Abstract
The TET (ten–eleven translocation) family of α-ketoglutarate (α-KG)-dependent dioxygenases catalyzes the sequential oxidation of 5-methylcytosine (5mC) to 5-hydroxymethyl-cytosine (5hmC), 5-formylcytosine and 5-carboxylcytosine, leading to eventual DNA demethylation. The TET2 gene is a bona fide tumor suppressor frequently mutated in leukemia, and TET enzyme activity is inhibited in IDH1/2-mutated tumors by the oncometabolite 2-hydroxyglutarate, an antagonist of α-KG, linking 5mC oxidation to cancer development. We report here that the levels of 5hmC are dramatically reduced in human breast, liver, lung, pancreatic and prostate cancers when compared with the matched surrounding normal tissues. Associated with the 5hmC decrease is the substantial reduction of the expression of all three TET genes, revealing a possible mechanism for the reduced 5hmC in cancer cells. The decrease of 5hmC was also observed during tumor development in different genetically engineered mouse models. Together, our results identify 5hmC as a biomarker whose decrease is broadly and tightly associated with tumor development.
Keywords: TET, 5-hydroxymethylation, DNA methylation, cancer biomarker
Introduction
Genomic DNA methylation, occurring predominantly on the 5th carbon atom of cytosine (5-methylcytosine (5mC)) in the context of CpG islands in mammals, has a broad and important role in normal development and tumor suppression (Jones and Baylin, 2007). The enzymes that catalyze DNA methylation, DNA methyltransferases, have been thoroughly studied, while the enzymes and mechanisms of DNA demethylation have remained elusive, especially in animals (Wu and Zhang, 2010). A recently discovered family of Fe(II)- and α-ketoglutarate (α-KG)-dependent dioxygenases, the TET (ten–eleven translocation) proteins, has the capacity to catalyze a three-sequential oxidation reactions: converting 5mC first to 5-hydroxymethylcytosine (5hmC), and then 5-formylcytosine, and finally 5-carboxylcytosine (5caC) (Tahiliani et al., 2009; Ito et al., 2010, 2011; He et al., 2011). A subsequent decarboxylation of 5caC, by either a thymine-DNA glycosylase or other DNA repair enzymes, could then lead to DNA demethylation.
Mammalian cells express three TET genes: TET1, TET2 and TET3. TET2 is mutationally inactivated in about 15% of myeloid cancers, including 22% of acute myeloid leukemia (AML) (Delhommeau et al., 2009; Langemeijer et al., 2009). Separately, AML (Mardis et al., 2009), as well glioma (Parsons et al., 2008), chondrosarcoma (Amary et al., 2011a), enchondroma (Pansuriya et al., 2011; Amary et al., 2011b), thyroid carcinomas (Hemerly et al., 2010; Murugan et al., 2010) also sustain frequent mutations in genes encoding for the metabolic enzymes isocitrate dehydrogenase (IDH1 and IDH2). IDH1 and IDH2 mutations have also been reported in a limited number of samples of several additional types of tumors at lower frequency. These mutations result in simultaneous loss and gain of activities in the production of α-KG and 2-hydroxyglutarate (2-HG), respectively (Dang et al., 2009; Zhao et al., 2009). 2-HG functions as an α-KG antagonist by binding to the same space in the catalytic site and competitively inhibiting the activity of α-KG-dependent dioxygenases, including TETs (Xu et al., 2011). These results provide a mechanistic interpretation for the mutually exclusive mutation patterns between IDH1/2 and TET2 genes in AML (Figueroa et al., 2010) and increased genomic methylation in IDH1-mutated gliomas (Noushmehr et al., 2010). They also highlight the important role of deregulated TET activity in tumorigenesis. Given these intrinsic links between decreased function of TET-catalyzed 5mC-to-5hmC conversion and tumorigenesis in both glioma and leukemia, we set to determine how broadly 5hmC decrease occurs in human tumors.
