Abstract
SnoN (Ski-novel protein) plays an important role in embryonic development, tumorigenesis and aging. Past studies largely focused on its roles in tumorigenesis. Recent studies of its expression patterns and functions in mouse models and mammalian cells have revealed that SnoN interacts with multiple signaling molecules at different cellular levels to modulate the activities of several signaling pathways in a tissue context and developmental stage dependent manner. These studies suggest that SnoN may have broad functions in the embryonic development and tissue morphogenesis.
Keywords: SnoN, Embryonic development, Morphogenesis
1. Introduction
SnoN (Ski novel protein) is a member of the Ski family of proteins and was initially identified as a nuclear proto-oncoprotein based largely on its close homology to v-ski, the transforming protein of the avian Sloan-Kettering retrovirus [1,2] and on its ability to induce anchorage-independent growth of chicken and quail embryo fibroblasts when overexpressed [3]. In addition to SnoN (684 amino acids), three alternatively spliced isoforms of SnoN, SnoN2 (638 amino acids), SnoA (415 amino acids) and SnoI (399 amino acids), all containing an identical N-terminal region of 366 amino acid residues, have been reported [4–6]. Apart from SnoN2, which is an abundant isoform in normal mouse tissues, the expression of SnoI and SnoA is limited to specific human cancer cell lines, and their functions in these cells have not been defined. The N-terminal 366 amino acid-residue region common to all isoforms is encoded by exon 1 of the snoN gene and shows a high level of sequence homology to Ski (Fig. 1). This ski homology region appears to mediate most if not all the known biological functions of Ski and SnoN. The carboxyl (C)-terminal halves of Ski and SnoN show little sequence homology, but may mediate homo- or hetero- dimerization with each other [7,8]. Although SnoN displays a certain degree of structural and functional redundancy with Ski, more recent studies suggest that SnoN also differs significantly from Ski in its functions and regulatory mechanisms [9–11]. SnoN is expressed ubiquitously throughout embryonic development and in all adult tissues and cells, suggesting that it may play a role in embryonic development and normal tissue morphogenesis and homeostasis. Most past studies on SnoN have focused on its actions in human cancers or cancer-derived cell lines as well as its regulatory and signaling mechanisms [9–11]. The physiological function of SnoN during embryonic and postnatal development has not been well characterized. In this review, we will focus on the recent progresses in understanding the functions of SnoN in embryogenesis and tissue morphogenesis with a brief summary of the mechanisms of SnoN signaling and expression. A more detailed summary of these mechanistic studies and the role of SnoN in tumorigenesis can be found in other reviews [10,11].
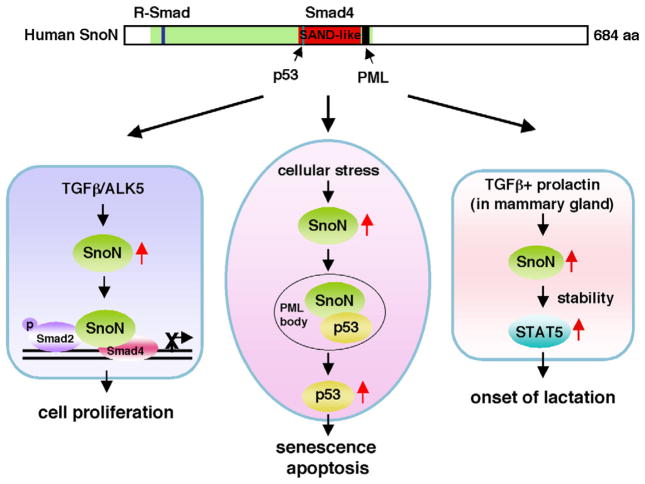
Fig. 1.
Domain structure and signaling activities of SnoN. SnoN is a protein of 684 amino acids residues with a conserved ski-homology domain (green), that contains the R-Smad-binding site (blue), a SAND (Sp100, AIRE1, NucP41/75, and DEAF1)-like domain (red) that mediates binding to Smad4. The p53 and PML binding regions are indicated by the arrowheads. Upon induction, SnoN can bind to the Smad proteins to antagonize the cytostatic activity of TGF-β. Upon induction by cellular stress signals, high levels of SnoN can be recruited to the PML nuclear bodies, where it interacts with p53, leading to its stabilization and activation. This ability to induce p53-dependent senescence and apoptosis responses underlies its anti-tumorigenic activity and its ability to promote aging. In the mammary gland, SnoN is induced by TGF-β and prolactin in a synergistic manner. The elevated SnoN enhances STAT5 stability, thereby sharply increasing the activity of prolactin signaling to trigger the onset of lactation.
2. Regulation of SnoN expression
The expression of SnoN in mammalian cells is dynamically regulated and under a precise temporal and spatial control. Regulation of SnoN expression occurs at multiple levels, including transcriptional regulation, protein degradation and subcellular localization [9–11]. SnoN expression can be induced during specific stages of tissue morphogenesis, by growth factors such as TGF-β, upon tissue injury, and in response to a variety of cellular stress signals including oxidative stress and chronic DNA damage [10,11].
