Abstract
Although molecular prognostics in breast cancer are among the most successful examples of translating genomic analysis to clinical applications, optimal approaches to breast cancer clinical risk prediction remain controversial. The Sage Bionetworks–DREAM Breast Cancer Prognosis Challenge (BCC) is a crowdsourced research study for breast cancer prognostic modeling using genome-scale data. The BCC provided a community of data analysts with a common platform for data access and blinded evaluation of model accuracy in predicting breast cancer survival on the basis of gene expression data, copy number data, and clinical covariates. This approach offered the opportunity to assess whether a crowdsourced community Challenge would generate models of breast cancer prognosis commensurate with or exceeding current best-in-class approaches. The BCC comprised multiple rounds of blinded evaluations on held-out portions of data on 1981 patients, resulting in more than 1400 models submitted as open source code. Participants then retrained their models on the full data set of 1981 samples and submitted up to five models for validation in a newly generated data set of 184 breast cancer patients. Analysis of the BCC results suggests that the best-performing modeling strategy outperformed previously reported methods in blinded evaluations; model performance was consistent across several independent evaluations; and aggregating community-developed models achieved performance on par with the best-performing individual models.
INTRODUCTION
Breast cancer is the leading female malignancy in the world (1) and is one of the first malignancies for which molecular biomarkers have exhibited promise for clinical decision making (2–5). Biomarkers can be used to divide the disease into predictive (the likelihood that a patient responds to a particular therapy) or prognostic (a patient’s risk for a defined clinical endpoint independent of treatment) subcategories. Such molecular subcategorization highlights the possibilities for precision medicine (6), in which biomarkers are leveraged to identify disease taxonomies that distinguish biologically relevant groupings beyond standard clinical measures and can potentially inform treatment strategies.
A decade after early achievements in the development of prognostic molecular classifiers (called signatures) of breast cancer based solely on gene expression analysis (4, 5, 7), a large number of signatures proposed as markers of clinical risk prediction either fail to surpass the performance of conventional clinical covariates or await meaningful prospective validation [for example, the MINDACT (8, 9) and TAILORx/RxPONDER (10) trials]. Although gene expression–based breast cancer prognostic tests have been successfully implemented in routine clinical use, the application of molecular data to guide clinical decision making remains controversial (11).
Slow progress to evolve useful molecular classifiers may relate to poor study design, inconsistent findings, or improper validation studies (12). Data and code that underlie a potential new disease classifier are often unavailable for diligence. In addition, the rigor and objectivity of assessing molecular models are confounded by the tendency of data generation, data analysis, and model validation to be combined within the same study. This leads to the “self-assessment trap,” in which the desire to demonstrate improved performance of a researcher’s own methodology may cause inadvertent bias in elements of study design, such as data set selection, parameter tuning, or evaluation criteria (13).
A potential response to such problems is leveraging the Internet, social media, and cloud computing technologies to make it possible for physically distributed researchers to share and analyze the same data in real time and collaboratively iterate toward improved models based on predefined objective criteria applied in blinded evaluations. The Netflix competition (14) and X-Prize (15) have successfully demonstrated that crowdsourcing novel solutions for data-rich problems and technology innovation is feasible when substantial monetary incentives are offered to engage the competitive instincts of a community of analysts and technologists. Alternatively, a game-like environment has been used to seek solutions to biological problems, such as protein (16) and RNA folding (http://eterna.cmu.edu/web), thereby appealing to a vast community of gamers. Others have demonstrated that both fee-for-service [Kaggle (17) and Innocentive (18)] and open-participation computational biology challenges (19) [CASP (20) and CAFA (21)] can be used as potential new research models for data-intensive science. Despite the wide breadth of areas covered by these competitions, a key finding is that the best models from a competition usually outperform analogous models generated using more traditional isolated research approaches (14, 17).
Building on the success of crowdsourcing efforts such as DREAM to solve important biomedical research problems, we developed and ran the Sage Bionetworks–DREAM Breast Cancer Prognosis Challenge (BCC) to determine whether predefined performance criteria, real-time feedback, transparent sharing of source code, and a blinded final validation data set could promote robust assessment and improvement of breast cancer prognostic modeling approaches. The BCC was designed to make use of the METABRIC data set, which contains nearly 2000 breast cancer samples with gene expression and copy number data and clinical information. The availability of such a large data set affords the statistical power required to assess the robustness of performance of many models evaluated in independent tests, but is subject to the trade-off of using (overall) survival time in a historical cohort as the clinical endpoint, rather than potential endpoints that could be driven by clinically actionable criteria, such as response to targeted therapies. Therefore, the aim of this Challenge was not direct clinical deployment of a full-fledged suite of complex biomarkers. Indeed, we expect this study to serve as a pilot that lays the groundwork for future breast cancer challenges designed at the outset to answer clinically actionable questions. With this in mind, the BCC resulted in the development of predictive models with better performance than standard clinicopathological measures for prediction of overall survival (OS). The performance of these models was highly consistent across multiple blinded evaluations, including a novel validation cohort generated specifically for this Challenge.
RESULTS
The BCC included 354 registered participants from more than 35 countries. Participants were tasked with developing computational models that predict OS of breast cancer patients based on clinical information (for example, age, tumor size, and histological grade; see Table 1), mRNA expression data, and DNA copy number data. The BCC used genomic and clinical data from a cohort of 1981 women diagnosed with breast cancer (the METABRIC data set) (22) and provided participants with authorized Web access to data from 1000 samples as a training data set, and held back the remaining samples as a test data set (see Materials and Methods for more details on the Challenge design). Participants used the data to train computational models on their own standardized virtual machine (donated to the Challenge by Google) and submitted their trained models to the Synapse computational platform (23) as an R binary object (24) and rerunnable source code, where they were immediately evaluated. The predictive value of each model was scored by calculating the concordance index (CI) of predicted death risk compared to OS in a held-out data set, and the CIs were posted on a real-time leaderboard (http://leaderboards.bcc.sagebase.org). The CI is a standard performance measure in survival analysis that quantifies the quality of ranking risk predictors with respect to survival (25). In essence, given two randomly drawn patients, the CI represents the probability that a model will correctly predict which of the two patients will experience an event before the other (for example, a CI of 0.75 for a model means that if two patients are randomly drawn, the model will order their survival correctly three of four times).
Table 1.
Clinical characteristics of METABRIC and OsloVal data sets.
| Categories | METABRIC | OsloVal |
|---|---|---|
| Cohort size | 1981 | 184 |
| Age, years (%) | ||
| ≤50 | 21.4 | 33.1 |
| 50–60 | 22.5 | 18.5 |
| ≥60 | 56.1 | 48.4 |
| Tumor size, cm (%) | ||
| ≤2 | 43.3 | 38.0 |
| 2–5 | 48.2 | 42.4 |
| ≥5 | 7.5 | 7.1 |
| NA | 1.0 | 12.5 |
| Node status (%) | ||
| Node negative | 52.3 | 49.5 |
| 1–3 nodes | 31.4 | 21.2 |
| 4–9 nodes | 11.4 | 9.8 |
| ≥10 nodes | 4.6 | 8.2 |
| NA | 0.3 | 11.3 |
| ER status (%) | ||
| ER+ | 76.3 | 60.9 |
| ER− | 23.7 | 39.1 |
| PR status (%) | ||
| PR+ | 52.7 | 21.2 |
| PR− | 47.3 | 78.8 |
| HER2 copy status (%) | ||
| HER2 amplification | 22.1 | 13.6 |
| HER2 neutral | 72.6 | 86.4 |
| HER2 loss | 5.0 | 0.0 |
| NA | 0.3 | 0.0 |
| Tumor grade (%) | ||
| 1 | 8.6 | 6.5 |
| 2 | 39.1 | 37.0 |
| 3 | 48.1 | 30.4 |
| NA | 4.2 | 26.1 |
NA, not available.
