SUMMARY
Topoisomerase I (TOP1) inhibitors are an important class of anticancer drugs. The cytotoxicity of TOP1 inhibitors can be modulated by replication fork reversal, in a process that requires PARP activity. Whether regressed forks can efficiently restart and the factors required to restart fork progression after fork reversal are still unknown. Here we combined biochemical and electron microscopy approaches with single-molecule DNA fiber analysis, to identify a key role for human RECQ1 helicase in replication fork restart after TOP1 inhibition, not shared by other human RecQ proteins. We show that the poly(ADPribosyl)ation activity of PARP1 stabilizes forks in their regressed state by limiting their restart by RECQ1. These studies provide new mechanistic insights into the roles of RECQ1 and PARP in DNA replication and offer molecular perspectives to potentiate chemotherapeutic regimens based on TOP1 inhibition.
INTRODUCTION
Topoisomerase I inhibitors are an important class of anticancer drugs that exert their function by perturbing DNA replication 1,2. The mechanism of tumor response to TOP1 inhibitors and the combination of TOP1 inhibitors with other drugs for more effective tumor treatment are areas of active investigation 3,4. One widely accepted mechanism for the cytotoxicity of TOP1 inhibitors has been their ability to create single-strand breaks (SSBs), which are converted to toxic DNA double-strand breaks (DSBs) during replication when the replication fork collides with a SSB 5. This notion has been recently challenged by the discovery that TOP1 inhibitors also impair TOP1 relaxation activity, inducing accumulation of positive supercoils ahead of the replication fork that may hamper fork progression and the conversion of SSBs to DSBs 1,6. Recent studies extended this observation by showing that replication forks rapidly slow down and undergo fork reversal upon treatment with clinically relevant doses of camptothecin (CPT), the prototype TOP1 inhibitor 7,8. This prevents DSB formation, and requires the activity of poly(ADP-ribose) polymerase 1 (PARP1), a well-known chromatin-associated enzyme that modifies various nuclear proteins by poly(ADP-ribosyl)ation, to accumulate regressed forks 7. However, the exact role of PARP1 in promoting fork reversal remained unexplained. In addition, other factors are likely to be involved in this process and the protein(s) required to restore and restart reversed replication forks, once the lesion is repaired, have not been identified.
RecQ helicases have been long proposed to assist replication forks in dealing with replication stress and have attracted considerable interest in recent years due to their connection to heritable human diseases associated with cancer predisposition 9,10. RecQ helicase enzymatic activities (helicase, branch migration, strand annealing) may play multiple roles during replication by virtue of their ability to interconvert numerous replication and recombination intermediates 11–13. Moreover, previous studies pointed to a potential role of RecQ helicases in fork reversal and restart by showing that two of the five human RecQ helicase family members, BLM and WRN, promote both regression and re-establishment of model replication forks in vitro 14–16. However, distinct roles or molecular functions for the five human RecQ helicases in replication stress and cancer remain to be defined 10,17.
In this work, we combined biochemical, single-molecule DNA fiber and electron microscopy approaches to investigate the function of the human RECQ1 helicase (also known as RECQL or RECQL1) during replication stress response. RECQ1 is the first of the human RecQ proteins to be discovered on the basis of its potent ATPase activity and the most abundant of the five human RecQ helicases in cells 18,19. However, little is known about its cellular functions to date. Here, we show that RECQ1 plays an essential role—not shared by other human RecQ helicases—in restoring active replication forks that have regressed as a result of TOP1 inhibition. Moreover, we provide a rationale for the requirement of the poly(ADPribosyl)ation activity of PARP in replication fork reversal. Our findings give new insight into a pivotal mechanism responsible for replication stress response and replication fork restart after chemotherapeutic drug damage. These findings have important clinical implications, as RECQ1 inactivation might affect the efficacy of combinatorial therapies that employ PARP inhibitors and DNA damaging agents, which are already in promising clinical trials.
RESULTS
RECQ1 interacts with PARP1 and poly(ADP-ribose)
In order to better define the role of human RECQ1 helicase in DNA replication and repair we first identified proteins associated with RECQ1, using a new integrated proteomic approach 20. We generated a stable, inducible HEK293 cell line expressing a doubly tagged-version of RECQ1, then isolated protein complexes containing RECQ1 by affinity purification (Supplementary Fig. 1a–d); the resulting complexes were characterized by mass spectrometry20. Among the most abundant co-purified proteins were PARP1; Ku70 and Ku80, key components of the DNA non-homologous end-joining (NHEJ) pathway; and several nucleosomal components (Supplementary Fig. 1e). Given recent reports indicating a role for PARP1 in replication stress response 7,21, we decided to focus our work on defining the role of RECQ1 interactions with PARP1.
We confirmed the RECQ1–PARP1 interaction by co-immunoprecipitation (Co-IP), using nuclear extracts from U-2 OS cells and an anti-RECQ1 antibody that recognizes the C-terminus of RECQ1 (aa 633–648). We obtained similar results using an anti-RECQ1 antibody that recognizes the N-terminus of the protein (data not shown). We also performed reciprocal co-IPs using an anti-PARP1 antibody (Fig. 1a and Supplementary Fig. 1f). All co-IPs were performed in the presence of EtBr or Benzonase, to ensure that DNA did not mediate the interactions. We obtained similar results with other cell lines (Supplementary Fig. 1f and data not shown), indicating that the association between RECQ1 and PARP1 is not cell type-specific. These observations are in agreement with a previous report showing that RECQ1 and PARP1 interact at viral replication origins, and with a recent study reporting an interaction between RECQ1 and PARP1 in human cells 22,23.
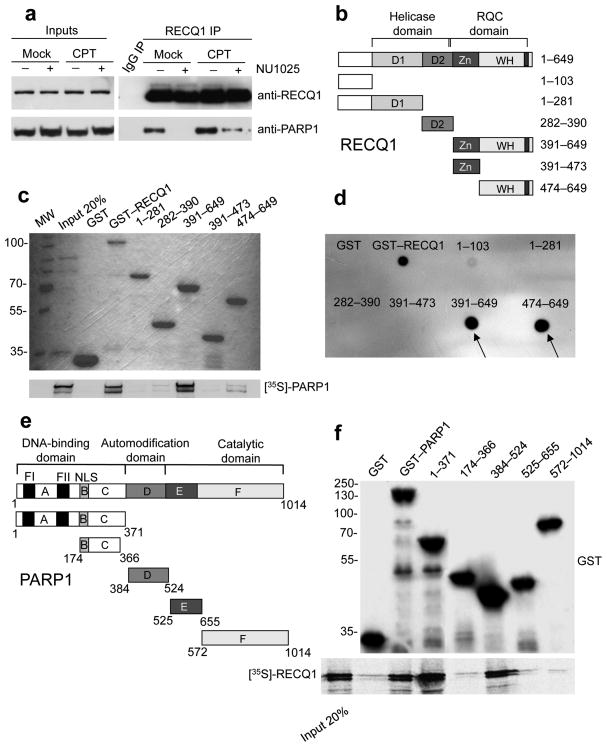
Figure 1. Analysis of the RECQ1-PARP1 interaction.
