Abstract
Lapatinib is an oral 4-anilinoquinazoline derivative that dually inhibits epidermal growth factor receptor and human epidermal growth factor receptor 2 (HER2). This drug is a mere decade old and has only been approved by the FDA for the treatment of breast cancer since 2007. Consequently, the intricacies of the pharmacokinetics are still being elucidated. In the work presented herein, we determined the biodistribution of orally administered lapatinib in mouse plasma, brain, heart, lung, kidney, intestine, liver, muscle and adipose tissue. Using this data, we subsequently developed a physiologically based pharmacokinetic (PBPK) model of lapatinib in mice that accurately predicted the tissue concentrations after doses of 30, 60 and 90 mg/kg. By taking into account interspecies differences in physiology and physiochemistry, we then extrapolated the mouse PBPK model to humans. Our model predictions closely reflected lapatinib plasma pharmacokinetics in healthy subjects. Additionally, we were also able to simulate the pharmacokinetics of this drug in the plasma of patients with solid malignancies by incorporating a decrease in liver metabolism into the model. Finally, our PBPK model also facilitated the estimation of various human tissue exposures to lapatinib, which harmonize with the organ-specific toxicities observed in clinical trials. This first-generation PBPK model of lapatinib can be further improved with a greater understanding of lapatinib absorption, distribution, metabolism and excretion garnered from subsequent in vitro and in vivo studies and expanded to include other pharmacokinetic determinants, including efflux transporters, metabolite generation, combination dosing, etc., to better predict lapatinib disposition in both mouse and man.
Keywords: Breast cancer, Lapatinib, Physiologically based pharmacokinetic modeling, Tyrosine kinase inhibitor
Introduction
Lapatinib is an oral 4-anilinoquinazoline derivative that dually inhibits epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor 2 (HER2) (estimated values of 3 and 13 nM, respectively) by competing with ATP [1]. Aberrant signaling of these tyrosine kinases is prevalent in various types of solid tumors, thus making them attractive therapeutic targets. Presently, lapatinib is approved by the US Food and Drug Administration (FDA) in combination with capecitabine for the treatment of HER2 positive metastatic breast cancer and in combination with letrozole for the treatment of hormone receptor positive, HER2 positive metastatic breast cancer. In addition, there are approximately 250 current clinical trials in cancer patients involving this drug [2].
Numerous preclinical studies and clinical trials have investigated the plasma pharmacokinetics of lapatinib [3–15]. However, none have elucidated the biodistribution of this compound in tissues other than blood. Based on adverse reactions reported in humans (including cardiac, hepatic, gastrointestinal and lung toxicities), it can be presumed that there are significant levels of drug in these organs.
To empirically determine both plasma and organ exposure to lapatinib, we developed a physiologically based pharmacokinetic (PBPK) model in mice and then scaled this model to humans. This type of pharmacologic modeling is a useful tool that facilitates the prediction of target tissue drug concentrations by incorporating mathematical descriptions of the uptake and disposition of chemicals based on quantitative interrelations among the critical determinants of physiological processes (i.e., absorption, metabolism, excretion and tissue solubility phenomena) [16]. Accordingly, PBPK models are comprised of compartments corresponding to discrete tissues or groupings of tissues with appropriate volumes, blood flows, and pathways for xenobiotic clearance including pertinent biochemical and physiochemical constants [17]. Each compartment in the model is described with a mass-balance differential equation whose terms mathematically represent biological processes; the set of equations is then solved by numerical integration to simulate tissue time-course concentrations of chemicals and their metabolites [17]. The PBPK model of lapatinib presented herein consisted of eight tissue compartments (plasma, brain, heart, lung, kidney, intestine, liver and slowly perfused tissues) and incorporated drug absorption, intestinal and hepatic metabolism and fecal elimination in both mouse and man.
Materials and methods
Chemicals
Lapatinib (GW572016) and GW572016AH were generously provided by GlaxoSmithKline. Hydroxypropyl methylcellulose and Tween® 80 were purchased from Sigma-Aldrich. All other reagents were of analytical grade.
Lapatinib pharmacokinetic studies in mice
Five to six-week-old female FVB mice were purchased from Taconic. Animals were housed in polycarbonate cages and kept on a 12 h light/dark cycle. Food and water were given ad libitum. All experimental procedures were approved by Colorado State University’s Animal Care and Use Committee and the Department of Defense US Army Medical Research and Material Command (USAMRMC) Animal Care and Use Review Office (ACURO).
Upon arrival, mice acclimated for a minimum of seven days prior to any experimentation. After acclimation, a time course distribution study of lapatinib was conducted at doses of 30, 60 and 90 mg/kg. Lapatinib was formulated as a suspension in 0.5 % hydroxypropyl methylcellulose: 0.1 % Tween® 80 in Milli-Q water and was administered via oral gavage as a single bolus dose. Subsequently, three mice were sacrificed at 0.25, 0.5, 1, 2, 4, 8, 12 and 16 h by cardiac stick exsanguination under isoflurane anesthesia. Plasma, brain, liver, proximal small intestine, kidney, heart, lung, muscle and adipose tissue were immediately collected, rinsed with phosphate buffered saline, frozen in liquid nitrogen and stored at −80 °C until analysis.
Lapatinib high-pressure liquid chromatography-tandem mass spectrometry analysis
Analysis of lapatinib in plasma and tissue was done using high-pressure liquid chromatography-tandem mass spectrometry (LC/MS/MS) analysis based on the method of Bai et al. [18], modified as follows. Briefly, lapatinib was extracted from plasma by adding 210 μL of acetonitrile and 10 μL of internal standard (17.2 pmol GW572016AH) to 100 μL of unknown sample plasma, vortexing for 10 min and centrifuging at 18,000×g for 10 min at 4 °C. An aliquot of 20 μL of the supernatant was injected into the LC/MS/MS system for analysis. Tissues (brain, liver, proximal small intestine, kidney, heart, lung, muscle and adipose) were homogenized at 100 mg/mL in water and 100 μL of the homogenates were extracted using the method for plasma detailed above. Standards and quality control samples were prepared in the appropriate matrix and analyzed as described above.
The HPLC system consisted of an Agilent 1200 Series binary pump SL, vacuum degasser, thermostatted column compartment SL (Agilent Technologies, Santa Clara, CA, USA) and a CTC Analytics HTC PAL System autosampler (Leap Technologies, Carrboro, NC, USA). The HPLC column was a Waters Sunfire C8 column (4.6 × 50 mm I.D., 2.5 μm bead size) (Waters Corporation, Milford, MA, USA) protected by a SecurityGuard™ C18 cartridge (4 × 2.0 mm I.D.) (Phenomenex, Torrance, CA, USA) and maintained at room temperature. The mobile phase consisted of an aqueous component (A) of 20 mM ammonium formate in MilliQ water, pH 2.2 (with formic acid) and an organic component (B) of acetonitrile with 1 % formic acid. The 3.5 min run consisted of the following linear gradient elution: 95 % A and 5 % B at 0 min, 95 % A and 5 % B at 0.25 min, 25 % A and 75 % B at 0.35 min, 25 % A and 75 % B at 3.0 min, 95 % A and 5 % B at 3.1 min and 95 % A and 5 % B at 3.5 min. The system operated at a flow-rate of 0.75 mL/min.
Mass spectrometric detection was performed on an API 3200™ triple quadrupole instrument (Applied Biosystems Inc., Foster City, CA, USA) using multiple reaction monitoring (MRM). Ions were generated in positive ionization mode using an electrospray interface. Lapatinib compound-dependent parameters were as follows: declustering potential (DP): 60 V; entrance potential (EP): 10 V; collision cell entrance potential (CEP): 21 V; collision energy (CE): 51 V and collision cell exit potential (CXP): 5.8 V. GW572016AH (internal standard) compound-dependent parameters were as follows: DP: 67 V; EP: 7.5 V; CEP: 23 V; CE: 49 V and CXP: 5.5 V. Source-dependent parameters were as follows: nebulizer gas (GS1): 50 psi; auxiliary (turbo) gas (GS2): 60 psi; turbo gas temperature (TEM): 500 °C; curtain gas [7]: 10 psi; collision-activated dissociation (CAD) gas (nitrogen): 6 psi; ionspray voltage (IS): 5,000 V and interface heater (IH): 500 °C. Peak areas ratios obtained from MRM of lapatinib (m/z 581 → 365.1) and GW572016AH (m/z 587 → 367) were used for quantification.
The lower limit of quantitation for this assay was 1 ng/mL for plasma and 5 ng/g for tissues. The accuracy for the assay was 95.61 ± 4.60 % in plasma and 95.83 ± 3.47 % in tissues. The precision of the assay was 1.97 % in plasma and 3.75 % in tissues.
PBPK model development
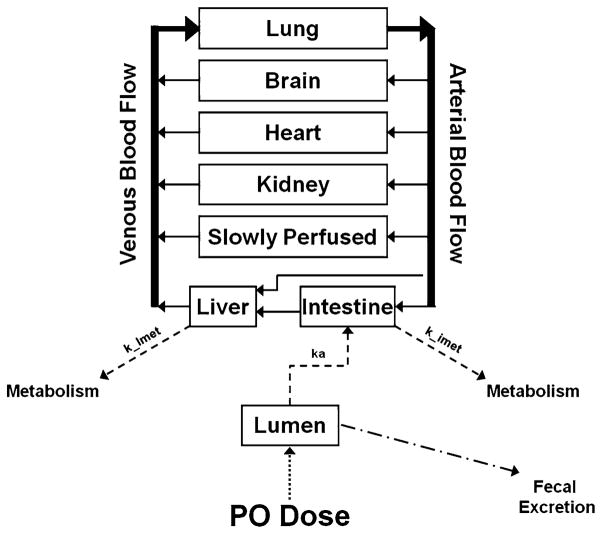
A PBPK model for lapatinib was developed incorporating absorption, intestinal and hepatic metabolism and fecal elimination. This flow-limited model was comprised of eight tissue compartments: plasma, brain, heart, lung, kidney, intestine, liver and slowly perfused tissue (Fig. 1). Physiological parameters (tissue volumes and tissue blood flows) were obtained from Brown et al. [19] and are shown in Table 1.
Fig. 1.
Schematic representation of a physiologically based pharmacokinetic (PBPK) model of lapatinib. Solid arrows represent blood flow. Dashed lines represent first-order rate constants for absorption from intestinal lumen (ka), hepatic metabolism (k_lmet) and intestinal metabolism (k_imet). The dotted line represents lapatinib input into the system via per os (p.o.) dosing. Drug remaining in the lumen is eliminated via fecal excretion
Table 1.