Throughout the 3-month orientation and training phases of the Challenge (phases 1 and 2, respectively), participants collectively submitted more than 1400 predictive models, 1400 of which successfully executed from the submitted binary object and were assigned CI scores in the test data set (Fig. 1). One unique characteristic of this Challenge with respect to previous biomedical research Challenges was that each participant’s source code was available for others to view and adapt in new models. At the end of the training phase, participants were given the opportunity to train five models each on all 1981 METABRIC samples. The overall Challenge was determined in the final phase (phase 3) by assessing up to five models per participant or team (see table S3 for a listing of those members of the Breast Cancer Challenge Consortium who submitted at least one model to phase 3 and requested to have their name and affiliation listed) against a newly generated breast cancer data set consisting of genomic and clinical data from 184 women diagnosed with breast cancer (the “OsloVal” data set) (see Materials and Methods for details on how to access the METABRIC and OsloVal data sets). The participant or team with the best-performing model in the new data set was invited to publish an article about the winning model in Science Translational Medicine, provided it exceeded the scores of preestablished benchmark models, including the first-generation 70-gene risk predictor (4, 7) and the best model developed by a group of expert researchers in a precompetition (26). This pioneering mechanism of Challenge-assisted peer review assessed performance metrics in a blinded validation test, and these results were the foremost criterion for publication in the journal.
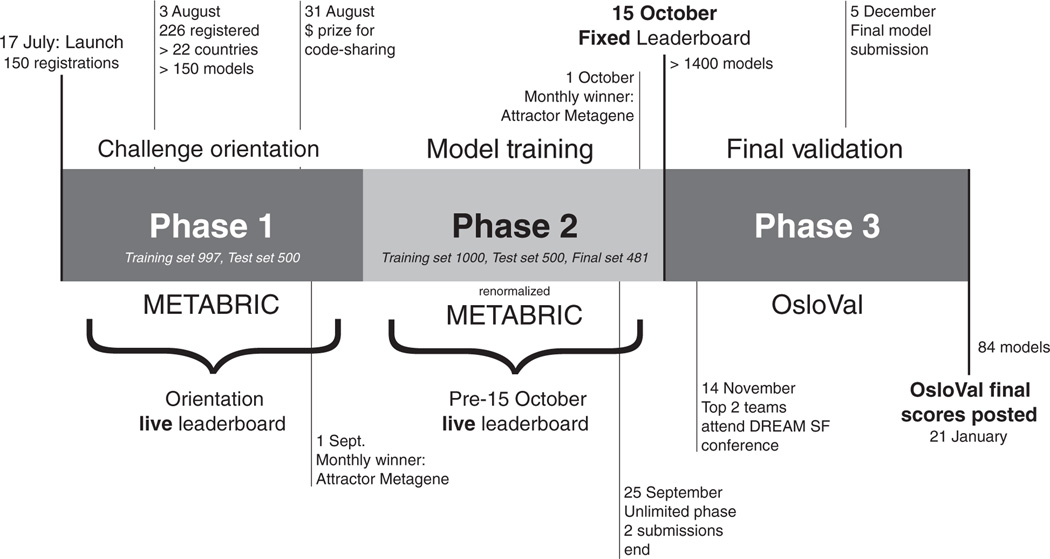
Fig. 1. Timeline and phases of BCC.
At initiation (phase 1), a subset of the METABRIC data set was provided along with orientation on how to use the Synapse platform for accessing data and submitting models and source code. Phase 2 provided a new randomization of samples, to eliminate biases in the distribution of clinical variables across training and test data, and renormalization of METABRIC mRNA expression and DNA copy number data, to reduce batch effects and harmonize data with the OsloVal data used in phase 3. During phase 2, there was a live “pre–15 October 2012” leaderboard that provided real-time scores for each submission against the held-out test set of 500 samples. At the conclusion of phase 2 on 15 October 2012, all models in the leaderboard were tested against the remaining held-out 481 samples. In the final validation round (phase 3), participants were invited to retrain up to five models on the entire METABRIC data set. Each model was then assigned a final CI score and consequently a rank based on the model’s performance against the independent OsloVal test set.
The METABRIC and the OsloVal data sets were comparable in terms of their clinical characteristics (Table 1). Roughly three-quarters of the patients were estrogen receptor–positive (ER+), and about half were lymph node–negative (LN−). The patients from the METABRIC and OsloVal cohorts received combinations of hormonal, radio-, and chemotherapy, and none were treated with more modern drugs such as trastuzumab, which specifically targets the human epidermal growth factor receptor 2/neu (HER2/neu) receptor pathway (27).
In phase 2 of the Challenge, participants trained on 1000 samples and obtained real-time feedback on model scores (CIs) in a held-out 500-sample test set (phase 1 results are displayed on the original leader-board and are not discussed here). A reference model using a random survival forest (28) trained on the clinical covariates and genomic data was provided as a baseline. Models submitted by Challenge participants quickly exceeded the performance of this baseline model and steadily improved over time (Fig. 2A), although we note that the improvement was modest compared to the baseline model (Fig. 2A, inset).
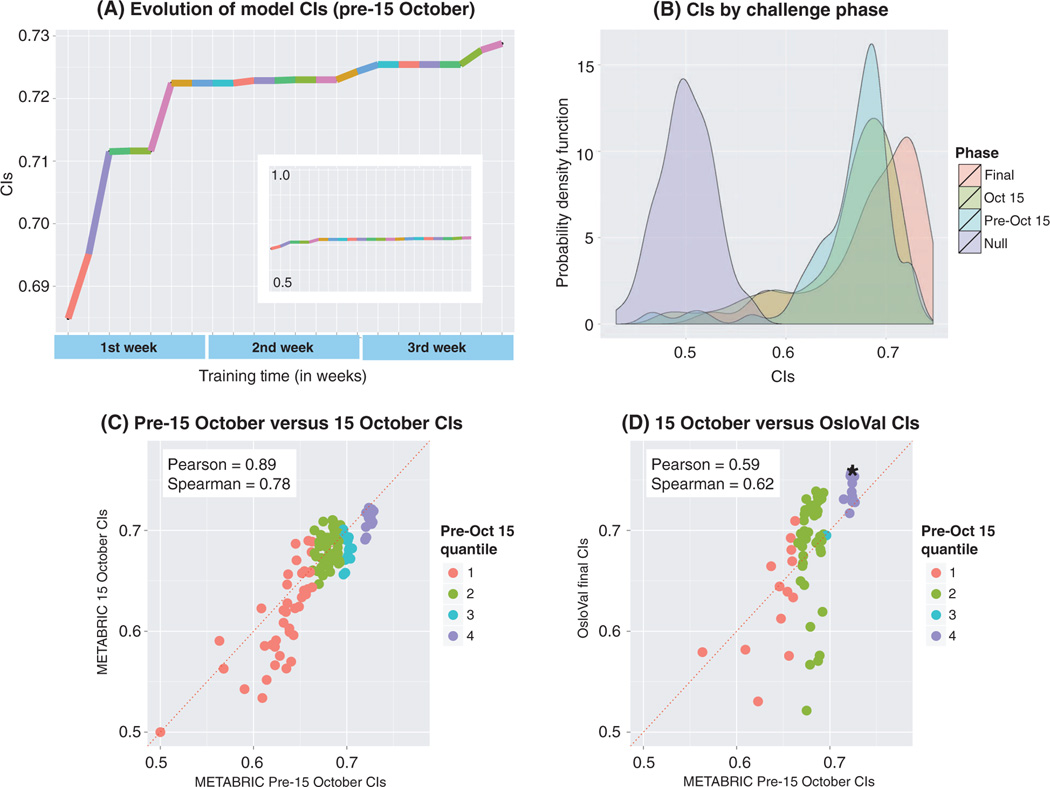
Fig. 2. BCC through time.
