Abstract
Protein domain family PF11267 (DUF3067) is a family of proteins of unknown function found in both bacteria and eukaryotes. Here we present the solution NMR structure of the 102-residue Alr2454 protein from Nostoc sp. PCC 7120, which constitutes the first structural representative from this conserved protein domain family. The structure of Nostoc sp. Alr2454 adopts a novel protein fold.
Keywords: Alr2454 protein, DUF3067, PF11267, Protein Structure Initiative, Solution NMR structure, Structural genomics
Introduction
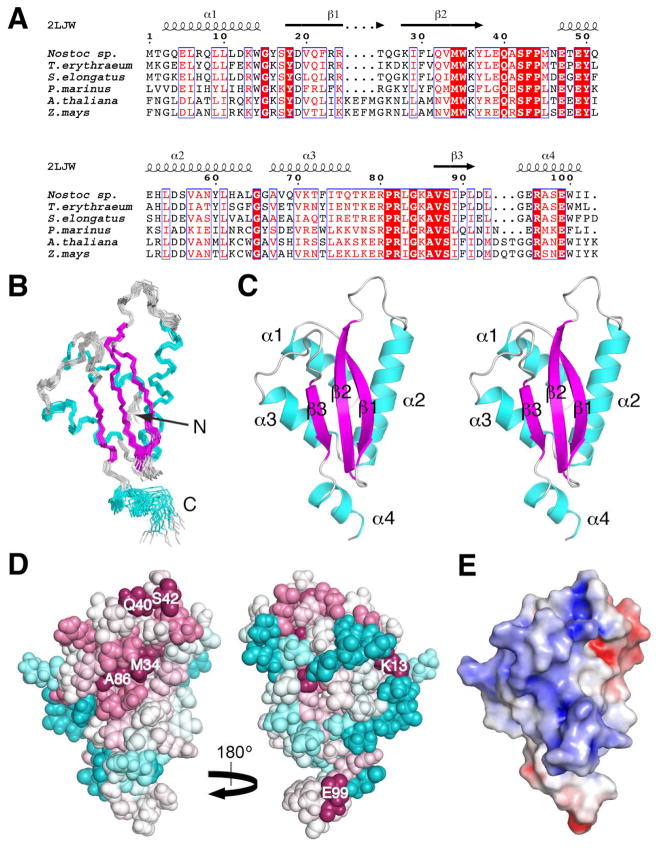
We present the solution NMR structure of the 102-residue Alr2454 protein from Nostoc sp. strain PCC 7120 (UniProtKB/TrEMBL ID, Q8YUA0_NOSS1; NESG ID, NsR264), a member of the functionally uncharacterized Pfam protein domain family PF11267 (DUF3067) comprised of bacterial and eukaryotic sequences, predominantly from cyanobacteria and viridiplantae (green plants), respectively (Fig. 1A) [1, 2]. This protein was selected for three-dimensional structure determination by the Northeast Structural Genomics Consortium (NESG) as part of the second phase of the Protein Structure Initiative (PSI-2) aimed at providing structural coverage of large, uncharacterized protein domain families [3]. Initial structural representatives of such families exhibit high homology modeling leverage [4], provide insights into protein evolution, and expand our knowledge of fundamental relationships between protein sequences, 3-dimensional structure, and protein function. The structure of Nostoc sp. Alr2454, the first structural representative from the PF11267 protein domain family, features a unique and, to the best of our knowledge, novel protein fold.
Fig. 1.
