Abstract
The Empirical Phase Diagram (EPD) technique is a vector-based multidimensional analysis method for summarizing large data sets from a variety of biophysical techniques. It can be used to provide comprehensive preformulation characterization of a macromolecule’s higher-order structural integrity and conformational stability. In its most common mode, it represents a type of stimulus-response diagram using environmental variables such as temperature, pH, and ionic strength as the stimulus, with alterations in macromolecular structure being the response. Until now EPD analysis has not been available in a high throughput mode because of the large number of experimental techniques and environmental stressor/stabilizer variables typically employed. A new instrument has been developed that combines circular dichroism, UV-absorbance, fluorescence spectroscopy and light scattering in a single unit with a 6-position temperature controlled cuvette turret. Using this multifunctional instrument and a new software system we have generated EPDs for four model proteins. Results confirm the reproducibility of the apparent phase boundaries and protein behavior within the boundaries. This new approach permits two EPDs to be generated per day using only 0.5 mg of protein per EPD. Thus, the new methodology generates reproducible EPDs in high-throughput mode, and represents the next step in making such determinations more routine.
Keywords: phase diagrams, formulation, stability, protein, vaccines, circular dichroism, fluorescence, absorbance, calorimetry, light scattering
INTRODUCTION
An Empirical Phase Diagram (EPD) is a data visualization tool used to assist in comparing alterations in macromolecular states due to environmental stress. The technique consists of applying Principal Components Analysis to a series of multidimensional biophysical measurements to find an optimal lower dimensional representation of these large data sets, then using this representation to construct a color diagram in which multidimensional differences in measurements are easily discernible. This approach may be thought of as intermediate between that of high resolution techniques such as X-Ray crystallography and NMR and individual biophysical analyses such as circular dichroism (CD), fluorescence spectroscopy, or static light scattering. The information content of EPDs is relatively high due to the use of stress variables to create what is effectively a stress-response diagram. Applications of EPDs have included the selection of excipient screening conditions and the determination of stabilizing solution conditions, as well as the comparison of proteins (e.g. mutants). The EPD technique has been used to assist in characterizing, stabilizing, and formulating proteins, viruses, virus-like particles (VLPs), and whole bacterial cells as pharmaceutical dosage forms for use as therapeutic drugs and vaccines.1,2
The time and effort required to generate an EPD for individual proteins have, however, somewhat limited the method's general applicability. It typically takes several days or up to 1–2 weeks to collect individual data sets with multiple biophysical techniques, process the large combined data set, and generate an EPD. Reducing the total time to a day or less would allow EPDs to be more routinely used during formulation development as a tool for excipient screening and to enable more reliable comparisons of the stability of higher-order macromolecular states.
Multimode instruments have recently become available that could significantly reduce the time to collect the experimental data used to generate an EPD. The Olis Multiscan (Bogart, GA) (also referred to as “The Protein Machine”) is a cuvette-based spectrophotometer that measures circular dichroism, UV-absorbance, fluorescence, turbidity and light scattering with high level photometric and wavelength accuracy and repeatability. In contrast to most plate readers, this instrument measures full spectra, in addition to incorporating a temperature controlled sample chamber capable of performing temperature studies from 0 to 100°C.
To automate data analysis, we have developed a software tool called the Declarative Array Transformer (DART, manuscript in preparation). In essence, this tool allows one to perform general mathematical, graphical, and file operations on data arrays without concern over the propagation and use of array axis information. DART is used to write data processing scripts in a declarative programming style, meaning that control flow operations such as looping and if/then statements are not necessary for most tasks. Scripts written in DART are self-explanatory and easily modified by non-expert programmers. Additionally, reproducibility of experiments and traceability of data are increased by saving data processing scripts along with their output data.
In this manuscript we describe for the first time the Olis Multiscan and its use to generate EPDs for four model proteins including aldolase, bovine serum albumin (BSA), α-chymotrypsin, and lysozyme. These proteins cover a range of molecular weights (~14 to 160 kDa), secondary structures (~10–67% alpha helical and ~10–49% beta sheet), and thermal stabilities (Tm values from ~44 to 74°C).3–8 These model proteins were characterized over a grid of environmental conditions consisting of solution pH values from 3 to 8 and temperatures from 10°C to 85°C. The characterization of each protein was performed over a 12 hour period. At each combination of temperature and pH, the following biophysical measurements were taken: CD at 217, 222, and 235 nm, absorbance from 238 to 343 nm (including optical density measurements from 320 to 340 nm), and intrinsic Trp fluorescence between 255 and 420 nm with 295 nm excitation. DART was then used to import and regularize the data, filter it, and generate EPDs. The resulting empirical phase diagrams have been interpreted in light of the original raw data, and the phase boundaries and protein behavior were found to be reproducible and similar to those obtained by independent measurements using separate instruments.