(a) IPs from U-2 OS cells using the anti-RECQ1 antibody ± PARP inhibitor (50 μM NU1025) and ± DNA damage (100 nM CPT for 2 hrs). (b) Schematic representation of the domain structure of RECQ1 and the GST-tagged RECQ1 fragments (D1 and D2 are the RecA-like domains). (c) Pull-down assays with GST-tagged RECQ1 fragments. Top: Coomassie stained gel of GST-RECQ1 fragments. Bottom: autoradiography of in vitro GST pull-down assay using 35S-labeled PARP1 protein. (d) Analysis of PAR binding in vitro. RECQ1 fragments (2 pmol) were dot-blotted onto a nitrocellulose membrane and incubated with 32P-labeled PAR. (e) Schematic representation of the domain structure of PARP1 and the GST-tagged PARP1 fragments (A, DNA binding domain; B, nuclear localization signal; D, BRCT–automodification domain; E, contains a WGR motif; F, catalytic domain. A third zinc-finger motif has been recently identified in domain C 36,37 in addition to the previously identified FI and FII zinc-finger motifs. NLS is a nuclear localization sequence. (f) Pull-down assays with GST-tagged PARP1 fragments. Bound proteins were revealed by autoradiography (bottom panel). Purified GST or GST–PARP1 proteins were detected with an anti-GST antibody (top panel). Input: 20% of the amount used in binding reactions.
The RECQ1–PARP1 interaction is regulated by PARP1 poly(ADP-ribosyl)ation activity: association of the two proteins was increased upon DNA damage, and sharply reduced upon inhibition of PARP1 activity with the PARP inhibitor NU1025 (Fig. 1a and Supplementary Fig. 1g). Using recombinant, purified PARP1 and RECQ1, we found that these two proteins interact directly, and the interaction was considerably stronger when using a poly(ADP-ribosyl)ated form of PARP1, indicating that the poly(ADP-ribose) (PAR) modification of PARP1 is important for the interaction with RECQ1 (Supplementary Fig. 2a,b). Indeed, we observed that RECQ1 interacted with PAR and binding to PAR was resistant to extensive washing with 1M salt, although no canonical PAR binding motifs could be identified in RECQ1 (Supplementary Fig. 2c) 24. As a control, we verified that NU1025 did not affect the interaction between recombinant RECQ1 and PARP1 in vitro, indicating that the reduced RECQ1–PARP1 interaction seen by co-IPs in the presence of this inhibitor is due to the inhibition of PARP1 poly(ADP-ribosyl)ation activity, rather than to a potential effect of NU1025 on PARP conformation (Supplementary Fig. 2b).
We next mapped the domains of RECQ1 that interact with PARP1 and PAR, using a series of GST-tagged RECQ1 fragments (Fig. 1b,c,d). Both PARP1 and PAR interacted with the C-terminal region of RECQ1 (amino acids 391–649) that contains the Zn-binding (Zn) and Winged Helix (WH) domains that form the so-called RecQ-C-terminal (RQC) domain, but not with the fragment 391–473 containing the Zn-binding domain alone (Fig. 1c). The WH domain alone (fragment 474–649) also bound PARP1, although more weakly than fragment 391–649. These results suggest that the region containing residues 391–473 might be important for the stability and/or conformation of the WH domain. Our data also suggest that the region 391–649 is poly(ADPribosyl)ated by PARP1 in vitro (Supplementary Fig. 2d), but it should be noted that RECQ1 does not seem to be poly(ADP-ribosylated) in vivo 22.
In order to determine the region(s) of PARP1 involved in RECQ1 interaction, we overexpressed PARP1 truncations fused to GST in HeLa cells (Fig. 1e,f). Only fragments 1-371 and 384–524 could efficiently pulldown RECQ1 in IP experiments, whereas fragment 174–366 did not pull-down RECQ1. These results indicate that the interaction with RecQ1 involves the first 173 N-terminal residues of PARP1 (containing the DNA binding domain), and residues 384–524 (containing the BRCT domain, which is also the automodification domain). These two PARP1 domains are also involved in homodimerization and in the binding of several partners, including WRN helicase 25,26.
RECQ1 catalyzes restoration of synthetic replication forks
Based on the recent discovery that PARP1 plays an important role in reversal of replication forks after CPT treatment 7, we decided to investigate whether RECQ1 is required for the cellular response to TOP1 inhibition. First, we confirmed previous observations that RECQ1 depletion leads to increased CPT sensitivity 27. Flow cytometric analysis showed that RECQ1 depleted cells are only mildly sensitive to most replication inhibitors or DNA damaging agents apart from CPT and etoposide (ETOP) (Supplementary Fig. 3a). These two drugs inhibit DNA replication by suppressing, respectively, the relaxation activity of the type IB and type IIA topoisomerases 2. Second, we confirmed that RECQ1 binds to replication forks in vivo and that the interaction increases upon CPT treatment by labeling newly replicated DNA with chlorodeoxyuridine (CldU), and investigating RECQ1 co-immunoprecipitation with CldU in the presence and absence of CPT (Supplementary Fig. 3b)
Next, we tested whether RECQ1 could mediate replication fork regression and/or restoration on synthetic DNA substrates, and whether PARP1 could affect RECQ1 activity. To measure these RECQ1 activities in vitro, we used a set of four oligonucleotides that can anneal into two alternative substrates that mimic model replication fork and “chicken-foot” structures, as previously described 14,28 (Supplementary Fig. 3c and Supplementary Table 1). We found that RECQ1 promotes model replication fork restoration very efficiently and in a concentration-dependent fashion: 50 nM RECQ1 converted >75 % of the chicken foot structure into the model replication fork after 20 min (Fig. 2a,b). In contrast, RECQ1 failed to catalyze the opposite reaction, fork regression: <2% of chicken foot fork regression structure was detected, even at the highest RECQ1 concentration. A variant of the same substrate lacking the 6 nt ssDNA gap on the leading strand template led to identical results, thus ruling out the possibility that the presence of the ssDNA gap prevented RECQ1-mediated fork regression (Supplementary Fig. 3d and 4a). Next, we confirmed that the ATPase activity of RECQ1 is essential to promote branch migration of the chicken foot structure and restoration of the active replication fork. We observed that the poorly hydrolysable ATP analog ATPγS, or the non-hydrolysable analog AMP-PNP, strongly inhibited the reaction, and two previously characterized ATPase-deficient RECQ1 mutants, K119R and E220Q, lacked fork restoration (Supplementary Fig. 4b). Additional experiments using Holliday junction (HJ) substrates with heterology regions of 1 and 4 bases, confirmed that RECQ1 has a strong branch migration activity, and that its helicase activity may be important to bypass regions of heterology. However, we observed a 50% reduction in the formation of the branch migration product when the heterology region was increased from 1 to 4 bases (Supplementary Fig. 5a,b).
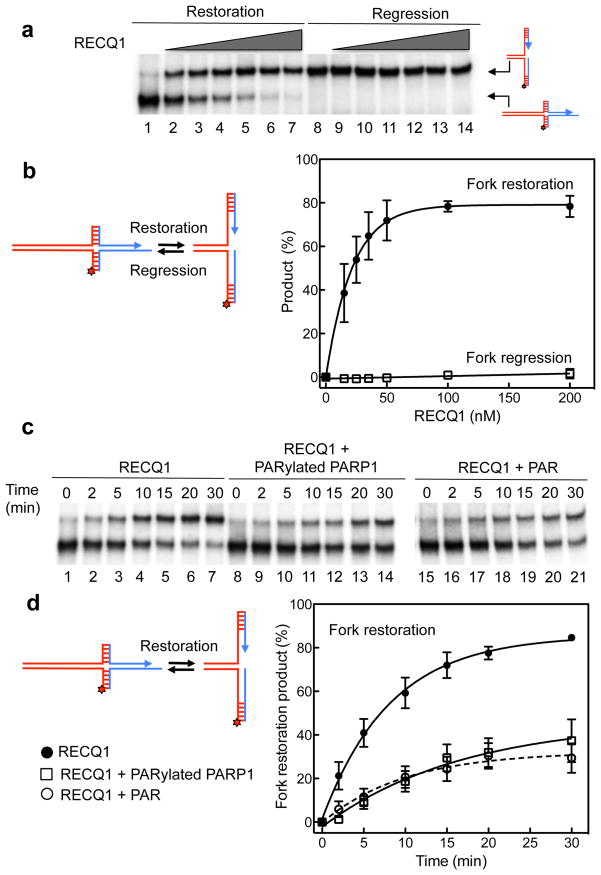
Figure 2. The in vitro fork restoration activity of RECQ1 is inhibited by PARylatedPARP1 and PAR.
