SUMMARY
Although autism spectrum disorder (ASD) is defined by core behavioral impairments, gastrointestinal (GI) symptoms are commonly reported. Subsets of ASD individuals display dysbiosis of the gut microbiota, and some exhibit increased intestinal permeability. Here we demonstrate GI barrier defects and microbiota alterations in a mouse model displaying features of ASD, maternal immune activation (MIA). Oral treatment of MIA offspring with the human commensal Bacteroides fragilis corrects gut permeability, alters microbial composition and ameliorates ASD-related defects in communicative, stereotypic, anxiety-like and sensorimotor behaviors. MIA offspring display an altered serum metabolomic profile, and B. fragilis modulates levels of several metabolites. Treating naïve mice with a metabolite that is increased by MIA and restored by B. fragilis causes behavioral abnormalities, suggesting that gut bacterial effects on the host metabolome impact behavior. Taken together, these findings support a gut-microbiome-brain connection in ASD and identify a potential probiotic therapy for GI and behavioral symptoms of autism.
INTRODUCTION
Autism spectrum disorder (ASD) is a serious neurodevelopmental condition characterized by stereotypic behavior and deficits in language and social interaction. The reported incidence of ASD has rapidly increased to 1 in 88 births in the United States as of 2008 (CDC, 2012), representing a significant medical and social problem. However, therapies for treating core symptoms of autism are limited. Much research on ASD has focused on genetic, behavioral and neurological aspects of disease, though the contributions of environmental risk factors (Hallmayer et al., 2011), immune dysregulation (Onore et al., 2012) and additional peripheral disruptions (Kohane et al., 2012) in the pathogenesis of ASD have gained significant attention.
Among several comorbidities in ASD, gastrointestinal (GI) distress is of particular interest, given its reported prevalence and correlation with symptom severity (Buie et al., 2010; Coury et al., 2012). While some issues remain regarding the standardized diagnosis of GI symptoms in ASD, abnormalities such as altered GI motility and increased intestinal permeability have been reported by several laboratories (Boukthir et al., 2010; D’Eufemia et al., 1996; de Magistris et al., 2010). Moreover, a recent multicenter study of over 14,000 ASD individuals reveals a higher prevalence of inflammatory bowel disease (IBD) and other GI disorders in ASD patients compared to controls (Kohane et al., 2012). The causes of autism-associated GI problems remain unclear, but may be linked to gut bacteria as a number of studies report that ASD individuals exhibit altered composition of the intestinal microbiota (Adams et al., 2011; Finegold et al., 2010; Finegold et al., 2012; Gondalia et al., 2012; Kang et al., 2013; Parracho et al., 2005; Williams et al., 2011; Williams et al., 2012). Though there is as yet no consistency in the specific species of microbes that are altered in ASD versus controls, three studies employing different methodologies report significantly elevated levels of Clostridium species in ASD individuals (Finegold et al., 2002; Parracho et al., 2005; Song et al., 2004). Altogether, evidence of GI complications and microbiota alterations in broadly defined ASD populations raises the intriguing question of whether such abnormalities can contribute to the clinical manifestations of ASD.
Dysbiosis of the microbiota is implicated in the pathogenesis of several human disorders, including IBD, obesity and cardiovascular disease (Blumberg and Powrie, 2012). Commensal bacteria also affect a variety of complex behaviors, including social, emotional and anxiety-like behaviors, and contribute to brain development and function in mice (Collins et al., 2012; Cryan and Dinan, 2012) and humans (Tillisch et al., 2013). Long-range interactions between the gut microbiota and brain underlie the ability of microbe-based therapies to treat symptoms of multiple sclerosis and depression in mice (Bravo et al., 2011; Ochoa-Reparaz et al., 2010) and the reported efficacy of probiotics in treating emotional symptoms of chronic fatigue syndrome and psychological distress in humans (Messaoudi et al., 2011; Rao et al., 2009).