Results and discussion
5hmC is decreased in multiple human cancers
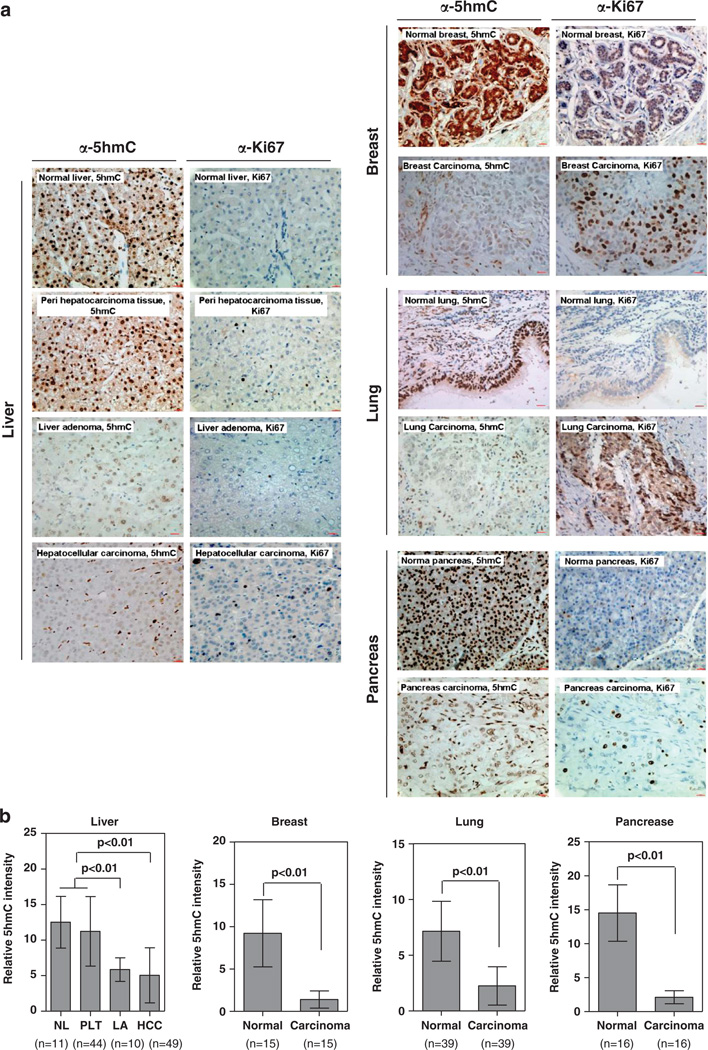
Immunohistochemistry (IHC) offers an efficient way for simultaneously examining the expression of a given antigen and histopathology of multiple samples in minimal quantity such as paraffin-embedded tissue sections. One critical reagent for reliable IHC is the quality of the antibody. To this end, we first verified the specificity of a polyclonal anti-5hmC antibody by antigen competition experiments. Using this antibody, we detected strong nuclear signals in normal human liver tissue, which were significantly diminished by preincubation of the antibody with the antigen dhmCTP (deoxyhydroxymethylcytidine triphosphate), but not by a similar preincubation with either dmCTP (deoxymethylcytidine triphosphate) or dCTP (deoxycytidine triphosphate) (Supplementary Figure S1). We then determined the levels and distribution of 5hmC in multiple types of human normal and cancer tissues, including liver, breast, lung, pancreas (Figure 1a), and prostate (Supplementary Figure S2). For liver samples, we assessed 11 normal liver (from liver transplantation), 44 peripheral liver, 10 liver adenoma and 49 HCC (hepatocellular carcinoma) samples. As shown normal liver and peripheral liver tissues displayed similar 5hmC levels. Notably, a significant decrease of 5hmC was observed in both liver adenoma and HCC as compared with normal liver and quantified via IHC (P < 0.01) (Figure 1b). In human breast tissue, we evaluated a total of 15 pairs of normal and carcinoma samples. Again, we found that the levels of 5hmC were dramatically reduced in human breast cancer when compared with the matched surrounding normal breast tissues (P < 0.01) (Figure 1b). In human lung, we evaluated a total of 39 samples and observed a profound reduction of 5hmC in the tumor samples when compared with normal lung tissues (P < 0.01) (Figure 1b). In human pancreas, we examined 16 cases and observed a similar reduction of 5hmC in both types of tumors (P < 0.01) (Figure 1b). Similar 5hmC reduction was also observed in human prostate tumors (P < 0.01) (Supplementary Figure S2). To verify the 5hmC staining in tumor samples, we performed IHC staining to detect the Ki67 protein, a cellular marker for proliferation, in the corresponding sections of normal and tumor samples. As compared with normal tissues, the Ki67 signal is stronger in tumor sections where the 5hmC signal is weak (Figure 1a). This provides evidence supporting that all the tumor samples have been properly prepared. Together, our IHC staining data demonstrate that the levels of 5hmC are dramatically decreased during human tumor development independently of cancer types.