(a) Lanes 1–7: fork restoration assays performed using increasing RECQ1 concentrations (0, 15, 25, 35, 50, 100, and 200 nM) and a fixed concentration of the chicken foot substrate (2 nM). Lanes 8–14: fork regression assays using increasing RECQ1 concentrations (0, 15, 25, 35, 50, 100, and 200 nM) and a fixed concentration of the replication fork structure (2 nM). The hatched regions indicate heterologous sequences that are included in the vertical arms to prevent complete strand separation. In addition, we inserted two mismatches and a single isocytosine modification to prevent spontaneous fork regression and restoration (see Supplementary Fig. 3 for more details). All the reactions were stopped after 20 min. (b) Left: reaction scheme. Right: Plot of the fork restoration and regression activities as a function of protein concentration. (c) Lanes 1–7: kinetic experiments performed using 40 nM RECQ1 and the chicken foot substrate (2 nM). Lanes 8–14: kinetic experiments performed in the presence of PARylatedPARP1 (40 nM). Lanes 15–21: kinetic experiments performed in the presence of PAR (100 nM). (d) Left: reaction scheme. Right: Plots of the fork restoration assays performed in the presence and absence of PARylatedPARP1 or PAR. The data points in a, b, c, and d represent the mean of three independent experiments. Error bars indicate standard error of the mean (s.e.m).
On the basis of our results that RECQ1 interacts with PARylatedPARP1 and previous observations that the poly(ADPribosyl)ation activity of PARP plays a key role in mediating the accumulation of regressed forks after DNA damage 7, we examined the effect of PARylatedPARP1 on the RECQ1 fork restoration activity. We found that PARylatedPARP1 strongly inhibited the fork restoration rates of RECQ1: 40 nM RECQ1 converted approximately 80 % of the chicken foot structure into a replication fork structure within 20 min. Addition of an equimolar concentration of PARylatedPARP1 reduced the fraction of restored fork structures to <30% (Fig. 2c,d). Experiments performed at increasing PARylatedPARP1 concentrations showed that a 2-fold excess of PARylatedPARP1 did not inhibit the reaction further indicating that equimolar concentrations are sufficient for maximal inhibition (Supplementary Fig. 4c). We observed a similar inhibition of RECQ1 activity in the presence of PARylatedPARP1 using the HJ (Supplementary Fig. 5c,d). To confirm that PARylatedPARP1 is also able to inhibit the DNA unwinding activity of RECQ1, we used and a fork duplex substrate with a duplex region of 20 bp (Supplementary Fig. 5e,f). In agreement with previous findings 29, EMSA experiments performed at increasing PARylatedPARP1 concentrations confirmed that PARylatedPARP1 binds DNA with low affinity indicating that the inhibitory effect of PARylatedPARP1 on RECQ1 activity is not due to a competition for DNA binding (Supplementary Fig. 6). Additional fork restoration assays performed with PAR, instead of PARylatedPARP1, supported this conclusion, confirming that the interaction of RECQ1 with PAR is responsible for the inhibition of the fork restoration activity (Fig. 2). Collectively, our biochemical data showed that RECQ1 has a strong fork restoration activity that could be responsible in restarting reversed forks associated with CPT-treatment. PARylatedPARP1 inhibits this activity of RECQ1 through a process not involving competition for DNA binding.
To test if other human RecQ helicases share the same activity we performed additional experiments with an exonuclease-deficient WRN-E84A mutant that allows to follow the branch migration reaction without possible complications arising from substrate digestion. WRN-E84A promoted both reactions with similar efficiency, with a slight bias toward fork restoration (Supplementary Fig. 7). Furthermore, the presence of PARylatedPARP1 did not inhibit the fork restoration activity of WRN-E84A, in agreement with previous studies performed using a different set of substrates 26. These results, along with previous observations made for BLM 14, showed that although other helicases are able to promote fork restoration and regression, RECQ1 has a striking preference to promote fork restoration versus regression, and its activity is uniquely regulated by PARylatedPARP1.
RECQ1 and PARP control CPT-induced replication fork slowing
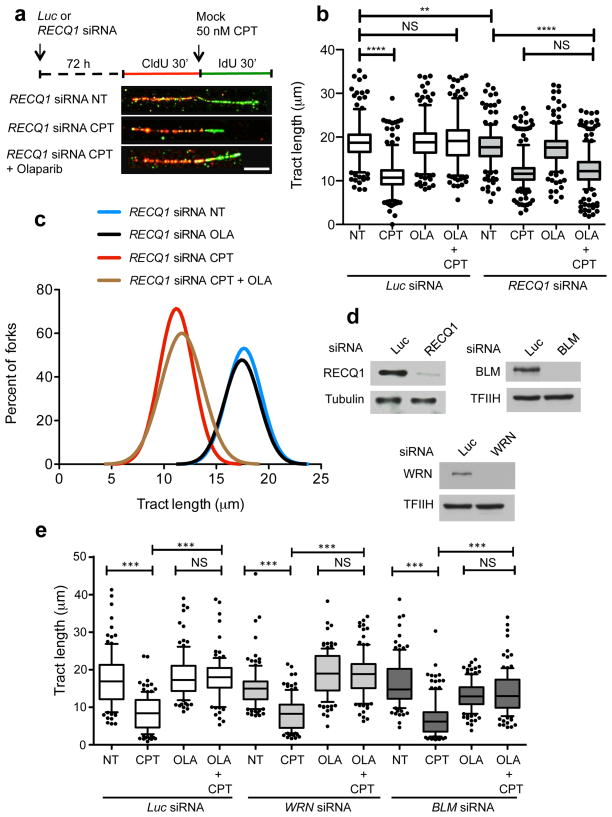
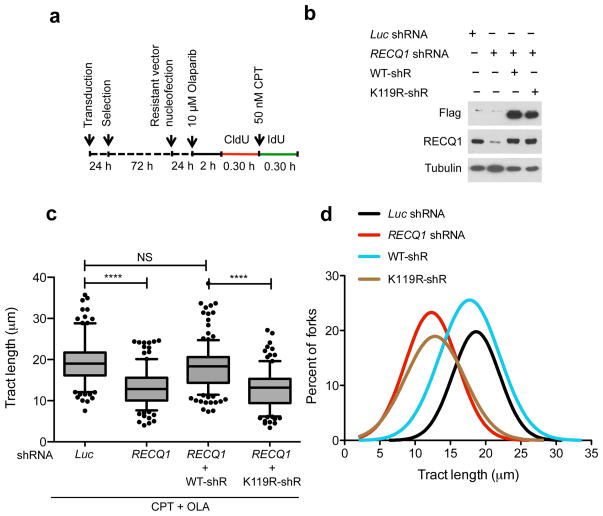
Next, we used genome-wide single-molecule DNA replication assays to test if RECQ1 depletion affects the rates of replication fork progression upon TOP1 inhibition in a cellular context. We pulse-labeled U-2 OS cells with the thymidine analog CIdU (red label) for 30 min, then treated cells with 50 nM CPT and concomitantly labeled with the second thymidine analog, ldU (green label), for an additional 30 min (Fig. 3). We then analyzed the IdU tract length distributions after CPT treatment with or without PARP inhibition. Using this approach, we initially confirmed the previous findings that replication forks rapidly slowed down upon treatment with low CPT doses (50 nM), and that this effect required the action of PARP1 7,8. This is consistent with the notion that PARP inactivation does not perturb normal fork progression, but prevents fork slowing after TOP1 inhibition. Then, we measured the rates of fork progression in RECQ1-depleted cells treated with CPT and the PARP inhibitor Olaparib. Our results showed that PARP inhibition did not rescue CPT-induced fork slowing in RECQ1-deficient cells. These results identify an essential role of RECQ1 in the control of fork progression upon TOP1 inhibition. RECQ1 downregulation using a lentiviral system and a different RNAi targeting sequence showed analogous results, supporting the notion that the observed effect was specifically associated with RECQ1 loss (data not shown). Additional DNA fiber experiments with BLM- and WRN-depleted cells showed that—in contrast to the results obtained with RECQ1-depleted cells—PARP inhibition was still able to rescue CPT-induced fork slowing in the absence of these two helicases. These results strongly support the notion that the identified role of RECQ1 in the control of fork progression upon TOP1 inhibition reflects a specific function of RECQ1 and not a more general role of the RecQ helicase family (Fig. 3e). Genetic knockdown-rescue experiments confirmed that complementation of RECQ1-depleted U-2 OS cells with shRNA resistant wild type RECQ1 abrogated the effect of RECQ1-depletion on replication fork progression upon TOP1 inhibition (Fig. 4). Moreover, complementation of RECQ1-depleted cells with the ATPase deficient RECQ1 mutant (K119R) confirmed that the ATPase activity of RECQ1 is essential for its role in replication fork progression upon TOP1 inhibition, as already inferred from the biochemical studies (Fig. 4). Interestingly, we observed a minor, but statistically significant, difference between the mean length of the replication tracts measured in the untreated RECQ1-depleted cells relative to the untreated control cells. This is in line with our previous studies where we observed that the replication tracts were slightly shorter in RECQ1-depleted cells versus controls in the absence of DNA damage 19. These data might reflect an additional role for RECQ1 in replication fork progression in unperturbed cells.