Methods
Materials
Albumin (from bovine serum), aldolase (from rabbit muscle), α-chymotrypsin (from bovine pancreas), and lysozyme (from chicken egg white) were obtained in the form of lyophilized powder from Sigma Life Sciences (St. Louis, MO). All chemicals were of reagent grade and purchased from Fisher Scientific (Pittsburg, PA).
Citrate-phosphate buffer was prepared at 20 mM at pH 3.0, 4.0, 5.0, 6.0, 7.0, and 8.0 from citric acid anhydrous and sodium phosphate dibasic anhydrous. The ionic strength of each buffer was controlled to I = 0.15 (dimensionless) by the addition of NaCl. For each pH, the lyophilized protein samples were dissolved into 2 mL of H2O and all protein solutions were dialyzed into between 1 and 2 L of citrate-phosphate buffer in Thermo Scientific Slide-a-Lyzer 0.5–3 mL 3500 Da MWCO dialysis cassettes (Waltham, MA). The concentration of each sample was obtained by absorbance spectroscopy with 1 cm path length at 280 nm using an Agilent Technologies 8453 spectrophotometer (Santa Clara, CA) and known extinction coefficients for each model protein. Samples were diluted to 0.2 mg/mL. The citrate-phosphate buffers used for protein dilution and as instrument controls were filtered with a Millipore Millex 0.45 µm syringe filter (Billerica, MA). Samples were stored at 4°C and measurements were taken within two weeks of reconstituting the lyophilized protein powders with the citrate-phosphate buffers (except for lysozyme which was used within 3 weeks).
High Throughput Spectroscopy
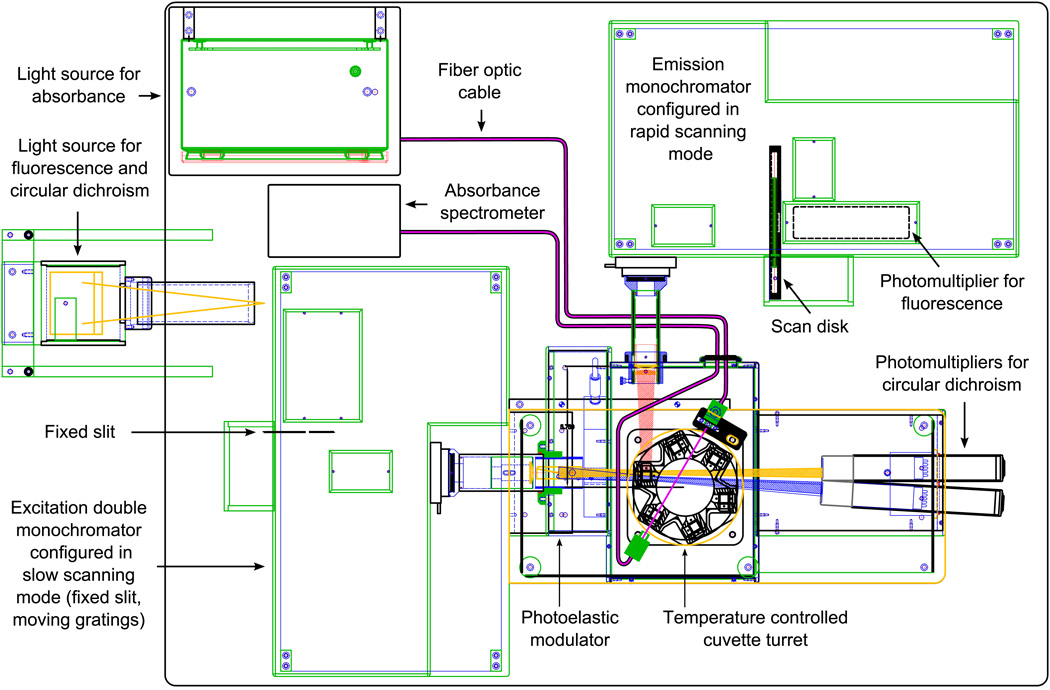
High throughput spectroscopy was performed with the Olis Multiscan (Bogart, GA) equipped with a Quantum Northwest peltier temperature controlled 6-position cuvette turret (Liberty Lake, WA). The Olis Multiscan uses a 150 W xenon arc lamp and dual grating Rapid Scanning Monochromators (RSM-1000) for circular dichroism (CD) and fluorescence excitation (see Figure 1). The RSM was set up in slow scanning mode, using a fixed 1.24 mm slit (corresponding to 1.6 nm band pass) and moveable gratings (for rapid scanning, the gratings would be fixed and a scan disk would be used instead of the fixed slit). Fluorescence signals are read at 90 degrees from the excitation source through a single grating RSM, using a rotating 1 mm slit (corresponding to a 6.3 nm band pass). The fluorescence monochromator collects a spectrum every 10 ms, and can measure an emission spectrum at each CD excitation wavelength. Absorbance is measured using a separate Avantes system (Eerbeek, The Netherlands) consisting of an Avalamp-DS deuterium light source and an AvaSpec-1024 photodiode array spectrometer that is built directly into the spectrometer system.