The best characterized regulator of SnoN expression is TGF-β. TGF-β regulates SnoN expression at both the level of protein stability and transcriptional activation [9–11]. Upon TGF-β stimulation,SnoN is rapidly degraded to allow the formation of a transcriptionally active Smad complex to activate the expression of TGF-β target genes [12]. This degradation of SnoN depends on ubiquitin-dependent proteasomes and requires the activity of several E3 ubiquitin ligases, including Smad ubiquitin regulatory factor 2 (Smurf2) [13], the Anaphase Promoting Complex (APC/C) [14,15] or Arkadia [16–18]. SnoN itself is also a transcription target of TGF-β. Transcription of SnoN is induced after 2 hours of TGF-β treatment by a direct binding of the Smad2/4 complex to the Smad-binding elements in the snoN promoter [12,19]. This upregulation of SnoN expression may function as a negative feedback mechanism to regulate the timing, duration and robustness of TGF-β signaling and may also participate in other cellular events [19]. SnoN transcription can also be induced by other growth factors, such as Hepatocyte growth factor (HGF) [20,21] and prolactin [22], upon tissue injury [23,24], and in response to oxidative stress, oncogene overexpression and chronic DNA damage (unpublished observations). The pathways and mechanisms involved in these regulations have not been fully defined.
SnoN was originally identified as a nuclear protein in established cell lines. However, more recent work suggests that in normal tissues and non-tumorigenic or primary epithelial cells, SnoN is predominantly expressed in the cytoplasm, while in cancer cell lines and some malignant tissues, it is nuclear [25,26]. Recently, we found that in mouse endothelial cells, a population of SnoN co-localizes with ALK1 on the plasma membrane to facilitate activation of Smad1/5/8 (unpublished data). Thus, SnoN localization may be regulated in a cell type specific manner and also subjected to regulation by the surrounding microenvironment and cell–cell interaction.
3. Mechanism of SnoN functions
SnoN has been shown to activate a number of signaling pathways, in some cases in a cell type-specific manner to regulate cell proliferation, apoptosis, senescence and differentiation (Fig. 1).
3.1. SnoN as a modulator in TGF-β signaling
SnoN is an important regulator of the Smad proteins of the TGF-β signaling pathways [9–11,27]. Members of TGF-β superfamily, including TGF-βs, activin, Nodal and BMPs (GDFs), play essential roles in embryonic development, organ formation, tissue homeostasis and cell–microenvironment interaction [28–32]. In particular, Nodal, the first TGF-β family member to be expressed in the embryo, is a morphogen whose concentration gradient is essential for gastrulation and establishment of anterior-posterior and left-right axes [33]. TGF-β1 ligand and its down-stream signaling components are critical for yolk sac and embryonic angiogenesis and are also important for the maturation and maintenance of vasculature in adult animals [34]. These growth factors signal through the cell surface type II and I receptor kinases and downstream Smad proteins. Binding of ligands to their specific type II receptors results in activation of the type I receptor kinases, which then phosphorylate and activate Smad (R-Smad) proteins. In mammals, seven type I receptors, ALK1-7, and five type II receptors function in combination to activate two major branches of R-Smads, Smad2/Smad3 and Smad1/Smad5/Smad8. TGF-β/Activin/Nodal signals mainly through Smad2/Smad3, while BMP/GDF predominantly activates Smad1/Smad5/Smad8 [27,35–38]. The phosphorylated R-Smads then form heteromeric complexes with the common partner Smad4 and translocate into the nucleus where they bind DNA, interact with various cellular co-factors and activate transcription of multiple target genes, leading to different cellular responses.
SnoN interacts with Smad2, Smad3 and Smad4 in both the cytoplasm and nucleus and represses their ability to activate downstream target genes by preventing their nuclear translocation, disrupting the functional heteromeric Smad complexes, recruiting transcription co-repressor complexes and blocking the binding of transcriptional co-activators to the Smads [9,12,39–41]. This ability of SnoN to antagonize Smad2, Smad3 and Smad4 implies that SnoN may function as a regulator of TGF-β, activin and Nodal pathways. Indeed, in cultured human and murine cell lines, SnoN has been shown to repress the expression of TGF-β target genes and block the cytostatic responses to TGF-β [12,42], and this is suggested to be responsible for the pro-oncogenic activity of SnoN in lung and breast cancer cell lines [43]. During embryogenesis in vivo, the ability of SnoN to regulate Smad2/Smad3 has been shown to be necessary for the TGF-β and Nodal-dependent developmental events (see below).
The effect of SnoN on the Smad proteins is not always inhibitory. In endothelial cells, SnoN also promotes activation of Smad1/Smad5 (Fig. 2 and see below for more details). In mink lung epithelial cells, reducing SnoN expression has been found to impair TGF-β-dependent transcription and cell cycle arrest [44,45], suggesting that at a lower expression level, SnoN may function to promote TGF-β signaling. Since this has only been observed in only one cell line, it is unclear how common this effect is and what the underlying mechanism is.