Based on the emerging appreciation of a gut-microbiome-brain connection, we asked whether modeling behavioral features of ASD in mice also causes GI abnormalities. Several mouse models of genetic and/or environmental risk factors are used to study ASD. We utilize the maternal immune activation (MIA) model, which is based on large epidemiological studies linking maternal infection to increased autism risk in the offspring (Atladottir et al., 2010; Gorrindo et al., 2012). This is further supported by many studies linking increased ASD risk to familial autoimmune disease (Atladottir et al., 2009; Comi et al., 1999) and elevated levels of inflammatory factors in the maternal blood, placenta and amniotic fluid (Abdallah et al., 2013; Brown et al., 2013; Croen et al., 2008). Modeling MIA in mice by injecting pregnant dams with the viral mimic poly (I:C) yields offspring that exhibit core behavioral symptoms of ASD, as well as a common autism neuropathology (Malkova et al., 2012; Shi et al., 2009). Furthermore, pregnant monkeys exposed to poly (I:C) yield offspring with cardinal symptoms of ASD(Bauman et al., 2013). Although several environmental and genetic risk factors for ASD have been investigated in animals, GI abnormalities have not been reported in preclinical models of ASD. We show herein that offspring of MIA mice, which display ASD-like behaviors, have defects in intestinal integrity and alterations in the composition of the commensal microbiota that are analogous to features reported in human ASD. To explore the potential contribution of GI complications to core ASD symptoms, we examine whether treatment with the gut bacterium Bacteroides fragilis, demonstrated to correct GI pathology in mouse models of colitis (Mazmanian et al., 2008) and to protect against neuroinflammation in mouse models of multiple sclerosis (Ochoa-Reparaz et al., 2010), impacts ASD-related GI and/or behavioral abnormalities in MIA offspring. Our findings suggest that targeting the microbiome may represent a novel approach for treating neurodevelopmental disorders such as autism.
RESULTS
Offspring of Immune-Activated Mothers Exhibit GI Symptoms of Human ASD
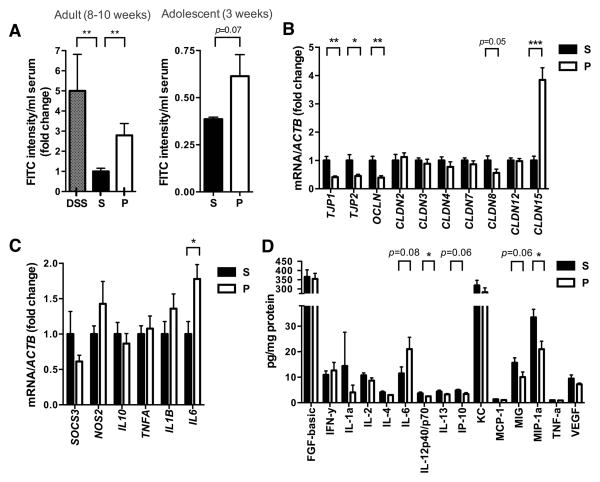
Subsets of ASD children are reported to display GI abnormalities, including increased intestinal permeability or “leaky gut” (D’Eufemia et al., 1996; de Magistris et al., 2010; Ibrahim et al., 2009). We find that adult MIA offspring, which exhibit cardinal behavioral and neuropathological symptoms of ASD (Malkova et al., 2012), also have a significant deficit in intestinal barrier integrity, as reflected by increased translocation of FITC-dextran across the intestinal epithelium, into the circulation (Figure 1A, left). This MIA-associated increase in intestinal permeability is similar to that of mice treated with dextran sodium sulfate (DSS), which induces experimental colitis (Figure 1A, left). Deficits in intestinal integrity are detectable in 3-week-old MIA offspring (Figure 1A, right), indicating that the abnormality is established during early life. Consistent with the leaky gut phenotype, colons from adult MIA offspring contain decreased gene expression of ZO-1, ZO-2, OCLN and CLDN8, and increased expression of CLDN15 (Figure 1B). Deficient expression of ZO-1 is also observed in small intestines of adult MIA offspring (Figure S1A), demonstrating a widespread defect in intestinal barrier integrity. Gut permeability is commonly associated with an altered immune response (Turner, 2009). Accordingly, colons from adult MIA offspring display increased levels of interleukin-6 (IL-6) mRNA and protein (Figures 1C and 1D) and decreased levels of the cytokines/chemokines IL-12p40/p70, IP-10, MIG and MIP-1α (Figure 1D). Small intestines from MIA offspring also exhibit altered cytokine/chemokine profiles (Figure S1C). Changes in intestinal cytokines are not accompanied by overt GI pathology, as assessed by histological examination of gross epithelial morphology from hematoxylin- and eosin-stained sections (data not shown). Overall, we find that adult offspring of immune-activated mothers exhibit increased gut permeability and abnormal intestinal cytokine profiles, features similar to those found in subsets of ASD.
Figure 1. MIA Offspring Exhibit GI Barrier Defects and Abnormal Expression of Tight Junction Components and Cytokines.
(A) Intestinal permeability assay, measuring FITC intensity in serum after oral gavage of FITC-dextran. Dextran sodium sulfate (DSS): n=6, S (saline+vehicle): adult n=16; adolescent n=4, P (poly(I:C)+vehicle): adult n=17; adolescent n=4. Data are normalized to saline controls.
(B) Colon expression of tight junction components relative to β-actin. Data for each gene are normalized to saline controls. n=8/group
(C) Colon expression of cytokines and inflammatory markers relative to β-actin. Data for each gene are normalized to saline controls. n=6–21/group
(D) Colon protein levels of cytokines and chemokines relative to total protein content. n=10/group
For each experiment, data were collected simultaneously for poly(I:C)+B. fragilis treatment group (See Figure 3). See also Figure S1.