Figure 3. Restoration of normal replication fork progression after TOP1 and PARP inhibition is impaired in RECQ1-depleted—but not in WRN- or BLM-depleted—U-2 OS cells.
(a) Schematic of single DNA fiber replication track analysis. U-2 OS cells were transfected with siRNA against luciferase (Luc) or RECQ1 before CldU or IdU labeling, as indicated. Red and green denote CldU- and IdU-containing tracts, respectively. 50 nM CPT has been added concomitantly with the second label. Representative DNA fiber tracts from Microfluidic-assisted replication tract analysis of RECQ1 depleted U-2 OS cells upon TOP1 and/or PARP inhibition are shown below. White scale bar represents 12.5 μm long. (b) Statistical analysis of IdU tract length measurements from Luc- or RECQ1-depleted cells. Relative length of IdU tracts (green) synthesized after mock (NT) or CPT treatment (50nM). At least 175 tracts were scored for each dataset. 10 μM Olaparip (OLA) was optionally added 2 hours before CldU labeling and maintained during labeling. Whiskers indicate the 10th and 90th percentiles. Statistical test according to Mann-Whitney, results are ns not significant, **p 0.006, **** p<0.0001. (c) The smoothed histogram shows the green (IdU) tract length distribution after TOP1 and/or PARP inhibition in RECQ1-downregulated cells. (d) Western blot for RECQ1, BLM, and WRN downregulation. (e) Statistical analysis of IdU tract length measurements from Luc-, WRN-, or BLM-depleted cells performed as described in b.
Figure 4. Genetic complementation of RECQ1-depleted cells with wild-type RECQ1, but not with the ATP-deficient K119R mutant, rescues the fork progression phenotype observed in RECQ1-depleted U-2 OS cells.
(a) Experimental scheme for the genetic knockdown-rescue experiments. U-2 OS cells were transduced with lentivirus to luciferase (Luc shRNA) or RECQ1 (RECQ1 shRNA). RECQ1-depleted cells were genetically complemented by nucleofection of RNAi resistant vectors, for the expression of wild-type (WT-shR) and ATPase-deficient (K119R-shR) shRNA resistant forms of RECQ1 before CldU labeling, as indicated. (b) Western blot analysis of RECQ1-depleted cells complemented with the shRNA resistant wild type RECQ1 or K119R mutant (both proteins are Flag-tagged). Tubulin was detected as a loading control. (c) Statistical analysis of IdU tract length measurements from a. Relative length of IdU tracts (green) synthesized after CPT treatment (50 nM). At least 175 tracts were scored for each dataset. 10 μM Olaparip was added 2 hours before CldU labeling and maintained during labeling. Whiskers indicate the 10th and 90th percentiles. Statistical test according to Mann-Whitney, results are ns not significant, **** p<0.0001. (d) The smoothened histogram shows green (IdU) tract length after Top1 and PARP inhibition in RECQ1-downregulated cells and RECQ1-downregulated cells genetically complemented with wild-type RECQ1 or the ATPase deficient K119R mutant.
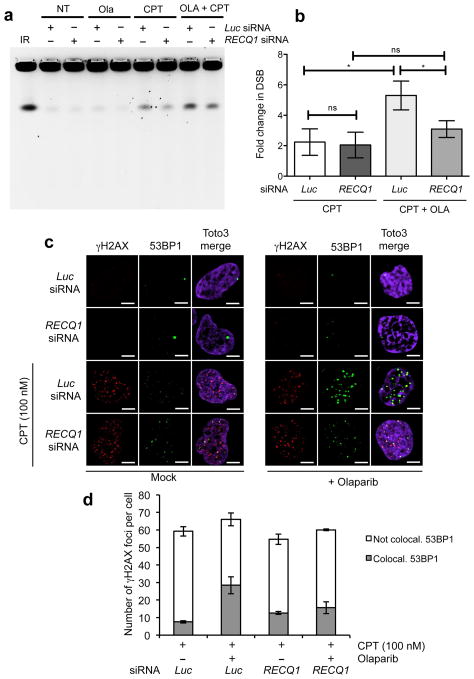
Previous work showed that CPT-induced fork slowing is uncoupled from DSB formation in human cells 7. To determine whether RECQ1 depletion also influenced DSB accumulation after CPT, we used a recently optimized Pulsed Field Gel Electrophoresis (PFGE) protocol 7,30. Our PFGE analysis confirmed that PARP inhibition in U-2 OS cells led to the induction of high levels of DSBs after CPT treatment (100nM) (Fig. 5a, b). These results are consistent with the notion that PARP-inhibited or defective cells do not slow or accumulate reversed forks after CPT treatment, leading to DSB formation even at low CPT doses 7. RECQ1 depletion, on the other hand, had the opposite effect: PARP1 inhibition did not prevent fork slowing after CPT and did not lead to increased DSB formation in RECQ1-depleted cells (Fig. 5a, b). As an alternative method to monitor DSB formation, we looked at γH2AX and 53BP1 foci colocalization under the same conditions used for the PFGE experiments. In agreement with previous findings, we found that only a minor fraction of γH2AX foci colocalized with 53BP1 upon 100 nM CPT treatment and that PARP inhibition led to a considerably higher degree of γH2AX and 53BP1 colocalization 7 (Fig. 5c, d). However, RECQ1 depletion reduced the fraction of colocalizing foci in the presence of Olaparib, supporting the notion that RECQ1-depletion prevents DSB formation following PARP inhibition. Collectively, these data indicate that RECQ1 regulates the rate of replication fork progression and that RECQ1 depletion makes PARP activity dispensable to prevent DSB accumulation after TOP1 inhibition.
Figure 5. PARP inactivation leads to DSB formation at low CPT doses in the presence, but not in the absence of RECQ1.