Figure 1.
Overhead line drawing of the Olis Multiscan (also referred to as “The Protein Machine”), a cuvette-based spectrophotometer that measures circular dichroism, absorbance, and fluorescence with high level photometric and wavelength accuracy and repeatability.
Samples were placed in 6Q Spectrosil cuvettes with a 2 mm path length in one direction and 1 cm path length in the other. The cuvettes were positioned so that the 2 mm path was parallel to the excitation beam and Avalamp absorbance beam (see Figure 1). Thermal stress was performed from 10–85°C in steps of 2.5°C. Excitation monochromator exit slit width and emission monochromator entrance slit width were both maximized to provide robust signals. These slits control light throughput and have no effect on band passes. Integration times were chosen to yield a cycle of 12 hours per experiment, for the convenience of experimenters. In those 12 hours, equal time was allotted to each of the 3 techniques.
The Olis Multiscan was operated in the following manner during temperature ramp experiments. After temperature equilibration of the sample chamber, the 6-position turret rotates to place one of the sample containing cuvettes into the excitation beam path. For each cuvette, the excitation monochromator scans through a series of excitation wavelengths, spending a user specified time at each wavelength while collecting one CD measurement and a full fluorescence spectrum. After scanning the excitation wavelengths, the next cuvette is rotated into the excitation beam, and this process repeats until each cuvette has been measured. The cuvettes are then scanned again, but this time they are placed into the beam path of the Avantes absorbance subsystem and UV absorbance spectra are measured. Finally, the temperature is raised to the next set point and the entire cycle is repeated.
One might ask whether the order of the operations could change the outcome, since the various measurements occur at different time delays after the temperature is increased. For example, CD and fluorescence are measured for the first cuvette shortly after the temperature is raised, whereas the absorbance spectrum of the last cuvette is measured roughly 20 minutes later. At all times, however, each cuvette has had ample time to equilibrate to the temperature before the current one. So the measured conformational state of the protein is somewhere between the equilibrated states at the last temperature and the current temperature. Thus, the order of operations can potentially affect the protein’s observed state and transition temperatures, but the effect is limited to an uncertainty equal to the size of the temperature step.
Circular Dichroism
Circular dichroism (CD) was used to measure molar ellipticity at the wavelengths of 217, 222, and 235 nm, chosen to correspond to typical peaks seen for alpha-helix and beta-structure secondary structures.9,10 Although Chymotrypsin has no CD peak at 222 nm, Chymotrypsin melting trends were similar for all 3 of the wavelengths chosen. Due to absorbance by the 20 mM citrate-phosphate buffer, far UV CD signals could not be monitored successfully at wavelengths below 215 nm for the majority of protein and pH combinations. Although shorter path lengths would have reduced absorbance by the buffer, they would result in a proportionally diminished fluorescence emission signal. Furthermore, the fluorescence emission measurements are taken parallel to the 1cm cuvette path length, making it difficult to use very short excitation beam path lengths due to reflection and alignment issues. To decrease noise in the data resulting from increased absorbance, CD data was integrated for 19 seconds at each wavelength.
Steady State Intrinsic Trp Fluorescence
The tertiary structure of all proteins was screened using intrinsic fluorescence. Proteins were excited at 295 nm to exclusively (>95%) excite tryptophan residues, as well as 300 nm to investigate red-edge shifts. Emission spectra were recorded between 255 and 420 nm with 18 seconds of integration time per excitation wavelength.
Each (temperature × wavelength) spectral melt matrix was filtered as follows. The matrix was first reconstructed using the top 5 singular vectors given by the Singular Value Decomposition. Each spectrum was then filtered with a third order, 4 nm radius Savitzky-Golay filter followed by a 4 nm radius Gaussian smooth. Then the temperature dimension of the matrix (i.e., the melt at each wavelength) was filtered with a third order, 6°C radius Savitzky-Golay filter followed by a 2.5°C radius Gaussian smooth.
Peak positions were determined from the filtered data using the spectral center of mass method. A red edge shift at each temperature and pH was determined by subtracting the emission peak position with 295 nm excitation from the peak position with 300 nm excitation.
Absorbance and Optical Density Measurements
Absorbance spectra were recorded between 238 and 343 nm with 46 seconds of integration time. The long integration time was required due to the low concentration and path length that were required for compatibility with far-UV CD measurements. The peak near-UV absorbance of BSA for instance, was approximately 0.029 absorbance units.