Figure 1.
5hmC is substantially reduced in multiple human tumors. IHC was performed using antibodies against 5hmC and Ki67 in human normal and cancer tissues from liver, breast, lung and pancreas (a). The mean values of the IHC quantification are shown (b). Scale bars are 20 µm. Data are represented as mean±s.d. Freshly frozen or paraffin-embedded human normal tissue and tumors of liver, breast, lung, prostate and pancreas were acquired from Department of Pathology, Huashan Hospital, Fudan University. The procedures relating to the acquisition of samples from human subjects were approved by the Ethics Committee of the Institutes of Biomedical Sciences (IBS), Fudan University. The paraffin-embedded specimens were cut into 5 µm thin sections, which were subsequently stained with hematoxylin and eosin (H/E). Subsequent separation of tumor and normal surrounding tissues were carried out microscopically by an experienced molecular pathologist and guided by the H/E staining. For IHC analysis, the labeled streptavidin-biotin (LSAB) method was applied using commercial kits (Dako Corporation, Santa Barbara, CA, USA). Briefly, paraffin sections were deparaffinized and rehydrated following standard protocols. Sections were incubated with 3% H2O2 to eliminate the endogenous peroxidase activity, and then blocked with 5% normal goat serum. After treatment with 2N HCl for 15 min at room temperature, sections were neutralized with 100mm Tris–HCl (pH 8.5) for 10 min and then washed three times with PBS, followed by incubation with a primary anti-5hmC antibody (Active Motif; Cat. 39769, dilution at 1:1000) at 37 °C for 1 h. A horseradish peroxidase (HRP)-conjugated secondary antibody (Dako Corporation) was then applied and incubated at 37 °C for 1 h. For Ki67 IHC staining, the LSAB method was used as described above, using a primary anti-Ki67 antibody (dilution 1:100; Abcam, Cambridge, MA, USA) and a HRP-conjugated secondary antibody (Dako Corporation). Sections were developed with DAB kit and stopped with water. To semiquantify the 5hmC-positive areas, five randomly selected fields (173 µm2 each) from each sample were randomly selected and microscopically examined. Images were captured using a charge-coupled device (CCD) camera and analyzed using Motic Images Advanced software (version 3.2, Motic China Group Co. Ltd). The relative 5hmC intensity was calculated by dividing the positively stained areas over the total area. All IHC image analysis values were calculated as mean ± s.d. Statistical analysis was performed using SPSS 11.5 statistical package. Statistical methods included Paired t-test and independent samples t-test. All statistical tests were considered significant at an α=0.05 (P < 0.05).