(a) PFGE of U-2 OS cells transfected with siRNA against Luc and RECQ1, and treated with either CPT (100 nM) and/or Olaparib (10 μM). (b) DSB signals were quantified by ImageJ and normalized to unsaturated signals of DNA retained in the wells. The values obtained from the different treatments were then normalized against their respective untreated controls to obtain the fold change in DSBs upon treatment with CPT. The graph integrates results from three independent experiments. Statistical analysis was done using paired t test (* represents significance level where p value <0.05, ns represents non-significance between the samples) (c) Immunofluorescence experiments of U-2 OS cells were treated with 100 nM CPT, as indicated, ± 10 μM Olaparib, and co-stained for γH2AX and 53BP1. A representative image is shown in each condition. (d) The plot shows the average number of γH2AX foci per cell and the average fraction (± s.e.m.) of γH2AX foci colocalizing with 53BP1.
RECQ1 is essential for fork restart upon TOP1 inhibition
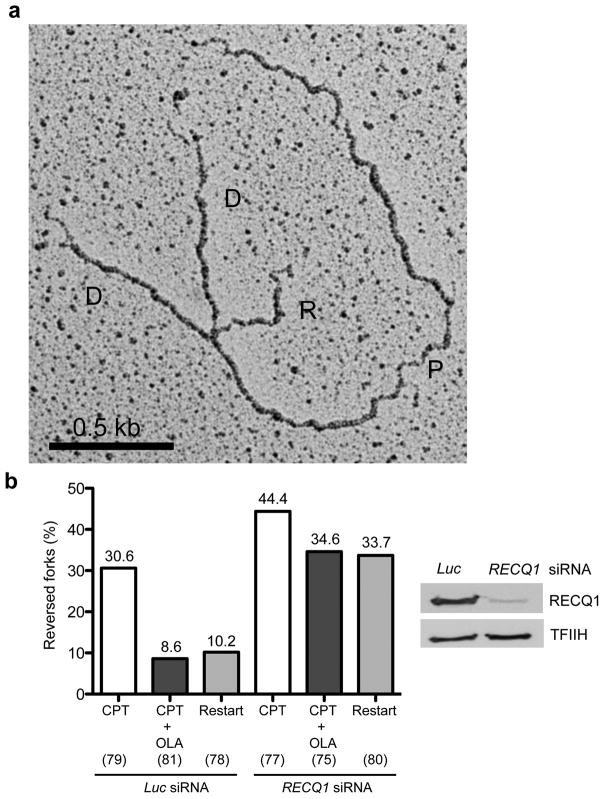
The fact that RECQ1 loss makes PARP activity dispensable to prevent fork slowing and DSB formation in CPT-treated cells suggests an accumulation of regressed forks in RECQ1-depleted cells. This is in agreement with our biochemical results pointing to a role for RECQ1 in replication fork restart. To provide more direct evidence for this idea we used electron microscopy (EM), one of the most powerful techniques to visualize the fine architecture of in vivo replication intermediates 31,32. Previous EM analysis of replication intermediates showed that replication forks undergo rapid fork reversal upon TOP1 inhibition 7. Furthermore, effective fork reversal required PARP1 activity, possibly by promoting the accumulation or stabilization of regressed replication forks and thus preventing fork collision with a CPT-induced lesion to generate a DSB. To test the hypothesis of an increased accumulation of reversed forks in RECQ1-depleted CPT-treated cells, we performed EM experiments using RECQ1-depleted CPT-treated U-2 OS cells with or without PARP inhibition (Fig. 6). Consistent with previous findings, we observed a high frequency of fork reversal (approximately 30% of molecules analyzed) in U-2 OS cells transfected with control Luc siRNA and treated with 25 nM CPT. The same experiments performed in the presence of Olaparib confirmed that PARP inhibition in control cells markedly decreased the fraction of reversed forks from 30% to less than 10%. RECQ1 depletion upon CPT treatment resulted in a higher frequency of fork reversal events (approximately 44%) than in control cells. Most importantly, PARP inactivation in RECQ1-depleted cells did not result in marked reduction in the fraction of regressed forks, suggesting that regressed forks fail to restart upon RECQ1 inactivation, even in the absence of PARP activity. To test this hypothesis directly, we performed recovery experiments and measured reversed fork frequency after CPT removal. While in control cells drug removal resulted in a marked decrease in the frequency of fork reversal (from 30 to 10%), RECQ1-depleted cells maintained a high frequency of reversed forks (approximately 33%) 3 hours after CPT withdrawal. These data strongly suggest that RECQ1 is essential to restart reversed forks and indicate that PARP requirement to observe CPT-induced reversed forks uniquely reflects its role in limiting RECQ1-mediated fork reactivation.
Figure 6. Reversed forks accumulate and are unable to restart in RECQ1-depleted cells after CPT treatment even if PARP is inhibited.
(a) Representative electron micrograph of a reversed fork observed on genomic DNA from CPT (25 nM) + Olaparib (10 μM) treated U-2 OS cells transfected with siRECQ1. The white arrow points to the four-way junction at the replication fork. D = Daughter strand, P = Parental strand, R = Reversed arm. (b) Frequency of fork reversal in U-2 OS cells transfected either with siLuc or siRECQ1 and treated with CPT ± Olaparib. Restart experiments were performed measuring the frequency of fork reversal 3 hours after CPT removal. In brackets (down), the number of analyzed molecules, and (up) the % of reversed forks.
DISCUSSION
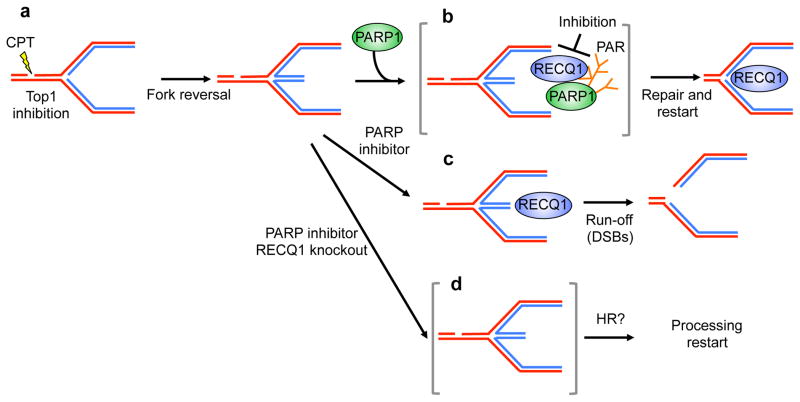
Replication fork regression is rapidly emerging as a pivotal response mechanism to replication stress induction. This notion is supported by the recent discovery that CPT damage with TOP1 inhibition induces replication fork slowing and reversal in a process that prevents double-strand break formation at clinically relevant CPT doses 7,8. A crucial cellular mediator required to accumulate or stabilize regressed forks upon TOP1 poisoning is PARP1. PARP1 itself is a target for anticancer therapies, most conspicuously in the case of BRCA-mutant breast and ovarian cancers. PARP inactivation prevents the accumulation of regressed forks without affecting the checkpoint response 7. However, the mechanism by which PARP activity promotes fork reversal is still unknown, and the requirements for the restart of reversed forks have not been defined. Our work provides novel insight into these mechanisms. First, our results show that regressed forks can restart in vivo and identify a key role for human RECQ1 in promoting efficient replication fork restart after TOP1 inhibition by virtue of its ATPase and branch migration activities (Fig. 7). Importantly, our results also show that this is a specific function of RECQ1 not shared by other helicases, such as BLM and WRN. Furthermore, our results provide new insight into the molecular role of PARP in fork reversal by showing that the poly(ADPribosyl)ation activity of PARP is important to regulate RECQ1 activity on replication forks after CPT treatment. An intriguing aspect of these data is that PARP activity is dispensable for the formation of reversed forks (Fig. 7a), but is required to “accumulate” them, i.e. to maintain or protect them from a counteracting activity (RECQ1) which would otherwise restart reversed forks untimely, leading to DSB formation (Fig. 7b, c). Indeed, we show that, differently from control cells, PARP activity is dispensable to accumulate reversed forks and to avoid CPT-induced DSB in RECQ1 depleted cells (Fig. 7d). We propose that PARP “signals” the presence of lesions on the template and inhibits locally RECQ1 thereby restraining the restart of reversed forks until TOP1 cleavage complex repair is complete (Fig. 7b). An important next step will be to identify factors that modulate RECQ1-catalyzed fork restart by PARP activity.
