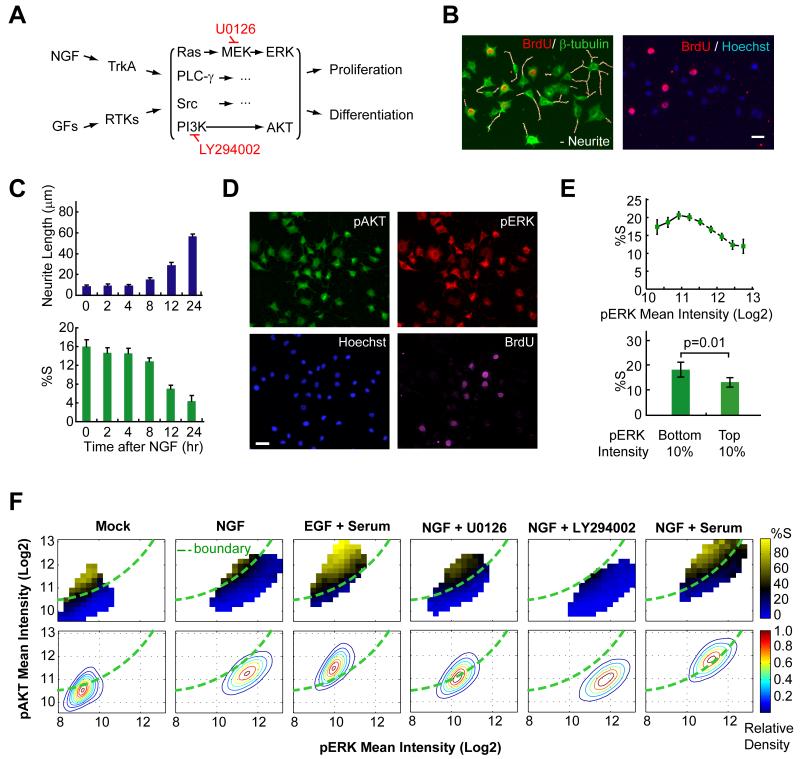
Previous studies with PC12 cells have shown that NGF stimulation of the TrkA receptor activates Ras, PI3K and a number of other signaling pathways to trigger neuronal differentiation (Huang and Reichardt, 2003) (Fig. 1A). The transition from a proliferative to a differentiated state occurs in most PC12 cells in the population within the first 24 hours of NGF stimulation (Fig. 1B). This switch can be tracked by the appearance of a neuron-like morphology (quantified by average neurite length, Fig. 1C, top) that is paralleled by the reduction of cells in S phase (monitored by BrdU incorporation, Fig. 1C, bottom).
Figure 1. Identification of a two dimensional pERK and pAKT signaling response map.
(A) Schematics of growth factors (GFs) induced receptor signaling. RTKs: receptor tyrosine kinases. Inhibitors used in the study are marked in red. (B) Automated image analysis of differentiation and proliferation after 24h of NGF treatment. Representative images used for the analysis are shown. (Left) Detected neurites (white) were superimposed over a merged tubulin and BrdU-stained image. (Right) Overlay of BrdU and DNA-stained image. Scale bar: 40μm. (C) Time courses of differentiation and proliferation following NGF stimulation (mean ± SD of triplicate wells). (D) Automated image analysis monitors pAKT, pERK and proliferation after 24h of NGF treatment. (E) Single cell analysis of pERK level versus the fraction of cells in S-phase shows only little correlation. The %S was calculated for equally-spaced bins of the ERK activity (top, mean ± 95% bootstrap confidence interval) or from the bottom and top 10 percentile of the ERK activity (bottom, mean ± SD of five replicate wells) after NGF stimulation for 24 hrs. (F) Heat map analysis of pERK-pAKT signaling and proliferation shows a clear boundary between proliferation and differentiation regions. Contour plots of cell density are shown in the lower panels. The %S was calculated for equally-spaced bins of the ERK and AKT activity and is marked in a color code. Cells were left untreated (mock) or stimulated as indicated for 24h before analysis. U0126 and LY294002 were added with NGF for 24h at 3.3μM and 6.3μM, respectively. The boundary (green line) was drawn across the black colored-bins on the NGF heat map and overlaid on top of other plots. Each panel contains ~40,000 cells. Note that due to day-to-day staining and imaging variations, the boundary position compares experiments done at the same time.
We measured the activity of the PI3K and Ras pathways by monitoring the phosphorylation status of their downstream targets, AKT and ERK, respectively (Fig. 1A). Using immunofluorescence and automated image analysis, we quantified the levels of pERK and pAKT and the proliferation status in single cells (Fig. 1D). Given the previous evidence that sustained pERK signals are important for the differentiation process (Marshall, 1995), we first tested whether the level of pERK in an individual cell correlates with the proliferation status when both are measured 24 hours after stimulation. Fig. 1E shows that the correlation between pERK and proliferation is statistically significant but quite small even when we selected the proliferation status from the bottom and top 10 percentile of pERK intensity for comparison.
It was striking that the proliferative state of each cell was much better defined by its location in a two dimensional pERK-pAKT plane (Fig. 1F, mock and NGF stimulus). This panel shows a probability map for proliferation of individual cells with different pERK and pAKT levels. The axes in this plot have a log base 2 and the proliferative state is represented by a color-coded heat map (from 0 to 80%). The region where cells have a high probability of proliferation is characterized by higher pAKT and lower pERK levels with a curved boundary separating the proliferating from non-proliferating cells (Fig. 1F).
Not only was the boundary between the two regions quite steep, it also remained at the same location when cells were stimulated with epidermal growth factor (EGF), NGF, low or high serum, or a combination with a low-dose of MEK or PI3K inhibitors (Fig. 1F). The existence of an invariant boundary between the two regions implied that each point in the pERK-pAKT response map can be reduced to a single parameter that reflects the distance from the boundary and predicts the proliferation probability.