MIA Offspring Display Dysbiosis of the Gut Microbiota
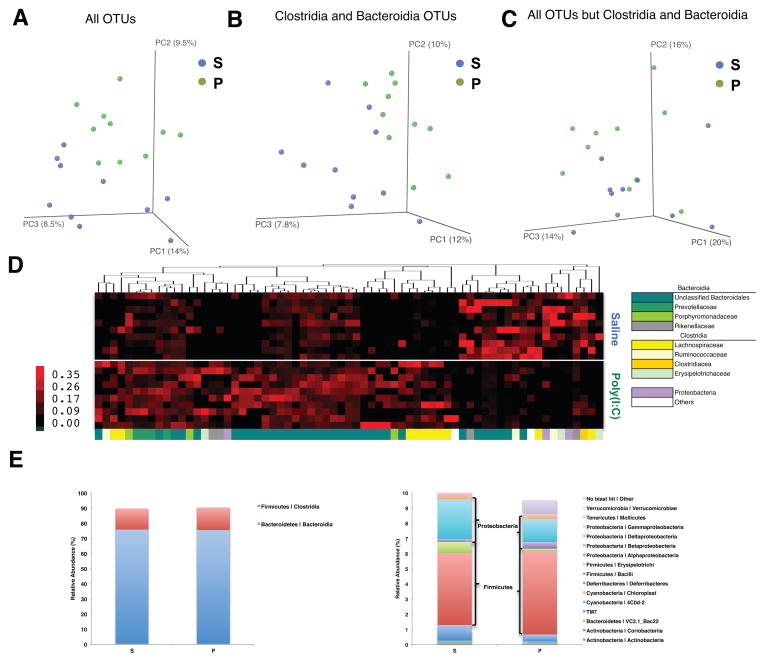
Abnormalities related to the microbiota have been identified in ASD individuals, including disrupted community composition (Adams et al., 2011; Finegold et al., 2010; Finegold et al., 2012; Gondalia et al., 2012; Parracho et al., 2005; Williams et al., 2011; Williams et al., 2012). although it is important to note that a well-defined ASD-associated microbial signature is lacking thus far. To evaluate whether MIA induces microbiota alterations, we surveyed the fecal bacterial population by 16S rRNA gene sequencing of samples isolated from adult MIA or control offspring. Alpha diversity, i.e., species richness and evenness, did not differ significantly between control and MIA offspring, as measured by several indices (Figures S2A and S2B). In contrast, unweighted UniFrac analysis, which measures the degree of phylogenetic similarity between microbial communities, reveals a strong effect of MIA on the gut microbiota of adult offspring (Figure 2). MIA samples cluster distinctly from controls by principal coordinate analysis (PCoA) and differ significantly in composition (Table S3, with ANOSIM R=0.2829, p=0.0030), indicating robust differences in the membership of gut bacteria between MIA offspring and controls (Figure 2A). The effect of MIA on altering the gut microbiota is further evident when sequences from the classes Clostridia and Bacteroidia, which account for approximately 90.1% of total reads, are exclusively examined by PCoA (R=0.2331, p=0.0070; Figure 2B), but not when Clostridia and Bacteroidia sequences are specifically excluded from PCoA of all other bacterial classes (R=0.1051, p=0.0700; Figure 2C). This indicates that changes in the diversity of Clostridia and Bacteroidia operational taxonomic units (OTUs) are the primary drivers of gut microbiota differences between MIA offspring and controls.
Figure 2. MIA Offspring Exhibit Dysbiosis of the Intestinal Microbiota.
(A) Unweighted UniFrac-based 3D PCoA plot based on all OTUs.
(B) Unweighted UniFrac-based 3D PCoA plot based on subsampling of Clostridia and Bacteroidia OTUs (2003 reads per sample).
(C) Unweighted UniFrac-based 3D PCoA plot based on subsampling of OTUs remaining after subtraction of Clostridia and Bacteroidia OTUs (47 reads per sample).
(D) Relative abundance of unique OTUs of the gut microbiota (bottom, x-axis) for individual biological replicates (right, y-axis), where red huesde note increasing relative abundance of a unique OTU.
(E) Mean relative abundance of OTUs classified at the class level for the most (left) and least (right) abundant taxa.
n=10/group. Data were simultaneously collected and analyzed for poly(I:C)+B. fragilis treatment group (See Figure 4). See also Figure S2 and Table S1.