5hmC is decreased in different genetically engineered mouse models
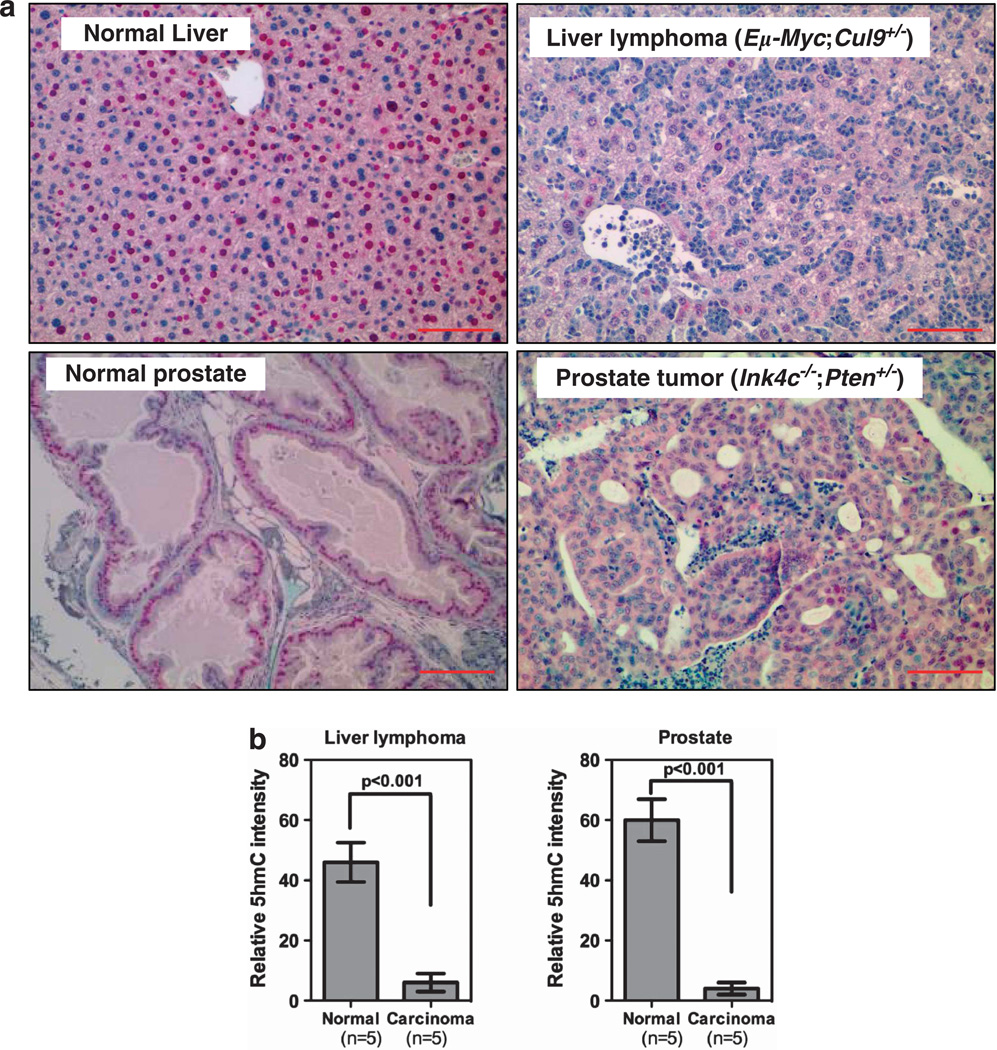
To determine whether a similar 5hmC reduction also occurs during mouse tumor development and whether the decrease is independent of specific genetic alterations, we performed IHC staining and determined the 5hmC levels in tumor generated from different genetically engineered mouse models that we have previously characterized including invasive lymphoma in the livers of Eµ-Myc;Cul9+/− mice, and lung tumors in p18Ink4c;Brca1+/−. In both types of mouse tumors, we found that 5hmC was significantly (P-value) decreased (Figure 2), supporting the notion that the reduction of 5hmC is a property common to different tumors in both humans and mice.
Figure 2.
5hmC levels are reduced in mouse tumors. IHC was performed using antibodies against 5hmC and Ki67 in two types of mouse tumors developed in different genetic backgrounds, a lymphoma invaded in liver and a prostate tumor, and counterstained with hematoxylin (a). The 5hmC IHC quantification is shown (b). Scale bars are 100 µm. Data are represented as mean ± s.d. (n=5).