NGF-induced signal variation spreads cells across a sharp boundary in the pERK-pAKT signaling plane
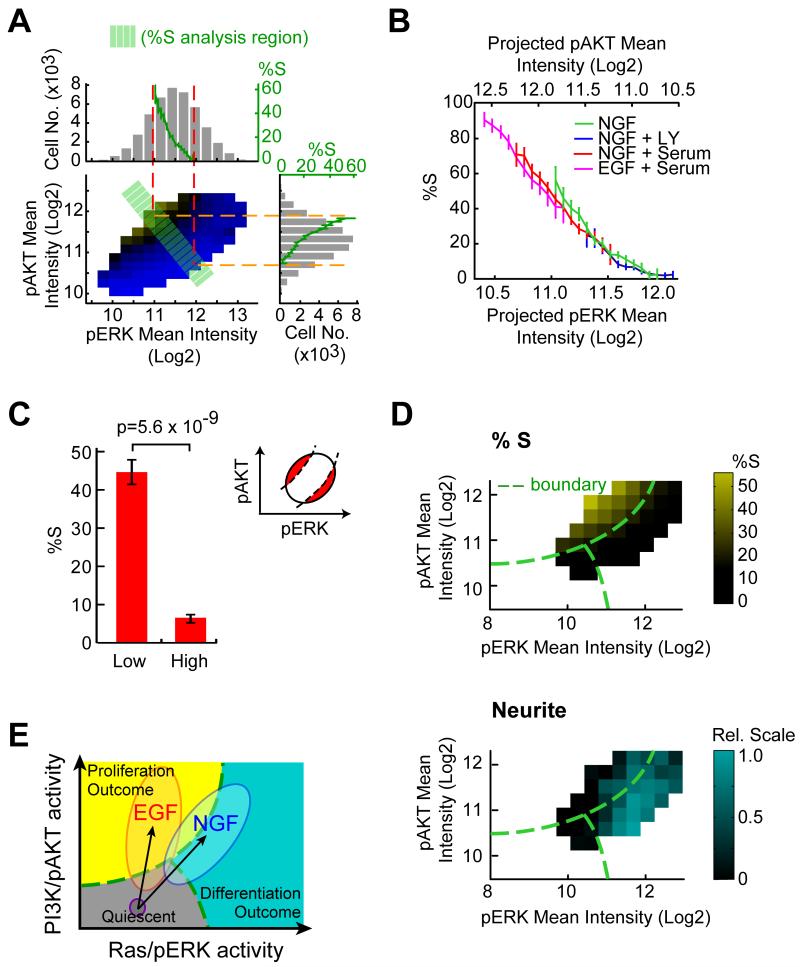
We noted that the same NGF stimulus induced a great variation in the pERK and pAKT signals even though we activated a homogenous cell population (Fig. 2A, subpanel top and right). The pERK and pAKT signal distribution shows a 4.2 and 3.1-fold difference between the bottom and top 5 percentile, respectively. This wide distribution allows cells to be spread over the narrow region boundary. We quantified the steepness of the proliferation probability change across the boundary (Fig. 2A; green band orthogonal to the boundary) by projecting the %S parameter onto the pERK and pAKT axis (Fig. 2A, green curves in subpanel top and right). Consistent with the visual impression, the boundary between the regions was very sharp with an approximately 30-fold increase of proliferating cells, from 2% to 60%, over only a 2-fold difference in the change in pERK or pAKT level.
Figure 2. A sharp boundary in the pERK-pAKT response map separates proliferating from differentiating cells.
(A) Quantitative analysis of NGF-triggered cell-to-cell signal variation and proliferation probabilities in the pERK-pAKT plane. The population distributions of pERK and pAKT are shown in the subpanel top and right (gray histograms). The same histogram includes a graph (green curves) of the %S calculated from cells located in the green band (orthogonal to the boundary shown in Fig. 1F). (B) Evidence of an invariant 2D signaling response map that determines proliferative cell fate. Proliferation changes were analyzed as shown in (A) from cells treated with different stimuli. The analysis only included cells located within the green band. In (A) & (B), data are mean ± 95% bootstrap confidence interval. (C) The proliferative status is better predicted by the 2D response map compared to pERK level shown in Fig. 1E (bottom). The %S was compared for the 10 percentile of cells farthest above (Low) and below (High) the boundary. Inset shows the schematic diagram of the analysis region. Data are mean ± SD of five replicate wells. (D) Heat map analysis showing proliferation (top) and differentiation (bottom) as a function of ERK and AKT activity at a single-cell level after 24h of NGF stimulation. %S was quantified as shown in Fig. 1F. Quantification of the integrated single-cell neurite parameter was achieved by measuring the presence of neurites proximal to the cell body of each cell and calculating mean neurite intensity for each cell as a function of pERK and pAKT levels. Each bin contains at least 300 cells. (E) Different directions and amplitudes of pERK-pAKT activity vectors correlate with cell fates. The schematic also shows a quiescent state for low pERK and pAKT levels. EGF and NGF not only trigger different amplitudes of signal activation but also have different directions of pERK-pAKT activity vector in the 2D plane.
When we extended the same analysis to cells treated with different stimuli, we found that the transition from proliferative to non-proliferative state was equally steep and indeed occurred at the same site in the pERK-pAKT plane independent of the stimulus and the position of the center of the pERK-pAKT distribution (Fig. 2B). In addition, when we compared the top and bottom 10 percentile of cells farthest away from the boundary as we did in the one dimensional case (Fig. 1E), the correlation with the cell fate was much higher (Fig. 2C).
Finally, we determined whether proliferation arrest and differentiation are indeed closely linked by performing a single cell analysis of neurite outgrowth as a marker of the differentiated state. The plot in Fig. 2D (bottom) shows that individual cells in a region below and to the right of the proliferation region have increased neuronal morphology. In this analysis, a third region in the plane can also be distinguished with cells that have low pERK and pAKT level that neither proliferate nor differentiate. This region likely reflects a quiescent G0/G1-state of the cell cycle.
Fig. 2E depicts the resulting working model that different regions in the pERK-pAKT plane are highly predictive as to whether a cell proliferates, differentiates or resides in a quiescent state. The usefulness of this response map analysis is particularly apparent when comparing an EGF stimulus, which primarily promotes proliferation and has an activation vector with higher relative pAKT, to the NGF stimulus, which promotes mostly differentiation and has a vector with higher relative pERK (Fig. 1F). As an added note, since the boundary between the cell fate regions is non-linear, simple linear combinations or ratios of the respective pERK and pAKT activities only partially correlate with the decision, implying that a 2D response map is needed for an optimal prediction of cell fate outcome.
An siRNA screen identifies regulators of cell fate and pERK-pAKT signals
To better understand how cells generate the pERK-pAKT response map, we performed a targeted siRNA screen to identify relevant molecular signaling components that we could use for subsequent perturbation studies. We made a rat siRNA library targeting 1308 signaling proteins using an in vitro Dicer cleavage strategy (Myers et al., 2003) and performed a screen as outlined in Fig. 3A with automated analysis of neurite extension and proliferation (Fig 1B). The 54 genes listed in Table S1 were hits that were validated by two sets of siRNAs against different regions of the mRNA with both siRNAs changing the cell fate outcome in the same direction (Fig. S1). When surveying the identified proteins for annotated processes, we found important regulators as well as known NGF signaling components among the hits (Table S1). The identified signaling regulators included for example the little characterized proteins Arf5 and Tao1 kinase which, when knocked down, greatly reduced or increased the differentiation state, respectively (Fig. 3B).
Figure 3. siRNA perturbation analysis validates the use of the signaling response map to predict cell fate.