(A) During phase 2, the highest-scoring model scores were recorded for each date until the leaderboard was closed. Each colored segment represents a top-scoring team at any given point for the period extending from late September 2012 until the final deadline of phase 2 (15 October 2012). The plot records only the times when there was an increase in the best score, whereas the teams that achieved this score are labeled with different colors. The sequence of colors highlights an important aspect of the real-time feedback, where teams were encouraged to improve their models after being bested on the leaderboard by another team. Inset, the same plot with a y-axis scale ranging from of 0.5 to 1.0 maximum CI. (B) Probability density function plots of model scores posted on the live pre–15 October leaderboard evaluated against (i) the first test set of 500 samples (blue), (ii) the second test set of 481 samples (yellow), and (iii) the OsloVal data set (red). The null hypothesis probability density, which corresponds to random predictions evaluated against the OsloVal data set, is shown in purple. (C) Scatter plot of pre–15 October 2012 model performance versus 15 October 2012 performance. Colors represent quantiles, meaning that the ordered data are divided into four equal groups numbered consecutively from the bottom-scoring models (1) to the top-scoring models (4) for pre–15 October model performance. (D) Scatter plot of pre–15 October 2012 model performance versus final OsloVal performance. Colors represent quantiles of pre–15 October model performance. Asterisk represents the highest-scoring submitted model.
At the end of phase 2, all models in the leaderboard were evaluated against 481 hidden validation samples from the METABRIC data set. Given that more than 1400 models were submitted, there was concern that improvement in model scores resulted from overfitting to the test set used to provide real-time feedback. However, the performance of most models in the new test set was consistent with the performance of the same models in the previous test set, with comparable score ranges (Fig. 2, B and C;Pearson correlation: 0.90).A similar outcome was observed for the five models from every team that were evaluated against the OsloVal data set, after being trained on the 1981 samples in the METABRIC data set. Again, there was little evidence of overfitting when compared to the previous METABRIC test scores (Fig. 2D; Pearson correlation: 0.59), especially for models ranked in the top quantile.
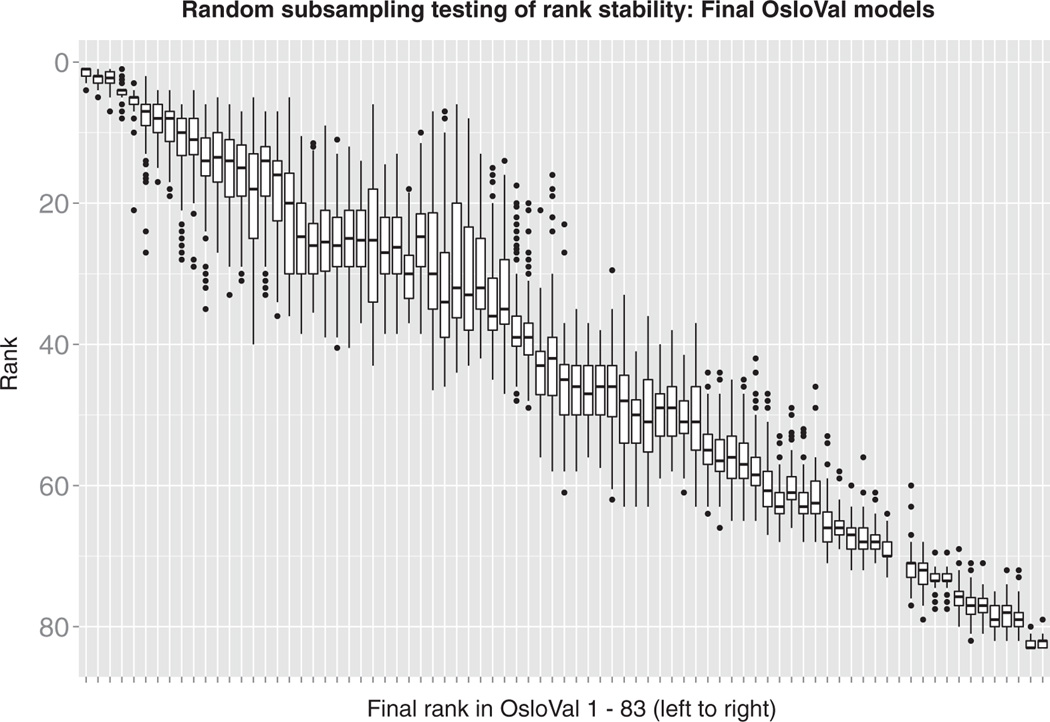
After participants trained and selected their final models, and after eliminating the ones that could not be evaluated because of run time errors, a total of 83 models were assessed and scored against the newly generated OsloVal data set. The winning team had three similar models that performed consistently better than all other scored models. The robustness of the final ranking was evaluated by sampling, without replacement, 80% of the validation set 100 times. Figure 3 shows box plots of the rankings obtained by each model across all trials ordered by the initial scores on all 184 OsloVal samples. The top three models belong to the same team and are ranked significantly better than the rest. The top model achieved a CI of 0.7561 [P = 5.1 × 10−28 compared to the fourth-ranked model, by Wilcoxon rank-sum test (29)].
Fig. 3. Rank stability of final models.
The OsloVal test data were randomly subsampled 100 times using 80% of the samples. Model rank was recalculated at each iteration. Models are ordered by their final posted leaderboard score (P values for the top three models, which were submitted by the same team, versus the fourth place model were as follows = 5.1 × 10−28, 1.8 × 10−22, and 1.7 × 10−20 by Wilcoxon rank-sum tests). With these box plots, the middle horizontal line represents the median, the upper whisker extends to the highest value within a 1.5× distance between the first and third quantiles, and the lower whisker extends to the lowest value within a 1.5× distance. Data beyond the ends of the whiskers are outliers plotted as points.
The BCC top-scoring model also significantly outperformed the top model developed during a pilot precompetition phase (26) in which BCC organizers tested 60 different models based on state-of-the-art machine learning approaches and clinical feature selection strategies. The best model from the precompetition used a random survival forest trained on the clinical feature data in addition to a genomic instability index derived from the copy number data.
The top-scoring model from the precompetition would have ranked as the sixth best model in the Challenge and achieved a CI of 0.7408 (Wilcoxon paired test, P = 4.33 × 10−1 compared to the winning model from the Challenge) when trained on the full METABRIC data set and evaluated in the OsloVal data. For comparison, a research version of a 70-gene risk signature (4) was also evaluated in the OsloVal cohort and achieved a CI of 0.60. In addition, two more test models were developed that included only the clinical covariates available for the two data sets (listed in tables S1 and S2). The first model was based on boosted regression (30) and achieved a CI of 0.7001 on the validation data set, whereas the second model used random forest regression (28) and achieved a score of 0.6964. The winning Challenge model achieved a score of 0.7562, significantly higher than the two clinical-only models (Wilcoxon paired test, P = 6.1 × 10−3 for both).