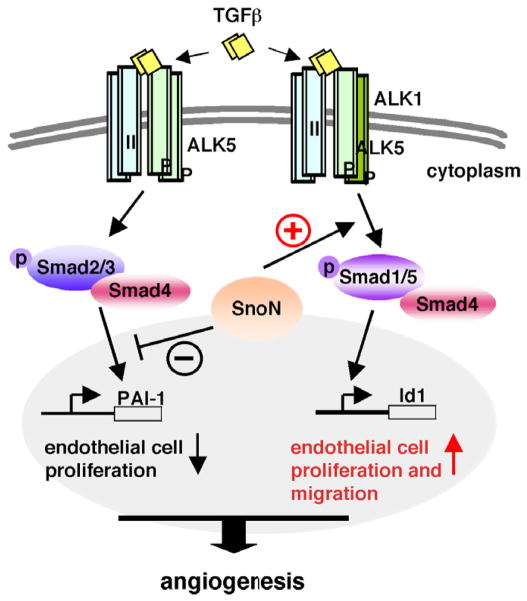
Fig. 2.
SnoN signaling in endothelial cells. SnoN plays a key role in angiogenesis through regulating the balance of two branches of TGF-β signaling in endothelial cells. SnoN enhances activation of Smad1/5 by ALK1 while represses the activity of the ALK5 branch, thus promoting endothelial cell proliferation and migration. Q. Zhu, K. Luo / FEBS Letters 586 (2012) 1971–1976 1973
3.2. SnoN as an activator of p53
Independent of its ability to antagonize TGF-β-induced growth inhibition, SnoN also functions to transduce signals in the cellular stress pathways by activating p53. SnoN expression is upregulated by a number of stress signals including chronic DNA damage, oxidative stress and oncogene overexpression (unpublished observations), and the elevated SnoN is recruited to the PML nuclear bodies through a direct physical interaction with the PML protein [46]. In the PML nuclear bodies, SnoN can bind directly to p53 and compete with Mdm2 for binding to p53, preventing p53 ubiquitination and degradation and facilitating p53 acetylation and phosphorylation, leading to p53 stabilization and activation [47]. Through this activity, overexpression of SnoN in both epithelial and fibroblast cells trigger senescence and increased apoptotic responses. This ability of SnoN to activate p53 to induce cell senescence is not only responsible for its anti-tumorigenic activity, but also results in accelerated aging [47]. Knockin mice expressing a form of SnoN that is accumulated to a high level are resistant to chemical carcinogen-induced tumorigenesis in a p53-dependent manner, are more sensitive to environmental stress and display premature aging in aspects that are known to be p53-dependent [47]. Thus, in adult cells, SnoN is not just an important negative regulator of TGF-β signaling, but plays a much broader role in co-coordinating the cellular stress responses by activating the p53 pathway to regulate cell cycle arrest, senescence and apoptosis.
3.3. Other cell or tissue-specific pathways activated by SnoN
SnoN has been shown to regulate a number of signaling pathways in a tissue-specific manner. In the mammary gland, SnoN is required for the onset of lactation by promoting prolactin signaling through inducing stabilization of STAT5 proteins [22]. SnoN-null mice show a dramatically reduced STAT5 expression and activation, leading to lactation failure [22]. In rat granule neurons, SnoN may function as a transcriptional activator to regulate the expression of a number of genes required for axon development [48]. For example, SnoN coordinates with p300 to activate the transcription of the actin-binding protein Ccd1 to promote axon growth [48]. It is likely that SnoN may be involved in crosstalk with more signaling pathways in specific cell types to regulate cell differentiation and tissue morphogenesis.
3.4. SnoN expression and functions in embryogenesis
SnoN is ubiquitously expressed in the mouse embryo, and its expression is selectively upregulated during specific stages of embryogenesis in tissues with a high degree of proliferative activity [5,26]. In mouse embryos, SnoN transcript is detected as early as E6.5 post coitum (p.c.) in the epiblast and the extramembryonic tissue [5]. Later it is highly expressed in the anterior endoderm, neuroectoderm and neural headfolds (unpublished observations). By E9.5 p.c., elevated SnoN mRNA is found in neural tissues and neural crest cells in addition to the developing vasculature, limb buds and mandibular arches. By E12.5, SnoN expression reaches the highest level in the central nervous system and remains elevated in the brain until birth. In addition, SnoN is induced from E11.5–E15.5 in skeletal muscles. The expression pattern of SnoN in the embryos suggests that it plays a role in the development of multiple lineages including vasculature, neuronal structures and skeletal muscles.
The functions of SnoN in embryonic development have not been well defined. Three snoN knockout mouse lines have been generated, all of which contain deletions of exon 1 and part of the snoN promoter region. One SnoN-null line shows embryonic lethality before E3.5 [49]. The other two SnoN−/− lines are viable and only show a moderate defect in T cell activation [50]. While the reason underlying this discrepancy in pheno-types is not clear, these knockout mice fail to be informative regarding the role of SnoN in embryogenesis. Given the important and complex roles that TGF-β family members play in embryogenesis, we reason that if SnoN is a crucial regulator of the Smad proteins, disruption of the SnoN–Smad interaction may lead to severe developmental outcomes. We therefore generated a knockin mouse (SnoNm/m) expressing a mutant SnoN defective in binding to the Smad proteins in the original snoN locus [46]. Analysis of this knockin mouse strain has revealed important functions of SnoN in mammalian embryogenesis.