PBPK model parameter values
| Parameter | Units | Mouse | Human |
|---|---|---|---|
| Lapatinib properties | |||
| Molecular weight | 581.06 g/mol | ||
| Percent unbound | 1 % | ||
| Tissue volumea | % of body weight | ||
| Blood | 4.90 | 7.9 | |
| Brain | 1.65 | 2.0 | |
| Heart | 0.50 | 0.5 | |
| Lung | 0.73 | 0.8 | |
| Kidney | 1.67 | 0.4 | |
| Intestine | 4.22 | 1.7 | |
| Liver | 5.49 | 2.6 | |
| Slowly perfusedb | 80.84 | 84.1 | |
| Tissue blood flowa | % of cardiac output | ||
| Brain | 3.3 | 11.4 | |
| Heart | 6.6 | 4.0 | |
| Lung | 100 | 100 | |
| Kidney | 9.1 | 17.5 | |
| Intestine | 14.1 | 18.1 | |
| Liver | 2.0 | 4.6 | |
| Slowly perfusedb | 64.9 | 44.4 | |
| Partition coefficientsc | Ratio | ||
| Brain:plasma | 10 (19) | 10 | |
| Heart:plasma | 215 (22) | 215 | |
| Lung:plasma | 1,643 (19) | 1,643 | |
| Kidney:plasma | 1,064 (18) | 1,064 | |
| Intestine:plasma | 531 (31) | 531 | |
| Liver:plasma | 12 (20) | 12 | |
| Slowly perfused:plasma | 65 (20) | 65 | |
| Absorption rate constantsd | h−1 | ||
| Lumen → Intestine | 0.237 (2) | 0.07 (6) | |
| Metabolism rate constants | h−1 | ||
| Liverd | 127 (13) | 75 (5) | |
| Intestine | 2.5e | 0.975f |
Physiological parameters obtained from Brown et al. [19]
Slowly perfused tissue parameters calculated as the remaining percent
Determined by parameter estimation optimized for observed plasma and tissue concentrations from mouse 60 mg/kg dose cohort. Data is parameter estimate (CV%)
First-order rate constants determined by parameter estimation optimized for observed plasma and tissue concentrations from mouse 60 mg/kg dose cohort for mouse model and observed plasma concentrations from healthy subject human data for human model. Data is parameter estimate (CV%)
Calculated as 2 % of liver metabolism
Calculated as 1.3 % of liver metabolism
The unbound fraction of drug in the plasma was set at 0.01 (1 %), as lapatinib is highly bound (>99 %) to albumin and alpha-1 acid glycoprotein [1]. The arterial blood drug concentration available to all tissues except liver was considered to be the unbound lapatinib concentration in the blood. Both unbound and bound lapatinib were available for uptake into the liver.
Tissue:plasma partition coefficients were determined by parameter estimation, optimizing the fit for the observed plasma and tissue concentrations from the mouse 60 mg/kg dose cohort. These fitted values were compared with values calculated as detailed in Chen and Gross [44] using our experimental data (Online Resource 1). For these calculations, we used our concentration–time data from the mouse 60 mg/kg dose study and considered the terminal elimination phase to include the 4, 8, 12 and 16 h time points. The tissue:plasma partition coefficients were calculated as , where and are the tissue and plasma intercepts (initial concentrations), respectively, from the concentration–time curves of the terminal elimination phase on a semilogarithmic plot. After the partition coefficients were determined, the values had to be adjusted because when we measured the plasma concentrations via LC/MS/MS, we analyzed both bound and unbound drug in the plasma. Thus, to correct the partition coefficients so they reflected only the unbound drug available for tissue uptake (1 % unbound), we multiplied the calculated value by 100. The Chen and Gross method [44] was applicable for the determination of kidney, lung and slowly perfused tissue (adipose and muscle were used as representative slowly perfused tissues) partition coefficients. However, for brain and heart partition coefficients, we were unable to utilize this method because the criteria for implementation of this equation were not met for these two tissues (K/Q was not ≪ 1, where K is the organ clearance and Q is the blood flow). Additionally, for liver and intestine partition coefficients, Chen and Gross [44] describes unique equations, which we could not use because we did not have the values for all necessary variables. Therefore, because we were only able to determine three of the seven tissue partition coefficients using the Chen and Gross equations [44], we chose to estimate all partition coefficients by fitting these parameters to the model. For kidney, lung and slowly perfused tissue partition coefficients, the fitted values were 37, 7 and 12 % different than the calculated values, respectively. For brain, heart, liver and intestine partition coefficients, the fitted values were 10, 42, 29 and 16 % different than the calculated values, respectively.
The first-order rate constants for absorption from intestinal lumen and hepatic metabolism were determined by parameter estimation. For the mouse model, the fit was optimized for the observed plasma and tissue concentrations from the mouse 60 mg/kg dose cohort. For the human model, the fit was optimized for the observed plasma concentrations from a single 100 mg dose study conducted by GlaxoSmithKline in healthy subjects (n = 21).
The first-order rate constant for intestinal metabolism was estimated as a constant percentage of hepatic metabolism based on the ratio of total liver:intestinal CYP3A, the major cytochrome P450 enzyme sub-family responsible for lapatinib metabolism [1]. In mice, the mean quantity of immunoreactive CYP3A is 2.24 and 0.64 pmol/mg microsomal protein in liver and intestinal microsomes, respectively [45]. The total amount of microsomal protein in a 20 g mouse liver (5.49 % body weight [19] = 1.098 g) and small intestine is 38.9 mg (35.4 mg hepatic microsomal protein/g liver [46] × 1.098 g) and 2.67 mg [47], respectively. Accordingly, the total amount of CYP3A in a 20 g mouse liver and small intestine is 87.136 and 1.709 pmol, respectively. As a result, we concluded that the first-order rate constant for intestinal metabolism in mice is 2 % that of liver metabolism. In humans, total hepatic and small intestine (duodenum, jejunum and ileum) CYP3A was calculated to be 5,490 and 70.5 nmol, respectively [48]. Therefore, we represented the first-order rate constant for human intestinal metabolism as 1.3 % that of liver metabolism. This ratio held true for the microsomal intrinsic clearance of midazolam, a CYP3A-specific substrate, which was 15800 mL/min and 213.7 mL/min (or 1.35 %) in human liver and small intestine, respectively [48].
Table 1 lists all parameter values for both the mouse and human PBPK models.
The rate of change of the amount of drug in a generic storage tissue compartment mass balance equation is as follows:
where AT is the amount of drug in the tissue compartment, t is time, QT is the blood flow to the tissue compartment, CA is the arterial blood drug concentration entering the tissue compartment and CVT is the venous blood drug concentration exiting the tissue compartment. Assuming venous equilibration, the drug concentration in the venous blood is:
where CT is the concentration of drug in the tissue compartment and PT is the tissue:plasma partition coefficient.
Assuming the volume of the tissue (VT) is constant, the drug concentration in the tissue is:
For metabolizing tissues (liver and intestine), the rate of change of the amount of drug metabolized (AM) is as follows:
where k is a first-order rate constant.
Computer simulation
For PBPK modeling, acslX Libero version 3.0.2.1 (The AEgis Technologies Group, Inc.) was used.
Pharmacokinetic analysis
Pharmacokinetic parameters were calculated using non-compartmental modeling performed with Microsoft Excel and standard equations for noncompartmental analysis.
Data analysis
The predictive capability of the model was evaluated by calculating the prediction error (PE%) as follows [20, 21]:
As a measure of the precision of the prediction, the median absolute prediction error (MAPE%) was calculated as follows:
As a measure of the bias of the prediction, the median prediction error (MPE%) was calculated as follows:
Sensitivity analysis
A normalized sensitivity analysis was performed as described in Loccisano et al. [22] to assess the influence of each PBPK model parameter on the simulated plasma area under the concentration–time curve (AUC) for both the mouse and human models. Briefly, sensitivity coefficients were calculated with the original parameters and for those resulting from a 1 % change in each parameter value. The following equation was used to calculate the normalized sensitivity coefficient (SC):
where A is the AUC resulting from the 1 % increase in the parameter value, B is the AUC resulting from the original parameter value, C is the parameter value increased by 1 % and D is the original parameter value.
Results
Lapatinib pharmacokinetics and model simulations in mice
A time course tissue distribution study of lapatinib was conducted in female FVB mice. Plasma and tissue concentrations were measured after single oral doses of 30, 60 and 90 mg/kg at 0.25, 0.5, 1, 2, 4, 8, 12 and 16 h post drug administration. These time points were chosen for sacrifice to provide multiple samplings during each pharmacokinetic phase (absorption, distribution and elimination).
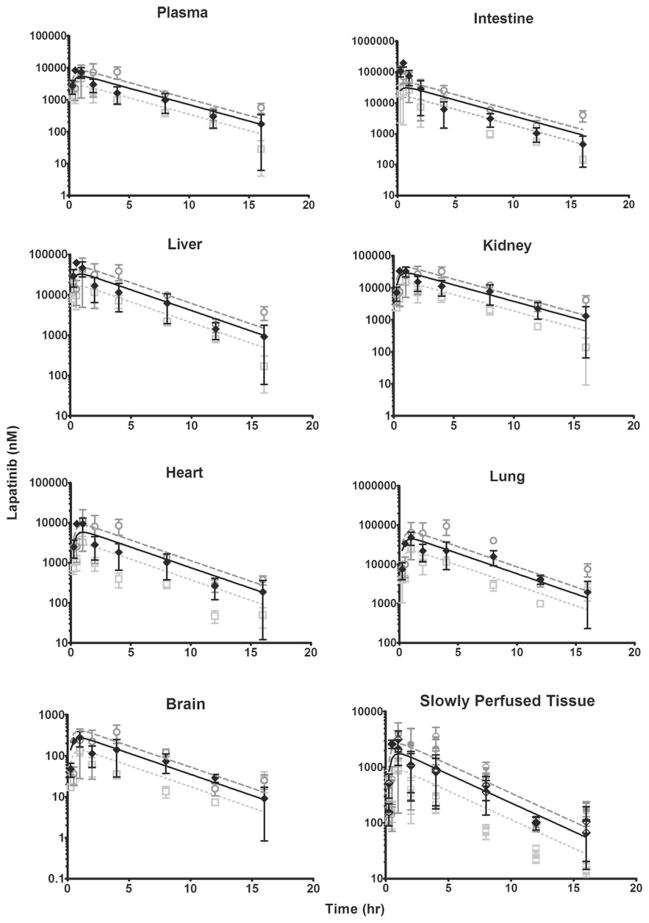
The mouse PBPK model development was based on the concentration–time data from the 60 mg/kg dose cohort; partition coefficients and first-order rate constants were determined by parameter estimation, optimizing the fit for the observed plasma and tissue concentrations from this study. The concentration–time profiles of lapatinib in plasma, intestine, liver, kidney, heart, lung, slowly perfused tissue and brain and the resulting PBPK model simulations are shown in Fig. 2. For all tissues except intestine, the PBPK model simulations closely mirrored the observed data.
Fig. 2.