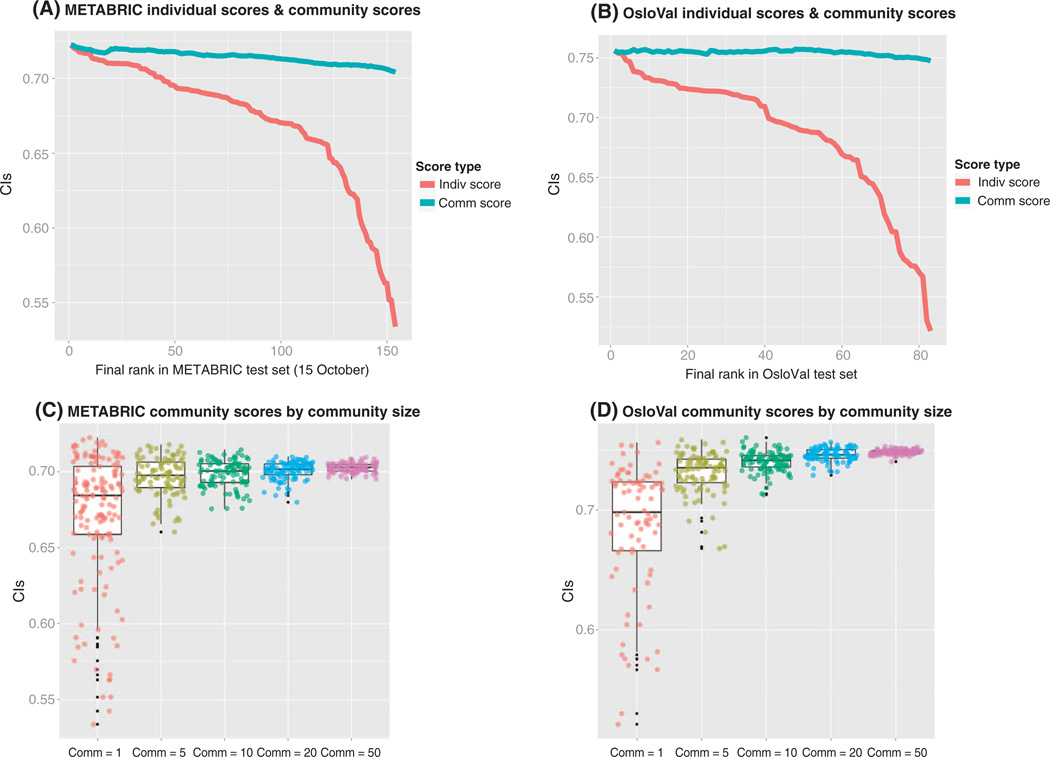
Meta-analyses of predictions submitted to past DREAM Challenges have systematically demonstrated (31–34) that the ensemble predictions resulting from the aggregation of the predictions of some or all the models usually perform similarly or even better than the best model. This phenomenon has been called the “wisdom of the crowds” and highlights one of the advantages of enabling research communities to work collaboratively to analyze the same data sets. The wisdom of the crowds was also at play in BCC (Fig. 4). For both cohorts, participants’ predictions were aggregated by calculating a community prediction formed by taking the average predicted rank of each patient across top n models for n = 1 … 83 (Fig. 4, A and B). Even when adding very poor predictions to the aggregate, the resulting score was robust and comparable with the top models. Robust predictors were also achieved by constructing community scores based on random subsamples of models (Fig. 4, C and D).
Fig. 4. Individual and community scores for METABRIC and OsloVal.
(A) Individual model scores are ordered by their rank on the pre–15 October 2012 METABRIC leaderboard (red line). For each model rank (displayed on the x axis), the blue line plots the aggregate model score based on combining all models less than or equal to the given rank. (B) Individual and aggregate model scores based on evaluation in the OsloVal data set. (C) Individual model scores (that is, community = 1) from the pre–15 October 2012 METABRIC leaderboard (http://leaderboards.bcc.sagebase.org/pre_oct15/index.html) are plotted alongside the community aggregate scores obtained when 5, 10, 20, and 50 randomly chosen models were considered. (D) Individual model scores (that is, community = 1) from the final OsloVal leaderboard (http://leaderboards.bcc.sagebase.org/final/index.html) are plotted alongside the community aggregate scores obtained when 5, 10, 20, and 50 randomly chosen predictions were considered. The colors correspond to community size: red = 1, yellow = 5, green = 10, blue = 20, purple = 50.
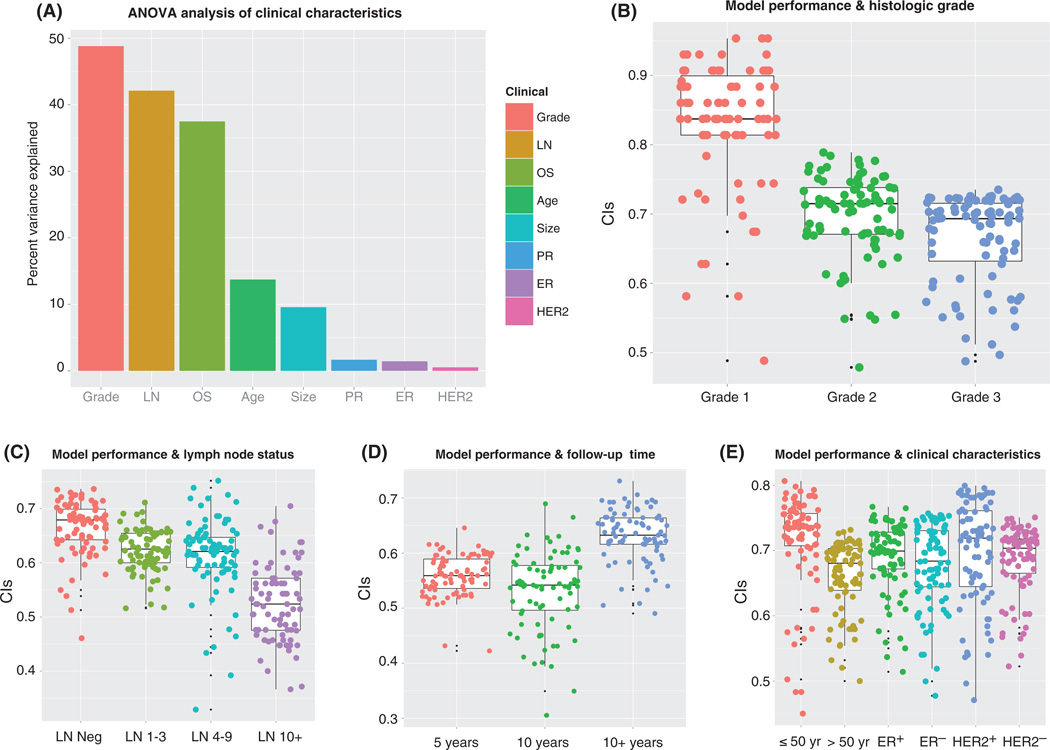
We evaluated model performance to determine whether there were specific clinical cases that are inherently more difficult to predict in terms of prognosis. We conducted analysis of variance (ANOVA) analyses by separating the patients from the OsloVal cohort based on the following clinical variables: age (≤50 years versus >50 years), tumor size (≤2 cm versus >2 cm), tumor grade (grades 1, 2, and 3), LN status (LN−, 1 to 3 positive LNs, 4 to 9 positive LNs, >9 positive LNs), ER status (ER+ versus ER−), progesterone receptor status (PR+ versus PR−), HER2/neu receptor amplification status (HER2+, HER2−), and OS (<5 years, 5 to 10 years, or >10 years). Of all variables considered (Fig. 5), tumor grade, LN status, OS, age, and tumor size were significantly associated with model performance (F test P values: 1.7 × 10−36, 1.1 × 10−38, 8.0 × 10−26, 9.0 × 10−7, and 5.0 × 10−5, respectively).
Fig. 5. Model performance and clinical characteristics.
(A) Percentage of CI variance explained by each clinical variable. (B) CIs were calculated for OsloVal models according to subsets of patients by histological grade. (C) CIs were calculated for OsloVal models according to subsets of patients by LN status. (D) CIs were calculated for OsloVal models according to subsets of patients by follow-up time (OS). (E) CIs were calculated for OsloVal models according to subsets of patients by age, ER status, and HER2 status. Patients were divided into subsets according to each of the above clinical characteristics. Individual model predictions were generated for patients belonging to each subset, and the CI was calculated by comparison with the actual survival for each patient.
Breast cancer patients with high-grade tumors and large numbers of positive LNs (>9) were associated with low CI scores (Fig. 5, B and C) and explain more than 40% of CI variance (Fig. 5A), suggesting that the OS of breast cancer patients with aggressive tumors is harder to predict. By contrast, there was no significant association between model performance and ER, PR, or HER2 status (Fig. 5E).