3.5. SnoN in regulation of angiogenesis
The SnoNm/m mice show partial embryonic lethality where approximately 33% of the embryos die before E12.5 [46], due mainly to defects in angiogenesis in both the yolk sac and embryo body (unpublished data). These SnoNm/m embryos display severe growth retardation, and their yolk sacs are pale and wrinkled with aberrant vasculature composed of dilated blood vessels surrounded by decreased numbers of vascular smooth muscle cells. The embryo bodies exhibit defects in secondary and tertiary vessel branching. They also exhibit dilated or collapsed dorsal aorta and pericardial effusion due to defects in both endothelial cell function and in recruitment and differentiation of vascular smooth muscle cells [51]. These phenotypes are very similar to those shown in mice lacking ALK1 or Smad5 [51–55], suggesting that SnoN regulates angiogenesis through modulating TGF-β signaling in endothelial cells.
In endothelial cells, TGF-β signals through two branches: the conventional ALK5-Smad2/3 branch and the endothelial specific ALK1-Smad1/5 branch [51,56]. Both branches are activated upon ligand binding but exert different effects on endothelial cell proliferation and migration. The ALK1 branch promotes endothelial cell proliferation and migration, while the ALK5 branch represses these processes, leading to the maturation phase of angiogenesis. The proper balance of the two signaling branches is necessary for angiogenesis.
Using mouse embryo endothelial cells (MEECs) isolated from the SnoNm/m embryos, we found that disruption of the SnoN–Smad interaction leads to not only elevated activation of the ALK5 branch, but also decreased ALK1 signaling activity (unpublished data). While WT SnoN can directly bind to ALK1 upon TGF-β treatment to promote its phosphorylation and activation of Smad1/5, mutant SnoN fails to bridge ALK1 with Smad1/5, resulting in decreased Smad1/5 activation (unpublished data). Thus, in endothelial cells, SnoN promotes Smad1/5 activation while repressing Smad2/3 activity to regulate the proper balance of overall TGF-β signaling (Fig. 2). This balance also controls the expression level of TGF-β1 itself, which in turn modulates the strength of angiogenic signals (unpublished data). Thus, SnoN is an integral part of the TGF-β signaling network and can act as an activator of the Smad proteins in addition to the reported negative regulatory role.
3.6. SnoN in regulation of forebrain development
Independent of the angiogenesis defects, approximately one third of the homozygous embryos that died before E12.5 and a fraction of the heterozygous embryos exhibit aberrant formation or patterning of the anterior neural structures with varying degrees of severity. These defects range from anterior truncations, absence of eye development to failure in neural tube closure and holoprosencephly (unpublished observations). Since the development of the midline structure and anterior patterning of the brain depend on optimal Nodal signaling activity [57], the above phenotypes suggest a role of SnoN in modulating Nodal-dependent developmental events. Indeed, SnoN can repress Nodal-dependent transcription in cells. Analysis of the expression of a number of Nodal target genes in individual embryos at E6.5 and E7.5 revealed that Nodal signaling in SnoNm/m embryos display a dramatic increased oscillation in comparison to that in WT embryos (unpublished observations), indicating that SnoN functions to limit the extent of signaling fluctuation and to maintain robustness of the Nodal signaling circuit.
Apart from angiogenesis and anterior forebrain development, a number of other phenotypes found in SnoNm/m embryos, such as neural tube closure, may be related to defective BMP signaling [58]. Although SnoN has not been found to directly antagonize BMP signaling through Smad1/5/8 proteins, it may indirectly affect BMP target gene expression in vivo through yet-to-be-identified mechanisms. Taken together, analysis of the SnoNm/m embryos suggests that SnoN can function as both an activator and repressor of Smad proteins dependent on tissue or cell types. Instead of inhibiting TGF-β signaling, SnoN is an integral part of the TGF-β superfamily signaling circuit and is necessary for maintaining the robustness of this signaling network.
4. SnoN expression and functions in adult tissue morphogenesis
SnoN is ubiquitously expressed in all adult cell types at a low level [2,5,6]. Recent study in normal human esophageal, ovarian, breast and pancreatic tissues indicate that it is found in both tissue epithelia and stroma and is predominantly cytoplasmic [26,59,60]. Given its ubiquitous expression pattern, SnoN is expected to function in normal tissue physiology likely in a tissue specific manner. However, very little is known about its function in these processes. Here we summarize the recent works that provide insights on SnoN function in mammary gland development and in axon growth.
4.1. SnoN in the mammary gland
The expression and functions of SnoN in mouse mammary gland development have been well characterized in recent years. SnoN is detected in the cytoplasm of luminal epithelial cells lining the ducts and epithelial cells of lobuli and terminal ducts. Its expression is low in the virgin gland, but sharply up regulated at late pregnancy and at the beginning of lactation [26]. The peak period of SnoN expression correlates with the structural and functional differentiation of the mammary alveoli, which polarize to form the secretory alveolar epithelium capable of milk production and secretion [61–64]. The high expression level of SnoN during this window suggests that it may play an important role in the structural and functional differentiation of the alveolar epithelial cells and that the expression of SnoN might be regulated by the pregnancy hormones.
