Abstract
We present a probabilistic data fusion framework that combines multiple computational approaches for drawing relationships between drugs and targets. The approach has special relevance to identifying surprising unintended biological targets of drugs. Comparisons between molecules are made based on 2D topological structural considerations, based on 3D surface characteristics, and based on English descriptions of clinical effects. Similarity computations within each modality were transformed into probability scores. Given a new molecule along with a set of molecules sharing some biological effect, a single score based on comparison to the known set is produced, reecting either 2D similarity, 3D similarity, clinical effects similarity or their combination. The methods were validated within a curated structural pharmacology database (SPDB) and further tested by blind application to data derived from the ChEMBL database. For prediction of off-target effects, 3D-similarity performed best as a single modality, but combining all methods produced performance gains. Striking examples of structurally surprising off-target predictions are presented.
Keywords: Molecular similarity, Surex-Sim, Patient Package Inserts, Off-Target Predictions
1. Introduction
In prior work, we introduced a methodological approach for data fusion which was used to predict the protein targets of small molecules based on molecular similarity.1 Given a test molecule and a set of small molecules with a known shared biological effect, the method produces a score corresponding to the likelihood that the test molecule will share the same activity. We showed that for predicting primary targets (i.e. targets modulating intended therapeutic effects) the performance advantage of a 3D similarity method over a 2D method was relatively small, due to the dominating effects of human 2D bias in drug design (i.e. “me-too” drugs).1,2 However, for predicting secondary targets (i.e. sources of side-effects) 3D similarity was much more effective than 2D topological comparisons. We also showed that clinical effects of drugs could be used as a surrogate for biochemical characterization,1 making use of common side effects of muscarinic antagonism as markers for the biochemical protein-ligand effect. It was possible using 3D chemical similarity to achieve strong separation of likely muscarinic modulators from those with no evidence of such effects.
In the current work, we expand the analysis to a much larger set of small molecule drugs, again making use of 2D and 3D chemical similarity computations. Additionally, computations involving structural similarity are augmented with clinical effects similarity, made possible by automating the extraction and weighting of relevant textual terms from drug package inserts. The top row of Figure 1 shows two highly similar first generation sulfonylureas, tolbutamide and tolazamide, each having highly similar pharmacological effects,3 with their therapeutic benefits deriving from identical mechanisms.4 Clinical effects similarity coincides here with high structural 2D and 3D similarity. Next, consider the two structurally dissimilar anticonvulsants on the bottom of Figure 1, carbamazepine and levetiracetam. Carbamazepine was one of the first anticonvulsants (approved in 1968), and its therapeutic benefit is attributed to stabilizing the inactivated state of voltage-gated sodium channels (Nav1.1).5 Levetiracetam is a newer anticonvulsant, believed to act through interaction with synaptic vesicle glycoprotein 2A (SV2A).6 As expected, the two package inserts have clinical effect terms in common due to shared indications. Given the high 3D structural similarity, our expectation is that these drugs do in fact share some molecular targets, as will be discussed later.
Fig. 1.
Relationship between small molecules based on molecular similarity, protein target modulation, and clinical effects. The optimal 3D superimposition (bottom) indicates high similarity, despite little topological commonality (green sticks correspond to regions of significant surface shape similarity and blue/red sticks correspond to regions of significant polar similarity).
The present study establishes a computational method to draw relationships between drugs based on the clinical effects present in Patient Package Inserts (PPI), whose utility for predicting drug target interactions has been shown previously.7 The present study makes three primary contributions. First, we introduce a method to extract and weight medically relevant terms from English clinical effects information. Second, we show that drug similarity computed from package inserts is directly correlated with drug similarity computed by molecular structure comparison. Third, we established that the combination of 2D, 3D, and PPI similarity yielded better off-target predictive performance over any single similarity computation. Recovery of roughly 40–50% of off-target annotations was possible with false positive rates of about 1–3%. The approach is generalizable to other computational modalities (e.g. docking of ligands to protein structures), and it is our hope that broad application of the methods will aid in identifying unexpected interactions between drugs and biological targets.
2. Methods and Data
The following describes the molecular data sets, computational methods, and specific computational procedures (see http://www.jainlab.org for additional details on software, data, and protocols).
2.1. Molecular Data Sets
In the present study two molecular data sets are used. The Structural Pharmacology Database (SPDB) is a deeply curated drug target database that is used as the basis to make predictions. A set of drug target annotations from ChEMBL that were not annotated in our database were used as a blind test set.