DISCUSSION
The BCC was an exercise in crowdsourcing that constitutes an open distributed approach to develop predictive models with the future potential to advance precision medicine applications. By creating a large, standardized breast cancer genotypic and phenotypic resource readily accessible via Web services and a common cloud computing environment, we were able to explore whether the open sharing of predictors of disease within a Challenge environment encourages the sharing of models and ideas before publication and whether decoupling of data creation, data analysis, and evaluation would minimize analytical and self-assessment biases.
Scientific conclusions from the Challenge
Improved model performance
The BCC results show that the best-performing model achieved significant CI improvements over currently available best-in-class methodologies, including the best model developed by a group of experts in a precompetition and a 70-gene risk signature. We note that the first-generation 70-gene risk signature used for comparison was designed as a binary risk stratifier for a specific patient subpopulation and should be viewed as a baseline and sanity check rather than a direct comparison. More significantly, the best-performing model (as well as slight variants of the same model submitted by the same BCC team) consistently outperformed all other approaches in three independent rounds of assessment and across multiple data sets. The top-scoring models used a methodology that minimized overfitting to the METABRIC training set by defining a “Metagene” feature space based on robust gene expression patterns observed in multiple external cancer data sets (35).
Robustness and generalizability of model performance
Our post hoc analysis suggests that the potential for training models overfit to the test set was not a significant confounding factor, even when participants were allowed to submit an unlimited number of models and obtain real-time scores for each submission. Comparison of the same models across multiple rounds of evaluation suggests a surprising degree of consistency between the unlimited submission phase and independent evaluations in the held-out METABRIC data and newly generated OsloVal data. This is especially remarkable because the OsloVal samples were collected from a different geographical location, by a different team, at a different time, and for a different purpose—in contrast to studies such as MAQC-II (36), in which the test sets and training sets were collected and processed by the same team and organization.
The distribution of CI scores improved with each round of evaluation, and the highest-scoring models in the OsloVal evaluation achieved higher CI scores than any of the 1400 models evaluated in either of the METABRIC phases. This apparent counterintuitive result can likely be explained by the fact that the average follow-up time for the OsloVal cohort is 4541 days, much longer than the average follow-up time of 2951 days for METABRIC. The rate of censored events is also lower in OsloVal (27.7%) than in METABRIC (55.2%); therefore, the proportion of long-term survivors scored in OsloVal is larger than that in the METABRIC validation set. Because the long-time survivors are better predicted (Fig. 5D) than the short-term survivors, the CIs associated with the METABRIC data are biased to shorter-time survivors and lower CIs.
The consistency of model scores across our three evaluations does not conclusively rule out some degree of overfitting, and, as with any scientific study, the generality of our findings will be continuously refined through sustained scrutiny in subsequent studies throughout the research community. However, the consistency of model performance across independent evaluations provides strong initial evidence that the findings of our study are likely to generalize to unseen data.
Robust performance of community models
Consistent with results of previous DREAM Challenges (31–34), the current study suggests that community models, constructed by aggregating predictions across many models submitted by participants, achieve performance on par with the highest-scoring individual models, and this high performance is remarkably robust to the inclusion of many low-scoring models into the ensemble. This result suggests that crowdsourcing a biomedical prediction question as a Challenge and using the community prediction as a solution is a fast and sound strategy to attain a model with strong performance and robustness. Intuitively, such an approach leverages the best ideas from many minds working on the same problem. More specifically, different approaches that accurately model biologically relevant signals are likely to converge on similar predictions related to the true underlying biology, whereas errant predictions resulting from erroneous approaches are less likely to be consistent. Thus, ensemble models may amplify the true signals that remain consistent across multiple approaches while decreasing the effects of less-correlated errant signals.
Contributions of BCC to the community
Challenge design
In designing BCC to incentivize crowdsourced solutions that might improve breast cancer prognostic signatures, our aims included continuous participant engagement throughout the Challenge, computational transparency, and rewarding of model generalizability. To accomplish these aims, respectively, we provided a framework that allowed users to make submissions and obtain realtime feedback on their performance, required submission of source code with each model, and provided multiple rounds of evaluation in independent data sets.
A common framework for comparing and sharing models
The model evaluation framework and CI scores calculated for models submitted during the Challenge provide a baseline set of model scores against which emerging tests, such as the PAM50 (37) risk of recurrence score and future prognostic models, may be compared.
The requirement for submission of publicly available source code provided an additional level of transparency compared to the typical Challenge design, which requires users to submit a prediction vector output from their training algorithm. We envision that sharing source code from multiple related predictive modeling Challenges will give rise to a community-developed resource of predictive models that is extended and applied across multiple projects and that might facilitate cumulative scientific progress, in which subsequent innovations build off of previous ones. It was encouraging to discover that participants learned from both well-performing and poorer-performing models. For example, the top-performing team consistently used the discussion forum to share with the other teams the prognostic ability of their Metagene features. Some of the other teams used aspects of the best-performing team’s code to improve their submissions, which in turn gave feedback to the best-performing team on the use of their methods by the other challenge participants.
Innovations from the community
Through the transparent code submission system and communication tools, such as a community discussion forum, the Challenge resulted in numerous examples of sharing and borrowing of scientific and technical insights between participants. At one point, we tested whether a cash incentive could be used to promote collaborative model improvement and offered a $500 incentive to any participant who could place atop the leader-board by borrowing code submitted by another participant (in addition to $500 to the participant whose code was borrowed). In less than 24 hours, a participant achieved the highest-scoring model by combining the computational approaches of the previously highest-scoring model with his clinical insight of modeling LN status as a continuous, rather than binary, variable. Unanticipated innovations also emerged organically from the community, including an online game within a game (http://genegames.org/cure), in which a player and a computer avatar successively select genes as input features to a predictive model, until one model is deemed statistically significantly superior to the other in predicting survival in the held-out data sets. This game attracted 120 players within 1 week, who played more than 2000 hands.
Limitations and extensions for future Challenges
The design of BCC included a number of simplifying assumptions intended to define a tractable prediction problem and evaluation criterion. However, it would be beneficial to account for biological and analytical complexities that were obscured by simplifications made in our experimental design.
First, we evaluated all models on the basis of a single metric, CI, which represents the most widely used statistic for evaluating survival models. However, a more complete assessment of advantages and disadvantages of each model would include additional criteria, such as model run time, trade-offs between sensitivity and specificity, or metrics more closely tied to the clinical relevance of a prognosticator.
Second, our choice to evaluate survival predictions across all samples in the cohort may obscure identification of models with advantages in particular breast cancer subtypes.
Third, by providing participants with normalized data, we tested only for modeling innovations given predefined input data, but did not assess different methods for data quality control and standardization that could contribute substantially to model improvements. A useful future Challenge design may allow participants to submit alternative methods for preprocessing raw data.
Fourth, we chose to evaluate models on the basis of OS. Other clinical endpoints, such as progression-free survival, are not currently available in the data sets used in BCC but may represent better evaluation endpoints if made available in the future. We chose OS over the other clinical endpoint available in the data set, disease-specific (DS) survival, to be consistent with decisions (38) by regulatory agencies such as the U.S. Food and Drug Administration and the European Medicines Agency. However, because of informal feedback we received from participants—that use of DS survival yielded more accurate models—we reevaluated all models and supported a separate exploratory leaderboard based on this metric. The model performance on this leaderboard suggested that using DS rather than OS as the clinical endpoint yielded improved CI scores, increased correlation with molecular features, and decreased correlation with confounding variables such as age. However, the best-performing models were consistent across both metrics.
Fifth, although prognostic models are one translational question, future Challenges that focus more directly on inferring predictive models of response to therapy may more directly affect clinical decision making. A laudable goal for future Challenges would be to directly engage the patient community and provide means to submit their own samples, help define questions, work alongside Challenge participants, and provide more direct feedback on how Challenge results can yield insights able to translate to improved patient care. In addition, providing large high-quality data sets (often prepublication) to a “crowd” of analysts is not common practice in academia and industry. However, such contributions by data generators would provide the necessary substrate for running such Challenges, with a potentially high impact on biomedical discovery.