Solution NMR structure of Alr2454 from Nostoc sp. PCC 7120 (PDB ID, 2LJW). (A) Sequence alignment of a representative subset of the Pfam PF11267 protein domain family [1] from cyanaobacteria (Nostoc sp., Q8YUA0_NOSS1; Trichodesmium erythraeum, Q114L5_TRIEI; Synechococcus elongatus, Q31PB2_SYNE7; Prochlorococcus marinus, A2BVC5_PROM5, residues 4 to 105) and plants (Arabidopsis thaliana, Q8LDI1_ARATH, residues 113 to 222; Zea mays, B4FUD0_MAIZE, residues 116 to 225). Sequences were aligned using Clustal W 2.0 [33] and the sequence alignment was rendered using ESPript [34]. Boxed residues represent identical (white font, highlighted in red) and similar (red font) amino acid conservation. Residue numbering for Nostoc sp. Alr2454 and secondary structural elements [26] from its solution NMR structure (PDB ID, 2LJW) are drawn above the alignment. (B) Superposition of the final ensemble of 20 conformers of Alr2454 (residues 1 to 102). The view of the β-face of the protein is shown; α-helices and β-strands are shown in cyan and magenta, respectively, and loops are colored grey. (C) Stereoview of the lowest energy (CNS) conformer of Alr2454. Same orientation and color scheme as in (B). Secondary structural elements are labeled. (D) ConSurf [27, 28] image showing the conserved residues in Alr2454 (residues 3 to 101). Residue coloring, reflecting the degree of residue conservation over the entire PF11267 protein domain family (Pfam 25.0 [1]; 80 sequences), ranges from magenta (highly conserved) to cyan (variable). (Left) β-sheet face of the protein. (Right) α-helical face of the protein. Selected highly conserved residues are labeled. (E) APBS [29, 30] solvent accessible electrostatic surface potential of the β-sheet face of Alr2454 (residues 1 to 102) showing negative (red), neutral (white), and positive (blue) charges. All structure figures were rendered using PyMOL (www.pymol.org).
Material and Methods
Protein purification
Isotopically enriched samples of Nostoc sp. Alr2454 ([U-13C,15N]- and [U-5%-13C,100%-15N]-Alr2454) for NMR spectroscopy were cloned, expressed, and purified following standard NESG protocols [5]. Briefly, the 102-residue coding sequence of the alr2454 gene from Nostoc sp. strain PCC 7120 was cloned into the pET21_NESG vector (Novagen) in frame with a C-terminal affinity tag (LEHHHHHH), transformed into codon-enhanced Escherichia coli BL21 (DE3) pMGK cells, and expressed in MJ9 minimal medium [6] containing U-(15NH4)2SO4 and U-13C-glucose as the sole nitrogen and carbon sources. Initial cell growth was carried out at 37 °C, and protein expression was induced at 17 °C by 1 mM IPTG at mid-log phase growth and continued overnight. Proteins were purified using an ÄKTAxpress system (GE Healthcare) with a two-step protocol consisting of HisTrap HP affinity chromatography followed by HiLoad 26/60 Superdex 75 gel filtration chromatography. The final yield of purified isotopically-enriched Alr2454 was approximately 40 mg/L of culture. Sample purity and molecular mass were confirmed by SDS-PAGE and MALDI-TOF mass spectrometry (MALDI-TOF mass of [U-13C,15N]-Alr2454 (Da): experimental, 13,733; expected, 13,724). Samples of [U-13C,15N]- and [U-5%-13C,100%-15N]-Alr2454 for NMR spectroscopy were concentrated by ultracentrifugation to 0.6 to 1.0 mM in 90% H2O/10% 2H2O solution containing 20 mM ammonium acetate, 100 mM NaCl, 10 mM DTT, 5 mM CaCl2, 50 μM DSS at pH 4.5. Analytical gel filtration followed by static light scattering (Suppl. Fig. S1) and 15N T1 and T2 relaxation data (Suppl. Fig. S2) demonstrate that Alr2454 is a monomer in solution under the conditions used in the NMR studies. The pET expression vector for Nostoc sp. Alr2454, (NESG NsR264-21.4), has been deposited in the PSI Materials Repository (http://psimr.asu.edu/).