The details of the SPDB and its relationship to other databases has been extensively described elsewhere.1,2,8 It has two features that are particularly important for the present study. First, “targets” are specific binding sites on proteins or protein complexes. This is a critical distinction in order to make inferences about small molecule activity based on structural similarity. Second, primary targets (those that are believed to be therapeutically beneficial) are distinguished from secondary targets (which mediate pharmacologically relevant off-target effects). By making this distinction, it is possible to explicitly quantify performance of methods for prediction of surprising effects. Of the roughly 1000 drugs within the SPDB, 602 met our criteria for inclusion based on PPI information (see below). Of the 257 primary and secondary targets of these 602 drugs, 91 had at least 5 annotated drugs and formed the basis of cross-validation experiments. These 91 targets were comprised of 83 human proteins, including 28 aminergic GPCRs, 19 ligand and voltage gated ion channels, 13 human enzymes, 7 nucleotide and short peptide GPCRs, 5 tyrosine kinases, 5 steroid receptors, 3 reuptake transporters, 2 ion transporters, and 1 transcription factor. The remaining 8 targets were bacterial, fungal, and viral proteins. To test the methodology, we employed ChEMBL version 14, which curates linkages between chemicals and biological targets.9 For each of the 602 drugs, corresponding ChEMBL compounds were identified based on direct structural equivalence. Equating the 91 SPDB target binding sites to ChEMBL bioactivities was done manually, yielding 65 corresponding ChEMBL targets. Significant bioactivity was defined as Kd, Ki, or IC50 values less than or equal to 1µM. There were 380 drug-target interactions present in ChEMBL that were missing from the SPDB matrix of 602 drugs and 91 targets. This set served as a blind test set and will be referred to as the ChEMBL set in what follows.
2.2. Patient Package Insert Similarity
We employed the well established vector space information retrieval approach10,11 to model patient package inserts (PPIs). Text documents are modeled as vectors in high dimensional space where each dimension corresponds to a term with an associated weight. Co-incidence of terms with high weight leads to high computed similarity between documents. The process to transform PPIs into weighted term vectors requires four steps. First, relevant sections are extracted, including: Indication, Contraindications, Precautions, Adverse Reactions, Drug Interactions, and Clinical Pharmacology. Second, term lists (up to five words each) are generated, with punctuation and short words like prepositions and articles removed. Third, to eliminate artifactual terms and enhance relevance, terms are identified that are part of two controlled vocabularies: Medical Subject Headings (MeSH, http://www.ncbi.nlm.nih.gov/mesh) and the low-level Medical Dictionary for Regulatory Activities (MedDRA, http://www.meddra.org). Last, term weights are assigned based on information richness (e.g. “generalized seizures” > “seizures”). Word frequencies from the Google Web 1T 5-gram Corpus (http://www.ldc.upenn.edu/Catalog/index.jsp, catalog number LDC2006T13) were used to compute term weights, with rare terms producing higher scores than common ones. For example, “seizures” produced a log odds weighting of 4.74, but the more specific term “generalized seizures” yielded 6.89. The final output for each drug is a vector composed of 6,591 term weights (the weight of the term if present and zero otherwise). From the PPI for carbamazepine, the Indication Section includes: “patients with the following seizure types: partial seizures with complex symptomatology (psychomotor, temporal lobe).” The unfiltered bigrams include both sensible ones such as “partial seizures” and useless ones such as “patients with” with the filtering process eliminating the latter. For carbamazepine, the two most heavily weighted terms were “failure liver” (8.83) and “syncope and collapse” (8.62). The term “partial seizures” scored 6.37, with many related terms (e.g. “grand mal”) scoring similarly.
| (1) |
Comparison of a pair of drug PPI vectors is quantified using the cosine similarity metric (Eq. 1). The metric has a range of 0–1, but its units are both arbitrary and counterintuitive. To employ such values in our data fusion framework, the raw similarity scores were normalized to p-values by generating a distribution of PPI similarity scores for unrelated molecule pairs. The unrelated pairs were identified based on having low 2D and low 3D similarity, quantified as described below with pairwise p-value comparisons ≥ 0.5 (we have previously shown that structurally unrelated drug pairs very infrequently share targets1). So, given a PPI similarity score S between a drug pair, the p-value is simply the proportion of occurrences of S or greater in the background set. For example, the raw PPI similarity between carbamazepine and levetiracetam was 0.286 (see Figure 1), and this corresponded to a p-value of 0.044. The most heavily weighted terms in the comparison included the following: pancytopenia (6.6), cytochrome p450 (6.6), grand mal (6.5), antiepileptic drugs (6.5), and partial seizures (6.4).