Sixth, although statistically significant, the BCC results show that the improvement of the best-performing model is moderate with respect to the score achieved by aggregating standard clinical information. Thus, whereas molecular prognostic models derived from BCC warrant further investigation into their clinical utility, our results also suggest a new benchmark for future predictive methods derived from incorporating clinical covariates into state-of-the-art ensemble methods such as boosting or random forests. Future Challenges also may investigate the use of additional types of genomic information.
Finally, the sharing of ideas enabled by requiring submissions as rerunnable source code may ironically inhibit the diversity of innovations, effectively encouraging a monoculture as the community converges on a local optimum, modifying and extending approaches with high performance in the early stages of feedback (39). Improvements for future Challenges may include short embargo periods before sharing source code, with release possibly associated with declaring winners in stages of sub-Challenges with slightly modified data or prediction criteria. In addition, future Challenges that promote code sharing should establish well-defined criteria for assigning proper attribution (for example, in publication of winning models resulting from the Challenge) to all participants who made material intellectual contributions that were incorporated into the final winning model. More generally, it is important to develop a reward system that favors collaborative research practices that balance the currently prevalent winner-takes-all reward system.
Our results reinforce the trend from efforts such as CASP, DREAM, and others that Challenges incentivize collaboration and rapid learning, create iterative solutions in which one Challenge may feed into follow-up Challenges, motivate the generation of new data to answer clinically relevant questions, and provide the means for objective criteria to prioritize approaches likely to translate basic research into improved benefit to patients. We envision that expanding such mechanisms to facilitate broad participation from the scientific community in solving impactful biomedical problems and assessing molecular predictors of disease phenotypes could be integral to building a transparent, extensible resource at the heart of precision medicine.
MATERIALS AND METHODS
Challenge timeline
The BCC comprised three phases spanning a period of 3 months: (i) an orientation phase, (ii) a training phase, and (iii) a validation phase.
In phase 1, participants were provided mRNA and copy number aberration (CNA) data from 997 samples from METABRIC alongside the clinical data to train predictive models of patient OS. Samples contained in this training set corresponded to those used as a training set in the original publication of the METABRIC data set (22). During this phase of the Challenge (17 July to 22 September 2012), models were evaluated in real time against a held-out data set of 500 METABRIC samples, and a leaderboard was developed to display CI scores for each model, based on predicted versus observed survival times in these 500 held-out samples.
In phase 2 (25 September to 15 October 2012), lessons learned from the previous phase were implemented: The clinical variable “LN status” was changed from binary (positive or negative) values to integer values, representing the number of affected LNs; the gene expression and CNA data were renormalized (as described below) to better correct for batch effects; and the full cohort was randomly split into a new training and a new testing set to correct for sample biases from the original split (for example, in the original split, nearly all missing clinical covariates were in the test set). During this phase, models were trained on a training set of 1000 samples and evaluated in real time against a held-out test data set of 500 samples. The resulting scores were posted on a new leaderboard and, at the end of phase 2, were evaluated against the remaining held-out 481 samples. The winner of this phase was determined on the basis of evaluation in this second test set. All official scoring was performed using the OS endpoint, although based on requests from Challenge participants, we also configured a leaderboard that allowed participants to assess their model scores against DS survival as a secondary “unofficial” evaluation.
Before submission of their final models in phase 3, participants were given the opportunity to retrain their models on the entire METABRIC data set of 1981 (for convenience, with clinical variables reduced to only those present in the OsloVal data set). Furthermore, participants were asked to select a maximum of five models per team for assessment against the OsloVal data set. All participants who submitted a model in phase 3 are listed in table S3. If a team did not choose their preferred five models, their five top-scoring models were selected by default. These models were scored on the basis of the CI predicted versus observed ranks of survival times in the OsloVal data set. The overall winner of the Challenge was determined by the top CI score in OsloVal. The significance of the top-scoring models compared to the rest was assessed on the basis of scores for multiple random subsamples without replacement of 80% of patients from the OsloVal cohort. This process was repeated 100 times, and the resulting rankings were compared with a Wilcoxon rank-sum test (29).
Data governance
The data generators’ institutional ethics committee approved use of the clinical data within the BCC. Expression and copy number data from METABRIC were made available to Challenge participants during the duration of phases 1 to 3. Data for clinical covariates from the METABRIC cohort had been made public previously (40). Each participant who accessed the data agreed to (i) use these data only for the purposes of participating in the Challenge and (ii) not redistribute the data. Data access permissions were revoked at the completion of each BCC phase, and participants were required to reaffirm their agreement to these terms to enter each phase.
The METABRIC and OsloVal data sets have been deposited in the Synapse database (https://synapse.prod.sagebase.org/#!Synapse:syn1710250) and will be available to readers for a 6-month “validation phase” of the BCC. Those interested in accessing these data for use in independent research are directed to the following links. Expression and CNA data from OsloVal are available through an open access mechanism. All of other data are available through a controlled access mechanism.
METABRIC: https://synapse.prod.sagebase.org/#!Synapse:syn1688369
OsloVal: https://synapse.prod.sagebase.org/#!Synapse:syn1688370
The final Challenge scoring was based on the CIs of models, which was the same metric used in the leaderboard at earlier phases of the Challenge. The CI was predefined from the beginning of the Challenge as the performance metric to be used at final scoring. For all final submissions, every effort was made by the Sage Bionetwork team to run the submitted code in the Synapse platform. However, because of different problems in the submitted code, some of the submissions did not run successfully. Each submitted model that could be successfully run yielded a final survival prediction for the OsloVal cohort. For each of these predictions, the CI was mathematically computed in an unambiguous way by a computer program, from which the performance ranking was generated in order of descending CI. All final models can be accessed at http://leaderboards.bcc.sagebase.org/final/index.html.
Data normalization
The Affymetrix Genome-Wide Human SNP 6.0 and Illumina HT12 Bead Chip data were normalized according to the supervised normalization of microarrays (snm) framework and Bioconductor package (41, 42). Following this framework, models were devised for each data set that expressed the raw data as functions of biological and adjustment variables. The models were built and implemented through an iterative process designed to learn the identity of important variables defined by latent factors identified via singular value decomposition. Once these variables were identified, we used the snm R package to remove the effects of the adjustment variables while controlling for the effects of the biological variables of interest.
For example, to normalize the METABRIC mRNA data, we used a model that included ER status as a biological variable and both scan date and intensity-dependent array effects as adjustment variables. The resulting normalized data consisted of the residuals from this model fit plus the estimated effects of ER status. For the relevant data sets, we list the biological and adjustment variables as follows: METABRIC mRNA: biological variable = ER status, adjustment variables = scan date and intensity-dependent array effects; METABRIC SNP: biological variable = none, adjustment variables = scan date and intensity-dependent array effects; OsloVal mRNA: biological variable = ER status, adjustment variables = Sentrix ID (43), intensity-dependent array effects; OsloVal SNP: biological variable = none, adjustment variables = scan date, intensity-dependent effects.
Summarization of probes to genes for the SNP6.0 copy number data was done as follows. First, probes were mapped to genes with information obtained from the pd.genomewidesnp.6 Bioconductor package (44). For genes measured by two probes, we defined the gene-level values as an unweighted average of the data from the two probes. For genes measured by a single probe, we defined the gene-level values as the data for the corresponding probe. For those measured by more than two probes, we devised an approach that weighted probes based on their similarity to the first eigengene as defined by taking a singular value decomposition of the probe-level data for each gene. The percent variance explained by the first eigengene was then calculated for each probe. The summarized values for each gene were then defined as the weighted mean, with the weights corresponding to the percent variance explained.