Figure 7. Schematic model of the combined roles of PARP1 and RECQ1 in response to TOP1 inhibition.
(a, b) PARP poly(ADPribosyl)ation activity is not required to form reversed forks, but it promotes the accumulation of regressed forks by inhibiting RECQ1 fork restoration activity, thus preventing premature restart of the regressed forks. (c) Inhibition of PARP activity leads to replication run-off and increased DSBs formation upon TOP1 inhibition since RECQ1 can restart reversed forks untimely. (d) PARP activity is no longer required in RECQ1-depleted cells were regressed forks accumulate because the cells lack the enzyme (RECQ1) necessary to promote fork restart. HR might be required to promote fork restart in the absence of RECQ1 and PARP activity.
These data provide new mechanistic insight to predict the efficiency of anticancer therapies that include both PARP and TOP1 inhibitors. These combinations are currently in clinical trials. Our results also suggest that RECQ1 itself might represent a new therapeutic target to be used in conjunction with TOP1. Inducing fork reversal (TOP1 poisons) and inhibiting reversed fork reactivation (RECQ1 depletion) should in principle synergize, explaining the observed CPT-sensitivity of RECQ1-depleted cells.
RecQ helicases are DNA unwinding enzymes essential for the maintenance of genome stability in many organisms. Why human cells should encode five RecQ homologs, while microorganisms like E. coli, S. cerevisiae, and S. pombe posses only one or two, remains unexplained. Our previous studies provided new insight by identifying important and distinct roles of RECQ1 and RECQ4 during DNA replication 19. These data, combined with previous observations that RECQ1 depletion leads to increased DNA damage and affects cellular proliferation 27,33,34, suggest that RECQ1 might play a distinct role in the stabilization and repair of replication forks. Our discovery that RECQ1 is required for replication fork restoration after TOP1 poisoning provides the first indication of a specific cellular function for this RecQ helicase. U-2OS cells lacking BLM or WRN do not show similar defects in replication fork restoration upon TOP1 poisoning, suggesting that RECQ1 is the RecQ helicase specifically responsible to promote replication fork restart upon CPT-induced fork reversal. Moreover, RECQ1 shows a striking preference for fork restoration versus regression and its activity is specifically regulated by poly(ADP-ribosyl)ation of PARP1, differently from WRN 26 (Supplementary Fig. 7). However, we cannot rule out the possibility as yet that other human RecQ helicases might be involved in different steps of the same process.
Important avenues for future studies will be to determine whether reversed forks are detected in response to genotoxic stress other than TOP1 inhibition and whether RECQ1 or other helicases, are implicated in replication fork reversal or restart depending on the type of DNA damage. In this regard, WRN- and BLM-deficient cells display increased sensitivity to selected genotoxic agents 35, while RECQ1-deficient cells are markedly sensitive to CPT and etoposide supporting the notion that these three RecQ helicases play distinct roles in replication stress response. The fact that RECQ1-depleted cells show increased sensitivity to etoposide opens the interesting possibility that a similar mechanism of fork reversal and restart might take place upon treatment with TOP2 poisons. EM analysis of replication intermediates after treatment with different classes of chemotherapeutic drugs will provide early clues to the most interesting combinations of drugs and RecQ helicases to pursue in future studies.
The discovery that RECQ1 is essential for fork restart upon TOP1 poisoning argues that RECQ1 may itself be a new therapeutic target, and could modulate the efficacy of combinatorial cancer therapies with PARP and TOP1 inhibitors that are already in clinical trials. A key experimental goal will be to determine the fate of regressed replication forks that accumulate in the absence of RECQ1. One intriguing possibility is that active replication forks are restored by the homologous recombination (HR) machinery in the absence of the fork restart activity of RECQ1 (Fig. 7d). Thus, RECQ1 depletion or inhibition might result in synthetic lethality in a HR-deficient background, thus providing a novel way to target and increase the efficacy of cancer therapies in the absence of efficient or inhibited HR repair.
ONLINE METHODS
Materials
The antibodies used were rabbit polyclonal anti-PARP1 (Enzo, ALX-210-302-R100) (1:2,000), mouse monoclonal anti-PARP1 (Santa Cruz, sc-8007) (1:1,000), mouse monoclonal anti-PAR (Enzo, ALX-804-220-R100, clone 10H) (1:2,000), rabbit polyclonal anti-PAR (Trevigen, 4336-BPC-100) (1:2,000), mouse monoclonal anti-Ku70 and -Ku86 (Santa Cruz, sc-5309 and sc-5280) (1:2,000), mouse monoclonal anti-tubulin (Sigma, T5168) (1:5,000), mouse monoclonal anti-WRN (BD laboratories, 611169) (1:1,000), rabbit polyclonal anti-BLM (Abcam, ab476) (1:1,000), rabbit polyclonal anti-TFIIH (Santa Cruz, sc293), rabbit polyclonal anti-RECQ1 raised against aa 1–110 (Santa Cruz, sc-25547) (1:2,000), and a custom made rabbit anti-RECQ1 polyclonal antibody against a synthetic peptide of a unique sequence in the last 16aa at the C-terminus of RECQ1 (Sigma) (1:2,000) 33. Camptothecin, and etoposide were from Sigma. The PARP1 inhibitors Olaparib and NU1025 were from Selleck Chemicals and Sigma, respectively.
Protein complex purification
To isolate protein complexes containing a RECQ1 bait protein, we prepared a HEK293 cell line expressing a double tagged-version of the human RECQ1 helicase by Flp recombinase-mediated integration. This system allows the generation of stable mammalian cell lines exhibiting tetracycline-inducible expression of a gene of interest from a single genomic location 20. The protein complexes containing RECQ1 were isolated and analyzed by mass spectrometry as already described 20 (Supplementary Fig. 1).
Immunoprecipitation
Human embryonic kidney 293T or human osteosarcoma U-2 OS cells were treated as indicated, washed 2 times with ice-cold PBS and resuspended in cytoplasmic extraction buffer [10 mM Tris·HCl (pH 7.9), 0.34 M sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 0.1 mM EDTA, 1 mM DTT, 20 mM NaF, 10 mM β-glycerophosphate, 0.2 mM Na3VO4, 0.5% Nonidet P-40 and protease inhibitors (Roche)] for 10 min at 4°C. Intact nuclei were pelleted by low-speed centrifugation, washed with cytoplasmic lysis buffer (without Nonidet P-40), lysed in nuclear lysis buffer [20 mM HEPES (pH 7.9), 150 mM KCl, 1.5 mM MgCl2, 20 mM NaF, 10 mM β-glycerophosphate, 0.2 mM Na3VO4 10% glycerol, 0.5% Nonidet P-40, and protease inhibitors] by homogenization and DNA and RNA in the suspension were digested with 50 U/μl Benzonase (Sigma) at 4°C for 1 hour. The nuclear soluble extract was clarified from insoluble material by centrifugation at 20,000 × g for 20 min, precleared with 50 μl protein A beads slurry (Santa Cruz) at 4°C for 1 hour and incubated overnight with anti-RECQ1 (Sigma), anti-PARP1 (Enzo), or a control IgG rabbit polyclonal antibody at 4°C. Immunocomplexes were captured by adding 50 μl of a protein A bead slurry for 2 hrs at 4°C. After extensive washing, proteins were eluted from beads with 2X Laemmli sample buffer at 95°C for 5 min, separated by SDS PAGE and detected by immunoblotting with the appropriate antibodies.