Development of a dot-blot hybridization method for sensitive and quantitative analysis of 5hmC
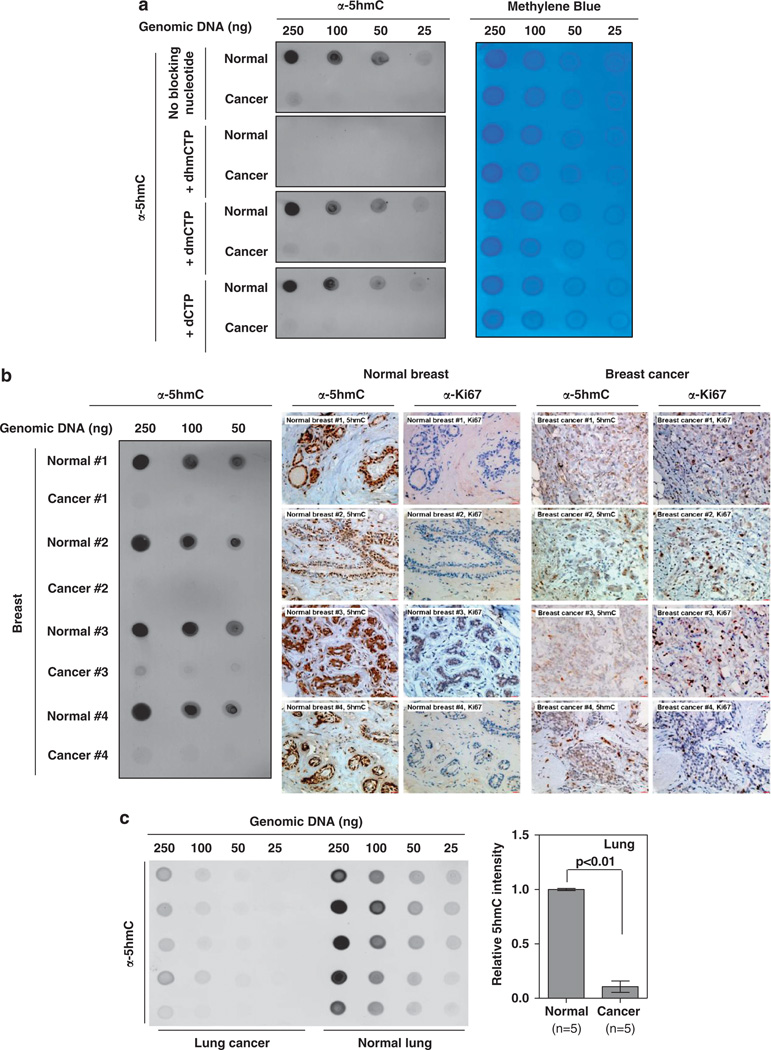
Although IHC can determine the tissue distribution of an antigen alone with histopathological examination, it offers only a semiquantitative measurement. Taking advantage that 5hmC is an antigen present in DNA, we developed a simple, sensitive and quantitative assay based on dot-blot hybridization for determining the levels of 5hmC. We first evaluated the specificity of the anti-5hmC antibody for the dot-blot by antigen nucleotide blocking experiments. We found that the anti-5hmC antibody could detect a clear signal in as little as 50 ng genomic DNA isolated from normal human breast tissue by dot-blot assay. Preincubation of the anti-5hmC antibody with the antigen dhmCTP, but not d5mC or dCTP, completely blocked the signal recognized by this antibody (Figure 3a). Prompted by this sensitivity and specificity of the antibody, we tested the feasibility of determining 5hmC by dot-blot assay using genomic DNA isolated from paraffin-embedded tissue sections. We found that as few as five sections (10 µm) of normal breast tissues could provide sufficient genomic DNA for the detection of 5hmC by the dot-blot assay. In 12 of 15 pairs of breast tissue samples, sufficient genomic DNA was harvested for dot-blot analysis. Most importantly, in all 12 paired samples that were examined, the levels of 5hmC were strikingly decreased even when the highest amount of DNA was used (250 ng; Figure 3b). In addition, Ki67 IHC staining results suggest that these breast cancer samples have been properly prepared (Figure 3b). An accurate determination on how many folds of reduction of 5hmC was exhibited in breast carcinoma samples was not reliable due to the very low levels of 5hmC in tumor genomic DNA, but the level of 5hmC in every tumor sample was at least ten fold lower than the matched normal tissue. Likewise, using this dot-blot assay, we also found that 5hmC is substantially reduced in human lung tumors when compared with their matched normal surrounding tissues (Figure 3c). Together, the IHC and dot-blot assay demonstrate that 5hmC levels are significantly reduced in multiple human tumors when compared with their matched normal surrounding tissues.