Each (temperature × wavelength) spectral melt matrix was filtered in the same manner as the fluorescence measurements. Second derivative spectra were then calculated with a third order, 4 nm radius, second derivative Savitzky-Golay filter.
The mean optical density from 320 to 340 nm was calculated from unfiltered spectra by averaging the optical density values in that range.
Construction of EPDs
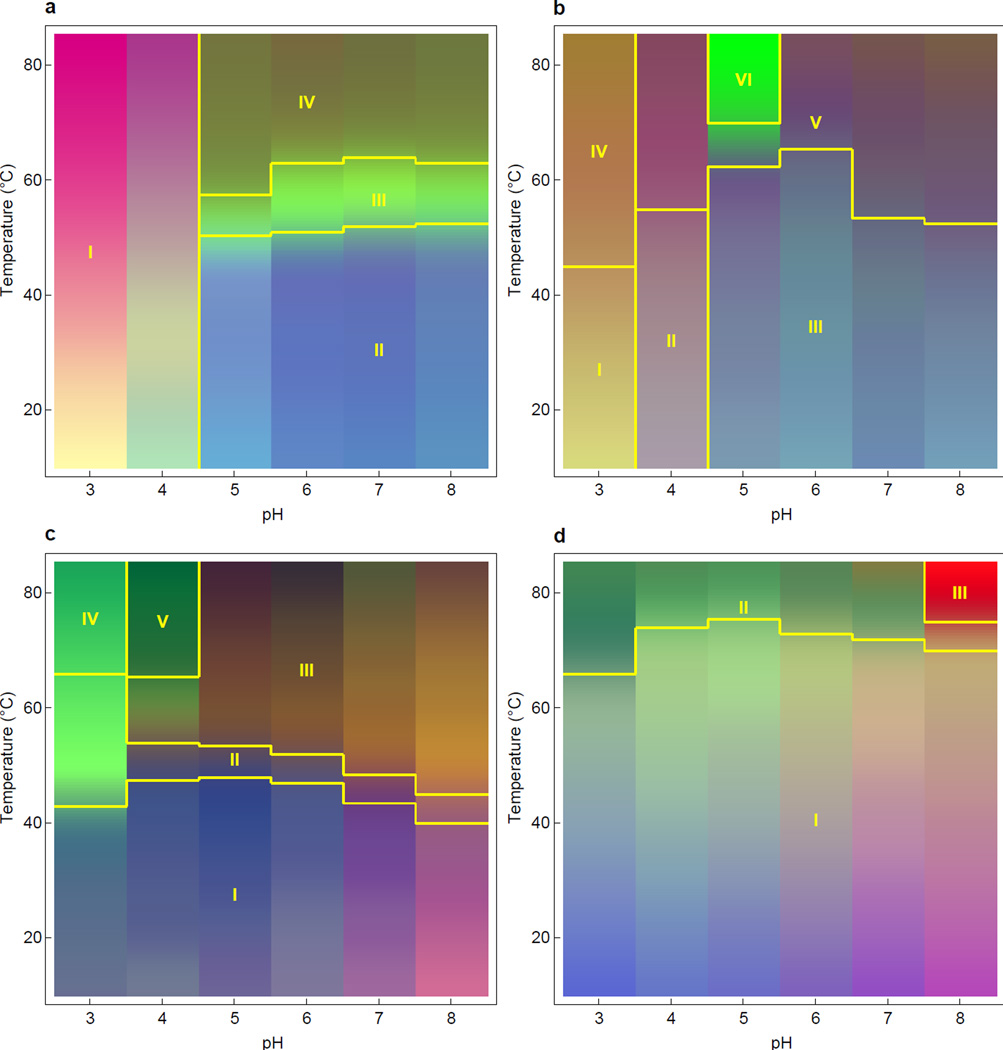
EPDs were constructed from each run, and were generated separately for each of the four model proteins. The EPDs resulting from the first 12 hour run of each protein are shown in Figure 3. The EPDs from further runs are virtually identical (data not shown). The following biophysical measurements were used: the second derivative near UV absorbance from 275 to 295 nm, mean optical density from 320 to 340 nm, far UV CD at 217, 222, and 235 nm, and fluorescence spectra from 315 to 370 nm. It should be noted that full UV absorbance and fluorescence spectra were used rather than selected peak positions and intensities. Before applying the EPD method, the data was interpolated in the temperature dimension from the original 2.5°C increments to smaller 0.5°C increments. For a detailed description of the EPD method, see Maddux et al.2
Figure 3.
Empirical phase diagrams of 4 model proteins: (a) aldolase, (b) BSA, (c) chymotrypsin, and (d) lysozyme. These EPDs summarize the representative biophysical data from Figure 2 (in conjunction with wavelength measurements using the full spectra; see text) and display protein structural responses to temperature and pH perturbations. The experimental techniques used were second derivative near UV absorbance from 275 to 295 nm (full spectra), fluorescence spectra from 315 to 370 nm (full spectra), far UV CD at 217, 222, and 235 nm, and the mean optical density from 320 to 340 nm.