NMR spectroscopy and resonance assignment
All NMR data were collected at 298 K on Bruker AVANCE 600 and 800 MHz spectrometers equipped with 1.7-mm TCI and 5-mm TXI cryoprobes, respectively, processed with NMRPipe [7], and visualized using SPARKY [8]. All spectra were referenced to internal DSS. Complete 1H, 13C, and 15N resonance assignments for Alr2454 were determined using conventional triple resonance NMR methods. Backbone resonance assignments were analyzed automatically using both PINE 1.0 [9] and AutoAssign 2.4.0 [10] software, based on peak lists for 2D 1H-15N HSQC and 3D HNCO, HN(CA)CO, HN(CO)CA, HNCA, CBCA(CO)NH and HNCACB spectra. The assigned 1H-15N HSQC spectrum of Alr2454 is provided as Suppl. Fig. S3, and a summary of the sequential connectivity and NOESY data used to determine these assignments as Suppl. Fig. S4. Side chain assignments were completed manually using 3D HBHA(CO)NH, HCCH-COSY, HCCH-TOCSY, and (H)CCH-TOCSY experiments. Stereospecific isopropyl methyl resonance assignments for all Val and Leu residues were determined from characteristic cross-peak fine structures in high resolution 2D 1H-13C HSQC spectra of [U-5%-13C,100%-15N]-Alr2454 [11]. Resonance assignments were validated using the Assignment Validation Suite (AVS) software package [12]. The final resonance assignments, NOESY spectral peak lists, and time domain data for Alr2454 were deposited in the BioMagResDB (BMRB accession number, 17965). 1H-15N heteronuclear NOE and 15N T1 and T2 relaxation measurements were made using gradient sensitivity-enhanced 2D heteronuclear NOE and 1D 15N T1 and T2 (CPMG) relaxation experiments, respectively [13].
Structure determination and refinement
The solution NMR structure of Alr2454 was calculated using CYANA 3.0 [14, 15] supplied with peak intensities from 3D 15N-edited NOESY (τm = 100 ms), 3D 13C-edited aliphatic NOESY (τm = 100 ms), and 3D 13C-edited aromatic NOESY (τm = 120 ms) spectra, together with broad dihedral angle constraints derived by TALOS+ [16] (φ, ψ ± 30°) for ordered residues with confidence scores of 10. The 20 structures with lowest target function out of 100 calculated in the final cycle were further refined by restrained molecular dynamics in explicit water using CNS 1.3 [17, 18] and the PARAM19 force field, supplied with the final NOE-derived distance and TALOS+ dihedral angle constraints. In this final stage of the structure determination, rotamer states of specific ordered residues were constrained (χ1, χ2 ± 20°) based on MolProbity [19, 20] and PROCHECK [21] analyses.
Structure validation and deposition
The final refined ensemble of 20 structures for Alr2454 (excluding the C-terminal His6 tag) was deposited into the Protein Data Bank (PDB ID, 2LJW). Structural statistics and global structure quality scores, including Verify3D [22], ProsaII [23], PROCHECK [21], and MolProbity [19, 20] raw and statistical Z-scores, were computed using the PSVS 1.4 software package [24]. The global goodness-of-fit of the final structure ensemble with the NOESY peak list data and resonance assignments was determined using the RPF analysis program [25].
Results and Discussion
The solution NMR structure of Nostoc sp. PCC 7120 Alr2454 features a unique mixed α+β fold comprised of four α-helices (α1, Gly3-Trp14; α2, Glu47-Leu64; α3, Ala67-Gln76; α4, Gly94-Ile101) and a sheet of three anti-parallel β-strands (β1, Tyr18-Thr25; β2, Lys28-Tyr37; β3, Val87-Leu91) arranged in a αββααβα topology (Fig. 1B, C) [26]. The three-stranded β-sheet packs against the first three helices (α1 to α3) to form a compact structure, whereas the C-terminal α-helix (α4) is somewhat poorly-defined. Structural statistics for Alr2454 are presented in Table 1.
Table 1.