2.3. Target Prediction using Patient Package Insert Similarity
We have previously reported a framework for data fusion which allows for the integration of similarity scores into a single value.1 Briey, given a molecule A and a set a molecules with a shared biological effect, Bn, the similarity between molecule A and each molecule Bi is computed. The similarity scores are normalized to p-values as detailed above by assessing score magnitude against score from a random background set. The multinomial distribution is then used to compute the likelihood, M, of observing the set of p-values and of the converse probabilities, M*. The log-odds score L is then computed by taking the log of the ratio of M and M* and inverting the sign. A detailed discussion of the computation and corresponding 2D and 3D similarity example can be found in the original publication.1 An attractive feature of our methodology is that it is able to integrate the results of different similarity computations into a single value. For example, the log-odds calculation for tolazamide interacting with PPAR-RXR yields single-modality values of 11.35 for PPI, 7.57 for 3D, and 5.49 for 2D. Combining the similarity methods gives a stronger prediction compared to using any single method alone with 3D+2D+PPI log-odds = 23.43.
2.4. Similarity and p-value Computation with Surex-Sim
The Surex-Sim 3D molecular similarity method and its use for virtual screening and off-target prediction has been extensively described in multiple publications.2,8,12,13 Briey, given two molecules in specific poses, a value from 0 to 1 is computed that reects the degree to which their molecular surfaces are congruent with respect to both shape and polarity. The function is based on the differences in distances from observer points surrounding the molecules to the closest points on their surfaces, including both the closest hydrophobic surface points and the closest polar surface points. So, two molecules that may have very different underlying chemical scaffolds may exhibit nearly identical surfaces to the observer points. These points are analogous to a protein binding pocket, which also “observes” ligands from the outside. Additional details regarding the theory and underlying algorithmic details can be found in the previously published work. In order to produce a log-odds value for a molecule against a list of molecules with a shared annotation, 3D similarity values must be computed against each annotated molecule, and these values must then be transformed into probabilities. Given the particulars of the conformational sampling density, 3D similarity optimization thoroughness, and empirical conversion of raw scores to p-values, the overall process required many hours for each comparison of one molecule to a typical set of annotated molecules.
In the current work, two improvements were made to support large-scale application of the methods. First, a new mode of pose optimization was developed in which diverse conformations of molecules are pre-generated prior to molecular comparison. Using this new mode, the optimal pose for one molecule onto a specific pose of another can be done quickly enough to process roughly 2 million drug-like molecules per day on a single computing core (compared with roughly 10,000 previously). Second, rather than using explicit computation of 1000 background similarity values for each molecule (as previously), we made use of the observation that these distributions were essentially always normally distributed. Given a molecule pair, only the particular mean and standard deviation for each need be estimated in order to derive a p-value rather than making use of the full empirical computation. Estimation of the distributional parameters was accomplished using simple linear regression models that made use of “molecular imprints” for each molecule.8 A molecular imprint is a vector of similarity values for a particular molecule against a fixed basis set of molecules (one pose each). Such vectors have precedent in predicting many molecular properties,14,15 and the conformational pre-search procedure was augmented to produce standard molecular imprints. So, given two pre-searched molecules, their mutual maximal 3D similarity can be rapidly calculated, and the p-value conversion is immediately derived from the estimated distributional parameters for each molecule. Taken together, the two improvements allow for typical 3D log-odds computations to be made in a few minutes for a given molecule against a target characterized by twenty known ligands. To test the accuracy of the faster method, we recomputed the p-values and log-odds values from our previous work. An all-by-all similarity of the 358 drugs from the original study yielded a Pearson’s correlation of 0.947 and Kendall’s tau of 0.814, both highly statistically significant. The full log-odds computation of 358 drugs against 44 targets yielded a Pearson’s correlation of 0.955 and Kendall’s Tau of 0.761 (again highly statistically significant).
For 2D molecular similarity computations, which make purely topological comparisons between molecules, we employed the previously described GSIM-2D method.1,2 This method is sufficiently efficient that empirical conversion of raw scores into p-values is possible, as we have previously described.1 For this method to yield high similarity, two molecules must be roughly the same size and contain similar subgraph compositions, especially for those subgraphs rooted at heteroatoms.