GST pull-down experiments with in vitro-translated PARP1
Pull-down assays with GST-RECQ1 fragments were performed as previously described 38. Briefly, 35S-methionine-labelled, in vitro- translated PARP1 was incubated with GST-fusion RECQ1 fragments bound to 10 μl of glutathione–Sepharose beads (Amersham) in binding buffer TNEN (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1.0 mM EDTA pH 8.0, 0.5 % NP-40, 1 mM DTT 1, mM PMSF) supplemented with 0.1 mg/ml ethidium bromide for 2 hrs at 4°C. The beads were subsequently washed two times in ethidium bromide-supplemented TNEN buffer, and three times with TNEN buffer. Bound proteins were eluted with SDS sample buffer, resolved by gel electrophoresis, and visualized by autoradiography. For the pull-downs with the GST-PARP1 fragments, 35S-Met-labelled, in vitro-translated RECQ1 was incubated with immobilized GST or GST–PARP1 domains. The GST-PARP fragments were expressed in HeLa cells as previously described 25,26. Cells were lysed 48 h later in 50 mM Tris–HCl, pH 8, 250 mM NaCl, 0.5% NP-40, 0.5 mM PMSF and protease inhibitors. Lysates were cleared by centrifugation and incubated for 2 hrs with glutathione sepharose beads. Beads were washed 3 times with lysis buffer, 2 times with lysis buffer supplemented with 1M NaCl and resuspended in GST binding buffer for pull-down experiments with the purified GST-PARP1 fragments.
PAR-binding assay
The PAR-binding assays were performed using 1M NaCl for the washing step as previously described 39.
Purification of recombinant proteins and in vitro poly(ADP-ribosyl)ation of PARP1
Recombinant RECQ1 and PARP1 were purified from insect cells, as previously described 40,41. For in vitro poly(ADP-ribosyl)ation of PARP1, recombinant PARP-1 was incubated in 20 μl of activity buffer (50 mM Tris, pH 7.5, 4 mM MgCl2, 50 mM NaCl, 200 μM DTT, 0.1 μg/μl BSA, 4 ng/μl DNaseI-activated calf thymus DNA, and 400 μM NAD+) for 10 min at 37°C.
In vitro fork regression and restart assays
The oligonucleotide sequences and the procedure used for the preparation of [γ-32P]-ATP-labeled substrate are in Supplementary Table 1 and Supplementary Fig. 3, respectively. Reactions were performed using the indicated protein concentrations and 2 nM DNA substrate in branch migration buffer (35 mM Tris-HCl, pH 7.5, 20 mM KCl. 5 mM MgCl2, 0.1 mg/ml BSA, 2 mM DTT, 15 mM phosphocreatine, 30 U/ml creatine phosphokinase, 5% glycerol) at 37°C for the indicated times. The reaction was started by the addition of 2 mM ATP. For the poly(ADPribosyl)ation experiments, the indicated concentrations of PARP1 and 200 μM NAD, or 100 nM purified PAR, were added to the reaction mixture without ATP and preincubated together with RECQ1 and the substrate at 37°C for 10 min. DNA substrates were deproteinized by adding 3X stop reaction (1.2% SDS, 30% glycerol supplemented with proteinase K (3mg/ml)) at room temperature for 10 min prior to being resolved on a native 8% polyacrylamide gel run in TBE buffer at 4°C.
Genetic knockdown-rescue assays
siRNA-mediated transient depletion of RECQ1 was achieved using an siRNA SMART pool against human RECQ1 (NM_032941, Dharmacon) in U-2 OS cells and a previously described protocol in which we established the specificity of the siRNA pool 19,33. siRNA-mediated depletion of WRN and BLM was achieved using the following siRNAs from Microsynth: WRN siRNA (5′-UAGAGGGAAACUUGGCAAAdTdT-3′) and BLM siRNA (5′-CCGAAUCUCAAUGUACAUAGA dTdT-3′). shRNA-mediated downregulation was achieved by cloning the sequence targeting RECQ1 (5′-GAGCTTATGTTACCAGTTA-3′) into the pLKO.1 lentiviral shRNA expression vector. Virus was generated by transient co-transfection of pLKO.1 and the packaging plasmids psPAX2 and pM2D.G into 293T cells. Viral supernatants were filtered through a 0.45 μM filter and transduced on U-2 OS cells for 24hrs, followed by selection with puromycin (8μg/ml) for 3 days. Control transductions were performed using the pLKO.1 vector expressing a shRNA targeting luciferase (5′-ACGCTGAGTACTTCGAAATGT-3′). The level of depletion was verified by western blot. For the complementation assays we cloned a RECQ1 RNAi-resistant open reading frame into a pIRES vector under the control of the CMV promoter. Specifically, the nucleotides targeted by the RNAi (5′-GAGCTTATGTTACCAGTTA-3′) were partially substituted without changing the amino acid sequence (5′-GTCACTATGCTATCAATTA-3′) by site-directed mutagenesis. Lentiviral depletion of endogenous RECQ1 was achieved using the protocol described above, and the resulting RECQ1-depleted cells were then nucleofected with an shRNA-resistant RECQ1 expression vector. Expression of the RNAi-resistant, Flag-tagged RECQ1 and K119R mutant was verified in control and RECQ1-depleted cells by western analysis 48 hrs after transfection.
Microfluidic–assisted DNA fiber stretching and replication fork progression analysis
Asynchronous U-2 OS cells were transiently transfected for 72 hrs with siRNA SMART pools (or specific shRNA) against RECQ1 or Luciferase as reported earlier 19,33. RECQ1- or Luciferase-depleted U-2 OS cells were labeled for 30 min each with 50μM CldU followed by 50 μM IdU. Cells were collected by trypsinization, and high molecular weight DNA from cells embedded in agarose plugs was isolated and stretched using a microfluidic platform as described earlier 42. For immunostaining, stretched DNA fibers were denatured with 2.5N HCl for 45 min, neutralized in 0.1 M Na borate pH 8.0 and PBS, and blocked with PBS/5% BSA/0.5% Tween20 for 30 min. Rat anti-CldU/BrdU (Abcam, ab6326), goat anti-rat Alexa 594 (Invitogen, A11007), mouse anti-IdU/BrdU (BD Biosciences, 347580), and goat anti-mouse Alexa 488 (Invitrogen, A11001) antibodies were used to reveal CldU and IdU labeled tracts, respectively. A Leica SP5X confocal microscope was used to visualize the labeled tracts, and tract lengths were measured using the Image J software. Statistical analysis of the tract length was performed using Graphpad Prism.
DSB detection by PFGE
DSB detection by PFGE was performed as previously described with minor modifications 7,30.
Immunofluorescence analyses
U-2 OS cells were grown on coverslips, fixed in 3.7% paraformaldehyde, permeabilized in 0.5% Triton X-100 and blocked in 3% BSA. Coverslips were then stained with rabbit polyclonal anti-53BP1 (Novus Biologicals, NB100-304) and mouse monoclonal anti-γH2AX (Millipore, 05-636), and detected by appropriate Alexa 488- and Alexa 594-conjugated secondary antibodies. Toto3 Iodide (Life Technologies, T3604) was used as a nuclear counterstain. Cells were imaged using a Zeiss LSM 510 Meta confocal microscope. Images were acquired using the LSM 5 software. Foci were counted with ImageJ “Analyze particles” function and “JACoP” plugin was used to calculate colocalization. The average number of foci was obtained from 3 independent experiments analyzing at least 35 cells per sample.
EM analysis of genomic DNA in mammalian cells
EM analysis of replication intermediates has been described in detail 31,32, including a description of the important parameters to consider specifically for the identification and the scoring of reversed forks 32.