Indeed we found that this sharp upregulation of SnoN expression at late pregnancy is induced by the synergistic actions of TGF-β and prolactin [22]. The elevated SnoN subsequently promotes STAT5 expression and activation by enhancing its stability, at thesametime relieving the repressive effects of TGF-β on prolactin signaling. This allows a rapid increase in prolactin/STAT5 signaling to trigger the onset of lactation. SnoN−/ − mice display severe defects in lactogenesis, and mammary epithelial cells from these mice fail to proliferate and undergo proper morphogenesis in a three-dimensional matrigel model [22]. These defects can be rescued by an active STAT5. Thus, SnoN is a critical player in the regulation of milk production and mammary alveologenesis through promoting STAT5 signaling.
In addition to its role at the onset of lactation, SnoN also stimulates mammary epithelial cell proliferation in the virgin gland. Transgenic mice overexpressing a SnoN fragment under the control of the mouse mammary tumor virus promoter show increased side-branching and lobular-alveolar proliferation in the virgin glands [26]. High levels of SnoN also accelerate post-lactational involution by inducing an increase in apoptosis and a reduction in survival cues. These activities of SnoN are likely dependent on its ability to repress TGF-β signaling since TGF-β has been shown to inhibit lobular alveolar proliferation, ductal growth [65], and regulate involution [66]. In addition, although overexpression of SnoN is insufficient to induce mammary tumorigenesis, elevated levels of SnoN cooperate with polyoma middle T antigen to accelerate breast adenocarcinoma development and pulmonary metastases [26]. Taken together, SnoN affects multiple stages of mammary gland development by regulating the proliferation, differentiation, polarity and apoptosis of mammary epithelial cells.
4.2. SnoN in the central nerve system
In the adult brain, snoN transcripts are found predominantly in the granule neurons of the cerebellum and Purkinje cells in the cerebellar cortex, in the hippocampus and in the cerebrum [5,67]. siRNA-based knockdown of SnoN in primary cerebellar granule and cortical neurons significantly reduces axonal growth [67]. In addition, reducing SnoN in the postnatal rat pups markedly impairs the morphogenesis of parallel fiber axons in the cerebellar cortex in vivo [48,68]. These results suggest that SnoN may play a critical role in axon development in the mammalian brain. Mechanistically, SnoN associates with p300/CBP to activate the expression of the signaling scaffolding protein Ccd1 to promote axon growth [48]. It can also associate with FOXO1 to repress the expression of the doublecortin (DCX) gene, and this repression of DCX expression appears to mediate SnoN regulation of primary neuron branching and granule neuron migration [69]. SnoN has also been found to function downstream of the Cdh1–anaphase promoting complex (Cdh1–APC) and Smad2, and can be degraded by the Cdh1–APC in neurons [70]. Clearly, more studies are needed to determine how these events are coordinated to allow SnoN activity and expression to be controlled in a precise manner to regulate neuronal specific functions.
5. SnoN in regulation of aging
SnoN can activate p53 in response to various cellular stress signals. In primary MEF cells, oxidative stress upregulates SnoN expression to trigger premature senescence in a p53-dependent manner [46]. Since activation of p53 is known to accelerate aging in mice, it is conceivable that SnoN may also contribute to progeria. Indeed, the SnoN knockin mice are more sensitive to environmental stress and exhibit accelerated aging phenotypes including a shortened lifespan, reduced body weight and reproductivity, kyphosis, osteoporosis, decreased wound-healing ability and a reduced regenerative capacity [47], similar to that found in mice expressing an active p53 [71]. Consistent with these aging phenotypes, tissues from the knockin mice contain an increased number of senescence and apoptotic cells than those from age-matched WT mice [47].
Thus, in adult cells, SnoN is not just an important negative regulator of TGF-β signaling, but plays a much broader role in coordinating the cellular stress responses by crosstalk with various intracellular signaling pathways to regulate cell cycle arrest, senescence and apoptosis. SnoN expression can be induced by a variety of stress signals and tissue injury. At times of prolonged cellular and tissue stress and injury, SnoN serves as an important cellular anticancer defense mechanism by integrating various cellular stress signals to the p53 anti-tumorigenic pathway. These stress responses protect the organism against cancer development, but at the price of accelerated aging.
6. Conclusion and future directions
Through the analyses of SnoN knockin and knockout mice as well as examination of primary cells isolated from these mice, a complex picture of SnoN functions in embryonic and adult tissue morphogenesis and homeostasis is emerging. An important concept that emerges from these analyses is that SnoN should not be considered an inhibitor of TGF-β signaling but as an integral and necessary component of the TGF-β signaling circuit. As a negative feedback regulator of Smad2/3, it functions to regulate the robustness of signaling. It also coordinates the activities of various signaling pathways of TGF-β signaling branches to ensure that proper developmental events occur. It is quite likely that our studies thus far have only uncovered a small fraction of the functions of SnoN in normal tissues. An important goal in the near future is to determine all the tissue specific functions of SnoN and to identify their underlying signaling mechanisms. Understanding these crucial facts would allow us to apply a systems biology approach to model the complex signaling network that SnoN regulates.