3. Results and Discussion
3.1. Relationship between Structural Novelty and Clinical Effects
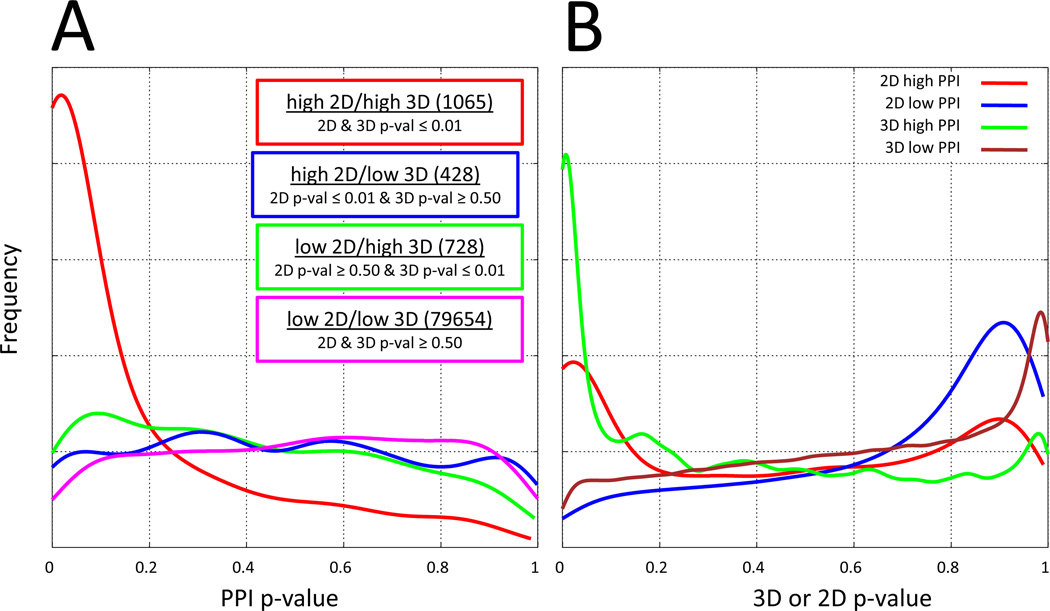
Previously, we quantified the effect of me-too drugs by showing that drug pairs with high 2D and high 3D similarity had four times more likelihood of having identical primary and secondary targets than drugs pairs where one was structurally novel.1 Here, this analysis has been extended to clinical effects by making use of the lexical similarity of package inserts. Both to establish the relevance of the PPI similarity metric and to quantify the degree to which structural novelty is related to changes in clinical effects, we computed the pairwise 2D, 3D, and PPI similarity of all 602 drugs. The drug pairs were separated into four categories based on chemical structural similarity: high 2D and 3D similarity, low 2D but high 3D, high 2D but low 3D, and low 2D and 3D. High similarity included pairs with p-values ≤ 0.01 and low similarity were those with p-values ≥ 0.5.
Figure 2A shows the histogram of the PPI p-value distributions for each of the four structural categories. It is clear that the “me-too” drug distribution (red line, drug pairs with high 2D and high 3D similarity) is different than the others. Toward the left side of the plot, where clinical effects similarity was high (PPI p-values ≤ 0.05), a large fraction of the me-too drug pairs had highly similar clinical effects. Structurally novel drug pairs (high 3D but low 2D similarity, green line) exhibited a significantly smaller fraction with highly concordant clinical effects but still showed some relationship between structural similarity and therapeutic profile. The high 2D and low 3D pairs had little signal (blue), and only a very small portion of structurally dissimilar drug pairs (low 2D and low 3D, magenta) shared clinically similar effects. Clearly, drug pairs with very high structural similarity (both by 2D and 3D methods) were much more likely to have closely shared clinical effects than molecule pairs of any other category, even those sharing high 3D similarity but low 2D similarity. The converse observations paralleled these observations. Figure 2B shows the corresponding histograms of 3D and 2D p-value distributions where molecule pair segregation was made based on clinical effect similarity. The 2D and 3D similarity p-value distributions for drug pairs with high PPI similarity (red and green lines) showed stronger enrichment for low p-values associated with high 3D structural similarity. As expected, drug pairs that had low PPI similarity (blue and brown lines) also had low 3D and 2D structural similarity.
Fig. 2.
Relationship between structural similarity and clinical effects similarity.
3.2. Internal SPDB Validation: Off Target Effects
An attractive aspect of the log-odds framework is that it allows us to combine different types of similarity computations into a single value. For each of the 602 drugs in our dataset, we computed the 2D, 3D, PPI, and combination log-odds scores of interacting with each of the 91 targets that had at least 5 drugs as ligands in the SPDB. In each case, any self/self comparisons were omitted from the calculations, making this exercise a leave-one-out cross-validation of the log-odds predictive methodology. The three methods were used independently and in combination to predict the log-odds of known primary and secondary target interactions. As we observed in our previous study, primary target predictions were dominated by the presence of me-too drugs, limiting the differences between any methods (data not shown). However, for prediction of secondary targets, i.e. those that mediate side-effects, significant differences appeared. Table 1 summarizes the true-positive rates observed for difference log-odds computations for secondary target prediction at different score thresholds.