(A) Protocol used to screen for siRNAs that change the fraction of proliferating and differentiating cells. (B) Knockdown images of selected genes identified in the siRNA screen of regulators of NGF-induced differentiation. Arf5 reduces differentiation and Tao1K increases differentiation. Scale bar: 40μm. (C) Perturbation analysis with 54 siRNAs showing the correlation between proliferation and the induction of differentiation (data were from duplicate wells; robust z-score: the median absolute deviation from the control median). (D) NGF signaling scheme and the corresponding secondary assays used to link different signaling processes to differentiation. (E) Perturbation parameter cross-correlation analysis showing that the 24h pERK is the most predictive parameter for neurite extension (Nrt) and proliferation (%S), both measured at 48h. All 54 siRNAs were used for the analysis. ERK5’, 1h and 24h denote measurements of pERK at 5 min, 1h and 24h after NGF stimulation. EGR1 is the induction of the early growth response 1 transcription factor. AKT24h represents AKT phosphorylation at 24h of NGF stimulation. Color bar represents the cross-correlation values (Pearson’s correlation coefficients). (F) Direct correlation analysis comparing the short-term (5 min, left) and long-term ERK (24h, right) signaling with differentiation (neurite length) for all 54 siRNAs. For (C) & (F), green lines are linear fits and R represents Pearson’s correlation coefficients. (G) Center of population distribution for all 54 siRNAs in the pERK-pAKT plane. Each dot represents the population median of the pERK and pAKT intensity after individual siRNA knockdown. Green line: region boundary that crosses 20% of S phase probability based on control knockdown (red circle). (H) Separation of genes that shift the boundary (x-axis) or move the population center away from the boundary of controls (y-axis) upon siRNA knockdown. The boundary shift was calculated by the sum of %S differences between specific siRNA knockdown and control per pERK-pAKT bin in the 2D plane. Movement of population relative to the boundary was represented as the orthogonal distance (Log2 unit) from the center of population distribution to the boundary (as shown in Fig. 3G). Knockdowns of cell cycle regulators (red circle) that shifted the boundary to the negative side include cyclin D1/D3, Cdks 1, 2, 4, 6 and Mdm4. The positive side contains the tumor suppressors Rb1, p21 and p16. SD represents the standard deviation calculated from all the siRNAs.
Shotgun perturbation analysis of the pERK-pAKT response map and cell fate
Since the 54 identified genes came from a relatively unbiased screen of signaling proteins, we used them as a signaling network perturbation tool. By comparing the effect of the different siRNAs on proliferation versus neurite growth, we confirmed that the coupling between differentiation and suppression of proliferation is indeed very close. Most siRNAs shifted proliferation and differentiation responses in opposite directions (Fig. 3C).
We next determined whether siRNA-mediated signaling changes measured at 5 min, 1 hour and 24 hours after NGF stimulation can predict the changes in the differentiation or proliferation outcomes 48 hours after NGF stimulation. The experiments shown before in Figs. 1 and 2 measured signaling as well as cell fate at 24 hours. We measured early pERK signaling at 5 minutes and at 1 hour, and the induction of an early response gene EGR1 at 1 hour after NGF stimulation. We also measured the late ERK and AKT signaling at 24 hours (Fig. 3D & Table S2). We then performed a correlation analysis between the changes induced by the 54 siRNAs (Fig. 3E). Interestingly, the siRNA-mediated changes in pERK signaling at 5 minutes had no significant predictive value either on the change of proliferation or differentiation at 48 hours (Fig. 3E & 3F, left panel). Similarly, the changes in pERK level and EGR-1 expression at 1 hour showed a higher but still small correlation with the later cell fates. In contrast, the siRNA-mediated changes of p-ERK signals 24 hours after stimulation provided a significantly higher predictive value for the neuronal differentiation and proliferation status at 48 hours (Fig. 3E & 3F, right panel). This is consistent with the interpretation derived from Figs. 1 and 2 that sustained pERK and pAKT signals control cell fate. This further implied that short term signals can be altered without changing the cell fate outcome as long as sustained signals are not affected.
The weaker but significant correlation of proliferation and differentiation with changes of pAKT at 24 hours showed that most of the siRNA impacted both pERK and pAKT signals in parallel, keeping the center of the respective population distributions close to the boundary. This can be seen more directly in a plot where the centers of the relative population distributions are shown for cells treated with different siRNAs (Fig. 3G). Two significant outliers are PTEN and Rasa2 which will be investigated in more detail below.
We hypothesized that siRNA perturbations can alter the 2D response map in at least two ways. One way is to move the pERK-pAKT signal distribution while leaving the boundary intact as we found before in Figs. 1 and 2. This can be represented as an orthogonal shift of the center of the population distribution relative to the boundary of control cells (y-axis in Fig. 3H). Another way would be to shift the decision boundary in the pERK-pAKT response map without altering the population distribution. We measured such a potential shift of the boundary position by using a comparison of S phase probability values between the siRNA knockdown cells and control cells (x-axis in Fig. 3H, Supplemental Material and Methods). We anticipated that knockdown of signaling steps downstream of pERK-pAKT might be of this second type. Markedly, among different siRNAs tested, the positive cell cycle regulators CDKs (1, 2, 4, and 6), MDM4 and cyclin D1/3 (Morgan, 2007) shifted the boundary towards the proliferating region (resulting in less proliferating cells) while the negative regulators p21, p16 and Rb (Morgan, 2007) shifted the boundary away from it (Fig. 3H), suggesting that they are indeed of this second type. For comparison, the different receptor stimuli as well as inhibition of PI3K and MEK belonged to the first type of perturbation that shifted the population distributions along the y-axis. In this group, PTEN and Rasa2 siRNAs stood out again, showing the strongest shift away from the boundary, an observation we examined further below.
Together, these siRNA perturbation studies showed that proliferation and differentiation are mutually exclusive (Fig. 3E) and that sustained pERK-pAKT signals are critical for the cell fate decision (Figs. 1, 2, 3F). Furthermore, downstream cell cycle effectors shift the decision boundary in the pERK-pAKT response map (Fig. 3H).
The cyclin D-mediated shift of the decision boundary involves pERK and pAKT regulation of protein stability
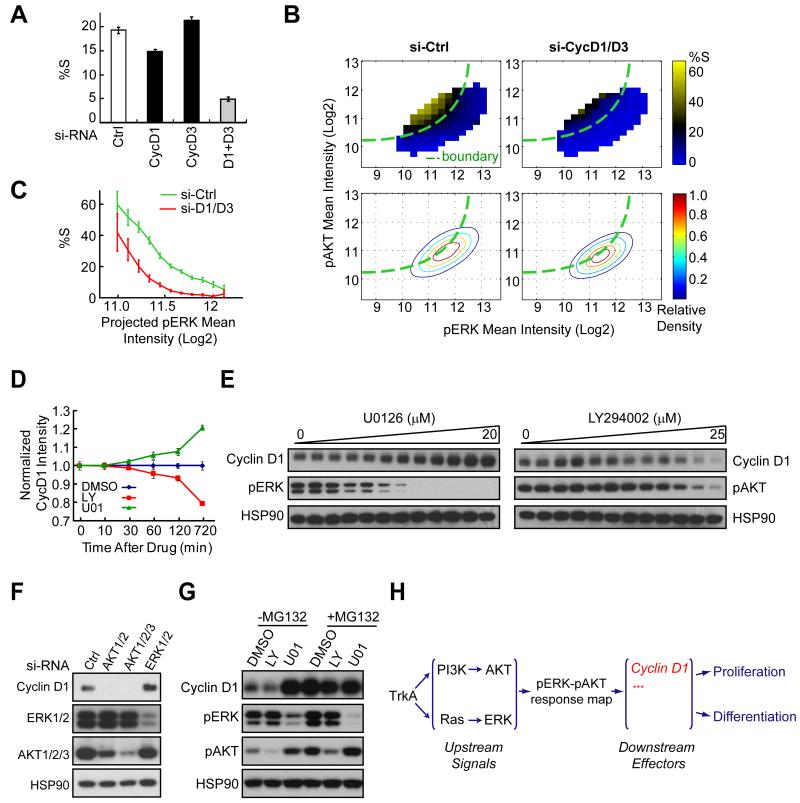
We were particularly interested in understanding the effect of the combined cyclin D1/3 knockdown since it strongly reduced proliferation (Fig. 4A) and caused the farthest shift of the boundary among the siRNAs (Fig. 3H, 4B). The shape of the shifted boundary is quantified in Fig. 4C. The knockdown of individual isoforms had only small effects (Fig. 4A), suggesting that cyclin D1 and D3 have redundant functions. Control experiments with the respective protein knockdowns are shown in Fig. S2A.