Observed and model-simulated lapatinib concentrations in mouse plasma, intestine, liver, kidney, heart, lung, brain and slowly perfused tissue after oral gavage dosing of 30, 60 and 90 mg/kg. In all graphs except slowly perfused tissue, open light gray squares, filled diamonds and open dark gray circles represent the observed data from the 30, 60 and 90 mg/kg cohorts, respectively. In the slowly perfused tissue graph, the observed data from the 30 mg/kg dose cohort is represented by the upper half-filled light gray squares (adipose tissue) and lower half-filled light gray squares (muscle tissue); the observed data from the 60 mg/kg dose cohort is represented by upper half-filled black diamonds (adipose tissue) and lower half-filled black diamonds (muscle tissue); and the observed data from the 90 mg/kg dose cohort is represented by upper half-filled dark gray circles (adipose tissue) and lower half-filled dark gray circles (muscle tissue). For all observed data, error bars symbolize the standard error of the mean (SEM). Light gray dottedlines, solid black lines, and dark gray dashed lines represent model simulations for the 30, 60 and 90 mg/kg dose cohorts, respectively
The model-predicted intestine concentrations for the first four time points (0.25, 0.5, 1 and 2 h) are significantly lower than the actual data. We suspect that the observed data is not an accurate measurement of the drug concentration in the intestinal epithelium. Instead, the measured values reflect both the lapatinib in the intestinal epithelium and unabsorbed lapatinib in the proximal intestinal lumen. As an attempt to circumvent this anticipated problem, we flushed the intestinal lumen with saline immediately after tissue collection; however, we still noted yellow aggregates of undissolved lapatinib within the lumen (resulting from administration of the drug as a suspension via oral gavage). Thus, the measured drug concentrations in the intestine are likely inflated due to the lapatinib suspension in the proximal intestinal lumen. After approximately 3 h, the model simulation accurately reflects the observed values. It is probable that the lapatinib suspension has moved through the intestinal lumen by this time, as the intestinal transit time in a mouse is approximately 3 h. Therefore, at these later time points, the measured drug is presumably only lapatinib that has been absorbed into the intestinal epithelium.
After developing the mouse PBPK model with the 60 mg/kg dose cohort as a training set, we employed the other two dose cohorts (30 and 90 mg/kg) as test sets. The concentration–time data and the corresponding model simulations for these dose cohorts are also presented in Fig. 2. Again, the model simulations approximated the observed data with the exception of the early time points in the intestine, likely a result of the same phenomenon as described previously for the 60 mg/kg dose cohort.
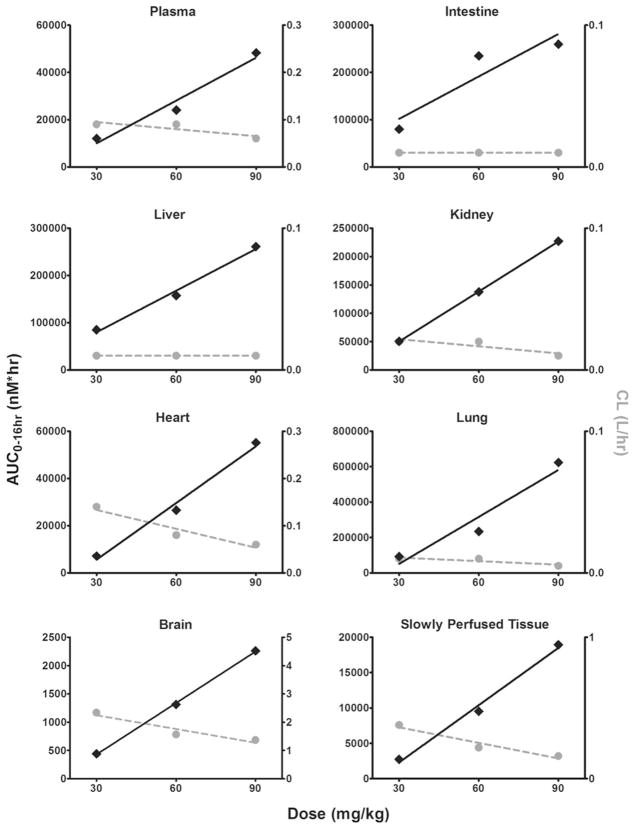
The area under the concentration–time curve from 0 to 16 h (AUC0–16 h), clearance (CL) and elimination half-life (t1/2) were calculated for both the observed and simulated data using noncompartmental analysis (Table 2). Lapatinib exhibits linear pharmacokinetics in all tissues within the 30–90 mg/kg dose range, as evidenced by a dose-dependent increase in AUC0–16 h and constant CL (Fig. 3). To compare the actual and predicted data, we determined the ratio of the observed to model-predicted values (Table 2). The mean AUC0–16 h ratio for all tissues was 1.00 and the range was 0.48 (heart from the 30 mg/kg dose cohort) to 1.81 (lung from the 90 mg/kg dose cohort), indicating that our model-predicted drug exposures reasonably mimicked the observed exposure for all tissues analyzed. As for CL, the model predictions also emulated the actual data; all ratios were between 0.45 (intestine from the 90 mg/kg dose cohort) and 2.10 (heart from the 30 mg/kg dose cohort), with the average ratio being 1.06. Lastly, all t1/2 ratios were within the range of 0.52 (slowly perfused tissue from the 90 mg/kg dose cohort) and 1.24 (brain from the 30 mg/kg dose cohort), with an average ratio of 0.90. Overall, the PK parameters derived from the PBPK model simulations accurately mirrored the observed mouse data.
Table 2.
Observed and model-simulated lapatinib pharmacokinetic (PK) parameters in micea
| PK parameter | Dose | Plasma | Intestine | Liver | Kidney | Heart | Lung | Brain | Slowly perfused | |
|---|---|---|---|---|---|---|---|---|---|---|
| AUC0–16 h (nM × h)b | 30 | Observed | 12,008 | 79,885 | 84,946 | 50,440 | 7,156 | 92,409 | 440.8 | 2,738 |
| Simulated | 13,972 | 78,143 | 83,505 | 74,392 | 15,022 | 114,792 | 698.6 | 4,544 | ||
| Ratio | 0.86 | 1.02 | 1.02 | 0.68 | 0.48 | 0.81 | 0.63 | 0.60 | ||
| 60 | Observed | 24,052 | 235,114 | 157,288 | 137,624 | 26,545 | 233,760 | 1,315 | 9,519 | |
| Simulated | 27,944 | 156,284 | 167,010 | 148,784 | 30,044 | 229,583 | 1,397 | 9,087 | ||
| Ratio | 0.86 | 1.50 | 0.94 | 0.92 | 0.88 | 1.02 | 0.94 | 1.05 | ||
| 90 | Observed | 48,230 | 259,808 | 261,237 | 227,184 | 55,179 | 623,277 | 2,261 | 18,930 | |
| Simulated | 41,916 | 234,426 | 250,515 | 223,176 | 45,066 | 344,375 | 2,096 | 13,631 | ||
| Ratio | 1.15 | 1.11 | 1.04 | 1.02 | 1.22 | 1.81 | 1.08 | 1.39 | ||
| CL (L/h)c | 30 | Observed | 0.09 | 0.01 | 0.01 | 0.02 | 0.14 | 0.01 | 2.34 | 0.38 |
| Simulated | 0.07 | 0.01 | 0.01 | 0.01 | 0.07 | 0.01 | 1.48 | 0.23 | ||
| Ratio | 1.16 | 0.98 | 0.98 | 1.47 | 2.10 | 1.24 | 1.58 | 1.66 | ||
| 60 | Observed | 0.09 | 0.01 | 0.01 | 0.02 | 0.08 | 0.01 | 1.57 | 0.22 | |
| Simulated | 0.07 | 0.01 | 0.01 | 0.01 | 0.07 | 0.01 | 1.48 | 0.23 | ||
| Ratio | 1.16 | 0.66 | 1.06 | 1.08 | 1.13 | 0.98 | 1.06 | 0.95 | ||
| 90 | Observed | 0.06 | 0.01 | 0.01 | 0.01 | 0.06 | 0.005 | 1.37 | 0.16 | |
| Simulated | 0.08 | 0.03 | 0.01 | 0.01 | 0.07 | 0.01 | 1.48 | 0.22 | ||
| Ratio | 0.78 | 0.45 | 0.96 | 0.98 | 0.79 | 0.55 | 0.93 | 0.73 | ||
| t1/2 (h)d | 30 | Observed | 2.73 | 2.79 | 2.48 | 2.53 | 2.59 | 2.39 | 3.70 | 3.88 |
| Simulated | 2.99 | 2.96 | 2.95 | 3.00 | 2.99 | 2.99 | 2.99 | 2.99 | ||
| Ratio | 0.91 | 0.94 | 0.84 | 0.84 | 0.87 | 0.80 | 1.24 | 1.30 | ||
| 60 | Observed | 3.42 | 3.05 | 2.75 | 3.57 | 3.05 | 3.04 | 3.12 | 2.57 | |
| Simulated | 2.99 | 2.96 | 2.95 | 3.00 | 2.99 | 2.99 | 2.99 | 2.99 | ||
| Ratio | 1.14 | 1.03 | 0.93 | 1.19 | 1.02 | 1.02 | 1.04 | 0.86 | ||
| 90 | Observed | 2.36 | 2.55 | 1.91 | 2.51 | 2.32 | 1.72 | 1.76 | 1.55 | |
| Simulated | 2.99 | 2.96 | 2.95 | 3.00 | 2.99 | 2.99 | 2.99 | 2.99 | ||
| Ratio | 0.79 | 0.86 | 0.65 | 0.83 | 0.78 | 0.58 | 0.59 | 0.52 |
PK parameters were calculated using noncompartmental modeling
AUC0–16 h is the area under the concentration–time curve from 0 to 16 h
CL is clearance
t1/2 is the half-life for elimination as calculated from linear regression of the terminal elimination phase
Fig. 3.
Area under the concentration–time curve calculated from 0 to 16 h (AUC0–16 h) and clearance (CL) for the mouse 30, 60 and 90 mg/kg dose cohorts in plasma, intestine, liver, kidney, heart, lung, brain and slowly perfused tissue. AUC0–16 h is presented on the left y axis and is represented by the solid black diamonds, with the corresponding linear regression trendline shown as the solid black line. CL is presented on the right y axis and is represented by the solid gray circles with the corresponding linear regression trendline shown as the dashed gray line. Both AUC0–16 h and CL were determined by noncompartmental analysis
To further assess the predictive performance of the mouse model, we calculated the median prediction error (MPE%) and the median absolute prediction error (MAPE%) for the concentrations, AUCs0–16 h and half-lives as measures of the bias and precision of the simulations, respectively (Table 3). Of these three variables, the concentrations were the most poorly predicted, with a mean MPE% of 28.6 and a mean MAPE% of 57.4. Although these prediction error assessments are not optimal, they are not surprising considering the large degree of variability in the data (mean concentration coefficient of variation of 78.6 %), likely due to the variable absorption of lapatinib when administered to unfasted animals. AUC0–16 h and t1/2 prediction errors were substantially better than the concentration prediction errors, feasibly because these parameters are derived from the cumulation of the concentration values and thus, the error of the individual points is muted. For AUC0–16 h, the average MPE% was 14.5 and the average MAPE% was 3.2. The MPE% for plasma and all tissue AUCs0–16 h was less than 28.0 and the MAPE% was less than 16.2. Regarding half-life, the average MPE% was 13.2 and the average MAPE% was 17.4, with no individual plasma or tissue MPE% and MAPE% being more than ±25.2 and 25.2 %, respectively.
Table 3.