Table 1.
SPDB Secondary Target Performance
| Log-Odds | 3D | 2D | PPI | 3D+2D | 3D+PPI | 2D+PPI | 3D+2D+PPI |
|---|---|---|---|---|---|---|---|
| 0 | 97 | 90 | 96 | 95 | 98 | 97 | 97 |
| 10 | 43 | 7 | 14 | 55 | 61 | 33 | 64 |
| 20 | 16 | 0 | 0 | 23 | 26 | 1 | 38 |
For all single methods and combinations of methods, the information present in the annotated drugs yielded positive information, evidenced by high true-positive rates at a log-odds threshold of 0. However, substantial differences among the methods appeared as higher log-odds thresholds were considered. At a threshold of 10, the 3D similarity approach showed a much higher retrieval rate than either of the other two single-mode methods. All combinations of methods showed synergy, with the most effective retrieval occurring with a combination of all three similarity methods to produce a single log-odds score. Roughly 60% of the true secondary target annotations could be recovered using the log-odds score from 3D+2D+PPI similarity computations. Note, however, that true positive rates without the context of false positive rates can be very misleading. The issue of estimating false positive rates is not straight-forward though. In our SPDB, a missing annotation between a drug and a target does not mean that the interaction does not occur. Authentic interactions within our 602 drug/91 target set may have been published after our curation or have yet to be biochemically characterized. Nonetheless, we expect that the large majority of unannotated interactions, in fact, represent true negative data. So, as a surrogate for a measurement of false positive rates for our similarity methods, we determined the number of drug/target predictions for interactions that were unannotated. At log-odds thresholds of 5, 10, and 20, predictions for non-existent SPDB annotations for both 3D similarity alone and 3D+2D+PPI were 3%, 1%, and 0.2%. These are upper limits of false positive predictions. As will be described below, the false positive rate was actually lower since many of the new predictions were validated as true by incorporating annotations from the ChEMBL database.
3.3. Prediction of New Drug-Target Pairs within ChEMBL
As discussed above, a missing annotation within the SPDB between a drug and a target does not necessarily mean that the interaction does not occur. For example, the drugs orphenadrine and mesoridazine showed high 3D log-odds against the muscarinic receptor but the interactions had been unannotated in the SPDB. Careful inspection of the literature revealed that the drugs were known to antagonize muscarinic receptors.1 Therefore, drug target annotations that are known but missing from our SPDB can serve as a blind set to test our methodology. To supplement annotations within the SPDB with a blind set for methodological testing, we searched ChEMBL and found 380 biochemically characterized drug/target interactions not present in the SPDB. We then investigated how well the methodology could identify the new ChEMBL annotations based only upon information within in the SPDB as the basis to compute the log-odds.
Table 2 shows the proportions correctly predicted at various log-odds using different methods and combinations. In general, the trends observed for the SPDB leave-one-out experiments were borne out. Among individual methods, 3D similarity strongly outperformed 2D- or PPI-based similarity, with the latter two having similar performance. However, the combination of the three methods, overall, yielded better performance than 3D alone. At log-odds thresholds of 10 and 20, using the full combination of methods, the percentage of recovered annotations within the SPDB test set was 22% and 4%, respectively. This compared with 16% and 2% using 3D similarity alone, and 3% and 0% using either 2D or PPI similarity alone. The enrichment ratios for the combination approach, using the upper-bound false positive rates discussed above, corresponded to 22-fold and 40-fold, respectively, at log-odds thresholds of 10 and 20.
Table 2.