Figure 3.
Quantification of 5hmC decrease in human breast cancer. (a) To validate the dot-blot assay, the anti-5hmC antibody was preincubated with equal amount (1mm) of dhmCTP, dmCTP or dCTP for 1 h at room temperature, and then was used for dot-blot hybridization. Note that the anti-5hmC antibody very specifically recognized 5hmC, and could be effectively blocked by the preincubation with dhmCTP, but not dmCTP or dCTP. In addition, the dot-blot membrane was hybridized with 0.02% methylene blue in 0.3M sodium acetate (pH 5.2) to stain DNA. (b) Genomic DNAs were extracted from 15 pairs of paraffin-embedded human breast carcinoma and the matched normal breast tissues. In 12 of the 15 paired samples, sufficient genomic DNA was harvested for dot-blot hybridization. Representative micrographs of dot-blot assay, 5hmC and Ki67 staining in four paired samples are shown. Dot-blot results of the other eight pairs of normal and cancer samples are shown in Supplemental Figure S3. (c) Genomic DNAs were extracted from five pairs of paraffin-embedded human lung carcinoma and the matched normal lung tissues and subjected to dot-blot hybridization to determine the levels of 5hmC. Genomic DNAs were extracted from paraffin-embedded sections from normal and tumor samples following standard protocols. In detail, paraffin-embedded sections (10 ìm thickness) were deparaffinized by xylene. The deparaffinized samples were treated with ribonuclease A (50 µg/ml; Takara) at 37 °C for 1 h, and then incubated with proteinase K (0.3–0.5 mg/ml, Takara) in a digestion buffer (100mm NaCl/10mm Tris–HCl, pH 8.0 and 25mm EDTA, pH 8.0/0.5% SDS) at 55 °C overnight. After complete RNase and proteases digestion, genomic DNA was extracted with the same volume of phenol/chloroform/isoamyl alcohol (25:24:1) (Sangon, Shanghai), and then precipitated with an equal volume of isopropanol. After centrifugation, DNA pellet was washed once with 75% ethanol, air dried and dissolved in 15 µl distilled water. The DNA concentration was measured by NanoDrop (Thermo Scientific). The procedure for the dot-blot assay was modified from a procedure described previously (Xu et al., 2011). Briefly, DNA was spotted on a nitrocellulose membrane (Whatman), which was placed under an ultraviolet lamp for 20 min to crosslink the DNA. Subsequently, the membrane was blocked with 5% milk in TBS-Tween for 1 h and incubated with the primary anti-5hmC antibody at 4 °C overnight. After incubation with a horse radish peroxidase-conjugated secondary antibody anti-rabbit IgG (GenScript) for 1 h at room temperature, the membrane was washed with TBS-Tween 20 for three times and then detected and scanned by a Typhoon scanner (GE Healthcare). The 5hmC intensity was quantified by Image-Quanta software (GE Healthcare).