Predictive performance for mouse PBPK model
| Concentrations
|
AUC0–16ha
|
t1/2b
|
||||
|---|---|---|---|---|---|---|
| MPE%c | MAPE%d | MPE%c | MAPE%d | MPE%c | MAPE%d | |
| Plasma | 28.53 | 43.36 | 16.18 | 16.18 | 9.57 | 12.64 |
| Intestine | −8.40 | 68.10 | 9.77 | −9.77 | 6.09 | 6.09 |
| Liver | 23.57 | 51.03 | 4.10 | −1.70 | 19.11 | 19.11 |
| Kidney | 45.40 | 52.90 | 8.11 | 8.11 | 18.57 | 18.57 |
| Heart | 52.81 | 73.34 | 18.33 | 13.18 | 15.41 | 15.41 |
| Lung | 4.45 | 59.25 | 24.22 | −1.79 | 25.14 | 25.14 |
| Slowly perfused | 28.77 | 47.40 | 27.99 | −4.53 | 16.24 | 22.96 |
| Brain | 53.30 | 63.65 | 7.30 | 6.24 | −4.29 | 19.16 |
AUC0–16 h is the area under the concentration–time curve from 0 to 16 h
t1/2 is the half-life for elimination as calculated from linear regression of the terminal elimination phase
MPE% is the median prediction error, which is a measure of the bias of the prediction
MAPE% is the median absolute prediction error, which is a measure of the precision of the prediction
Lapatinib pharmacokinetics and model simulations in humans
The mouse PBPK model developed using the 60 mg/kg dose cohort was scaled to humans by using human parameters for tissue volumes and tissue blood flows and fitting the first-order rate constants for absorption and liver metabolism to the observed plasma concentrations from a single 100 mg dose study conducted by GlaxoSmithKline in healthy subjects (n = 21) (Table 1). The first-order rate constant for intestinal metabolism was set as 1.3 % that of liver metabolism as explained previously.
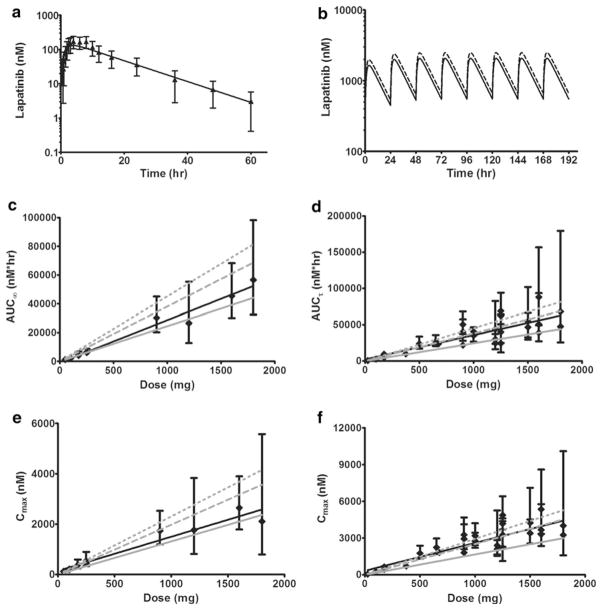
The concentration–time profiles of lapatinib in actual human plasma and the resulting PBPK model simulation are shown in Fig. 4a. The PBPK model prediction closely parallels the observed plasma concentration data. The MPE% and MAPE% for the lapatinib concentrations were − 8.17 and 11.69, respectively. Regarding the actual and simulated plasma pharmacokinetic parameters, AUCs0–60 h were 2,698 and 2,409 nM × h, CLs were 63.8 and 71.4 L/h and half-lives were 9.5 and 10.0 h, respectively. The AUCs0–60 h for plasma and all tissues in the model are shown in Table 4. From largest to smallest, exposure to lapatinib ranked as follows: intestine, lung, liver, kidney, heart, plasma, slowly perfused tissue and brain.
Fig. 4.
Observed and model-simulated lapatinib concentrations, area under the concentration–time curve (AUC) and maximum concentration (Cmax) in human plasma. a Single oral dose of 100 mg. Filled black triangles represent the observed data with error bars symbolizing the standard deviation (SD). The solid black line represents the model simulation. b Multiple doses (q24 h) for 8 days. Solid black line represents the model simulation for daily dosing of 1,250 mg. Dashed black line represents the model simulation for daily dosing of 1,500 mg. c After a single dose of lapatinib, solid black diamonds represent observed AUCs∞ (calculated from time 0 to infinity) with error bars symbolizing the 95 % confidence intervals and the solid black line is the corresponding linear regression trendline. The solid gray line represents the model-predicted AUCs∞. The dashed gray line represents simulated AUCs∞ from the model with moderate hepatic impairment. The dotted gray line represents simulated AUCs∞ from the model with severe hepatic impairment. d After multiple doses (q24 h) of lapatinib, solid black diamonds represent observed steady-state AUCsτ (calculated within the dosing interval from time 0 to 24 h) with error bars symbolizing the 95 % confidence intervals and the solid black line is the corresponding linear regression trendline. The solid gray line represents the model-predicted AUCs∞. The dashed gray line represents simulated AUCsτ from the model with moderate hepatic impairment. The dotted gray line represents simulated AUCsτ from the model with severe hepatic impairment. e After a single dose of lapatinib, solid black diamonds represent observed Cmax with error bars symbolizing the 95 % confidence intervals and the black line is the corresponding linear regression trendline. The solid gray line represents the model-predicted Cmax. The dashed gray line represents simulated Cmax from the model with moderate hepatic impairment. The dotted gray lines represents simulated Cmax from the model with severe hepatic impairment. f After multiple doses (q24 h) of lapatinib, solid black diamonds represent observed Cmax with error bars symbolizing the 95 % confidence intervals and the solid black line is the corresponding linear regression trendline. The solid gray line represents the model-predicted Cmax. The dashed gray line represents simulated Cmax from the model with moderate hepatic impairment. The dotted gray line represents simulated Cmax from the model with severe hepatic impairment
Table 4.
Human tissue AUCs for single and multiple (q24 h) lapatinib doses
| 100 mg single dose AUC0–60 ha(nM × h) | 1,250 mg multiple dose q24 h AUCτb (nM × h) | 1,500 mg multiple dose q24 h AUCτb(nM × h) | |
|---|---|---|---|
| Plasma | 2,409 | 30,631 | 36,757 |
| Intestine | 21,884 | 277,286 | 332,744 |
| Liver | 14,470 | 183,723 | 220,468 |
| Kidney | 12,817 | 162,959 | 195,550 |
| Heart | 2,590 | 32,929 | 39,514 |
| Lung | 19,792 | 251,637 | 301,964 |
| Brain | 121 | 1,532 | 1,838 |
| Slowly perfused | 783 | 9,955 | 11,946 |
AUC0–60 h is the area under the concentration–time curve from 0 to 60 h
AUCτ is the steady-state area under the concentration–time curve within the dosing interval (0–24 h)
Clinically, the recommended dose of lapatinib is 1,250 or 1,500 mg orally once daily continuously with either capecitabine (for advanced or metastatic breast cancer) or letrozole (for hormone receptor positive, HER2 positive metastatic breast cancer), respectively [1]. Thus, we modified our original model to incorporate multiple dosing of lapatinib. The resulting simulations of 1,250 and 1,500 mg doses of lapatinib q24 h for 8 days are shown in Fig. 4b. The steady-state area under the concentration–time curves calculated within the dosing interval from 0 to 24 h (AUCτ) for plasma and all tissues in the model are shown in Table 4.
To further assess the predictive performance of the human model, we were not able to accrue concentration–time data for any other subjects/patients so we compared our model-predicted AUC, half-life, maximum concentration (Cmax) and time of maximum concentration (Tmax) values with those found in the literature for both healthy subjects [4, 24] and patients with solid tumors [5–13, 15]. The results along with the subject/patient characteristics (disease state, fasted or not fasted when administered lapatinib, liver function and age) are presented in Table 5 (single dose lapatinib) and Tables 6, 7 (multiple dose lapatinib). Graphically, observed and predicted AUCs are depicted in Fig. 4c, d. For the single dose comparison, all of the prediction errors were less than ±27.2 %, with a MPE% of 0.29 and a MAPE% of 7.7. The single dose prediction errors were smaller for the area under the concentration–time curve calculated from time 0 to infinity (AUCs∞) of healthy subjects (MPE% of 1.5 and MAPE% of 2.5, n = 6 studies) than for the AUCs∞ of patients with solid tumors (MPE% of −17.8 and MAPE% of 17.8, n = 4 studies), which was not surprising given that our model was developed with data from healthy subjects who presumably cleared (metabolized) lapatinib more efficiently than the patients with advanced solid malignancies, as they were both younger and had normal liver function. Thus, our model tended to underpredict the AUC∞ for the patients with solid tumors, as indicated by the negative value of the MPE%. For the multiple dose lapatinib study, the prediction errors were larger, with a MPE% of −29.9 and a MAPE% of 29.9. Again, the negative MPE% was the result of our model simulations underpredicting lapatinib exposure, likely due to impaired hepatic function related to the age and disease state of the test population (n = 24 studies with cancer patients and only three studies with healthy subjects) versus the healthy training population used to develop the PBPK model.
Table 5.
Single dose lapatinib observed and predicted human AUC∞ and subject characteristics
| Dose (mg) | Observed AUC∞ (nM × h) | Predicted AUC∞ (nM × h) | AUC∞ PE%c | Subjects | Food | Bilirubind | ASTd | ALTd | Age (years)e | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 1,170 (756–1,817)a | 1,225 | 4.7 | Healthy | Fasted | Normal | Normal | Normal | 22 (18–53) | [4] |
| 100 | 2,096 (1,492–2,941)a | 2,450 | 16.9 | Healthy | Fasted | Normal | Normal | Normal | 22 (18–53) | [4] |
| 100 | 2,459 (2,062–2,933)b | 2,450 | −0.4 | Healthy | Not fasted | Normal | Normal | Normal | 28 (20–47) | [14] |
| 175 | 4,206 (2,588–6,834)a | 4,288 | 1.9 | Healthy | Fasted | Normal | Normal | Normal | 22 (18–53) | [4] |
| 250 | 6,313 (4,550–8,758)a | 6,126 | −3.0 | Healthy | Fasted | Normal | Normal | Normal | 22 (18–53) | [4] |
| 250 | 6,068 (4,970–7,411)b | 6,126 | 1.0 | Healthy | Not fasted | Normal | Normal | Normal | 29 (20–48) | [14] |
| 900 | 30,250 (20,328–45,011)a | 22,054 | −27.1 | Cancer | Fasted | ≤1.5× ULN | ≤2.5× ULN | ≤2.5× ULN | 60 (37–73) | [13] |
| 1,200 | 26,574 (12,753–55,375)a | 29,405 | 10.7 | Cancer | Fasted | ≤1.5× ULN | ≤2.5× ULN | ≤2.5× ULN | 60 (37–73) | [13] |
| 1,600 | 45,367 (30,150–68,263)a | 39,207 | −13.6 | Cancer | Fasted | ≤1.5× ULN | ≤2.5× ULN | ≤2.5× ULN | 60 (37–73) | [13] |
| 1,800 | 56,519 (32,499–98,293)a | 44,108 | −22.0 | Cancer | Fasted | ≤1.5× ULN | ≤2.5× ULN | ≤2.5× ULN | 60 (37–73) | [13] |
AUC∞ is the geometric mean (95 % confidence interval) of the area under the concentration–time curve calculated from time 0 to infinity
AUC∞ is the geometric mean (range) of the area under the concentration–time curve calculated from time 0 to infinity
PE% is the prediction error
Bilirubin, aspartate transaminase (AST) and alanine transaminase (ALT) are measures of liver function. ULN is upper limit of normal
Median age (range)
Table 6.