ChEMBL Prediction Performance
| Log-Odds | 3D | 2D | PPI | 3D+2D | 3D+PPI | 2D+PPI | 3D+2D+PPI |
|---|---|---|---|---|---|---|---|
| 5 | 43 | 14 | 13 | 42 | 41 | 19 | 41 |
| 10 | 16 | 3 | 3 | 20 | 18 | 8 | 22 |
| 20 | 2 | 0 | 0 | 3 | 1 | 0 | 4 |
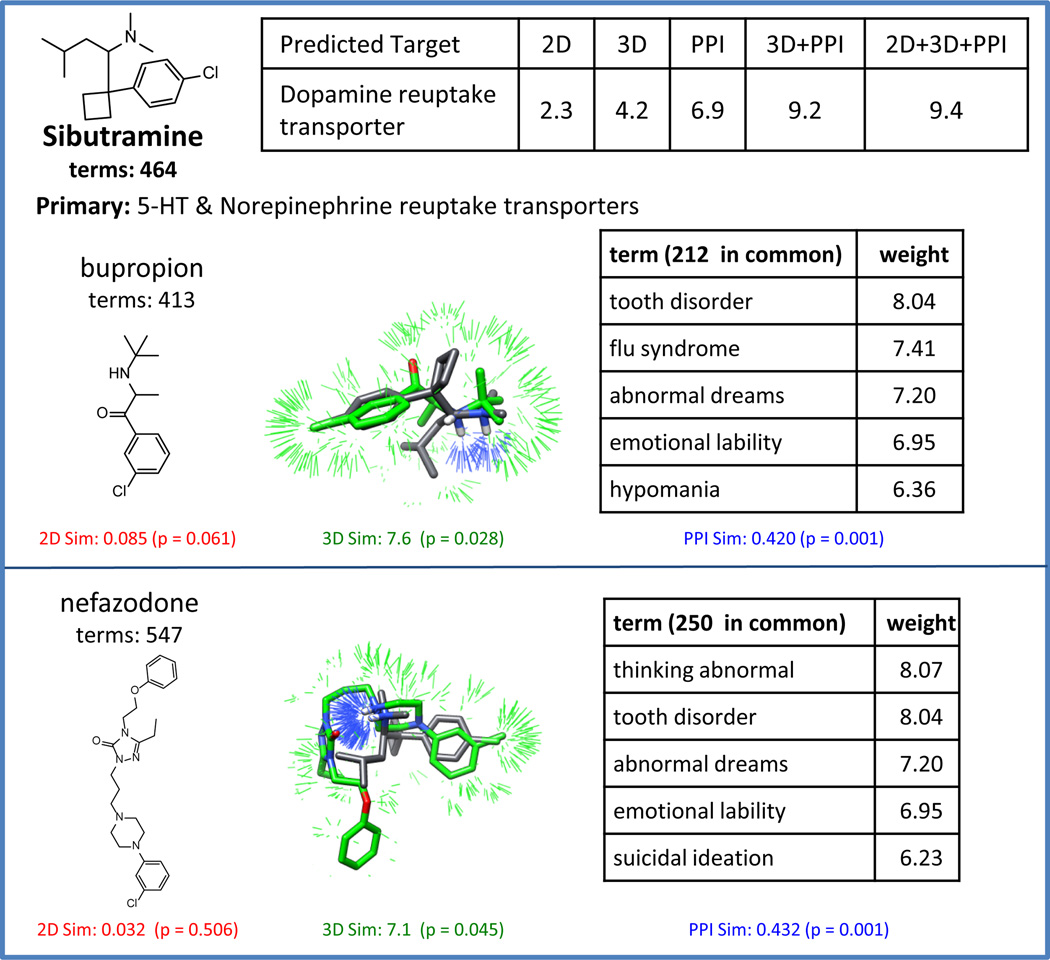
Figure 3 shows a typical example of a drug/target interaction not annotated in the SPDB where the combination similarity approach confidently identified a pharmacologically relevant target. Sibutramine is an anorexic annotated in the SPDB as a ligand of the serotonin and norepinephrine reuptake transporters. However, it has been shown that sibutramine also interacts with the dopamine reuptake transporter and that this interaction contributes to the therapeutic benefit (indicated in the Meridia package insert, http://www.rxabbott.com/pdf/meridia.pdf). Computing the similarity between sibutramine and 11 other dopamine reuptake transporter inhibitors (two are shown in Figure Figure 3), the log-odds were 2.3, 4.2, and 6.9 using 2D, 3D, and PPI, respectively. These predictions were strengthened by combining all three methods, with corresponding log-odds of 9.4. The pairwise PPI similarities between sibutramine and bupropion and nefazodone are highly significant as are the individual 3D similarities. Clinical effects can be sufficient to infer off-targets, but combining similarity methods generally adds confidence to predictions.
Fig. 3.
ChEMBL example showing that combination similarity effectively predicts a drug target interaction not covered within the SPDB. Shown are the 2D structures, 3D overlays, and common clinical terms between sibutramine and two dopamine reuptake transporter inhibitors, bupropion and nefazodone.
Note, however, that the numerical performance on the ChEMBL set was lower than for the SPDB set in terms of pure true positive recovery rates (see Tables 1 and 2). This stemmed from an increase in structural diversity for molecules within ChEMBL compared to those molecules within the SPDB for the target identified by the ChEMBL annotation. To quantify structural novelty, we performed a nearest-neighbor analysis. For each drug within ChEMBL, the most similar 2D representative from the SPDB was identified (based on p-value) from within the collection of drugs having the same target annotation. An analogous leave-one-out computation was performed for each drug target annotation within the SPDB. Figure 4 shows a histogram of the distributions of p-values for the ChEMBL (red line) and SPDB (green line) sets. Within the SPDB set, there were substantially more cases with extremely low p-values than for the ChEMBL set. The nearest structural neighbor for each ChEMBL test molecule were generally more divergent. Two examples are highlighted from the ChEMBL set where the nearest neighbor had poor 2D p-values relative to the much more significant 3D p-values which provided support for high log-odds scores.