Figure 4. The cell fate decision is in part mediated by pERK and pAKT-control of cyclin D1 protein stability.
(A) Quantitative analysis of the effect of cyclin D1/D3 single and co-knockdown on proliferation. siRNA treated cells were stimulated with NGF for 24h before analysis (mean ± SD of triplicate wells). (B) Heat map analysis of the cyclinD1/D3 knockdown effect on pERK-pAKT signaling and proliferation. The knockdown (right) shifted the boundary to the top-left between the differentiation and proliferation regions without significantly changing the pERK and pAKT distribution itself. Assays were performed as described in Fig. 1D & 1F. (C) Evidence of the boundary shift with cyclin D1/D3 co-knockdown. Proliferation changes were calculated from cells located in the region orthogonal to the boundary as shown in Fig. 2A (mean ± 95% bootstrap confidence interval). (D) Time courses of the effects of PI3K (LY294002) and MEK inhibition (U0126) on cyclin D1 protein levels. 12.5μM LY294002 or 10μM U0126 was added at 24h after NGF stimulation for different lengths of time as indicated before immunostaining. Cyclin D1 levels were measured by automated image analysis (mean ± SD of triplicate wells). (E) Dose effects of U0126 and LY294002 on cyclin D1 protein level changes. Cells were treated with increasing doses of U0126 or LY294002 together with NGF for 4 hours. (F) Knockdown of AKT or ERK mimics the LY294002 and U0126 drug effects on cyclin D1 protein level changes. Knockdown cells were subjected to 24h of NGF stimulation before analysis. (G) The opposing regulation of cyclin D1 protein level by LY294002 and U0126 is proteasome-dependent. Cells were stimulated with NGF for 4 hours with the drug combination as indicated. MG132 was used at 50μM. LY294002 and U0126 were used at 12.5μM and 10μM, respectively. (H) Schematics of signaling diagram showing cyclin D1 as one of the downstream mediators linking the pERK-pAKT response map to cell fates.
Cyclin D is a key cell cycle as well as differentiation regulator (Sherr and Roberts, 2004) whose concentration is known to be controlled by growth factor inputs acting through the Ras and PI3K pathways (Diehl et al., 1998; Shao et al., 2000). This made us examine how the position in the pERK-pAKT response map is translated into a change in cyclin D1 concentration. Inhibition of MEK increased cyclin D1 protein level whereas inhibition of PI3K or AKT decreased cyclin D1 (Figs. S2B, S2C, 4D, 4E), a finding that was also confirmed by combined knockdowns of the isoforms AKT1-3 (targets of PI3K and PIP3) or ERK1-2 (targets of MEK) (Fig. 4F). When cells were treated with proteasome inhibitor (MG132), the differential effects of LY294002 and U0126 on cyclin D1 protein level were lost (Fig. 4G & S2C). Furthermore, the opposing ERK-AKT regulation on cyclin D1 levels remained intact when protein translation was inhibited with cycloheximide (CHX) (Fig. S2D). Together, this shows that the control of cyclin D1 stability is a rate-limiting regulatory step that translates a decision made at the pERK-pAKT integration point into cell fates (Fig. 4H). Thus, the observed shift of the decision boundary in cyclin D knockdown cells (Fig. 4B) reflects a requirement for higher pAKT and lower pERK signals to increase cyclin D protein stability and thereby make up for the reduced cyclin D translation to restore a proliferating subpopulation.
Evidence that Rasa2 mediates a negative feedback from PI3K to Ras signaling
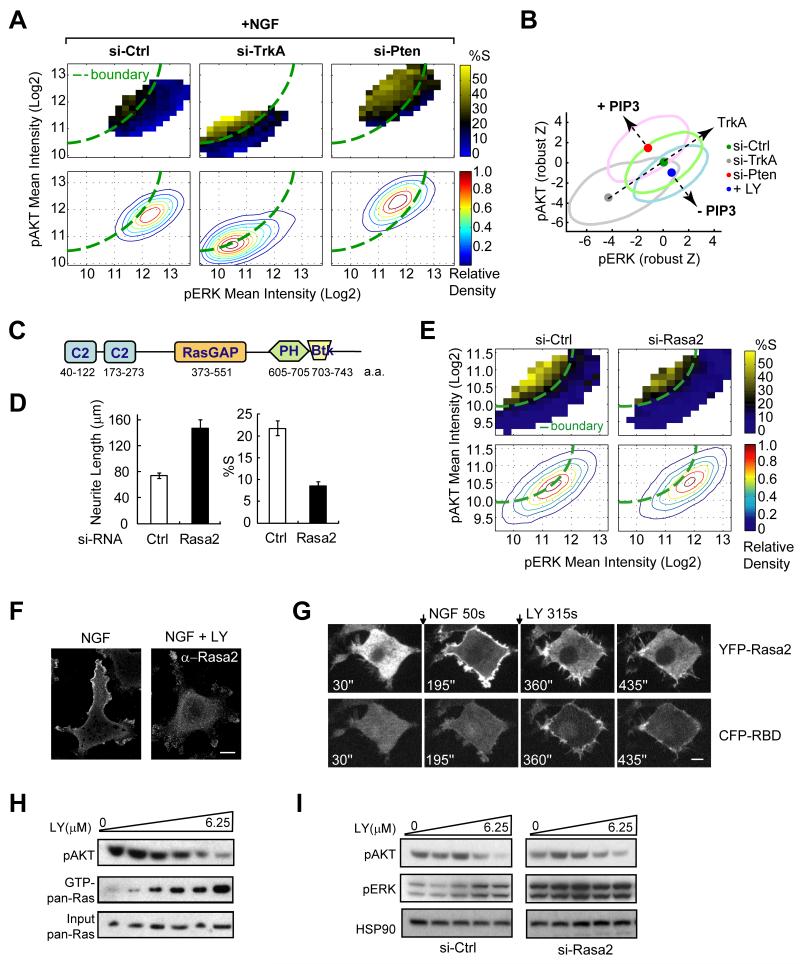
When we first generated the pERK-pAKT response map in Fig 1F, we were intrigued by the observation that a subpopulation of cells continues to proliferate upon NGF stimulation. We considered that maintaining such a subpopulation enables more cells to differentiate over longer time periods. An effective way to control the size of the proliferating subpopulation would be to shift the activation vector orthogonal to the boundary, towards the top left or bottom right. A candidate orthogonal regulator was the lipid second messenger PIP3 that is generated by activation of PI3K. This hypothesis was based on the observation that PTEN knockdown or PI3K inhibition both shifted the pERK-pAKT vector along this orthogonal line but in opposite directions (Figs. 5A, 5B, 3G, 3H and 1F). We found that cells with knocked down PTEN, a PIP3 lipid phosphatase that lowers PIP3 levels, not only suppresses AKT signaling (Carracedo and Pandolfi, 2008) but also lowers the average pERK signal response (Fig. 5B). Similarly, PI3K inhibition reduces PIP3 with a concomitant increase in pERK signals (Figs. 1F, 5B). This explains how PIP3 changes can create an orthogonal shift of the pERK-pAKT response compared to the shift mediated by knockdown of the NGF receptor TrkA. This raised the question of how PIP3 reduces ERK phosphorylation.