Multiple dose lapatinib (25–1,200 mg) observed and predicted human AUCτ and subject characteristics
| Dose (mg) | Observed AUCτ (nM × h)a | Predicted AUCτ (nM × h) | AUCτ PE%b | Subjects | Food | Bilirubinc | ASTc | ALTc | Age (years)d | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 515 (329–809) | 613 | 19.1 | Healthy | Fasted | Normal | Normal | Normal | 22 (19–38) | [4] |
| 100 | 25,68 (1,594–4,137) | 2,450 | −4.6 | Healthy | Fasted | Normal | Normal | Normal | 22 (19–38) | [4] |
| 175 | 4,869 (3,313–7,154) | 4,288 | −11.9 | Healthy | Fasted | Normal | Normal | Normal | 22 (19–38) | [4] |
| 175 | 9,431 | 4,288 | −54.5 | Cancer | Fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 61 (25–80) | [15] |
| 375 | 9,878 | 9,189 | −7.0 | Cancer | Fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 62 (25–81) | [15] |
| 500 | 23,922 (17,107–33,731) | 12,252 | −48.8 | Cancer | Not fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 56 (28–74) | [5] |
| 650 | 27,020 (20,480–35,797) | 15,928 | −41.1 | Cancer | Not fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 60 (37–82) | [5] |
| 675 | 23,578 | 16,540 | −29.8 | Cancer | Fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 63 (25–82) | [15] |
| 900 | 21,857 | 22,054 | 0.9 | Cancer | Fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 64 (25–83) | [15] |
| 900 | 40,099 (28,052–57,481) | 22,054 | −45.0 | Cancer | Not fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 57 (34–82) | [5] |
| 900 | 50,377 (37,204–68,217) | 22,054 | −56.2 | Cancer | Fasted | ≤1.5× ULN | ≤2.5× ULN | ≤2.5× ULN | 60 (37–73) | [13] |
| 1,000 | 40,615 (35,452–46,639) | 24,504 | −39.7 | Cancer | Not fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 53 (43–59) | [5] |
| 1,000 | 35,280 (26,847–46,295) | 24,504 | −30.5 | Cancer | Fasted | Adequate | Adequate | Adequate | 53 (30–80) | [9] |
| 1,200 | 24,610 (16,212–37,518) | 29,405 | 19.5 | Cancer | Not fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 54 (37–67) | [5] |
| 1,200 | 44,195 (23,626–82,673) | 29,405 | −33.5 | Cancer | Fasted | ≤1.5× ULN | ≤2.5× ULN | ≤2.5× ULN | 60 (37–73) | [13] |
| 1,200 | 29,773 | 29,405 | −1.2 | Cancer | Fasted | ≤2 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 65 (25–84) | [15] |
AUCτ is the geometric mean (95 % confidence interval) of the steady-state area under the concentration–time curve calculated within the dosing interval (0–24 h)
PE% is the prediction error
Bilirubin, aspartate transaminase (AST) and alanine transaminase (ALT) are measures of liver function. ULN is upper limit of normal
Median age (range)
Table 7.
Multiple dose lapatinib (1,250–1,800 mg) observed and predicted human AUCτ and subject characteristics
| Dose (mg) | Observed AUCτ (nM × h) | Predicted AUCτ (nM × h) | AUCτ PE%c | Subjects | Food | Bilirubind | ASTd | ALTd | Age (years)e | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 1,250 | 62,300 (40,271–96,376)a | 30,631 | −50.8 | Cancer | Not fasted | ≤1.5 mg/dL | ≤2.0× ULN | ≤2.0× ULN | 58.3 (2–79) | [7] |
| 1,250 | 68,840 (50,425–94,138)b | 30,631 | −55.5 | Cancer | Not fasted | Significant dysfunction excluded | 58 (34–78) | [8] | ||
| 1,250 | 24,438 (11,789–50,941)b | 30,631 | 25.3 | Cancer | Fasted | ≤1.5 mg/dL | ≤2.0× ULN | ≤2.0× ULN | 59 (19–74) | [10] |
| 1,250 | 40,099 (24,954–64,193)b | 30,631 | −23.6 | Cancer | Not fasted | ≤2.0× ULN | ≤5.0× ULN | ≤5.0× ULN | 57.5 (33–74) | [12] |
| 1,500 | 46,639 (32,699–66,430)b | 36,757 | −21.2 | Cancer | Not fasted | ≤1.5 mg/dL | ≤2.0× ULN | ≤2.0× ULN | 59.5 (39–73) | [6] |
| 1,500 | 54,900 (29,601–101,833)b | 36,757 | −33.0 | Cancer | Fasted | ≤1.5 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 57 (31–73) | [11] |
| 1,600 | 50,597 (27,364–93,450)b | 39,207 | −22.5 | Cancer | Not fasted | ≤2.0 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 55 (38–70) | [5] |
| 1,600 | 87,941 (49,348–156,717)b | 39,207 | −55.4 | Cancer | Fasted | ≤1.5× ULN | ≤2.5 × ULN | ≤2.5× ULN | 60 (37–73) | [13] |
| 1,600 | 39,067b | 39,207 | 0.4 | Cancer | Fasted | ≤2.0 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 66 (25–85) | [15] |
| 1,800 | 47,671b | 44,108 | −7.5 | Cancer | Fasted | ≤2.0 mg/dL | ≤3.0× ULN | ≤3.0× ULN | 67 (25–86) | [15] |
| 1,800 | 67,895 (25,658–179,656)b | 44,108 | −35.0 | Cancer | Fasted | ≤1.5× ULN | ≤2.5 × ULN | ≤2.5× ULN | 60 (37–73) | [13] |
AUCτ is the mean (90 % confidence interval) of the steady-state area under the concentration–time curve calculated within the dosing interval (0–24 h)
AUCτ is the geometric mean (95 % confidence interval) of the steady-state area under the concentration–time curve calculated within the dosing interval (0–24 h)
PE% is the prediction error
Bilirubin, aspartate transaminase (AST) and alanine transaminase (ALT) are measures of liver function. ULN is upper limit of normal
Median age (range)
Previously, lapatinib pharmacokinetics were assessed in subjects with moderate or severe hepatic impairment (Child-Pugh scores of 7–9, or greater than 9, respectively) and in 8 healthy control subjects; after a single oral dose of 100 mg, the lapatinib AUC increased approximately 56 and 85 % in subjects with moderate and severe hepatic impairment, respectively [25]. To imitate this liver dysfunction in our model, we decreased the first-order rate constant for liver metabolism by 35 and 45 % and, accordingly, achieved AUC increases of 56 and 85 %, respectively. Decreased liver metabolism of this magnitude has been observed in aged patients; a review of 16 cytochrome P450 (CYP) 3A substrates showed an average 37.2 % reduction in the clearance of these substrates by elderly versus young volunteers or patients [26]. The resulting AUC predictions from our modified model are graphed in Fig. 4c, d. The AUCs∞ resulting from hepatic impairment in the single dose studies both overpredicted exposure, conceivably because 60 % of the studies were done in healthy subjects. In contrast, the moderately impaired liver function simulation more correctly reflected the observed AUCsτ from the multiple dose lapatinib clinical trials in which 86 % of the studies were done in cancer patients. Thus, decreasing the liver metabolism in our model improves the lapatinib exposure predictions for cancer patients.
In addition to actual and simulated human lapatinib exposures, we also wanted to evaluate concentration–time curve shape parameters. Accordingly, we compared observed and predicted half-life, Cmax and Tmax (Tables 8, 9, 10). For single dose lapatinib, the model-predicted and mean observed (n = 10 studies) half-lives were 10.0 and 10.3 h, respectively. For multiple dose lapatinib, the model predicted and mean observed (n = 6 studies) half-lives were 10.2 and 16.6 h, respectively. Overall, half-life MPE% was −8.1 and MAPE% was 28.1. In healthy subjects, the model overpredicted the half-life in 78 % of the studies (MPE% of 14.6) and in cancer patients, the model underpredicted the half-life in all studies (MPE% of −38.0).
Table 8.
Single dose lapatinib observed and predicted human half-life (t1/2), maximum concentration (Cmax) and time of maximum concentration (Tmax)
| Dose (mg) | Observed t1/2 (h) | Predicted t1/2 (h) | T1/2 PE%c | Observed Cmax (nM) | Predicted Cmax (nM) | Cmax PE%c | Observed Tmax (h) | Predicted Tmax (h) | Tmax PE%c | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 6.0a (4.8–7.5) | 10.0 | 66.7 | 124 (88–177)d | 66 | −46.7 | 3.0 (2.0–6.0)f | 3.75 | 25.0 | [4] |
| 100 | 6.3a (5.6–7.0) | 10.0 | 58.7 | 213 (148–308)d | 132 | −38.1 | 4.0 (2.5–5.9)f | 3.75 | −6.3 | [4] |
| 100 | 9.6a (8.5–10.7) | 10.0 | 4.2 | 198 (174–224)e | 132 | −33.3 | 4.0 (2.5–8.0)f | 3.75 | −6.3 | [14] |
| 175 | 8.2a (6.7–9.9) | 10.0 | 22.0 | 380 (241–599)d | 231 | −39.3 | 3.0 (2.0–4.0)f | 3.75 | 25.0 | [4] |
| 250 | 8.8a (6.6–11.7) | 10.0 | 13.6 | 546 (330–902)d | 329 | −39.7 | 4.0 (3.0–6.0)f | 3.75 | −6.3 | [4] |
| 250 | 10.2a (9.24–11.3) | 10.0 | −2.0 | 449 (360–563)e | 329 | −26.8 | 4.0 (2.5–6.0)f | 3.75 | −6.3 | [14] |
| 900 | 12.9b (10.1–18.3) | 10.0 | −22.5 | 1,740 (1,194–2,533)d | 1,185 | −31.9 | 4.0 (2.0–6.0)g | 3.75 | −6.3 | [13] |
| 1,200 | 11.5b (10.1–19.5) | 10.0 | −13.0 | 1,767 (816–3,833)d | 1,581 | −10.5 | 3.5 (2.1–6.0)g | 3.75 | 7.1 | [13] |
| 1,600 | 13.9b (9.6–18.0) | 10.0 | −28.1 | 2,647 (1,793–3,903)d | 2,107 | −20.4 | 4.0 (2.0–8.0)g | 3.75 | −6.3 | [13] |
| 1,800 | 15.7b (11.0–133.1) | 10.0 | −36.3 | 2,112 (800–5,579)d | 2,371 | 12.3 | 3.9 (3.0–8.0)g | 3.75 | −3.8 | [13] |
t1/2 is the terminal half-life geometric mean (95 % confidence interval)
t1/2 is the terminal half-life median (95 % confidence interval)
PE% is the prediction error
Is the geometric mean (95 % confidence interval) of Cmax
Is the geometric mean (range) of Cmax
Is the median (range) of Tmax
Is the median (95 % confidence interval) of Tmax
Table 9.