5hmC decrease is associated with a substantial reduction of TET gene expression in human tumors
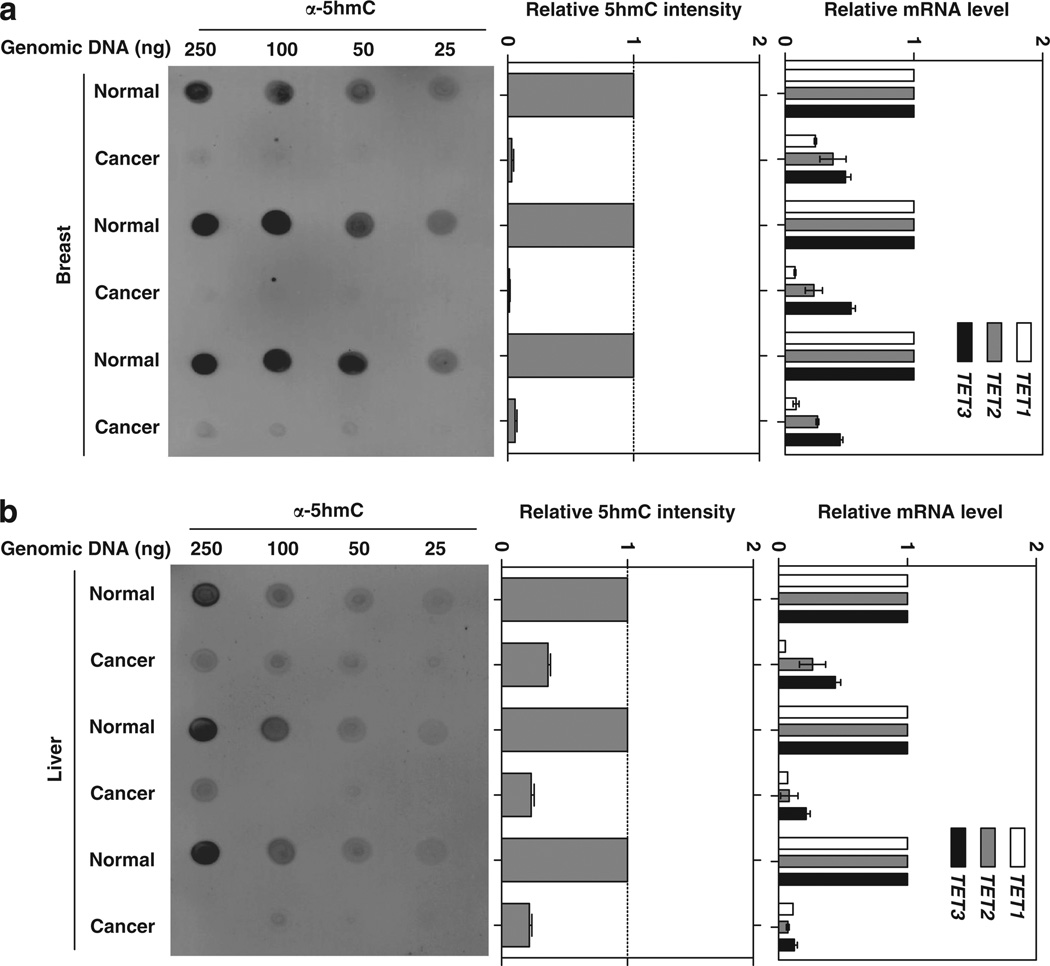
Two mechanisms have been reported that would lead to decreased levels of 5hmC in human tumors—loss-of-functional mutations targeting TET2 gene and inhibition of TET activity by the reduction of α-KG and accumulation of 2-HG resulting from IDH1/2 mutations (Delhommeau et al., 2009; Mullighan, 2009; Figueroa et al., 2010; Xu et al., 2011). However, neither TET nor IDH1/2 gene has thus far been reported to be mutated in liver, breast, lung, pancreas and prostate tumors. To investigate the possible mechanism underlying 5hmC decrease in these cancers, we determined the mRNA expression of three TET genes in both human breast and liver cancers. Interestingly, we found that the expressions of all three TET genes were significantly and uniformly reduced in both types of human tumors when compared with their matched normal tissues (Figures 4a and b). Of note, the decrease of three TET genes varied, with TET1 reduced most significantly, followed by TET2 and TET3. These observations provide a potential molecular mechanism for the observed reduction of 5hmC in human cancers.