Compute resources
A total of 2000 computational cores in the Google Cloud were provided to participants for the ~5-month duration of the Challenge, corresponding to a maximum of 7.5 million core hours if used at capacity. Specifically, each participant was provisioned an 8-core, 16-GB RAM machine, preconfigured and tested with an R computing environment and required libraries used in the Challenge. Compute resources were provisioned to each participant for dedicated use throughout the Challenge, and once capacity was reached, resources assigned to inactive users were recycled to new registrants such that all active users were provisioned compute resources. Users were also allowed to work on their own computing resources.
OsloVal data generation
The OsloVal cohort consisted of fresh-frozen primary tumors from 184 breast cancer patients collected from 1981 to 1999 (148 from 1981 to 1989 and 36 from 1994 to 1999) at the Norwegian Radium Hospital. Tumor material collection, clinical characterization, and DNA and mRNA extraction methods are described in the Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank I. R. Bergheim, E. U. Due, A. H. T. Olsen, C. Pedersen, M. La. Skrede, and P. Vu (Department of Genetics, Institute for Cancer Research) and L. I. Håseth (Department of Pathology, Oslo University Hospital, The Norwegian Radium Hospital) for their great efforts in the laboratory in collecting, cataloging, preparing, and analyzing the breast cancer tissue samples in a very short time; I. S. Jang for help on data analysis; H. Dai for assistance with the 70-gene signature evaluation; and L. Van’t Veer for helpful discussions in formulating the Challenge. The Breast Cancer Challenge Consortium members (listed in table S3) are each acknowledged for their submission of at least one computational model to phase 3 of the BCC.
Funding: This work was funded by NIH/National Cancer Institute grant 5U54CA149237 and Washington Life Science Discovery Fund grant 3104672. Generation of the OsloVal data set was funded by the Avon Foundation (Foundation for the NIH grant FRIE12PD), The Norwegian Cancer Society, and The Radium Hospital Foundation. Computational resources provided to Challenge participants were donated by Google.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/5/181/181re1/DC1
Materials and Methods
Table S1. Univariate and multivariate Cox regression statistics for the clinical covariates in the METABRIC data set.
Table S2. Univariate and multivariate Cox regression statistics for the clinical covariates in the OsloVal data set.
Table S3. The Breast Cancer Challenge Consortium: Challenge participants who submitted a model to phase 3 of the BCC.
Author contributions: A.A.M., E.B., E.H., T.C.N., H.K.M.V., O.M.R., J.G., V.N.K., S.H.F., G.S., S.A., C. Caldas, and A.-L.B.-D. conceived and designed the study. A.A.M., E.B., E.H., T.C.N., S.H.F., and G.S. drafted the manuscript. A.A.M., E.B., and E.H. performed data analysis. L.O., H.K.M.V., H.G.R., D.P., V.O.V., and A.-L.B.-D. organized clinical data and samples for the OsloVal data set. B.H.M. performed data normalization. B.S., M.R.K., M.D.F., N.A.D., B.H., and X.S. developed software infrastructure. L.M.M. organized data governance. T.P., L.Y., C. Citro, and J.H. developed Google compute resources. C. Curtis, S.A., and C. Caldas developed the METABRIC data set.
REFERENCES AND NOTES
- 1.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int. J. Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Rijn Mvan de S.S. Jeffrey, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijver van de MJ, He YD, van’t Veer LJ, Dai H, Hart AAM, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 5.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: National Academies Press; 2011. http://dels.nas.edu/Report/Toward-Precision-Medicine-Building-Knowledge/13284. [PubMed] [Google Scholar]
- 7.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 8.Rutgers E, Piccart-Gebhart MJ, Bogaerts J, Delaloge S, Van’t Veer L, Rubio IT, Viale G, Thompson AM, Passalacqua R, Nitz U, Vindevoghel A, Pierga JY, Ravdin PM, Werutsky G, Cardoso F. The EORTC 10041/BIG 03-04 MINDACT trial is feasible: Results of the pilot phase. Eur. J. Cancer. 2011;47:2742–2749. doi: 10.1016/j.ejca.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Cardoso F, Piccart-Gebhart M, Van’t Veer L. E. Rutgers; TRANSBIG Consortium, The MINDACT trial: The first prospective clinical validation of a genomic tool. Mol. Oncol. 2007;1:246–251. doi: 10.1016/j.molonc.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sparano JA. TAILORx: Trial assigning individualized options for treatment (Rx). Clin. Breast Cancer. 2006;7:347–350. doi: 10.3816/CBC.2006.n.051. [DOI] [PubMed] [Google Scholar]
- 11.Marchionni L, Wilson RF, Wolff AC, Marinopoulos S, Parmigiani G, Bass EB, Goodman SN. Systematic review: Gene expression profiling assays in early-stage breast cancer. Ann. Intern. Med. 2008;148:358–369. doi: 10.7326/0003-4819-148-5-200803040-00208. [DOI] [PubMed] [Google Scholar]
- 12.Simon R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J. Clin. Oncol. 2005;23:7332–7341. doi: 10.1200/JCO.2005.02.8712. [DOI] [PubMed] [Google Scholar]
- 13.Norel R, Rice JJ, Stolovitzky G. The self-assessment trap: Can we all be better than average? Mol. Syst. Biol. 2011;7:537. doi: 10.1038/msb.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell RM, Koren Y. Lessons from the Netflix prize challenge. ACM SIGKDD Explor. Newsl. 2007;9:75–79. [Google Scholar]
- 15.Kedes L, Liu ET. The Archon Genomics X PRIZE for whole human genome sequencing. Nat. Genet. 2010;42:917–918. doi: 10.1038/ng1110-917. [DOI] [PubMed] [Google Scholar]
- 16.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, Beenen M, Leaver-Fay A, Baker D, Popović Z, Players F. Predicting protein structures with a multiplayer online game. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter J. May the best analyst win. Science. 2011;331:698–699. doi: 10.1126/science.331.6018.698. [DOI] [PubMed] [Google Scholar]
- 18.Allio RJ. CEO interview: The InnoCentive model of open innovation. Strategy Leadership. 2004;32:4–9. [Google Scholar]
- 19.Marbach D, Costello JC, Küffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, Kellis M, Collins JJ, Stolovitzky G Consortium DREAM5. Wisdom of crowds for robust gene network inference. Nat. Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23:ii–v. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- 21.Radivojac P, Clark WT, Oron TR, Schnoes AM, Wittkop T, Sokolov A, Graim K, Funk C, Verspoor K, Ben-Hur A, Pandey G, Yunes JM, Talwalkar AS, Repo S, Souza ML, Piovesan D, Casadio R, Wang Z, Cheng J, Fang H, Gough J, Koskinen P, Törönen P, Nokso-Koivisto J, Holm L, Cozzetto D, Buchan DWA, Bryson K, Jones DT, Limaye B, Inamdar H, Datta A, Manjari SK, Joshi R, Chitale M, Kihara D, Lisewski AM, Erdin S, Venner E, Lichtarge O, Rentzsch R, Yang H, Romero AE, Bhat P, Paccanaro A, Hamp T, Kaβner R, Seemayer S, Vicedo E, Schaefer C, Achten D, Auer F, Boehm A, Braun T, Hecht M, Heron M, Hönigschmid P, Hopf TA, Kaufmann S, Kiening M, Krompass D, Landerer C, Mahlich Y, Roos M, Björne J, Salakoski T, Wong A, Shatkay H, Gatzmann F, Sommer I, Wass MN, Sternberg MJE, Skunca N, Supek F, Bošnjak M, Panov P, Džeroski S, Smuc T, Kourmpetis YAI, van Dijk ADJ, Ter Braak CJF, Zhou Y, Gong Q, Dong X, Tian W, Falda M, Fontana P, Lavezzo E, Camillo BDi, Toppo S, Lan L, Djuric N, Guo Y, Vucetic S, Bairoch A, Linial M, Babbitt PC, Brenner SE, Orengo C, Rost B, Mooney SD, Friedberg I. A large-scale evaluation of computational protein function prediction. Nat. Methods. 2013;10:221–227. doi: 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S Group METABRIC. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derry JMJ, Mangravite LM, Suver C, Furia MD, Henderson D, Schildwachter X, Bot B, Izant J, Sieberts SK, Kellen MR, Friend SH. Developing predictive molecular maps of human disease through community-based modeling. Nat. Genet. 2012;44:127–130. doi: 10.1038/ng.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R language definition. http://lib.stat.cmu.edu/R/CRAN/doc/manuals/R-lang.html.