Figure 5. Rasa2 increases the number of proliferating cells after NGF stimulation by adding a negative feedback from PI3K to Ras and ERK signaling.
(A) Heat map analysis of PTEN and TrkA siRNA effects on pERK-pAKT signaling and proliferation. Assays were performed as described in Fig. 1F. (B) Changes in PIP3 levels cause a shift of the activation vector orthogonal to NGF activation. Data from Fig. 1F & Fig. 5A were normalized to their respective control and plotted together with robust z-score units. The large ovals represent the population distributions and the small filled circles represent the centroids of each population. (C) Domain structure of Rasa2. (D) Quantitative analysis of the effect of Rasa2 knockdown on reducing proliferation and increasing differentiation (mean ± SD of triplicate wells). (E) Heat map analysis of the Rasa2 siRNA-mediated shift of the population distribution towards higher pERK levels. Assays were performed as described in Fig. 1F. The boundary was drawn according to control cells. (F) Membrane localization of endogenous Rasa2. Cells after 24h of NGF stimulation were left untreated (left) or treated with PI3K inhibitor (LY294002 at 25μM) for 5 min before subjected to Rasa2 antibody staining. Scale bar: 10μm. (G) Time series of images showing YFP-Rasa2 (top) and CFP-RBD (bottom) translocation after NGF stimulation and subsequently, after PI3K inhibitor (LY294002) addition. Cells were co-transfected with YFP-Rasa2, CFP-RBD (Raf) and H-Ras. CFP and YFP confocal images were taken from the same representative cell. NGF and LY (100μM) were added as indicated. Ras activity was monitored using the relative plasma membrane translocation of CFP-RBD. Scale bar: 5μm. (H) Ras pull-down followed by western blotting showing that inhibition of PI3K is paralleled by an increase of GTP-bound Ras level. Cells were treated with increasing dose of PI3K inhibitor (0 to 6.25μM, 2-fold dilution from the right) for 15 min at 24h post NGF stimulation. (I) Analysis of control and Rasa2 siRNA effect on ERK activity changes in response to PI3K inhibition. Cells were treated as described in (H) and assayed by western blotting.
Our siRNA screen identified Rasa2 as a strong enhancer of proliferation and suppressor of differentiation (Fig. 3G, 3H, 5C, 5D & Fig. S3A). Rasa2 has a RasGAP domain specific for Ras (Maekawa et al., 1994) that could explain the observed effect on Erk signaling. It further provided a potential link from PIP3 to pERK since PIP3 has been shown to bind to its PH-domain and recruit Rasa2 to the plasma membrane (PM) (Lockyer et al., 1999). Nevertheless, these studies did not determine whether its RasGAP activity is regulated by PIP3.
We first showed that Rasa2 knockdown significantly enhanced neurite growth and decreased proliferation. Furthermore, it shifted the pERK-pAKT activity vector to the right as expected for a knockdown of a RasGAP (Fig. 5E). The specificity and effectiveness of Rasa2 siRNA knockdowns were confirmed by two different siRNAs, quantitative RT-PCR and western blotting as well as by Rasa2 expression rescue experiments (Fig. S3B, S3C). We further confirmed that Rasa2 knockdown enhanced the level of GTP-bound Ras (Fig. S3D). Moreover, neurite extension induced by a constitutively active (CA) Ras (Bar-Sagi and Feramisco, 1985) could not be further enhanced by Rasa2 knockdown while control knockdown of the downstream ERK could (Fig. S3E). Thus, Rasa2 functions as a RasGAP and the observed increase in pERK and differentiation following Rasa2 knockdown is due to increased Ras activity.
We then confirmed that the PM localization of endogenous Rasa2 and the expressed YFP-Rasa2 are both dependent on PI3K activity (Figs. 5F, 5G, top; Fig. S3F). To determine whether PIP3 regulates Rasa2 activity, we co-expressed a CFP-tagged Ras binding RBD domain, a biosensor for active Ras (Chiu et al., 2002), together with YFP-Rasa2. The Ras-GTP biosensor showed initially only a weak PM localization and translocated more strongly to the PM after PI3K inhibition, demonstrating that the PIP3-mediated PM translocation of Rasa2 activates its RasGAP activity and that the dissociation of Rasa2 from the PM rapidly increases the concentration of Ras-GTP in the PM (Fig. 5G, bottom). Furthermore, the strength of the initial CFP-RBD translocation was markedly enhanced when we expressed instead a mutant Rasa2 defective in PIP3 binding (R629C) (Lockyer et al., 1999), arguing that PIP3 binding is critical for its GAP activity (Fig. S3I-K).
The same result was obtained using a biochemical analysis of Ras, pERK and pAKT signaling. Increasing doses of a PI3K inhibitor led to a progressive decrease in pAKT that was paralleled by a marked increase in Ras and ERK activity (Fig. 5H, 5I). Notably, this dose-dependent pERK increase was not observed in Rasa2 knockdown cells, demonstrating that Rasa2 provides the main link from PI3K to the suppression of ERK signaling (Fig. 5I). Taken together, this shows that PIP3 induces PM translocation and activation of Rasa2 creates a negative feedback by which PI3K activation suppresses Ras and ERK signaling.