Acknowledgments
Weapologize to the authors whose workcould not be cited in this review due to space constraints. Research in the authors’ laboratory was supported by NIH RO1 CA101891, RO1 DK090347 to K. Luo.
References
- 1.Li Y, Turck CM, Teumer JK, Stavnezer E. Unique sequence, ski, in Sloan-Kettering avian retroviruses with properties of a new cell-derived oncogene. J Virol. 1986;57:1065–1072. doi: 10.1128/jvi.57.3.1065-1072.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nomura N, Sasamoto S, Ishii S, Date T, Matsui M, Ishizaki R. Isolation of human cDNA clones of ski and the ski-related gene, sno. Nucleic Acids Res. 1989;17:5489–5500. doi: 10.1093/nar/17.14.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer PL, Colmenares C, Stavnezer E, Hughes SH. Sequence and biological activity of chicken snoN cDNA clones. Oncogene. 1993;8:457–466. [PubMed] [Google Scholar]
- 4.Pearson-White S. SnoI, a novel alternatively spliced isoform of the ski protooncogene homolog, sno. Nucleic Acids Res. 1993;21:4632–4638. doi: 10.1093/nar/21.19.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelzer T, Lyons GE, Kim S, Moreadith RW. Cloning and characterization of the murine homolog of the sno proto-oncogene reveals a novel splice variant. Dev Dyn. 1996;205:114–125. doi: 10.1002/(SICI)1097-0177(199602)205:2<114::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 6.Pearson-White S, Crittenden R. Proto-oncogene Sno expression, alternative isoforms and immediate early serum response. Nucleic Acids Res. 1997;25:2930–2937. doi: 10.1093/nar/25.14.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heyman HC, Stavnezer E. A carboxyl-terminal region of the ski oncoprotein mediates homodimerization as well as heterodimerization with the related protein SnoN. J Biol Chem. 1994;269:26996–27003. [PubMed] [Google Scholar]
- 8.Cohen SB, Zheng G, Heyman HC, Stavnezer E. Heterodimers of the SnoN and Ski oncoproteins form preferentially over homodimers and are more potent transforming agents. Nucleic Acids Res. 1999;27:1006–1014. doi: 10.1093/nar/27.4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo K. Ski and SnoN: negative regulators of TGF-beta signaling. Curr Opin Genet Dev. 2004;14:65–70. doi: 10.1016/j.gde.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Jahchan NS, Luo K. SnoN in mammalian development, function and diseases. Curr Opin Pharmacol. 2010;10:670–675. doi: 10.1016/j.coph.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deheuninck J, Luo K. Ski and SnoN, potent negative regulators of TGF-beta signaling. Cell Res. 2009;19:47–57. doi: 10.1038/cr.2008.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 13.Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- 14.Stroschein SL, Bonni S, Wrana JL, Luo K. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 2001;15:2822–2836. doi: 10.1101/gad.912901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan Y, Liu X, Kirschner MW. The anaphase-promoting complex mediates TGF-beta signaling by targeting SnoN for destruction. Mol Cell. 2001;8:1027–1039. doi: 10.1016/s1097-2765(01)00382-3. [DOI] [PubMed] [Google Scholar]
- 16.Nagano Y, Mavrakis KJ, Lee KL, Fujii T, Koinuma D, Sase H, Yuki K, Isogaya K, Saitoh M, Imamura T, Episkopou V, Miyazono K, Miyazawa K. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. J Biol Chem. 2007;282:20492– 20501. doi: 10.1074/jbc.M701294200. [DOI] [PubMed] [Google Scholar]
- 17.Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol Cell Biol. 2007;27:6068–6083. doi: 10.1128/MCB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Scolan E, Zhu Q, Wang L, Bandyopadhyay A, Javelaud D, Mauviel A, Sun L, Luo K. Transforming growth factor-beta suppresses the ability of Ski to inhibit tumor metastasis by inducing its degradation. Cancer Res. 2008;68:3277–3285. doi: 10.1158/0008-5472.CAN-07-6793. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Q, Pearson-White S, Luo K. Requirement for the SnoN oncoprotein in transforming growth factor beta-induced oncogenic transformation of fibroblast cells. Mol Cell Biol. 2005;25:10731–10744. doi: 10.1128/MCB.25.24.10731-10744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Dai C, Liu Y. A novel mechanism by which hepatocyte growth factor blocks tubular epithelial to mesenchymal transition. J Am Soc Nephrol. 2005;16:68–78. doi: 10.1681/ASN.2003090795. [DOI] [PubMed] [Google Scholar]
- 21.Tan R, Zhang X, Yang J, Li Y, Liu Y. Molecular basis for the cell type specific induction of SnoN expression by hepatocyte growth factor. J Am Soc Nephrol. 2007;18:2340–2349. doi: 10.1681/ASN.2007010128. [DOI] [PubMed] [Google Scholar]