Summary of structural statisticsa
| Alr2454 | ||
|---|---|---|
| Completeness of resonance assignments b: | ||
| Backbone (%) | 99.4 | |
| Side chain (%) | 98.3 | |
| Aromatic (%) | 96.6 | |
| Stereospecific methyl (%) | 100 | |
| Conformationally-restricting constraintsc: | ||
| Distance constraints | ||
| Total | 2478 | |
| intra-residue (i = j) | 688 | |
| sequential (|i−j| = 1) | 619 | |
| medium range (1 < |i − j| < 5) | 462 | |
| long range (|i − j| ≥ 5) | 709 | |
| Dihedral angle constraints | 162 | |
| Hydrogen bond constraints | 0 | |
| No. of constraints per residue | 25.4 | |
| No. of long range constraints per residue | 6.8 | |
| Residual constraint violationsc: | ||
| Average no. of distance violations per structure: | ||
| 0.1 – 0.2 Å | 8.75 | |
| 0.2 – 0.5 Å | 1.85 | |
| > 0.5 Å | 0 | |
| Average no. of dihedral angle violations per structure: | ||
| 1 – 10° | 8.75 | |
| > 10° | 0 | |
| Model Qualityc: | ||
| RMSD backbone atoms (Å)d | 0.6 | |
| RMSD heavy atoms (Å)d | 0.9 | |
| RMSD bond lengths (Å) | 0.018 | |
| RMSD bond angles (°) | 1.1 | |
| MolProbity Ramachandran statisticsc,d | ||
| most favored regions (%) | 96.8 | |
| allowed regions (%) | 3.1 | |
| disallowed regions (%) | 0.1 | |
| Global quality scores (Raw/Z-score)c | ||
| Verify3D | 0.40 | −0.96 |
| ProsaII | 0.66 | 0.04 |
| Procheck (phi-psi)d | −0.15 | −0.28 |
| Procheck (all)d | −0.03 | −0.18 |
| MolProbity clash score | 12.51 | −0.62 |
| RPF Scorese | ||
| Recall/Precision | 0.976 | 0.934 |
| F-measure/DP-score | 0.955 | 0.817 |
| Model Contents: | ||
| Ordered residue ranged | 1–100 | |
| BMRB accession number: | 17965 | |
| PDB ID: | 2LJW | |
Structural statistics computed for the ensemble of 20 deposited structures.
Computed using AVS software [12] from the expected number of resonances, excluding: highly exchangeable protons (N-terminal, Lys, and Arg amino groups, hydroxyls of Ser, Thr, Tyr), carboxyls of Asp and Glu, non-protonated aromatic carbons, and the C-terminal His6 tag.
Calculated using PSVS 1.4 [24]. Average distance violations were calculated using the sum over r−6.
Based on ordered residue ranges [S(phi) + S(psi) > 1.8].
RPF scores [25] reflecting the goodness-of-fit of the final ensemble of structures (including disordered residues) to the NOESY data and resonance assignments.
ConSurf [27, 28] and electrostatic surface potential [29, 30] analyses of the Alr2454 structure reveal that amino acid residues conserved across the PF11267 protein domain family are primarily clustered on the somewhat basic βsheet face of the protein, particularly in the loops between secondary structural elements (Fig. 1D, E). On the basis of Skan [31] and Dali [32] structural alignment analyses, the structure of Nostoc sp. Alr2454 shows no significant similarity to any protein structure reported to date (Dali Z-scores < 3). Hence, we conclude that Alr2454 adopts a unique protein fold. Moreover, the structure of Alr2454 is sufficiently remote from other known protein structures to preclude a useful prediction of the function of this protein domain family.
An important goal in the PSI program has been to provide novel modeling leverage [4]; i.e., the number of proteins for which homology models can be made using a subject protein structure as a template, that could not be made given the structures in the PDB on the date the subject structure was deposited. Based on criteria for homology modeling described by Liu et al. [4], the Alr2454 structure reported here has a novel homology modeling leverage of 95 structures, including all members of Pfam domain family PF11267.
In summary, the solution NMR structure of Nostoc sp. Alr2454 reported here provides the first structural representative from the Pfam PF11267 (DUF3067) protein domain family of unknown function. In addition, the structure represents a novel protein fold. However, uncovering the exact function of the PF11267 protein domain family awaits further study.