Rasa2 and TrkA expression is upregulated by NGF stimulation
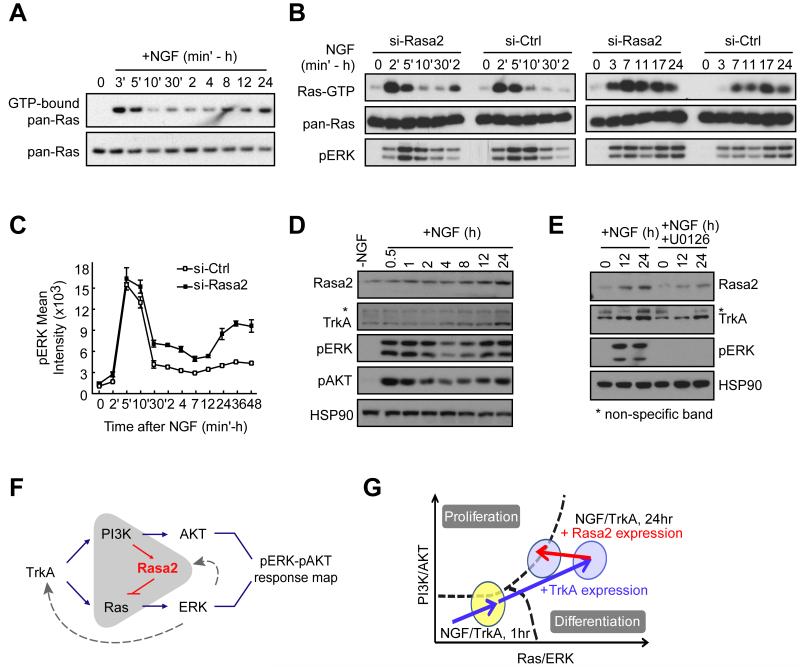
We then examined when Rasa2 acts to regulate cell fate. We observed two waves of Ras activation following NGF stimulation (Fig. 6A). The first wave happened immediately after NGF stimulation (2-5 min) and the second wave occurred 7-24 hours later, coinciding with the onset of differentiation (Fig. 1C). Knocking down Rasa2 increased the second wave of both Ras and pERK signaling but had only a small effect on the initial pERK activity peak (Fig. 6B, C). We were able to explain this delayed role of Rasa2 by the finding that Rasa2 expression significantly increased during the second wave of ERK activation (Fig. 6D), arguing that Rasa2 is important during the time window when the cell fate decision is made but plays a minor role in regulating short term Ras and ERK signaling. Nevertheless, these findings left unanswered what creates the second peak in Ras, pERK and pAKT activities (Fig 6A-D).
Figure 6. NGF-triggered expression of Rasa2 and TrkA directs the pERK-pAKT activation vector close to the boundary.
(A) NGF stimulation triggers two waves of Ras activation. (B) Knockdown of Rasa2 enhances Ras and pERK activities during the second wave. For (A) & (B), Ras pull-down assays were performed at the indicated time and assayed by western blotting. (C) Knockdown of Rasa2 selectively enhances a second wave of pERK activation with little effect on the first peak (mean ± SD of triplicate wells). (D) Time course analysis of Rasa2 and TrkA expression compared to pERK and pAKT activation. Cells were assayed by western blotting. HSP90 was shown as a protein loading control. (E) TrkA and Rasa2 upregulation is partially dependent on MEK signaling. U0126 was used at 10μM. (F) Schematic representation of the feedback between PI3K, Rasa2 and Ras. (G) Schematic model of the roles of the positive TrkA expression feedback, which increases the amplitude of the activation vector, and the negative Rasa2 expression feedback that turns the activation vector closer to the proliferation boundary.
Differentiated neurons have been shown to have increased expression of Trk family growth factor receptors (Deppmann et al., 2008; Zhou et al., 1995). We considered that such an upregulation of TrkA may contribute to the delayed increase in ERK activity. We show that TrkA expression is indeed induced by NGF in PC12 cells with a time course that paralleled the increase in Ras, pERK and pAKT activities as well as Rasa2 expression (Fig. 6D). We would like to note that both, the pERK and the pAKT increase have the same kinetics, consistent with the interpretation that the second activation peak is mediated by the induced expression of TrkA. Nevertheless, since Rasa2 is upregulated in parallel with TrkA (Fig. 6D), the amplitude of the second activation peak of pERK results from a competition between TrkA upregulation, which amplifies NGF-triggered PI3K and Ras signaling, and Rasa2 upregulation, which suppresses Ras/ERK activities. We further showed that both Rasa2 and TrkA expression are at least partially dependent on MEK activity since inhibition of MEK reduced their expression (Fig. 6E). Thus, Rasa2 functions as a negative feedback regulator that begins to lower Ras activity a few hours after NGF stimulation when Rasa2 expression increases (Fig. 6F and 6G). As an added note, given the unimodal population distributions shown in Fig. 2A, the positive feedback resulting from NGF upregulating its own TrkA receptor is primarily a mechanism to amplify the long term NGF signaling response rather than creating a bistable switch for ERK and AKT activation.
Role of Rasa2 in expanding the number of cells during differentiation
The feedback mediated by Rasa2 changes the direction of the pERK-pAKT activity vector by reducing pERK signals as PI3K signaling increases. This leaves a significantly larger fraction of cells in the proliferation region by forcing the activation vector to turn and stay closer to the boundary as TrkA expression increases during the 7-24 hour time window (Fig. 6G). We hypothesized that cells benefit from having the center of the pERK-pAKT vector close to the boundary by maintaining a balance between cell number expansion (proliferation) and differentiation.
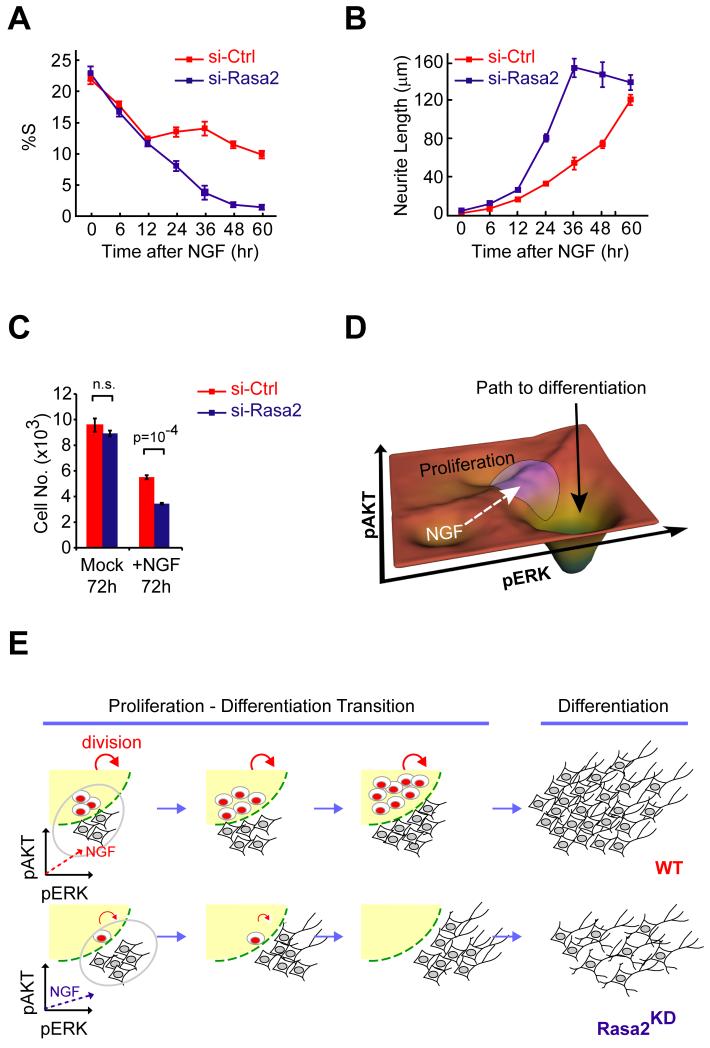
Indeed, we found that Rasa2 has a role in maintaining a pool of proliferating cells when we monitored the fraction of proliferating cells over a 60 hour period after NGF stimulation. Rather than observing a near complete drop in the number of proliferating cells by 36 hours as observed in Rasa2 knockdown cells, control cells maintained a fraction of proliferating cells for more than 60 hours (Fig. 7A). The continued proliferation comes at a small cost since cells that upregulate Rasa2 expression take longer to differentiate (Fig. 7B). However, the continued proliferation provides a benefit since a larger number of differentiated cells are generated after this period in control cells compared to Rasa2 knockdown cells (Fig. 7C). Thus, by positioning the population near the proliferation boundary, Rasa2 maintains proliferation competent precursor cells to create more differentiated cells.