Fig. 4.
A near-neighbor analysis for each of 602 drugs (SPDB in green, ChEMBL in red) based on target annotations from the SPDB.
Fluoxetine (blue box) is a selective serotonin reuptake inhibitor which mediates its therapeutic benefit through inhibition of the 5-HT reuptake transporter. The ChEMBL data indicated that uoxetine also interacts with the muscarinic M3 receptor. The nearest-neighbor molecule sharing this annotation was methadone (2D p-value = 0.041). Considering all of the muscarinic M3 receptor ligands (38 total), the 2D, 3D, and PPI log-odds were 1.2, 7.7, and 3.7 respectively. Combining all of the methods gave a score of 8.2.
Apomorphine (red box) is indicated to treat Parkinson’s disease and its therapeutic benefit is thought to be primarily due to activating dopamine D2 receptors. However, apomorphine was indicated within ChEMBL to also interact with the dopamine D3 receptor (which is also known to play a role in the beneficial effects for other anti-Parkinsonian drugs). The nearest-neighbor drug within the D34 ligands was ropinirole (2D p-value = 0.210), which is structurally distinct in a topological sense in Figure 4. As in the previous case, when considering all 11 dopamine D3 ligands, the 3D comparisons provide primary support for a positive log-odds score. The 2D, 3D, and PPI log-odds were −1.1, 7.7, and 1.2 respectively. The combination of all three comparison types yielded a score of 3.3. Here, the 3D molecular similarity information was the most reliable predictor.
4. Conclusion
In the present study, we report a means to combine chemical similarity between molecules with information derived from computing similarity based upon lexical analysis of patient package inserts (PPI). As expected based on our prior work, drugs that were highly structurally similar (both by 2D and 3D comparison) were much more likely to have significant overlap of their clinical effects compared to drugs that were structurally different (low 2D similarity but high 3D similarity). Our prior work illustrated a similar effect with respect to specifically annotated molecular targets: me-too drugs tend to have nearly identical target profiles.1 The correlation between lexical and chemical similarity also served to validate the lexical comparison methodology.
We extended a probabilistic data fusion method to include observations from both molecular and clinical effects similarity and reported performance on predicting protein targets of small molecules. This was done both by leave-one-out cross-validation on our internal database of drug-target interactions (the SPDB) as well as on a blind test on new interactions present in ChEMBL. For off-target prediction within the SPDB, 3D similarity was the most effective single information source. However, combining the methods predicted a larger proportion of secondary targets than any of the individual methods, while maintaining a similar nominal false positive rate. On the test against previously unseen ChEMBL drug-target linkages, again 3D similarity was the single most effective predictor, but gains were derived from combining the different data sources. We note that the method supports the integration of any method that produces scores relating molecules to targets (e.g. docking), and that inclusion of additional information sources is likely to produce further benefits. It is also important to understand that this framework is similar in character to virtual screening methods, in that while enrichment for compounds with the predicted effects occurs, the actual potencies of the effects are not predicted. This point is discussed at length in a prior study.16
In contemplating the problem of off-target prediction for drugs, the problem of molecular design ancestry can confuse the issue of methodological validation. For example, ligands of aminergic GPCRs offer troublesome test case, owing to the established promiscuity of such drugs among numerous targets.17 Returning to Figure 1 (bottom), we see the example of levetiracetam, an anticonvulsant believed to have a unique mechanism of action when compared with most existing anticonvulsants. The established CNS targets of the major classes of anticonvulsant drugs include the GABAA receptor (for barbiturates such as pentobarbital) and neuronal voltage-gated sodium channels (for drugs such as carbamazepine and phenytoin). These drugs have been recently shown to modulate voltage-gated potassium channels as part of their anti-epileptic effects.18–21 Levetiracetam, having a novel scaffold, has been proposed to work through an entirely new mechanism of action due to high binding affinity to the synaptic vesicle protein SV2A (which is not a known therapeutic target of any drug).6,22,23 Our methods strongly predict that levetiracetam is a voltage-gated sodium channel modulator with 3D log-odds alone of 14.5 (the combination log-odds was 21.4). Levetiracetam has been shown to inhibit voltage-gated potassium currents,22 leading to the suggestion that this drug, like other anti-epileptics, acts at least in part through potassium channels. Considering that many antiepileptics modulate both sodium and potassium channels,23 our prediction supports the notion that levetiracetam shares a similar mechanism of action, perhaps in addition to the interaction with SV2A.