Supplementary Material
Acknowledgments
The authors thank Roberto Tejero, Barry Honig, Bomina Yu, Colleen Ciccosanti, and G.V.T. Swapna for helpful discussions. This research was supported by National Institute of General Medical Sciences Protein Structure Initiative (PSI-Biology) program grants U54-GM074958 and U54-GM094597.
Abbreviations
- DSS
2,2-Dimethyl-2-silapentane-5-sulfonic acid
- DTT
Dithiothreitol
- HSQC
Heteronuclear single quantum coherence
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- NOE
Nuclear Overhauser effect
- NESG
Northeast Structural Genomics Consortium
- PDB
Protein Data Bank
- RMSD
Root-mean-square-deviation
Contributor Information
James M. Aramini, Email: jma@cabm.rutgers.edu, Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, and the Northeast Structural Genomics Consortium, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA
Donald Petrey, Department of Biochemistry and Medical Biophysics, Center for Computational Biology and Bioinformatics, and the Northeast Structural Genomics Consortium, Columbia University, New York, NY 10032, USA.
Dong Yup Lee, Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, and the Northeast Structural Genomics Consortium, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Haleema Janjua, Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, and the Northeast Structural Genomics Consortium, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Rong Xiao, Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, and the Northeast Structural Genomics Consortium, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Thomas B. Acton, Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, and the Northeast Structural Genomics Consortium, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA
John K. Everett, Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, and the Northeast Structural Genomics Consortium, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA
Gaetano T. Montelione, Email: guy@cabm.rutgers.edu, Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, and the Northeast Structural Genomics Consortium, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA. Department of Biochemistry, Robert Wood Johnson Medical School, University of Medicine and Dentistry of New Jersey, Piscataway, NJ 08854, USA
References
- 1.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman A, Coggill P, Finn RD. Acta Crystallogr F. 2010;66:1148–1152. doi: 10.1107/S1744309110001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dessailly BH, Nair R, Jaroszewski L, Fajardo JE, Kouranov A, Lee D, Fiser A, Godzik A, Rost B, Orengo C. Structure. 2009;17:869–881. doi: 10.1016/j.str.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Montelione GT, Rost B. Nat Biotechnol. 2007;25:849–851. doi: 10.1038/nbt0807-849. [DOI] [PubMed] [Google Scholar]
- 5.Acton TB, Xiao R, Anderson S, Aramini J, Buchwald WA, Ciccosanti C, Conover K, Everett J, Hamilton K, Huang YJ, Janjua H, Kornhaber G, Lau J, Lee DY, Liu G, Maglaqui M, Ma L, Mao L, Patel D, Rossi P, Sahdev S, Shastry R, Swapna GVT, Tang Y, Tong S, Wang D, Wang H, Zhao L, Montelione GT. Methods Enzymol. 2011;493:21–60. doi: 10.1016/B978-0-12-381274-2.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, Nilsson B. J Biomol NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- 7.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 8.Goddard TD, Kneller DG. SPARKY. Vol. 3. University of California; San Francisco: 2006. [Google Scholar]
- 9.Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. PLoS Comput Biol. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moseley HN, Monleon D, Montelione GT. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- 11.Neri D, Szyperski T, Otting G, Senn H, Wüthrich K. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 12.Moseley HN, Sahota G, Montelione GT. J Biomol NMR. 2004;28:341–355. doi: 10.1023/B:JNMR.0000015420.44364.06. [DOI] [PubMed] [Google Scholar]
- 13.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 14.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann T, Güntert P, Wüthrich K. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, Delaglio F, Cornilescu G, Bax A. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 19.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 20.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 22.Lüthy R, Bowie JU, Eisenberg D. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 23.Sippl MJ. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya A, Tejero R, Montelione GT. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 25.Huang YJ, Powers R, Montelione GT. J Am Chem Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 27.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 28.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. Nucleic Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrey D, Fischer M, Honig B. Proc Natl Acad Sci U S A. 2009;106:17377–17382. doi: 10.1073/pnas.0907971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Rosenström P. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 34.Gouet P, Courcelle E, Stuart DI, Métoz F. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.