- 22.Jahchan NS, Wang D, Bissell MJ, Luo K. SnoN regulates mammary gland alveologenesis and onset of lactation by promoting prolactin/ STAT5 signaling. 2012. Paper submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macias-Silva M, Li W, Leu JI, Crissey MA, Taub R. Up-regulated transcriptional repressors SnoN and Ski bind Smad proteins to antagonize Q. Zhu, K. Luo / FEBS Letters 586 (2012) 1971–1976 1975 transforming growth factor-beta signals during liver regeneration. J Biol Chem. 2002;277:28483–28490. doi: 10.1074/jbc.M202403200. [DOI] [PubMed] [Google Scholar]
- 24.Mayoral R, Valverde AM, Llorente Izquierdo C, González-Rodríguez A, Boscá L, Martín-Sanz P. Impairment of transforming growth factor beta signaling in caveolin-1-deficient hepatocytes: role in liver regeneration. J Biol Chem. 2010;285:3633–3642. doi: 10.1074/jbc.M109.072900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krakowski AR, Laboureau J, Mauviel A, Bissell MJ, Luo K. Cytoplasmic SnoN in normal tissues and nonmalignant cells antagonizes TGF-beta signaling by sequestration of the Smad proteins. Proc Natl Acad Sci USA. 2005;102:12437–12442. doi: 10.1073/pnas.0504107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jahchan NS, You YH, Muller WJ, Luo K. Transforming growth factor-beta regulator SnoN modulates mammary gland branching morphogenesis, postlactational involution, and mammary tumorigenesis. Cancer Res. 2010;70:4204–4213. doi: 10.1158/0008-5472.CAN-10-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 28.Massagué J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mummery CL. Transforming growth factor beta and mouse development. Microsc Res Tech. 2001;52:374–386. doi: 10.1002/1097-0029(20010215)52:4<374::AID-JEMT1022>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 30.Derynck R, Miyazono K. TGF-β and the TGF-β family. In: Derynck R, Miyazono K, editors. The TGF-β Family. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. pp. 29–44. [Google Scholar]
- 31.Wu MY, Hill CS. TGF-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19:71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- 34.ten Dijke P, Arthur HM. Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol. 2007;8:857–869. doi: 10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
- 35.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 36.Feng XH, Derynck R. Specificity and versatility in TGF-signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 37.Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 38.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 39.Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. C-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. J Biol Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- 40.Wu JW, Krawitz AR, Chai J, Li W, Zhang F, Luo K, Shi Y. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell. 2002;111:357– 367. doi: 10.1016/s0092-8674(02)01006-1. [DOI] [PubMed] [Google Scholar]
- 41.He J, Tegen SB, Krawitz AR, Martin GS, Luo K. The transforming activity of Ski and SnoN is dependent on their ability to repress the activity of Smad proteins. J Biol Chem. 2003;278:30540–30547. doi: 10.1074/jbc.M304016200. [DOI] [PubMed] [Google Scholar]
- 42.Sun Y, Liu X, Ng-Eaton E, Lodish HF, Weinberg RA. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proc Natl Acad Sci USA. 1999;96:12442–12447. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Q, Krakowski AR, Dunham EE, Wang L, Bandyopadhyay A, Berdeaux R, Martin GS, Sun L, Luo K. Dual role of SnoN in mammalian tumorigenesis. Mol Cell Biol. 2007;27:324–339. doi: 10.1128/MCB.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarker KP, Wilson SM, Bonni S. SnoN is a cell type-specific mediator of transforming growth factor-beta responses. J Biol Chem. 2005;280:13037–13046. doi: 10.1074/jbc.M409367200. [DOI] [PubMed] [Google Scholar]
- 45.Sarker KP, Kataoka H, Chan A, Netherton SJ, Pot I, Huynh MA, Feng X, Bonni A, Riabowol K, Bonni S. ING2 as a novel mediator of transforming growth factor-beta-dependent responses in epithelial cells. J Biol Chem. 2008;283:13269–13279. doi: 10.1074/jbc.M708834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan D, Zhu Q, Luo K. SnoN functions as a tumour suppressor by inducing premature senescence. EMBO J. 2009;28:3500–3513. doi: 10.1038/emboj.2009.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan D, Zhu Q, Conboy MJ, Conboy IM, Luo K. SnoN directly activates p53 to regulate aging and tumorigenesis. 2012. Paper submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikeuchi Y, Stegmüller J, Netherton S, Huynh MA, Masu M, Frank D, Bonni S, Bonni A. A SnoN-Ccd1 pathway promotes axonal morphogenesis in the mammalian brain. J Neurosci. 2009;29:12–21. doi: 10.1523/JNEUROSCI.0126-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinagawa T, Dong HD, Xu M, Maekawa T, Ishii S. The sno gene, which encodes a component of the histone deacetylation complex, acts as a tumor suppressor in mice. EMBO J. 2000;19:2280–2291. doi: 10.1093/emboj/19.10.