Supplementary Material
Acknowledgments
We are grateful to A. Mazin for sharing information regarding the substrate preparation. We thank P. Janscak (Institute of Molecular Cancer Research, University of Zurich, Zurich, Switzerland) and D. Orren (University of Kentucky College of Medicine, Lexington, Kentucky, USA) for providing aliquots of their purified WRN and WRN-E84A proteins, respectively. We thank G. de Murcia (Unité 9003 du Centre National de la Recherche Scientifique, École Supérieure de Biotechnologie de Strasbourg, Illkirch, France) for providing the constructs for the production of the PARP1 fragment. We thank Yuna Ayala for critical discussions and Gianluca Triolo for his help in recombinant protein production. This work was supported by startup funding from the Doisy Department of Biochemistry and Molecular Biology and from the Saint Louis University Cancer Center to A.V., by grants from the President’s Research Fund of Saint Louis University and from the Associazione Italiana per la Ricerca sul Cancro, Italy (AIRC: AIRC10510) to A.V., by NIH grant CA77852 to R.J.M., Jr., by the Swiss National Science Foundation grants PP0033-114922 and PP00P3-135292 to M.L., and by a contribution from Fonds zur Förderung des akademischen Nachwuchses (FAN) of the Zürcher Universitätsverein (ZUNIV), Switzerland, to M.L. and A.R.C. We thank members of Nano Research Facility (NRF), a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation, USA, under Grant No. ECS-0335765 and part of the School of Engineering and Applied Science at Washington University in St.Louis for microfabrication and the use of clean room facility. We also thank the Center for Microscopy and Image Analysis of the University of Zurich for technical assistance with electron microscopy.
Footnotes
AUTHOR CONTRIBUTIONS
M.B. conducted the immunoprecipitations, GST-pulldown, Far Western, and dot-blot experiments and the in vitro fork regression and restoration studies with the synthetic DNA substrates. A.R.C. conducted the PFGE and the EM analysis. S.T. conducted the single-molecule DNA replication assays. S.G. expressed the recombinant proteins and contributed to the in vitro fork regression and restoration assays. S.K. performed the immunofluorescence experiments and contributed to the cell survival assays. M.K. conducted the single-molecule DNA replication assays with the BLM and WRN depleted cells. F.O. performed the protein complex purification experiments. T.G. contributed to the design of the proteomic experiments and performed the mass spectrometry analysis. S.G. performed the cell survival assays. R.M.M. contributed to the production of the GST-tagged fragments used in the GST-pulldown assays. B.L. expressed the RECQ1 mutants and contributed to the optimization of protocols for RECQ1 expression. V.B. expressed the recombinant PARP1 protein. M.G. contributed to the design and supervision of the proteomic experiments. R.A. supervised the proteomic experiments. J.S. contributed to the establishment of the single-molecule DNA replication assays in Vindigni’s lab. R.J.M. contributed in the establishment of the single-molecule DNA replication assays in Vindigni’s lab and assisted A.V. in finalizing the manuscript. M.L. panned, designed and supervised the PFGE and EM experiments and assisted A.V. in finalizing the manuscript. A.V. planned and supervised the project and wrote the manuscript.
References
- 1.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–7. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chemistry & biology. 2010;17:421–33. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nature reviews Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Galindo C, et al. Clinical use of topoisomerase I inhibitors in anticancer treatment. Medical and pediatric oncology. 2000;35:385–402. doi: 10.1002/1096-911x(20001001)35:4<385::aid-mpo1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 5.Pommier Y, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutation research. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 6.Koster DA, Crut A, Shuman S, Bjornsti MA, Dekker NH. Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell. 2010;142:519–30. doi: 10.1016/j.cell.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray Chaudhuri A, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nature structural & molecular biology. 2012;19:417–23. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 8.Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. The Journal of cell biology. 2008;183:1203–12. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nature reviews Cancer. 2009;9:644–54. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 11.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–33. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 12.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends in biochemical sciences. 2008;33:609–20. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vindigni A, Marino F, Gileadi O. Probing the structural basis of RecQ helicase function. Biophysical chemistry. 2010;149:67–77. doi: 10.1016/j.bpc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Bugreev DV, Rossi MJ, Mazin AV. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic acids research. 2011;39:2153–64. doi: 10.1093/nar/gkq1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machwe A, Karale R, Xu X, Liu Y, Orren DK. The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) holliday junctions to functional replication forks. Biochemistry. 2011;50:6774–88. doi: 10.1021/bi2001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machwe A, Lozada E, Wold MS, Li GM, Orren DK. Molecular cooperation between the Werner syndrome protein and replication protein A in relation to replication fork blockage. The Journal of biological chemistry. 2011;286:3497–508. doi: 10.1074/jbc.M110.105411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickson ID. RecQ helicases: caretakers of the genome. Nature reviews Cancer. 2003;3:169–78. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 18.Seki M, et al. Purification of two DNA-dependent adenosinetriphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. Journal of biochemistry. 1994;115:523–31. doi: 10.1093/oxfordjournals.jbchem.a124369. [DOI] [PubMed] [Google Scholar]
- 19.Thangavel S, et al. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Molecular and cellular biology. 2010;30:1382–96. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Molecular systems biology. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer research. 2012 doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- 22.Sharma S, Phatak P, Stortchevoi A, Jasin M, Larocque JR. RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA repair. 2012 doi: 10.1016/j.dnarep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Li H, Tang Q, Maul GG, Yuan Y. Kaposi’s sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: involvement of host cellular factors. Journal of virology. 2008;82:2867–82. doi: 10.1128/JVI.01319-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleine H, Luscher B. Learning how to read ADP-ribosylation. Cell. 2009;139:17–9. doi: 10.1016/j.cell.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Schreiber V, et al. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. The Journal of biological chemistry. 2002;277:23028–36. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 26.von Kobbe C, et al. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic acids research. 2004;32:4003–14. doi: 10.1093/nar/gkh721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma S, Brosh RM., Jr Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PloS one. 2007;2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–3. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 29.Ferro AM, Olivera BM. Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. The Journal of biological chemistry. 1982;257:7808–13. [PubMed] [Google Scholar]
- 30.Hanada K, et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nature structural & molecular biology. 2007;14:1096–104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 31.Lopes M. Electron microscopy methods for studying in vivo DNA replication intermediates. Methods in molecular biology. 2009;521:605–31. doi: 10.1007/978-1-60327-815-7_34. [DOI] [PubMed] [Google Scholar]
- 32.Neelsen KJ, Ray Chaudhuri A, Follonier C, Herrador R, Lopes M. Visualization and interpretation of eukaryotic DNA replication intermediates by electron microscopy in vivo. Methods in molecular biology. doi: 10.1007/978-1-62703-706-8_15. (in press) [DOI] [PubMed] [Google Scholar]
- 33.Mendoza-Maldonado R, et al. The human RECQ1 helicase is highly expressed in glioblastoma and plays an important role in tumor cell proliferation. Molecular cancer. 2011;10:83. doi: 10.1186/1476-4598-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma S, et al. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Molecular and cellular biology. 2007;27:1784–94. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer research. 2010;70:6548–55. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. The Journal of biological chemistry. 2008;283:4105–14. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 37.Tao Z, Gao P, Hoffman DW, Liu HW. Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry. 2008;47:5804–13. doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- 38.Lucic B, et al. A prominent beta-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic acids research. 2011;39:1703–17. doi: 10.1093/nar/gkq1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahel D, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–3. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui S, et al. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic acids research. 2004;32:2158–70. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muzzolini L, et al. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS biology. 2007;5:e20. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidorova JM, Li N, Schwartz DC, Folch A, Monnat RJ., Jr Microfluidic-assisted analysis of replicating DNA molecules. Nature protocols. 2009;4:849–61. doi: 10.1038/nprot.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.