EPD segmentation
The EPD method assists in the determination of regions of conserved behavior, but does not itself determine transition regions. Instead, human visual assessments have traditionally been used to perform the task of separating regions of the same color on an EPD. A similar classification can also be performed mathematically by the use of cluster analysis. Since it is automatic, cluster analysis may be a valuable tool for high-throughput protein stability characterization using EPDs.
Cluster analysis was performed on the average of 3 runs, separately for each protein, using the same combination of measurements used to generate EPDs. The standardization step in the EPD method results in a list of multidimensional vectors, with one vector for each combination of temperature and pH. K-means clustering was applied to these vectors to find a natural categorization in the high dimensional space, thus dividing the temperature-pH plane into regions of similar measurements. For a description of clustering in general and the K-means algorithm in particular, see Jain 2006.11
The phases and transitions temperatures given by K-means clustering did not, however, always match the phases and transitions perceived by visual assessments of the EPDs. To address this issue, the EPDs were segmented using a different method. For each phase in an EPD, a characteristic point in the temperature-pH plane was visually selected to represent that phase. Then for each point in the EPD the nearest characteristic point was determined, where the distance utilized was the Euclidean distance between measurement vectors. The boundaries in the resulting segmented EPD were used to find transition temperatures by averaging the temperature above and below a boundary. These were then averaged over the 3 runs to determine the transition temperatures given below. The error in a transition temperature was calculated by adding (in quadrature) the standard deviation over 3 runs and the quantization error of 2.5°C.
RESULTS AND DISCUSSION
The major result of this work is the direct demonstration of the small amount of protein sample needed and short period of time required to generate high resolution EPDs for biophysical characterization of protein conformational stability as a function of solution pH and temperature. Each EPD (containing data from triplicate runs) required approximately only 1.8 mg of protein, about 8 hours of sample preparation time, ~72 hours of instrument time, and minutes of data analysis time. (The 1.8 mg of protein is the amount used in 3 spectroscopic runs, not including planned overages. In practice the amount of protein required may vary, depending on the cost of the analyte and skill of the experimentalist.) The instrument time could be further reduced since data was collected in triplicate and buffers were measured for every pH and run. The buffer spectra could be measured for only one pH condition using one of the six turret positions. With this experimental setup, an EPD across 5 pH values could be generated using all 3 experimental techniques with only ~12 hours of instrument time and ~0.5 mg of protein. If CD measurements were not used as part of the EPD analysis, the protein concentration and cuvette path length could be increased and instrument integration times could be decreased, allowing perhaps up to 4 EPDs to be generated per day.
If separate instruments had been used, the data collection run described in this paper would have required 3 times as much protein, because new samples would be needed for each instrument (absorbance, fluorescence, and circular dichroism spectrophotometers). Preparation time would also increase due to the need to prepare more samples and cuvettes for multiple experimental runs. Total instrument time, however, would not increase due to the use of separate instruments, since the Olis Multiscan collects data from the 3 experimental measurements separately (i.e., the Far UV CD measurements use wavelengths in the 200 to 240 nm range, the fluorescence measurements use excitation wavelengths of 295 and 300 nm, and the absorbance measurements are not simultaneous with CD or fluorescence). In fact, total instrument time would probably be reduced somewhat by using separate instruments, because path lengths and concentrations could be optimized separately for each instrument.
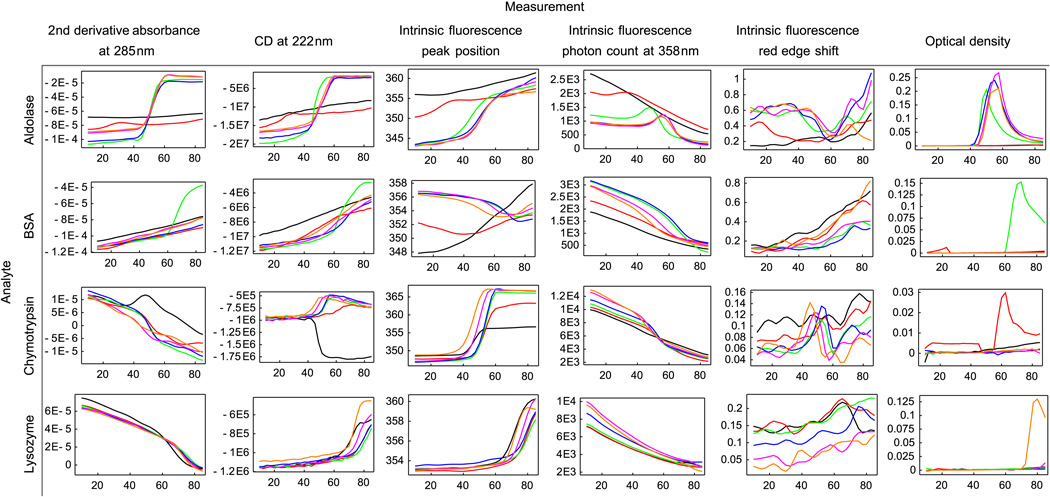
Representative examples of measurements as a function of pH and temperature from each of the biophysical measurements for the four model proteins are shown in Figure 2. These results are averaged over 3 runs as a function of temperature and solution pH. For error bars associated with these measurements, see Supplementary Figures S1-S4. Figure 3 shows EPDs summarizing the biophysical data presented in Figure 2. Note that the EPD's were generated using data collected from all wavelengths, not just the representative wavelengths shown in Figure 2. As explained elsewhere, in an EPD the colors themselves have no absolute meaning.2 Instead, differences in color alone assist in the determination of protein behavior and behavioral transitions.