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearson-White S, McDuffie M. Defective T-cell activation is associated with augmented transforming growth factor Beta sensitivity in mice with mutations in the Sno gene. Mol Cell Biol. 2003;23:5446–5459. doi: 10.1128/MCB.23.15.5446-5459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pardali E, Goumans MJ, ten Dijke P. Signaling by members of the TGF-beta family in vascular morphogenesis and disease. Trends Cell Biol. 2010;20:556–567. doi: 10.1016/j.tcb.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 52.Seki T, Hong KH, Oh SP. Nonoverlapping expression patterns of ALK1 and ALK5 reveal distinct roles of each receptor in vascular development. Lab Invest. 2006;86:116–129. doi: 10.1038/labinvest.3700376. [DOI] [PubMed] [Google Scholar]
- 53.Urness LD, Sorensen LK, Li DY. Arteriovenous malformations in mice lacking activin receptor-like kinase-1. Nat Genet. 2000;26:328–331. doi: 10.1038/81634. [DOI] [PubMed] [Google Scholar]
- 54.Seki T, Yun J, Oh SP. Arterial endothelium-specific activin receptor-like kinase 1 expression suggests its role in arterialization and vascular remodeling. Circ Res. 2003;93:682–689. doi: 10.1161/01.RES.0000095246.40391.3B. [DOI] [PubMed] [Google Scholar]
- 55.Yang X, Castilla LH, Xu X, Li C, Gotay J, Weinstein M, Liu PP, Deng CX. Angiogenesis defects and mesenchymal apoptosis in mice lacking SMAD5. Development. 1999;126:1571–1580. doi: 10.1242/dev.126.8.1571. [DOI] [PubMed] [Google Scholar]
- 56.Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Balancing the activation state of the endothelium via two distinct TGF-beta type I receptors. EMBO J. 2002;21:1743–1753. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 58.Levine AJ, Brivanlou AH. Proposal of a model of mammalian neural induction. Dev Biol. 2007;308:241–256. doi: 10.1016/j.ydbio.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang F, Lundin M, Ristimaki A, Heikkila P, Lundin J, Isola J, Joensuu H, Laiho M. Ski-related novel protein N (SnoN), a negative controller of transforming growth factor-beta signaling, is a prognostic marker in estrogen receptor-positive breast carcinomas. Cancer Res. 2003;63:5005–5010. [PubMed] [Google Scholar]
- 60.Zhang X, Egawa K, Xie Y, Ihn H. The expression of SnoN in normal human skin and cutaneous keratinous neoplasms. Int J Dermatol. 2009;48:579–583. doi: 10.1111/j.1365-4632.2009.03685.x. [DOI] [PubMed] [Google Scholar]
- 61.Neville MC, McFadden TB, Forsyth I. Hormonal regulation of mammary differentiation and milk secretion. J Mammary Gland Biol Neoplasia. 2002;7:49–66. doi: 10.1023/a:1015770423167. [DOI] [PubMed] [Google Scholar]
- 62.Oakes SR, Hilton HN, Ormandy CJ. The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res. 2006;8:207. doi: 10.1186/bcr1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richert MM, Schwertfeger KL, Ryder JW, Anderson SM. An atlas of mouse mammary gland development. J Mammary Gland Biol Neoplasia. 2000;5:227–241. doi: 10.1023/a:1026499523505. [DOI] [PubMed] [Google Scholar]
- 64.Anderson SM, Rudolph MC, McManaman JL, Neville MC. Key stages in mammary gland development. Secretory activation in the mammary gland: it’s not just about milk protein synthesis! Breast Cancer Res. 2007;9:204. doi: 10.1186/bcr1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 66.Muraoka-Cook RS, Shin I, Yi JY, Easterly E, Barcellos-Hoff MH, Yingling JM, Zent R, Arteaga CL. Activated type I TGFbeta receptor kinase enhances the survival of mammary epithelial cells and accelerates tumor progression. Oncogene. 2006;25:3408–3423. doi: 10.1038/sj.onc.1208964. [DOI] [PubMed] [Google Scholar]
- 67.Stegmüller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1–APC target SnoN. Neuron. 2006;50:389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 68.Pot I, Ikeuchi Y, Bonni A, Bonni S. SnoN: bridging neurobiology and cancer biology. Curr Mol Med. 2010;10:667–673. doi: 10.2174/156652410792630616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huynh MA, Ikeuchi Y, Netherton S, de la Torre-Ubieta L, Kanadia R, Stegmüller J, Cepko C, Bonni S, Bonni A. An isoform-specific SnoN1-FOXO1 repressor complex controls neuronal morphogenesis and positioning in the mammalian brain. Neuron. 2011;69:930–944. doi: 10.1016/j.neuron.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stegmüller J, Huynh MA, Yuan Z, Konishi Y, Bonni A. TGFbeta- Smad2 signaling regulates the Cdh1–APC/SnoN pathway of axonal morphogenesis. J Neurosci. 2008;28:1961–1969. doi: 10.1523/JNEUROSCI.3061-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Hee Park S, Thompson T, Karsenty G, Bradley A, Donehower LA. P53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]