- 25.Harrell FE. Regression Modeling Strategies. New York: Springer; 2001. p. 600. http://www.amazon.com/Regression-Modeling-Strategies-Frank-Harrell/dp/0387952322. [Google Scholar]
- 26.Bilal E, Dutkowski J, Guinney J, Jang IS, Logsdon BA, Pandey G, Sauerwine BA, Shimoni Y, Vollan HKM, Mecham BH, Rueda OM, Tost J, Curtis C, Alvarez MJ, Kristensen VN, Aparicio S, Børresen-Dale AL, Caldas C, Califano A, Friend SH, Ideker T, Schadt EE, Stolovitzky GA, Margolin AA. Improving breast cancer survival analysis through competition-based multidimensional modeling. PLoS Comput. Biol. 2013;9:e1003047. doi: 10.1371/journal.pcbi.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garnock-Jones KP, Keating GM, Scott LJ. Trastuzumab: A review of its use as adjuvant treatment in human epidermal growth factor receptor 2 (HER2)-positive early breast cancer. Drugs. 2010;70:215–239. doi: 10.2165/11203700-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 28.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann. Appl. Stat. 2008;2:841–860. [Google Scholar]
- 29.Bauer D. Constructing confidence sets using rank statistics. J. Am. Stat. Assoc. 1972;67:687–690. [Google Scholar]
- 30.Friedman JH. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002;38:367–378. [Google Scholar]
- 31.Prill RJ, Saez-Rodriguez J, Alexopoulos LG, Sorger PK, Stolovitzky G. Crowdsourcing network inference: The DREAM predictive signaling network challenge. Sci. Signal. 2011;4:mr7. doi: 10.1126/scisignal.2002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marbach D, Prill RJ, Schaffter T, Mattiussi C, Floreano D, Stolovitzky G. Revealing strengths and weaknesses of methods for gene network inference. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6286–6291. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prill RJ, Marbach D, Saez-Rodriguez J, Sorger PK, Alexopoulos LG, Xue X, Clarke ND, Altan-Bonnet G, Stolovitzky G. Towards a rigorous assessment of systems biology models: The DREAM3 challenges. PLoS One. 2010;5:e9202. doi: 10.1371/journal.pone.0009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stolovitzky G, Prill RJ, Califano A. Lessons from the DREAM2 challenges. Ann. N. Y. Acad. Sci. 2009;1158:159–195. doi: 10.1111/j.1749-6632.2009.04497.x. [DOI] [PubMed] [Google Scholar]
- 35.Cheng WY, Yang THO. D. Anastassiou, Development of a prognostic model for breast cancer survival in an open challenge environment. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3005974. [DOI] [PubMed] [Google Scholar]
- 36.Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, Shaughnessy JDJ, Oberthuer A, Thomas RS, Paules RS, Fielden M, Barlogie B, Chen W, Du P, Fischer M, Furlanello C, Gallas BD, Ge X, Megherbi DB, Symmans WF, Wang MD, Zhang J, Bitter H, Brors B, Bushel PR, Bylesjo M, Chen M, Cheng J, Cheng J, Chou J, Davison TS, Delorenzi M, Deng Y, Devanarayan V, Dix DJ, Dopazo J, Dorff KC, Elloumi F, Fan J, Fan S, Fan X, Fang H, Gonzaludo N, Hess KR, Hong H, Huan J, Irizarry RA, Judson R, Juraeva D, Lababidi S, Lambert CG, Li L, Li Y, Li Z, Lin SM, Liu G, Lobenhofer EK, Luo J, Luo W, McCall MN, Nikolsky Y, Pennello GA, Perkins RG, Philip R, Popovici V, Price ND, Qian F, Scherer A, Shi T, Shi W, Sung J, Thierry-Mieg D, Thierry-Mieg J, Thodima V, Trygg J, Vishnuvajjala L, Wang SJ, Wu J, Wu Y, Xie Q, Yousef WA, Zhang L, Zhang X, Zhong S, Zhou Y, Zhu S, Arasappan D, Bao W, Lucas AB, Berthold F, Brennan RJ, Buness A, Catalano JG, Chang C, Chen R, Cheng Y, Cui J, Czika W, Demichelis F, Deng X, Dosymbekov D, Eils R, Feng Y, Fostel J, Fulmer-Smentek S, Fuscoe JC, Gatto L, Ge W, Goldstein DR, Guo L, Halbert DN, Han J, Harris SC, Hatzis C, Herman D, Huang J, Jensen RV, Jiang R, Johnson CD, Jurman G, Kahlert Y, Khuder SA, Kohl M, Li J, Li M, Li QZ, Li S, Li Z, Liu J, Liu Y, Liu Z, Meng L, Madera M, Martinez-Murillo F, Medina I, Meehan J, Miclaus K, Moffitt RA, Montaner D, Mukherjee P, Mulligan GJ, Neville P, Nikolskaya T, Ning B, Page GP, Parker J, Parry RM, Peng X, Peterson RL, Phan JH, Quanz B, Ren Y, Riccadonna S, Roter AH, Samuelson FW, Schumacher MM, Shambaugh JD, Shi Q, Shippy R, Si S, Smalter A, Sotiriou C, Soukup M, Staedtler F, Steiner G, Stokes TH, Sun Q, Tan PY, Tang R, Tezak Z, Thorn B, Tsyganova M, Turpaz Y, Vega SC, Visintainer R, Frese Jvon, Wang C, Wang E, Wang J, Wang W, Westermann F, Willey JC, Woods M, Wu S, Xiao N, Xu J, Xu L, Yang L, Zeng X, Zhang J, Zhang L, Zhang M, Zhao C, Puri RK, Scherf U, Tong W, Wolfinger RD. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat. Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guidance for industry clinical trial endpoints for the approval of cancer drugs and biologics. http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm071590.pdf. [Google Scholar]
- 39.Williams D, Davidson JW, Hiser JD, Knight JC, Nguyen-Tuong A. Security through diversity: Leveraging virtual machine technology. IEEE Security Privacy Mag. 2009;7:26–33. [Google Scholar]
- 40.Stephens PJ, Tarpey PS, Davies H, Loo PVan, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerød A, Lee MTM, Shen CY, Tee BTK, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van’t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SAJR, Salomon AV, Børresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mecham BH, Nelson PS, Storey JD. Supervised normalization of microarrays. Bioinformatics. 2010;26:1308–1315. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gene expression profiling with Sentrix focused arrays. http://www.illumina.com/Documents/products/techbulletins/techbulletin_rna.pdf.
- 44.Carvalho B. pd.genomewidesnp.6: Platform design info for Affymetrix GenomeWideSNP_6. http://www.bioconductor.org/packages/2.12/data/annotation/html/pd.genomewidesnp.6.html.
- 45.Tavassoéli FA, Devilee P, editors. World Health Organization: Tumours of the Breast and Female Genital Organs (WHO/IARC Classification of Tumours) Lyon: IARC Press; 2003. The International Agency for Research on Cancer; p. 432. http://www.amazon.com/World-Health-Organization-Tumours-Classification/dp/9283224124. [Google Scholar]
- 46.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of his-tological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology. 2002;41:151–152. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.