Figure 4.
Reduced 5hmC is associated with the substantial reduction of TET gene expression in human cancers. Genomic DNA and mRNA were extracted from three pairs of frozen human breast carcinoma and the matched normal breast tissues (a) and three pairs of frozen human HCC and the matched normal liver tissues (b). The levels of 5hmC and mRNAs of three TET genes were determined by dot-blot assay and quantitative real-time PCR, respectively. Data are represented as mean ± s.d. RNA was extracted using QIAamp RNA Mini Kit following the manufacturer’s instructions (Qiagen). Quantitative real-time PCR was performed using an Applied Biosystems 7500 Sequence Detection System with SYBR green labeling with β-actin as an endogenous control. Primer sequences are listed in Supplementary Table S1.
In this paper, we demonstrate that 5hmC, a newly discovered modification of genomic DNA whose level is inversely correlated with that of 5mC, is substantially reduced in multiple types of human cancer. During the preparation of this paper, Haffner et al. (2011) reported that that 5hmC is substantially decreased in three types of human cancers, including breast, colon and prostate. Our study supports the link between 5hmC reduction and tumor development by expanding the finding to three additional types of cancer; liver, lung and pancreases. In every sample examined, remarkably, the tumor cells always have reduced 5hmC than the corresponding normal cells with no exception. Together, these studies suggest that 5hmC reduction is broadly and tightly linked with tumorigenesis. Moreover, our study shows that detection of 5hmC could be valuable biomarker for diagnosis of many cancer types. The tight inverse correlation between 5hmC and tumorigenesis suggests a potentially fundamental role of 5hmC and cytosine epigenetic modification in cancer development.
Our study also provides novel insights into the decrease of 5hmC levels in human tumors. First, we found that TET gene expression is significantly reduced in the tumors we examined, providing a potential mechanism underlying the 5hmC reduction. This result indicates that beside mutations targeting either TET2 or IDH1/2 genes, there are additional mechanisms, such as transcriptional inactivation of TET gene expression, which can block the 5mC-to-5hmC conversion and thus DNA demethylation. It will be important to determine how TET gene expression is silenced broadly in so many different types of tumors. Second, we showed that in different mouse tumor models that 5hmC was also decreased, indicating a gene-independent 5hmC reduction during tumor development. Lastly and importantly, we have developed a sensitive, reliable and quantitative dot-blot assay for determining the change in 5hmC levels using genomic DNA isolated from paraffin-embedded tissue sections. This assay should have broad applicability for clinical diagnosis.
Supplementary Material
Acknowledgements
We thank the members of the Fudan MCB laboratory for discussions and support throughout this study, and Eric Oermann for reading the manuscript. This work was supported by MOST 973 (No. 2009CB918401, No. 2011CB910600), NSFC (Grant No. 30600112, 30871255, 31071192). This work was also supported by the 985 Program, Shanghai key project (Grant No. 09JC1402300), the Shanghai Leading Academic Discipline Project (project number B110), and NIH grants (to YX and KLG).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011a;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011b;43:1262–1265. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della VV, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly JP, Bastos AU, Cerutti JM. Identification of several novel non-p.R132 IDH1 variants in thyroid carcinomas. Eur J Endocrinol. 2010;163:747–755. doi: 10.1530/EJE-10-0473. [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG. TET2 mutations in myelodysplasia and myeloid malignancies. Nat Genet. 2009;41:766–767. doi: 10.1038/ng0709-766. [DOI] [PubMed] [Google Scholar]
- Murugan AK, Bojdani E, Xing M. Identification and functional characterization of isocitrate dehydrogenase 1 (IDH1) mutations in thyroid cancer. Biochem Biophys Res Commun. 2010;393:555–559. doi: 10.1016/j.bbrc.2010.02.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansuriya TC, van Eijk R, d’Adamo P, van Ruler MA, Kuijjer ML, Oosting J, et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet. 2011;43:1256–1261. doi: 10.1038/ng.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.