Figure 7. Function of the pERK-pAKT response map in balancing cell number expansion and differentiation.
(A & B) Time course analysis of proliferation (A) and neurite extension (B) in control and Rasa2 knockdown cells following NGF stimulation (mean ± SD of four replicate wells). In (A), subpopulations of control cells stay proliferative over a period of 60h while Rasa2 knockdown cells cease to proliferate after 48h of NGF stimulation. (C) Quantification of Rasa2 knockdown effect on cell number expansion following NGF stimulation. Cells transfected with control or Rasa2 siRNA were treated with Mock or NGF for 3 days before counting cell number (mean ± SD of four replicate wells). (D) Landscape scheme of the 2D pERK-pAKT response map emphasizes the boundary between the two regions that predict the proliferation and differentiation outcomes. The purple circle depicts the variation of the NGF-induced signaling response that spreads the population of cells across the boundary. The white dashed arrow reflects the NGF-induced shift of the activation vector and the black solid arrow depicts the path to differentiation. (E) Schematic showing how Rasa2 maintains a balance between cell number expansion and differentiation.
DISCUSSION
Significance of a pERK-pAKT signaling code
When we initiated our studies, we considered that the level of sustained ERK activation alone might predict the decision between differentiation and proliferation (Marshall, 1995). Our study showed instead that pERK and pAKT are both critical intermediate signaling steps and single cell measurements are needed to reveal the relationship between signaling and cell fate. We found that the position of the activation vector relative to the boundary in the pERK-pAKT response map determines whether a particular cell differentiates or proliferates. This boundary idea can be applied to the probability response map because the transition from non-proliferating to proliferating cells is steep (Figs. 2A and 2B). Our model of a response map that defines the paths to differentiation or proliferation is schematically shown in a landscape representation in Fig. 7D. This concept shares some similarity to the idea that complex systems use hubs to process information (Albert, 2005; Barabasi and Oltvai, 2004).
Using different stimuli, small molecule inhibitors and siRNA knockdown of signaling proteins, we found that the curved boundary between the regions was independent of the cellular signaling processes that activate cells (Fig. 1F & 2B), arguing for a separation of upstream and downstream components from pERK and pAKT signals. We showed that the decision of a cell in the pERK-pAKT response map is in part translated into a cell fate by regulation of cyclin D1 stability. Knocking down cyclin D or other downstream cell cycle regulators shifted the decision boundary. While cyclin D1 is well known to be induced by mitogens (Sherr and Roberts, 2004) and separate studies have shown that its stability can be regulated by Ras (Diehl et al., 1998) or PI3K signaling (Diehl et al., 1998; Shao et al., 2000), our study provides evidence that both pathways regulate cyclin D1 in concert with opposing positive and negative regulation by pAKT and pERK, respectively. Given that AKT and ERK also regulate other cell cycle as well as differentiation-related proteins (Chambard et al., 2007; Manning and Cantley, 2007; von Kriegsheim et al., 2009), it is likely that cyclin D is a key player among a broader set of effectors that cooperate to translate the pERK-pAKT decision into a cell fate.
The existence of a pERK-pAKT response map highlights the dual roles of PI3K and Ras signaling in regulating proliferation and differentiation cell fates. Indeed, other than their well-known roles in proliferation, Ras and PI3K have been implicated in a variety of differentiation processes including those in neuronal, myeloid, muscle and adipocyte cells (Crespo and Leon, 2000; Katso et al., 2001). In the case of PC12 cell fate decisions, a number of studies have shown that the temporal differences in pERK kinetics correlate with cell fates with EGF-mediated transient ERK activity promoting proliferation and NGF-mediated sustained ERK activity triggering differentiation outcomes (Marshall, 1995; Santos et al., 2007; Sasagawa et al., 2005). Our results using the response map analysis and the systemic siRNA perturbation experiments suggest that changes of the amplitude of the first wave of ERK signaling (2-5 minutes) have no significant impact on the cell fate decision while the sustained pERK and pAKT activity is predictive (Fig. 3E, 3F). The relevance of long term PI3K/AKT signaling was demonstrated using multiple perturbation experiments that shifted the NGF-regulated proliferation and differentiation balance, further arguing that PI3K as well as Ras signaling jointly control the cell fate decision (Figs. 1F, 3H, 4).
pERK-pAKT signal variation enables differentiation and proliferation decisions for the same NGF receptor stimulus
Single cell intrinsic noise in protein expression typically alters protein levels by over 30% (Blake et al., 2003; Niepel et al., 2009). Such variation can make it difficult for cells to execute reliable signal transduction processes (Arias and Hayward, 2006). Most studies in eukaryotic cells have therefore focused on mechanisms that make signaling less variable and outcomes more robust (Acar et al., 2010; Colman-Lerner et al., 2005). However, noise can also be important in cellular decision making (Balazsi et al., 2011; Blake et al., 2006; Spencer et al., 2009). Our study shows that signal variation generated by stimulation of a uniform cell population can be used to generate two subpopulations with different cell fates. This required a cell fate boundary in the pERK-pAKT plane that is narrower (< factor of 2) than the NGF induced signal variation (~ factor of 4), allowing individual cells to distribute into the proliferating and differentiating regions, respectively (Fig. 2A). Given the multiple feedbacks and signaling processes involved in regulating long-term AKT and ERK activity (Campbell et al., 1998; Carracedo and Pandolfi, 2008), the relatively large variation in the pERK-pAKT response map can plausibly be explained by the sum of smaller relative expression differences of multiple upstream regulatory proteins.
A Rasa2-mediated feedback controls differentiation along with cell number expansion
The analysis of Rasa2 regulation on cell number and differentiation in Fig. 7A and 7B showed that the combination of a response map and signal variation has a potential advantage in the execution of an effective differentiation response. Instead of simply differentiating all cells, the same stimulus can keep some cells proliferating which eventually increases the total number of terminally differentiated cells. Fig. 7E illustrates this design, whereby each cycle of proliferation splits off a pool of differentiating cells. This process does not require a predefined differentiation program for each cell but rather relies on stochastic signal variation to create the two subpopulations. Thus, by combining a signaling network with 1) a response map where the decision is made, 2) sufficient single cell signal variation to spread the signal over a sharp boundary as well as 3) a feedback mechanism that positions the activity vector near the boundary, cells gain tight control over the fraction of terminally differentiated cells. In an in vivo context, we envision that this pERK-pAKT signaling code enables cells to maintain a homeostatic balance of precursor and differentiated cells since excessive proliferation and premature differentiation are both harmful to multicellular organisms.