Figure 2.
Representative biophysical measurements of 4 model proteins collected with the Olis Multiscan. Data include second derivative near UV absorbance, circular dichroism, intrinsic Trp fluroscence and optical density measurements for the model proteins aldolase, BSA, chymotrypsin, and lysozyme. For error bars, see the supplemental figures. The units for the Y-axis of each column are, respectively: absorbance unit/nm2 , deg·cm2/dmol, nm, number of photons, emission peak shift per excitation wavelength change (nm/nm), and optical density units. The line colors are black-pH 3, red-pH 4, green-pH 5, blue-pH 6, purple-pH 7, and orange-pH 8. Each figure has temperature (°C) as the horizontal axis.
The EPD for aldolase displays 4 structural states as a function of pH and temperature stress (Figure 3a). At pH 3 and pH 4, the protein was unfolded, as shown by a lack of transitions (color changes) in the EPD and by inspection of the biophysical data itself. At pH 5 to 8, the protein manifested structural changes near 50°C. This apparent phase change is characterized by the melting of secondary and tertiary structure as shown by transitions in absorbance, CD and fluorescence.9,10,12–18 A few degrees after the onset of melting, the protein self-associated, as shown by increases in optical density. The protein initiated structural changes again at pH values 5 to 8 near 60°C. Although this phase may be partially unfolded, some structure remains, as indicated by the slight recovery in the red shift between 60°C and 80°C.19,20 The EPD in Figure 3a for aldolase is lacking a structural transition near 30°C at pH 3 and 4, which was found by Hu et al.21 This difference is probably due to the fact that the EPD in this work does not include near-UV CD data.
The EPD for BSA also displays multiple structural states as a function of pH and temperature stress (Figure 3b). At pH 3, BSA is partially unfolded by the acidic conditions, as indicated by the weak transitions visible in both the EPD and biophysical data. At pH 4, the same phenomenon is observed, though to a lesser extent. The protein had the highest melting temperature at pH 6, with a structural transition observed in the EPD near 65.6 ± 2.6°C. From pH 5 to 8, the first transition is characterized by melting of the secondary structure as indicated by CD, melting of the tertiary structure indicated by fluorescence changes, and protein association reflected by a slight blue shift in the fluorescence peak position and a rising red edge shift. At pH 5 the protein undergoes a transition near 68.9 ± 2.6°C in which secondary structure melts more completely, as seen in CD measurements, and the protein more strongly self-associates, as indicated by increases in optical density.
The EPD for chymotrypsin also displays multiple structural states as a function of pH and temperature stress (Figure 3c). Unlike aldolase and BSA, however, chymotrypsin exhibits the same structural state across the entire pH range of 3 to 8 at lower temperatures. At pH 3, the protein’s tertiary structure begins to change near 35°C, as shown in the UV second derivative absorbance plot. The secondary structure then begins to change to a more helical state with an onset near 42.8 ± 2.5°C, as suggested by the CD measurements and the EPD. Although chymotrypsin does not display a CD peak at the wavelength of 222 nm used for the plots in Figure 2, the melting trends are the same at all CD wavelengths monitored (data not shown). At pH 4 to 8, the protein’s secondary and tertiary structure alter simultaneously near 42.5°C, as shown by CD and fluorescence peak position measurements. During the transition, the red edge shift increases temporarily, consistent with protein association. At pH 4 near 65.6 ± 2.9°C, the protein changes to a state characterized by strong association behavior, as indicated by OD measurements. The color variations visible at lower temperatures (20 to 40°C) result from noise in the data, as they were different for each run (EPDs for each run are not shown).