Identification of off-target activities of drugs is a difficult problem, particularly in cases where the drug in question has a non-obvious structural relationship with the known ligands of a given target. Our hope is that methods that make use of multiple information sources will help to identify clinically important and unexpected effects.
References
- 1.Yera E, Cleves A, Jain A. Journal of Medicinal Chemistry. 2011;54:6771. doi: 10.1021/jm200666a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cleves AE, Jain AN. Journal of Computer-Aided Molecular Design. 2008;22:147. doi: 10.1007/s10822-007-9150-y. [DOI] [PubMed] [Google Scholar]
- 3.Wright J, Willette R. Journal of Medicinal Chemistry. 1962;5:815. doi: 10.1021/jm01239a016. [DOI] [PubMed] [Google Scholar]
- 4.Nagashima K, Takahashi A, Ikeda H, Hamasaki A, Kuwamura N, Yamada Y, Seino Y. Diabetes Research and Clinical Practice. 2004;66:S75. doi: 10.1016/j.diabres.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Ragsdale DS, Avoli M. Brain Research. 1998;26:16. doi: 10.1016/s0165-0173(97)00054-4. [DOI] [PubMed] [Google Scholar]
- 6.Lynch B, Lambeng N, Nocka K, Kensel-Hammes P, Bajjalieh S, Matagne A, Fuks B. PNAS. 2004;101:9861. doi: 10.1073/pnas.0308208101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campillos M, Kuhn M, Gavin A, Jensen L, Bork P. Science. 2008;321:263. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 8.Cleves AE, Jain AN. Journal of Medicinal Chemistry. 2006;49:2921. doi: 10.1021/jm051139t. [DOI] [PubMed] [Google Scholar]
- 9.Gaulton A, Bellis L, Bento A, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B. Nucleic Acids Research. 2012;40:D1100. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salton G, McGill MJ. Introduction to Modern Information Retrieval. New York, NY, USA: McGraw-Hill, Inc.; 1986. [Google Scholar]
- 11.Han J, Kamber M, Pei J. Data Mining: Concepts and Techniques, Second Edition (Morgan Kaufmann Series in Data Management Systems. 2 edn. Morgan Kaufmann; 2006. [Google Scholar]
- 12.Jain AN. Journal of Computer-Aided Molecular Design. 2000;14:199. doi: 10.1023/a:1008100132405. [DOI] [PubMed] [Google Scholar]
- 13.Jain AN. Journal of Medicinal Chemistry. 2004;47:947. doi: 10.1021/jm030520f. [DOI] [PubMed] [Google Scholar]
- 14.Ghuloum A, Carleton R, Jain A. Journal of Medicinal Chemistry. 1999;42:1739. doi: 10.1021/jm980527a. [DOI] [PubMed] [Google Scholar]
- 15.Mount J, Ruppert J, Welch W, Jain A. Journal of Medicinal Chemistry. 1999;42:60. doi: 10.1021/jm970775r. [DOI] [PubMed] [Google Scholar]
- 16.Jain A, Cleves A. J Comput Aided Mol Des. 2012;26:57. doi: 10.1007/s10822-011-9530-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keiser MJ, Roth BL, N. Armbruster B, Ernsberger P, Irwin JJ, Shoichet BK. Nature Biotechnology. 2007;25:197. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 18.Zona C, Tancredi V, Palma E, Pirrone G, Avoli M. Canadian Journal of Physiology and Pharmacology. 1990;68:545. doi: 10.1139/y90-079. [DOI] [PubMed] [Google Scholar]
- 19.Bloom F, Kupfer D, Bunney B. Psychopharmacology: The Fourth Generation of Progress. Raven Press; 1995. [Google Scholar]
- 20.Nobile M, Vercellino P. British Journal of Pharmacology. 1997;120:647. doi: 10.1038/sj.bjp.0700969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambrósio A, Soares-da Silva P, Carvalho C, Carvalho A. Neurochem. Res. 2002;27:121. doi: 10.1023/a:1014814924965. [DOI] [PubMed] [Google Scholar]
- 22.Madeja M, Georg Margineanu D, Gorji A, Siep E, Boerrigter P, Klitgaard H, Speckmann E. Neuropharmacology. 2003;45:661. doi: 10.1016/s0028-3908(03)00248-x. [DOI] [PubMed] [Google Scholar]
- 23.Surges R, Volynski K, Walker M. Therapeutic Advances in Neurological Disorders. 2008;1:13. doi: 10.1177/1756285608094212. [DOI] [PMC free article] [PubMed] [Google Scholar]