Multiple dose lapatinib (25–1,200 mg) observed and predicted human half-life (t1/2), maximum concentration (Cmax) and time of maximum concentration (Tmax)
| Dose (mg) | Observed t1/2 (h) | Predicted t1/2 (h) | t1/2 PE%c | Observed Cmaxd (nM) | Predicted Cmax (nM) | Cmax PE%c | Observed Tmax (h) | Predicted Tmax (h) | Tmax PE%c | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 7.9a (6.4–9.8) | 10.2 | 29.1 | 55 (38–81) | 42 | −23.7 | 2.7 (2.5–4.0)e | 3.5 | 29.6 | [4] |
| 100 | 8.9a (6.1–12.9) | 10.2 | 14.6 | 251 (150–420) | 166 | −33.9 | 3.0 (2.0–6.0)e | 3.5 | 16.7 | [4] |
| 175 | 11.1a (7.3–16.8) | 10.2 | −8.1 | 429 (270–678) | 290 | −32.3 | 4.0 (3.0–6.0)e | 3.5 | −12.5 | [4] |
| 175 | ND | 10.2 | NA | 637 | 290 | −54.5 | ND | 3.5 | NA | [15] |
| 375 | ND | 10.2 | NA | 706 | 622 | −11.8 | ND | 3.5 | NA | [15] |
| 500 | ND | 10.2 | NA | 1,755 (1,308–2,375) | 830 | −52.7 | ND | 3.5 | NA | [5] |
| 650 | ND | 10.2 | NA | 2,237 (1,687–2,960) | 1,078 | −51.8 | ND | 3.5 | NA | [5] |
| 675 | ND | 10.2 | NA | 1,824 | 1,120 | −38.6 | ND | 3.5 | NA | [15] |
| 900 | ND | 10.2 | NA | 1,807 | 1,493 | −17.4 | ND | 3.5 | NA | [15] |
| 900 | ND | 10.2 | NA | 2,926 (2,082–4,130) | 1,493 | −49.0 | ND | 3.5 | NA | [5] |
| 900 | 23.1b (9.8–38.2) | 10.2 | −55.8 | 3,261 (2,270–4,683) | 1,493 | −54.2 | 4.0 (3.0–6.0)f | 3.5 | –12.5 | [13] |
| 1,000 | ND | 10.2 | NA | 3,184 (2,409–4,216) | 1,659 | −47.9 | ND | 3.5 | NA | [5] |
| 1,000 | ND | 10.2 | NA | 2,754 (2,203–3,442) | 1,659 | –39.8 | ND | 3.5 | NA | [9] |
| 1,200 | ND | 10.2 | NA | 2,100 (1,549–2,840) | 1,990 | −5.2 | ND | 3.5 | NA | [5] |
| 1,200 | 16.9b (15.1–34.3) | 10.2 | −39.6 | 2,952 (1,661–5,246) | 1,990 | −32.6 | 3.6 (3.0–7.9)f | 3.5 | −2.8 | [13] |
| 1,200 | ND | 10.2 | NA | 2,392 | 1,990 | −16.8 | ND | 3.5 | NA | [15] |
ND not determined, NA not applicable
t1/2 is the terminal half-life geometric mean (95 % confidence interval)
t1/2 is the terminal half-life median (95 % confidence interval)
PE% is the prediction error
Is the geometric mean (95 % confidence interval) of Cmax
Is the median (range) of Tmax
Is the median (95 % confidence interval) of Tmax
Table 10.
Multiple dose lapatinib (1,250–1,800 mg) observed and predicted human half-life (t1/2), maximum concentration (Cmax) and time of maximum concentration (Tmax)
| Dose (mg) | Observed t1/2 (h)a | Predicted t1/2 (h) | t1/2 PE%b | Observed (nM) | Predicted Cmax (nM) | Cmax PE%b | Observed Tmax (h) | Predicted Tmax (h) | Tmax PE%b | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 1,250 | ND | 10.2 | NA | 4,182 (2,702–6,488)c | 2,073 | −50.4 | 3.5 (2.0–10)e | 3.5 | 0 | [7] |
| 1,250 | ND | 10.2 | NA | 4,870 (3,700–6,419)d | 2,073 | −57.4 | ND | 3.5 | NA | [8] |
| 1,250 | ND | 10.2 | NA | 2,220 (1,119–4,389)d | 2,073 | −6.6 | 3.0 (1.5–8.0)f | 3.5 | 16.7 | [10] |
| 1,250 | ND | 10.2 | NA | 3,253 (2,289–4,612)d | 2,073 | −36.3 | 3.0 (2.6–8.0)f | 3.5 | 16.7 | [12] |
| 1,500 | ND | 10.2 | NA | 3,390 (2,547–4,526)d | 2,488 | −26.6 | 3.4 (0.0–6.0)f | 3.5 | 3.2 | [6] |
| 1,500 | ND | 10.2 | NA | 4,251 (2,530–7,108)d | 2,488 | −41.5 | 3.0 (0.0–12.2)f | 3.5 | 16.7 | [11] |
| 1,600 | ND | 10.2 | NA | 3,666 (2,341–5,765)d | 2,654 | −27.6 | ND | 3.5 | NA | [5] |
| 1,600 | 26.2 (12.9–48.3) | 10.2 | −61.1 | 5,354 (3,334–8,598)d | 2,654 | −50.4 | 5.1 (0.9–8.0)g | 3.5 | −31.4 | [13] |
| 1,600 | ND | 10.2 | NA | 3,304d | 2,654 | −19.7 | ND | 3.5 | NA | [15] |
| 1,800 | ND | 10.2 | NA | 3,253d | 2,986 | −8.2 | ND | 3.5 | NA | [15] |
| 1,800 | 21.8 (18.5–104.5) | 10.2 | −53.2 | 4,015 (1,595–10,102)d | 2,986 | −25.6 | 3.9 (3.0–7.9)g | 3.5 | −10.3 | [13] |
ND not determined, NA not applicable
t1/2 is the terminal half-life median (95 % confidence interval)
PE% is the prediction error
Is the mean (90 % confidence interval) of Cmax
Is the geometric mean (95 % confidence interval) of Cmax
Is the median (90 % confidence interval) of Tmax
Is the median (range) of Tmax
Is the median (95 % confidence interval) of Tmax
For single dose lapatinib, our model-predicted Tmax to be at 3.75 h post administration and the average observed Tmax was 3.7 h. The MPE% and MAPE% were −6.3 and 9.6, respectively. For multiple dose lapatinib, our model-predicted steady-state Tmax was 3.5 h and the mean observed Tmax was 3.5 h. The MPE% and MAPE% were 1.6 and 14.6, respectively.
Regarding Cmax, the actual values versus our model-simulated values are graphically shown in Fig. 4e, f. The single dose predictions directly paralleled the actual Cmax (MPE% and MAPE% of −28.8 and 28.8, respectively). For the multiple dose predictions, our model underestimated steady-state Cmax (MPE% and MAPE% of −33.9 and 33.9, respectively). However, when we decreased liver metabolism to mimic hepatic impairment (as we did with AUC), the predicted steady-state Cmax for moderate liver dysfunction closely mirrored the observed data.
Overall, our PBPK model properly predicted lapatinib pharmacokinetic parameters from actual populations. As our model was developed with data from healthy subjects, the predictions were better for studies which were conducted in healthy subjects versus patients with solid tumors. To improve our model simulations for cancer patients, we altered our liver metabolism parameter to reflect hepatic impairment resulting from disease and/or age. With this modification, the model more precisely reproduced actual AUCs and Cmax from patients with solid tumors.
Sensitivity analysis
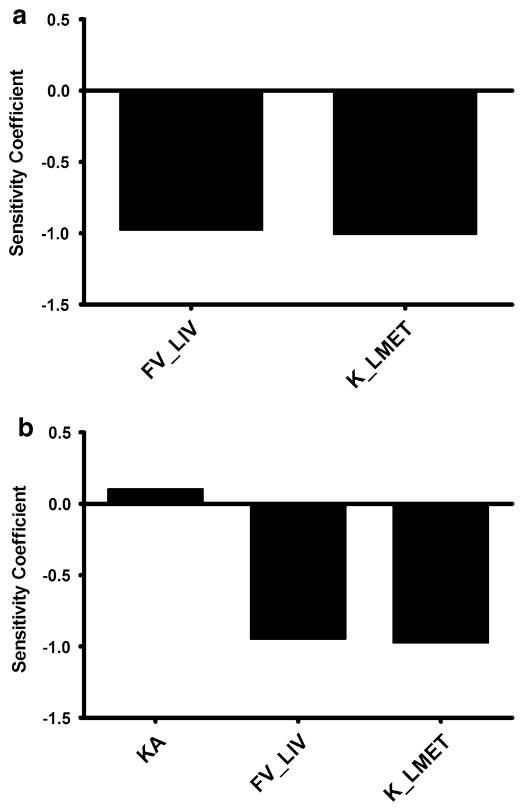
The normalized sensitivity coefficients for the mouse (60 mg/kg dose) and human (100 mg dose) PBPK models with respect to plasma AUC are shown in Fig. 5. Only parameters with sensitivity coefficients greater than 0.1 are shown. In both models, no normalized sensitivity coefficient was greater than ±1, indicating that there are no amplified parameter errors.
Fig. 5.
Calculated sensitivity coefficients for PBPK model parameters with respect to plasma AUC for the (a) mouse model and (b) human single dose model. Only parameters with sensitivity coefficients>0.1 are shown. FV_LIV fractional volume of liver, K_LMET first-order rate constant for liver metabolism; and KA first-order rate constant for absorption from intestinal lumen
Discussion
Physiologically based pharmacokinetic models have been developed for numerous antineoplastic agents including methotrexate [27, 28], cisplatin [29], actinomycin-D [30], 5-fluorouracil [31], capecitabine [32], 1-β-D-arabinofuranosylcytosine [33], adriamycin [34–36], topotecan [37] and docetaxel [38]. The need for these types of pharmacokinetic models for chemotherapeutics is great because of the challenges presented by this class of pharmaceutical compounds, specifically the narrow therapeutic index which is governed by drug distribution in the body. With PBPK modeling, the dynamics of drug distribution can be predicted using basic information on physiochemical properties, transport, biotransformation and excretion, thus leading to a better understanding of target tissue exposure resulting in either a therapeutic or toxic effect.
We have successfully developed a first-generation PBPK model for the dual EGFR/HER2 tyrosine kinase inhibitor lapatinib. This drug is a mere decade old and has only been approved by the FDA for the treatment of breast cancer since 2007. Consequently, the intricacies of the pharmacokinetics are still being elucidated. To our knowledge, the details of mouse tissue distribution of lapatinib have been limited to plasma and brain [39, 40] whereas, in humans, only plasma concentrations have been determined [3–15]. The tissue distribution of [14C] lapatinib was resolved by whole-body autoradiography in rats with detectable amounts quantified in the blood, brain, cerebrospinal fluid, harderian gland, heart, kidney, liver and muscle [41]. Our mouse data demonstrated tissue:blood concentration ratios that were comparable to those presented by Polli et al. [41], indicating that lapatinib exhibits similar distribution dynamics in these two rodents. Considering the autoradiography data [41] and the work presented herein, we now have a comprehensive assessment of the biodistribution of lapatinib in rats and mice.