The EPD for lysozyme (Figure 3d), exhibits a single apparent phase from pH 3–8 and 10–60°C. The protein is most stable at pH 4 to 6, undergoing a transition in secondary and tertiary structure near 73°C. The color variations visible at lower temperatures (20 to 60°C) result from a constant slope in the data, visible in Figure 2. At pH 8, the protein begins to strongly self-associate with an onset near 75 ± 2.7°C, as indicated by OD measurements. Analysis of the same four model proteins was conducted by measurements in separate biophysical instruments and virtually identical results were obtained.
CONCLUSION
A combined multifunctional spectrometer (the Olis Multiscan) is capable of collecting a variety of protein biophysical data with a single instrument for the construction of EPDs at a much higher throughput than has previously been possible, while maintaining reproducibility and good signal to noise levels. A newly developed software analysis package was combined with the Olis Multiscan instrument to rapidly produce EPDs in a high throughput fashion with minimal sample requirements. The utility of this new methodology was demonstrated by evaluating the conformational stability of four model proteins as a function of solution pH and temperature. The interpretation of protein structural phases observed in the EPDs is the only remaining non-automated step in the EPD method. As noted earlier, the EPD colors themselves have no absolute meaning and the focus is on evaluating color changes which represent structural changes in the protein. The generation of EPD's containing colors with real physical (i.e., protein structural) meaning is equivalent to automated interpretation, since one would enable the other. In the near future, we plan to use the systems described in this paper to assist in the development of analysis techniques for automated interpretation of protein characterization data sets.
Supplementary Material
Supplemental Figure 1S - Error bars associated with biophysical measurements of Aldolase collected with the Olis Multiscan. The units are for each column are, respectively: absorbance unit/nm2 , deg·cm2/dmol, nm, number of photons, emission peak shift per excitation wavelength change (nm/nm), and optical density units. Each figure has temperature (°C) as the horizontal axis.
Supplemental Figure 2S- Error bars associated with biophysical measurements of BSA collected with the Olis Multiscan. The units are for each column are, respectively: absorbance unit/nm2 , deg·cm2/dmol, nm, number of photons, emission peak shift per excitation wavelength change (nm/nm), and optical density units. Each figure has temperature (°C) as the horizontal axis.
Supplemental Figure 3S - Error bars associated with biophysical measurements of Chymotrypsin collected with the Olis Multiscan. The units are for each column are, respectively: absorbance unit/nm2 , deg·cm2/dmol, nm, number of photons, emission peak shift per excitation wavelength change (nm/nm), and optical density units. Each figure has temperature (°C) as the horizontal axis.
Supplemental Figure 4S - Error bars associated with biophysical measurements of Lysozyme collected with the Olis Multiscan. The units are for each column are, respectively: absorbance unit/nm2 , deg·cm2/dmol, nm, number of photons, emission peak shift per excitation wavelength change (nm/nm), and optical density units. Each figure has temperature (°C) as the horizontal axis.
ACKNOWLEDGEMENTS
This work has been funded by NIH grant #NIH48811, project 5T32AI070089, titled “Graduate training program in multidimensional vaccinogenesis”.
REFERENCES
- 1.Kueltzo LA, Ersoy B, Ralston JP, Middaugh CR. Derivative absorbance spectroscopy and protein phase diagrams as tools for comprehensive protein characterization: A bGCSF case study. J Pharm Sci. 2003;92:1805–1820. doi: 10.1002/jps.10439. [DOI] [PubMed] [Google Scholar]
- 2.Maddux NR, Joshi SB, Volkin DB, Ralston JP, Middaugh CR. Multidimensional methods for the formulation of biopharmaceuticals and vaccines. J Pharm Sci. 2011;100:4171–4197. doi: 10.1002/jps.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beernink PT, Tolan DR. Subunit interface mutants of rabbit muscle aldolase form active dimers. Protein Science. 1994;3:1383–1391. doi: 10.1002/pro.5560030904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byler DM, Susi H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Sturtevant JM. The observed change in heat capacity accompanying the thermal unfolding of proteins depends on the composition of the solution and on the method employed to change the temperature of unfolding. Biochemistry. 1996;35:3059–3062. doi: 10.1021/bi952198j. [DOI] [PubMed] [Google Scholar]