Conclusions
Our study introduces a two-dimensional signaling code that describes the NGF-induced proliferation and differentiation decision of PC12 cells. Given the broad importance of PI3K and Ras signaling, our study likely exemplifies a general principle whereby pERK-pAKT response maps represent key integration points where subpopulations of cells are specified to different cell fates.
EXPERIMENTAL PROCEDURES
Cell culture, transfection, antibodies and plasmids
A PC12 subline, Neuroscreen-1 (here referred to as PC12 cells) (Dijkmans et al., 2008), was used for the study due to its reduced tendency toward cell aggregation (Cellomics, Inc). Unless otherwise noted, cells were induced to differentiate in low serum-containing media (F12K supplemented with 0.5% horse serum (HS)) while high-serum media (Fig. 1F) contained 5% HS and 0.8% FBS. siRNA (10-20 nM) and DNA transfection (100 ng per 96-well) were carried out with Lipofectamine 2000 (Invitrogen) according to manufacturer’s protocol. For expression of multiple constructs, PC12 were electroporated with the Amaxa system following manufacturer’s instructions (Lonza). Sources for antibodies, reagents, qRT-PCR primers and other constructs used in this study are provided in the Supplemental Methods.
Image Analysis
Images for 2D response map experiments were analyzed using custom made MATLAB image analysis programs (Salmeen et al., 2010). Briefly, nuclear centroids were identified in images of Hoechst stain. A nucleus mask was generated for each cell by expansion from the centroid to reach 30% of maximum intensity. A cell mask was then generated by expansion of the nucleus mask 5μm to include both the nucleus and the perinuclear region. After local background subtraction, the pERK (cell mask), pAKT (cell mask) and BrdU (nucleus mask) mean intensity were measured. The threshold level used to determine BrdU-positive cells was set using a k-means clustering algorithm on a well-to-well basis. Detailed analysis related to the 2D response map was described in the Supplemental Methods.
Statistical analysis
Error bars represent the standard deviation, standard error of the mean or 95% bootstrap confidence interval as indicated in the legends. Statistical comparisons (p values) were obtained from two-sided t tests. The Pearson’s correlation coefficients (R) were calculated as indicated.
Supplementary Material
Highlights.
A two dimensional ERK-AKT code decides between proliferation and differentiation.
Single-cell signal variation creates two cell fates in an identical cell population.
Different growth factor inputs are integrated at the level of ERK and AKT.
Rasa2 enhances proliferation over differentiation in a population of PC12 cells.
Acknowledgements
We would like to thank J.H. Chen for the graphic design and Dr. A. Salmeen for help with image analysis. We are also grateful for Dr. S. Spencer and A. Winans for comments. This work was supported by a Stanford Graduate Fellowship to JYC, a Department of Defense (Breast Cancer Research Program) Predoctoral Fellowship (W81XWH-09-1-0026) to JRL, NIH grant ES016486 to KAC and NIH grant MH64801 to TM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acar M, Pando BF, Arnold FH, Elowitz MB, van Oudenaarden A. Science. Vol. 329. New York, N.Y: 2010. A general mechanism for network-dosage compensation in gene circuits; pp. 1656–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R. Scale-free networks in cell biology. Journal of cell science. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nat Rev Genet. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sagi D, Feramisco JR. Microinjection of the ras oncogene protein into PC12 cells induces morphological differentiation. Cell. 1985;42:841–848. doi: 10.1016/0092-8674(85)90280-6. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Blake WJ, M KA, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Campbell SL, Khosravi-Far R, Rossman KL, Clark GJ, Der CJ. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- Chambard JC, Lefloch R, Pouyssegur J, Lenormand P. ERK implication in cell cycle regulation. Biochim Biophys Acta. 2007;1773:1299–1310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, 2nd, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nature cell biology. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- Crespo P, Leon J. Ras proteins in the control of the cell cycle and cell differentiation. Cell Mol Life Sci. 2000;57:1613–1636. doi: 10.1007/PL00000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppmann CD, Mihalas S, Sharma N, Lonze BE, Niebur E, Ginty DD. Science. Vol. 320. New York, N.Y: 2008. A model for neuronal competition during development; pp. 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkmans TF, van Hooijdonk LW, Schouten TG, Kamphorst JT, Vellinga AC, Meerman JH, Fitzsimons CP, de Kloet ER, Vreugdenhil E. Temporal and functional dynamics of the transcriptome during nerve growth factor-induced differentiation. Journal of neurochemistry. 2008;105:2388–2403. doi: 10.1111/j.1471-4159.2008.05338.x. [DOI] [PubMed] [Google Scholar]
- Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007;128:225–231. doi: 10.1016/j.cell.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci U S A. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annual review of cell and developmental biology. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockyer PJ, Wennstrom S, Kupzig S, Venkateswarlu K, Downward J, Cullen PJ. Identification of the ras GTPase-activating protein GAP1(m) as a phosphatidylinositol-3,4,5-trisphosphate-binding protein in vivo. Curr Biol. 1999;9:265–268. doi: 10.1016/s0960-9822(99)80116-x. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Li S, Iwamatsu A, Morishita T, Yokota K, Imai Y, Kohsaka S, Nakamura S, Hattori S. A novel mammalian Ras GTPase-activating protein which has phospholipid-binding and Btk homology regions. Mol Cell Biol. 1994;14:6879–6885. doi: 10.1128/mcb.14.10.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Morgan DO. The cell cycle: principles of control. New Science Press; Sinauer Associates; London Sunderland, MA: 2007. [Google Scholar]
- Myers JW, Jones JT, Meyer T, Ferrell JE., Jr. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nature biotechnology. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- Niepel M, Spencer SL, Sorger PK. Non-genetic cell-to-cell variability and the consequences for pharmacology. Current opinion in chemical biology. 2009;13:556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. 2003;3:317–330. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- Salmeen A, Park BO, Meyer T. The NADPH oxidases NOX4 and DUOX2 regulate cell cycle entry via a p53-dependent pathway. Oncogene. 2010;29:4473–4484. doi: 10.1038/onc.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nature cell biology. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- Sasagawa S, Ozaki Y, Fujita K, Kuroda S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nature cell biology. 2005;7:365–373. doi: 10.1038/ncb1233. [DOI] [PubMed] [Google Scholar]
- Shao J, Sheng H, DuBois RN, Beauchamp RD. Oncogenic Ras-mediated cell growth arrest and apoptosis are associated with increased ubiquitin-dependent cyclin D1 degradation. J Biol Chem. 2000;275:22916–22924. doi: 10.1074/jbc.M002235200. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kriegsheim A, Baiocchi D, Birtwistle M, Sumpton D, Bienvenut W, Morrice N, Yamada K, Lamond A, Kalna G, Orton R, et al. Cell fate decisions are specified by the dynamic ERK interactome. Nature cell biology. 2009;11:1458–1464. doi: 10.1038/ncb1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Valletta JS, Grimes ML, Mobley WC. Multiple levels for regulation of TrkA in PC12 cells by nerve growth factor. Journal of neurochemistry. 1995;65:1146–1156. doi: 10.1046/j.1471-4159.1995.65031146.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.