By incorporating the mouse tissue distribution data into a PBPK model, we were able to effectively predict lapatinib concentrations in mouse plasma, brain, heart, lung, kidney, intestine, liver and slowly perfused tissue after oral doses of 30, 60 and 90 mg/kg. Subsequently, by taking into account interspecies differences in physiology and physiochemistry, we extrapolated this PBPK model to humans. To validate the human model, we were only able to compare our model simulations with observed plasma lapatinib concentrations and pharmacokinetic parameters, as there is no data in the literature regarding human tissue levels. Our model correctly predicted plasma exposure [23], Cmax, Tmax and half-life following single doses of lapatinib ranging from 50 to 1,800 mg and following multiple doses of lapatinib ranging from 25 to 1,800 mg. After taking the clinical trial subject/patient characteristics into consideration, it was evident that our model predictions were more accurate for healthy subjects than for patients with solid tumors (whose AUCs and Cmax were consistently underpredicted). This was not surprising given that our human PBPK model was developed with data from healthy subjects. In addition to the absence or presence of solid malignancies, the other major biological differences between these two populations were age and liver function. Both most likely contribute to hepatic impairment which results in a decrease in lapatinib clearance via metabolism and a subsequent increase in tissue exposure. When we altered our PBPK model to mimic hepatic impairment by decreasing the first-order rate constant for liver metabolism, the simulations for moderate hepatic impairment (incorporated as a 35 % decrease in liver metabolism) closely reflected the observed AUC and Cmax in cancer patients. Thus, our model can not only predict lapatinib plasma pharmacokinetics in healthy subjects but, with a minor metabolic alteration, can also predict the pharmacokinetics of this drug in the plasma of patients with solid malignancies.
The human PBPK model additionally facilitates the estimation of tissue levels of lapatinib. There is incredible utility in this application of the model, as it is not feasible to collect actual tissue concentration data from humans. Based on the adverse reactions to lapatinib observed in clinical trials, we can speculate as to the organ distribution of this drug. It is probable that the heart, liver, intestine and lung are exposed to significant levels of lapatinib as patients administered this compound have experienced decreased left ventricular ejection fraction, QT prolongation, hepatotoxicity, diarrhea and interstitial lung disease/pneumonitis. From largest to smallest, our multiple dose (1,250 mg q24 h) model-predicted ratios of lapatinib tissue:plasma AUCsτ were intestine (9.1), lung (8.2), liver (6.0), kidney (5.3), heart (1.1), slowly perfused tissue (0.3) and brain (0.05). Thus, for all organs in which adverse reactions to lapatinib have been noted, our model predicted tissue:plasma AUC ratios greater than 1, indicating substantial distribution into these tissues. Regarding brain, our model predicted low levels of lapatinib, which is consistent with the poor central nervous system (CNS) penetration observed in mice, owing to ABCB1- and ABCB2-mediated efflux [39]. Despite low lapatinib exposure in normal brain tissue, this drug has been shown to reduce the burden of metastatic breast cancer cells in the brains of mice [42] and have a modest CNS antitumor activity in human patients with brain metastases from HER2-positive breast cancer [43].
In summary, we have been able to successfully develop a PBPK model of lapatinib in mice, scale this model to humans and accurately predict the pharmacokinetics of this drug in human plasma over a wide range of doses. Additionally, our model also facilitated the estimation of various tissue exposures to lapatinib, which harmonize with the organ-specific toxicities documented in clinical trials. We acknowledge that this is a first-generation PBPK model which can be further improved with a greater understanding of lapatinib absorption, distribution, metabolism and excretion garnered from subsequent in vitro and in vivo studies. Moreover, our base model can be expanded to include other pharmacokinetic determinants, including efflux transporters, metabolite generation, combination dosing, etc., to make this PBPK model even more beneficial for the prediction of lapatinib disposition in both mouse and man.
Supplementary Material
Acknowledgments
We are grateful to Jerry L. Campbell (Center for Human Health Assessment, The Hamer Institutes for Health Sciences, Research Triangle Park, Durham, NC, USA), Conrad Housand (The AEgis Technologies Group, Oshawa, ON, USA) and Robin McDougall (The AEgis Technologies Group, Oshawa, ON, USA) for all of their help and guidance with this project. This work was supported in part by Grant number W81XWH-09-1-0457 from the Department of Defense (DOD) Breast Cancer Research Program (BCRP) of the Office of the Congressionally Directed Medical Research Programs (CDMRP).
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Electronic supplementary material The online version of this article (doi:10.1007/s10928-012-9295-8) contains supplementary material, which is available to authorized users.
References
- 1.GlaxoSmithKline. [Accessed 01 July 2012];Tykerb prescribing information. 2012 http://us.gsk.com/products/assets/us_tykerb.pdf.
- 2.www.clinicaltrials.gov
- 3.Gaul MD, Guo Y, Affleck K, Cockerill GS, Gilmer TM, Griffin RJ, Guntrip S, Keith BR, Knight WB, Mullin RJ, Murray DM, Rusnak DW, Smith K, Tadepalli S, Wood ER, Lackey K. Discovery and biological evaluation of potent dual ErbB-2/EGFR tyrosine kinase inhibitors: 6-thiazolylquinazolines. Bioorg Med Chem Lett. 2003;13(4):637–640. doi: 10.1016/s0960-894x(02)01047-8. [DOI] [PubMed] [Google Scholar]
- 4.Bence AK, Anderson EB, Halepota MA, Doukas MA, DeSimone PA, Davis GA, Smith DA, Koch KM, Stead AG, Mangum S, Bowen CJ, Spector NL, Hsieh S, Adams VR. Phase I pharmacokinetic studies evaluating single and multiple doses of oral GW572016, a dual EGFR-ErbB2 inhibitor, in healthy subjects. Invest New Drugs. 2005;23(1):39–49. doi: 10.1023/B:DRUG.0000047104.45929.ea. [DOI] [PubMed] [Google Scholar]
- 5.Burris HA, III, Hurwitz HI, Dees EC, Dowlati A, Blackwell KL, O’Neil B, Marcom PK, Ellis MJ, Overmoyer B, Jones SF, Harris JL, Smith DA, Koch KM, Stead A, Mangum S, Spector NL. Phase I safety, pharmacokinetics, and clinical activity study of lapatinib (GW572016), a reversible dual inhibitor of epidermal growth factor receptor tyrosine kinases, in heavily pretreated patients with metastatic carcinomas. J Clin Oncol. 2005;23(23):5305–5313. doi: 10.1200/JCO.2005.16.584. [DOI] [PubMed] [Google Scholar]
- 6.Siegel-Lakhai WS, Beijnen JH, Vervenne WL, Boot H, Keessen M, Versola M, Koch KM, Smith DA, Pandite L, Richel DJ, Schellens JH. Phase I pharmacokinetic study of the safety and tolerability of lapatinib (GW572016) in combination with oxaliplatin/fluorouracil/leucovorin (FOLFOX4) in patients with solid tumors. Clin Cancer Res. 2007;13(15 Pt 1):4495–4502. doi: 10.1158/1078-0432.CCR-07-0004. [DOI] [PubMed] [Google Scholar]
- 7.Chu QS, Schwartz G, de Bono J, Smith DA, Koch KM, Versola MJ, Pandite L, Arya N, Curtright J, Fleming RA, Ho PT, Rowinsky EK. Phase I and pharmacokinetic study of lapatinib in combination with capecitabine in patients with advanced solid malignancies. J Clin Oncol. 2007;25(24):3753–3758. doi: 10.1200/JCO.2007.11.1765. [DOI] [PubMed] [Google Scholar]
- 8.Midgley RS, Kerr DJ, Flaherty KT, Stevenson JP, Pratap SE, Koch KM, Smith DA, Versola M, Fleming RA, Ward C, O’Dwyer PJ, Middleton MR. A phase I and pharmacokinetic study of lapatinib in combination with infusional 5-fluorouracil, leucovorin and irinotecan. Ann Oncol. 2007;18(12):2025–2029. doi: 10.1093/annonc/mdm366. [DOI] [PubMed] [Google Scholar]
- 9.Storniolo AM, Pegram MD, Overmoyer B, Silverman P, Peacock NW, Jones SF, Loftiss J, Arya N, Koch KM, Paul E, Pandite L, Fleming RA, Lebowitz PF, Ho PT, Burris HA., 3rd Phase I dose escalation and pharmacokinetic study of lapatinib in combination with trastuzumab in patients with advanced ErbB2-positive breast cancer. J Clin Oncol. 2008;26(20):3317–3323. doi: 10.1200/JCO.2007.13.5202. [DOI] [PubMed] [Google Scholar]
- 10.LoRusso PM, Jones SF, Koch KM, Arya N, Fleming RA, Loftiss J, Pandite L, Gadgeel S, Weber BL, Burris HA., III Phase I and pharmacokinetic study of lapatinib and docetaxel in patients with advanced cancer. J Clin Oncol. 2008;26(18):3051–3056. doi: 10.1200/JCO.2007.14.9633. [DOI] [PubMed] [Google Scholar]
- 11.Chu QS, Cianfrocca ME, Goldstein LJ, Gale M, Murray N, Loftiss J, Arya N, Koch KM, Pandite L, Fleming RA, Paul E, Rowinsky EK. A phase I and pharmacokinetic study of lapatinib in combination with letrozole in patients with advanced cancer. Clin Cancer Res. 2008;14(14):4484–4490. doi: 10.1158/1078-0432.CCR-07-4417. [DOI] [PubMed] [Google Scholar]
- 12.Molina JR, Kaufmann SH, Reid JM, Rubin SD, Galvez-Peralta M, Friedman R, Flatten KS, Koch KM, Gilmer TM, Mullin RJ, Jewell RC, Felten SJ, Mandrekar S, Adjei AA, Erlichman C. Evaluation of lapatinib and topotecan combination therapy: tissue culture, murine xenograft, and phase I clinical trial data. Clin Cancer Res. 2008;14(23):7900–7908. doi: 10.1158/1078-0432.CCR-08-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakagawa K, Minami H, Kanezaki M, Mukaiyama A, Minamide Y, Uejima H, Kurata T, Nogami T, Kawada K, Mukai H, Sasaki Y, Fukuoka M. Phase I dose-escalation and pharmacokinetic trial of lapatinib (GW572016), a selective oral dual inhibitor of ErbB-1 and -2 tyrosine kinases, in Japanese patients with solid tumors. Jpn J Clin Oncol. 2009;39(2):116–123. doi: 10.1093/jjco/hyn135. [DOI] [PubMed] [Google Scholar]
- 14.Smith DA, Koch KM, Arya N, Bowen CJ, Herendeen JM, Beelen A. Effects of ketoconazole and carbamazepine on lapatinib pharmacokinetics in healthy subjects. Br J Clin Pharmacol. 2009;67(4):421–426. doi: 10.1111/j.1365-2125.2009.03370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burris HA, III, Taylor CW, Jones SF, Koch KM, Versola MJ, Arya N, Fleming RA, Smith DA, Pandite L, Spector N, Wilding G. A phase I and pharmacokinetic study of oral lapatinib administered once or twice daily in patients with solid malignancies. Clin Cancer Res. 2009;15(21):6702–6708. doi: 10.1158/1078-0432.CCR-09-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan K, Loizou GD, Spendiff M, Lipscomb JC, Andersen ME. PBPK modeling: a primer. In: Krishnan K, Andersen ME, editors. Quantitative modeling in toxicology. Vol. 17. Wiley; Chichester: 2010. p. 485. [Google Scholar]
- 17.Andersen ME, Yang RSH, Clewell HJ, III, Reddy MB. Introduction: a historical perspective of the development and applications of PBPK models. In: Reddy MB, Yang RSH, Clewell HJ III, Andersen ME, editors. Physiologically based pharmacokinetic modeling: science and applications. Vol. 19. Wiley-Interscience; Hoboken: 2005. p. 420. [Google Scholar]
- 18.Bai F, Freeman BB, III, Fraga CH, Fouladi M, Stewart CF. Determination of lapatinib (GW572016) in human plasma by liquid chromatography electrospray tandem mass spectrometry (LC-ESI-MS/MS) J Chromatogr B Analyt Technol Biomed Life Sci. 2006;831(1–2):169–175. doi: 10.1016/j.jchromb.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 19.Brown RP, Delp MD, Lindstedt SL, Rhomberg LR, Beliles RP. Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health. 1997;13(4):407–484. doi: 10.1177/074823379701300401. [DOI] [PubMed] [Google Scholar]
- 20.Sheiner LB, Beal SL. Some suggestions for measuring predictive performance. J Pharmacokinet Biopharm. 1981;9(4):503–512. doi: 10.1007/BF01060893. [DOI] [PubMed] [Google Scholar]
- 21.Wu G. Calculating predictive performance: a user’s note. Pharmacol Res. 1995;31(6):393–399. doi: 10.1016/1043-6618(95)80095-6. [DOI] [PubMed] [Google Scholar]
- 22.Loccisano AE, Campbell JL, Jr, Butenhoff JL, Andersen ME, Clewell HJ., III Comparison and evaluation of pharmacokinetics of PFOA and PFOS in the adult rat using a physiologically based pharmacokinetic model. Reprod Toxicol. 2012;33(4):452–467. doi: 10.1016/j.reprotox.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Castellino S, O’Mara M, Koch K, Borts DJ, Bowers GD, MacLauchlin C. Human metabolism of lapatinib, a dual kinase inhibitor: implications for hepatotoxicity. Drug Metab Dispos. 2012;40(1):139–150. doi: 10.1124/dmd.111.040949. [DOI] [PubMed] [Google Scholar]
- 24.Kwara A, Lartey M, Sagoe KW, Rzek NL, Court MH. CYP2B6 (c.516G–>T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol. 2009;67(4):427–436. doi: 10.1111/j.1365-2125.2009.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GlaxoSmithKline. [Accessed 01 July 2012];Tyverb prescribing information. 2010 http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_Product_Information/human/000795/WC500044957.pdf.