- 6.Lozano P, Diego T, Iborra JL. Dynamic Structure/Function Relationships in the α-Chymotrypsin Deactivation Process by Heat and pH. Eur J Biochem. 1997;248:80–85. doi: 10.1111/j.1432-1033.1997.00080.x. [DOI] [PubMed] [Google Scholar]
- 7.Murayama K, Tomida M. Heat-induced secondary structure and conformation change of bovine serum albumin investigated by Fourier transform infrared spectroscopy. Biochemistry. 2004;43:11526–11532. doi: 10.1021/bi0489154. [DOI] [PubMed] [Google Scholar]
- 8.Sawyer L, Fothergill-Gilmore LA, Freemont PS. The predicted secondary structures of class I fructose-bisphosphate aldolases. Biochem J. 1988;249:789. doi: 10.1042/bj2490789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly SM, Price NC. The application of circular dichroism to studies of protein folding and unfolding. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1997;1338:161–185. doi: 10.1016/s0167-4838(96)00190-2. [DOI] [PubMed] [Google Scholar]
- 10.Venyaminov SY, Yang J. Determination of protein secondary structure. In: Fasman GD, editor. Circular dichroism and the conformational analysis of biomolecules. 1 ed. New York: Plenum Press; 1996. pp. 69–107. [Google Scholar]
- 11.Jain AK. Data clustering: 50 years beyond K-means. Pattern Recogn Lett. 2010;31:651–666. [Google Scholar]
- 12.Kueltzo LA, Middaugh CR. Ultraviolet absorption spectroscopy. In: Jiskoot W, Crommelin D, editors. Methods for structural analysis of protein pharmaceuticals. 1 ed. Arlington, Virginia: 2005. pp. 1–25. [Google Scholar]
- 13.Mach H, Volkin D, Burke C, Middaugh C. Ultraviolet absorption spectroscopy. Methods Mol Biol. 1995;40:91–114. doi: 10.1385/0-89603-301-5:91. [DOI] [PubMed] [Google Scholar]
- 14.Mach H, Thomson JA, Middaugh CR, Lewis RV. Examination of phenylalanine microenvironments in proteins by second-derivative absorption spectroscopy. Arch Biochem Biophys. 1991;287:33–40. doi: 10.1016/0003-9861(91)90384-u. [DOI] [PubMed] [Google Scholar]
- 15.Nakanishi K, Berova N, Woody RW. Circular dichroism: principles and applications. 1 ed. New York: VCH; 1994. [Google Scholar]
- 16.Sreerama N, Manning MC, Powers ME, Zhang JX, Goldenberg DP, Robert W. Tyrosine, phenylalanine, and disulfide contributions to the circular dichroism of proteins: circular dichroism spectra of wild-type and mutant bovine pancreatic trypsin inhibitor. Biochemistry. 1999;38:10814–10822. doi: 10.1021/bi990516z. [DOI] [PubMed] [Google Scholar]
- 17.Jiskoot W, Visser A, Herron J, Sutter M. Fluorescence spectroscopy. In: Jiskoot W, Crommelin D, editors. Methods for structural analysis of protein pharmaceuticals. 1 ed. Arlington, Virginia: AAPS Press; 2005. pp. 27–82. [Google Scholar]
- 18.Lakowicz JR, Masters BR. Principles of fluorescence spectroscopy. Journal of Biomedical Optics. 2008;13:029901. [Google Scholar]
- 19.Demchenko A. Red-edge-excitation fluorescence spectroscopy of single-tryptophan proteins. European Biophysics Journal. 1988;16:121–129. doi: 10.1007/BF00255522. [DOI] [PubMed] [Google Scholar]
- 20.Demchenko AP. The red edge effects: 30 years of exploration. Luminescence. 2002;17:19–42. doi: 10.1002/bio.671. [DOI] [PubMed] [Google Scholar]
- 21.Hu L, Olsen CM, Maddux NR, Joshi SB, Volkin DB, Middaugh CR. Investigation of Protein Conformational Stability Employing a Multimodal Spectrometer. Anal Chem. 2011;83:9399–9405. doi: 10.1021/ac201995c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1S - Error bars associated with biophysical measurements of Aldolase collected with the Olis Multiscan. The units are for each column are, respectively: absorbance unit/nm2 , deg·cm2/dmol, nm, number of photons, emission peak shift per excitation wavelength change (nm/nm), and optical density units. Each figure has temperature (°C) as the horizontal axis.
Supplemental Figure 2S- Error bars associated with biophysical measurements of BSA collected with the Olis Multiscan. The units are for each column are, respectively: absorbance unit/nm2 , deg·cm2/dmol, nm, number of photons, emission peak shift per excitation wavelength change (nm/nm), and optical density units. Each figure has temperature (°C) as the horizontal axis.
Supplemental Figure 3S - Error bars associated with biophysical measurements of Chymotrypsin collected with the Olis Multiscan. The units are for each column are, respectively: absorbance unit/nm2 , deg·cm2/dmol, nm, number of photons, emission peak shift per excitation wavelength change (nm/nm), and optical density units. Each figure has temperature (°C) as the horizontal axis.
Supplemental Figure 4S - Error bars associated with biophysical measurements of Lysozyme collected with the Olis Multiscan. The units are for each column are, respectively: absorbance unit/nm2 , deg·cm2/dmol, nm, number of photons, emission peak shift per excitation wavelength change (nm/nm), and optical density units. Each figure has temperature (°C) as the horizontal axis.