- 26.Cotreau MM, von Moltke LL, Greenblatt DJ. The influence of age and sex on the clearance of cytochrome P450 3A substrates. Clin Pharmacokinet. 2005;44(1):33–60. doi: 10.2165/00003088-200544010-00002. [DOI] [PubMed] [Google Scholar]
- 27.Bischoff KB, Dedrick RL, Zaharko DS. Preliminary model for methotrexate pharmacokinetics. J Pharm Sci. 1970;59(2):149–154. doi: 10.1002/jps.2600590203. [DOI] [PubMed] [Google Scholar]
- 28.Bischoff KB, Dedrick RL, Zaharko DS, Longstreth JA. Methotrexate pharmacokinetics. J Pharm Sci. 1971;60(8):1128–1133. doi: 10.1002/jps.2600600803. [DOI] [PubMed] [Google Scholar]
- 29.Evans WE, Crom WR, Tsiatis A, Green AA, Hayes FA, Pratt CB. Pharmacokinetic modeling of cisplatin disposition in children and adolescents with cancer. Cancer Chemother Pharmacol. 1982;10(1):22–26. doi: 10.1007/BF00257231. [DOI] [PubMed] [Google Scholar]
- 30.Lutz RJ, Galbraith WM, Dedrick RL, Shrager R, Mellett LB. A model for the kinetics of distribution of actinomycin-D in the beagle dog. J Pharmacol Exp Ther. 1977;200(3):469–478. [PubMed] [Google Scholar]
- 31.Collins JM, Dedrick RL, King FG, Speyer JL, Myers CE. Nonlinear pharmacokinetic models for 5-fluorouracil in man: intravenous and intraperitoneal routes. Clin Pharmacol Ther. 1980;28(2):235–246. doi: 10.1038/clpt.1980.156. [DOI] [PubMed] [Google Scholar]
- 32.Tsukamoto Y, Kato Y, Ura M, Horii I, Ishitsuka H, Kusuhara H, Sugiyama Y. A physiologically based pharmacokinetic analysis of capecitabine, a triple prodrug of 5-FU, in humans: the mechanism for tumor-selective accumulation of 5-FU. Pharm Res. 2001;18(8):1190–1202. doi: 10.1023/a:1010939329562. [DOI] [PubMed] [Google Scholar]
- 33.Dedrick RL, Forrester DD, Ho DH. In vitro-in vivo correlation of drug metabolism—deamination of 1-β-D-arabinofuranosylcytosine. Biochem Pharmacol. 1972;21(1):1–16. doi: 10.1016/0006-2952(72)90245-6. [DOI] [PubMed] [Google Scholar]
- 34.Harris PA, Gross JF. Preliminary pharmacokinetic model for adriamycin (NSC-123127) Cancer Chemother Rep. 1975;59(4):819–825. [PubMed] [Google Scholar]
- 35.Chan KK, Cohen JL, Gross JF, Himmelstein KJ, Bateman JR, Tsu-Lee Y, Marlis AS. Prediction of adriamycin disposition in cancer patients using a physiologic, pharmacokinetic model. Cancer Treat Rep. 1978;62(8):1161–1171. [PubMed] [Google Scholar]
- 36.Gustafson DL, Rastatter JC, Colombo T, Long ME. Doxorubicin pharmacokinetics: macromolecule binding, metabolism, and excretion in the context of a physiologic model. J Pharm Sci. 2002;91(6):1488–1501. doi: 10.1002/jps.10161. [DOI] [PubMed] [Google Scholar]
- 37.Sung C, Blaney SM, Cole DE, Balis FM, Dedrick RL. A pharmacokinetic model of topotecan clearance from plasma and cerebrospinal fluid. Cancer Res. 1994;54(19):5118–5122. [PubMed] [Google Scholar]
- 38.Bradshaw-Pierce EL, Eckhardt SG, Gustafson DL. A physiologically based pharmacokinetic model of docetaxel disposition: from mouse to man. Clin Cancer Res. 2007;13(9):2768–2776. doi: 10.1158/1078-0432.CCR-06-2362. [DOI] [PubMed] [Google Scholar]
- 39.Polli JW, Olson KL, Chism JP, John-Williams LS, Yeager RL, Woodard SM, Otto V, Castellino S, Demby VE. An unexpected synergist role of P-glycoprotein and breast cancer resistance protein on the central nervous system penetration of the tyrosine kinase inhibitor lapatinib (N-{3-chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methylsulfonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine; GW572016) Drug Metab Dispos. 2009;37(2):439–442. doi: 10.1124/dmd.108.024646. [DOI] [PubMed] [Google Scholar]
- 40.Taskar KS, Rudraraju V, Mittapalli RK, Samala R, Thorsheim HR, Lockman J, Gril B, Hua E, Palmieri D, Polli JW, Castellino S, Rubin SD, Lockman PR, Steeg PS, Smith QR. Lapatinib distribution in HER2 overexpressing experimental brain metastases of breast cancer. Pharm Res. 2012;29(3):770–781. doi: 10.1007/s11095-011-0601-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polli JW, Humphreys JE, Harmon KA, Castellino S, O’Mara MJ, Olson KL, John-Williams LS, Koch KM, Serabjit-Singh CJ. The role of efflux and uptake transporters in [N-{3-chloro-4-[(3-fluo-robenzyl)oxy]phenyl}-6-[5-({[2-(methylsulfonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine (GW572016, lapatinib) disposition and drug interactions. Drug Metab Dispos. 2008;36(4):695–701. doi: 10.1124/dmd.107.018374. [DOI] [PubMed] [Google Scholar]
- 42.Gril B, Palmieri D, Bronder JL, Herring JM, Vega-Valle E, Feigenbaum L, Liewehr DJ, Steinberg SM, Merino MJ, Rubin SD, Steeg PS. Effect of lapatinib on the outgrowth of metastatic breast cancer cells to the brain. J Natl Cancer Inst. 2008;100(15):1092–1103. doi: 10.1093/jnci/djn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin NU, Dieras V, Paul D, Lossignol D, Christodoulou C, Stemmler HJ, Roche H, Liu MC, Greil R, Ciruelos E, Loibl S, Gori S, Wardley A, Yardley D, Brufsky A, Blum JL, Rubin SD, Dharan B, Steplewski K, Zembryki D, Oliva C, Roychowdhury D, Paoletti P, Winer EP. Multicenter phase II study of lapatinib in patients with brain metastases from HER2-positive breast cancer. Clin Cancer Res. 2009;15(4):1452–1459. doi: 10.1158/1078-0432.CCR-08-1080. [DOI] [PubMed] [Google Scholar]
- 44.Chen HS, Gross JF. Estimation of tissue-to-plasma partition coefficients used in physiological pharmacokinetic models. J Pharmacokinet Biopharm. 1979;7(1):117–125. doi: 10.1007/BF01059446. [DOI] [PubMed] [Google Scholar]
- 45.Perloff MD, Von Moltke LL, Greenblatt DJ. Differential metabolism of midazolam in mouse liver and intestine microsomes: a comparison of cytochrome P450 activity and expression. Xenobiotica. 2003;33(4):365–377. doi: 10.1080/0049825031000066259. [DOI] [PubMed] [Google Scholar]
- 46.Hietanen E, Vainio H. Interspecies variations in small intestinal and hepatic drug hydroxylation and glucuronidation. Acta Pharmacol Toxicol (Copenh) 1973;33(1):57–64. doi: 10.1111/j.1600-0773.1973.tb01507.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhang QY, Dunbar D, Kaminsky LS. Characterization of mouse small intestinal cytochrome P450 expression. Drug Metab Dispos. 2003;31(11):1346–1351. doi: 10.1124/dmd.31.11.1346. [DOI] [PubMed] [Google Scholar]
- 48.Paine MF, Khalighi M, Fisher JM, Shen DD, Kunze KL, Marsh CL, Perkins JD, Thummel KE. Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther. 1997;283(3):1552–1562. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.