67 of the 1,474 OTUs detected across any of the samples discriminate between treatment groups, including those assigned to the bacterial families Lachnospiraceae, Ruminococcaceae, Erysipelotrichaceae, Alcaligenaceae, Porphyromonadaceae, Prevotellaceae and Rikenellaceae, and unclassified Bacteroidales (Figure 2D and Table S1). Of these 67 discriminatory OTUs (relative abundance: 13.3±1.65% control, 15.93±0.62% MIA), 19 are more abundant in the control samples and 48 are more abundant in MIA samples. Consistent with the PCoA results (Figures 2A–C), the majority of OTUs that discriminate MIA offspring from controls are assigned to the classes Bacteroidia (45/67 OTUs or67.2%; 12.02±1.62% control, 13.48±0.75% MIA) and Clostridia (14/67 OTUs or20.9%; 1.00±0.25% control, 1.58±0.34% MIA). Interestingly, Porphyromonadaceae, Prevotellaceae, unclassified Bacteriodales (36/45 discriminatory Bacteroidial OTUs or 80%; 4.46±0.66% control, 11.58±0.86% MIA), and Lachnospiriceae (8/14 discriminatory Clostridial OTUs or 57%; 0.28±0.06% control, 1.13±0.26% MIA) were more abundant in MIA offspring. Conversely, Ruminococcaceae (2 OTUs), Erysipelotrichaceae (2 OTUs), and the beta Proteobacteria family Alcaligenaceae (2 OTUs) were more abundant in control offspring (Figure 2D and Table S1; 0.95±0.31% control, 0.05±0.01% MIA). This suggests that specific Lachnospiraceae, along with other Bacteroidial species, may play a role in MIA pathogenesis, while other taxa may be protective. Importantly, there is no significant difference in the overall relative abundance of Clostridia (13.63 ± 2.54% vs 14.44 ± 2.84%; p=0.8340) and Bacteroidia (76.25 ± 3.22% vs 76.22 ± 3.46%; p=0.9943) between MIA offspring and controls (Figure 2E, left), indicating that alterations in the membership of Clostridial and Bacteroidial OTUs drive major changes in the gut microbiota between experimental groups.
Overall, we find that MIA leads to dysbiosis of the gut microbiota, driven primarily by alterations in specific OTUs of the bacterial classes Clostridia and Bacteroidia. Changes in OTUs classified as Lachnospiraceae and Ruminococcaceae of the order Clostridiales parallel select reports of increased Clostridium species in the feces of subjects with ASD (Finegold et al., 2012). Altogether, modeling MIA in mice induces not only behavioral and neuropathological features of ASD, but also microbiome changes as described in subsets of ASD individuals.
Bacteroides fragilis Improves Gut Barrier Integrity in MIA Offspring
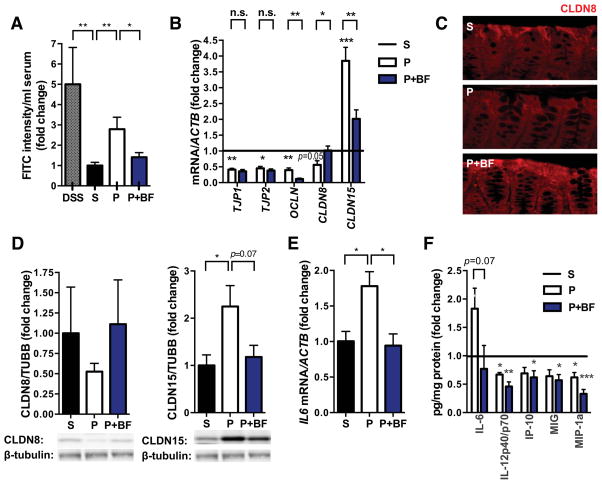
Gut microbes play an important role in the development, maintenance and repair of the intestinal epithelium (Turner, 2009). To determine whether targeting the gut microbiota could impact MIA-associated GI abnormalities, we treated mice with the human commensal B. fragilis at weaning, and tested for GI abnormalities at 8 weeks of age. B. fragilis has previously been shown to ameliorate experimental colitis (Mazmanian et al., 2008; Round and Mazmanian, 2010). Remarkably, B. fragilis treatment corrects intestinal permeability in MIA offspring (Figure 3A). In addition, B. fragilis treatment ameliorates MIA-associated changes in expression of CLDNs 8 and 15 (Figure 3B). Similar changes are observed in protein levels of CLDNs8 and 15 in the colon, with restoration by B. fragilis treatment (Figures 3C and 3D). No effects of B. fragilis on tight junction expression are observed in the small intestine (Figure S1B), consistent with the fact that Bacteroides species are predominantly found in the colon. Finally, the presence of GI defects prior to probiotic administration (Figure 1A, right) suggests that B. fragilis may treat, rather than prevent, this ASD-related pathology in MIA offspring.
Figure 3. B. fragilis Treatment Corrects GI Deficits in MIA offspring.
(A) Intestinal permeability assay, measuring FITC intensity in serum after oral gavage of FITC-dextran. Data are normalized to saline controls. Data for DSS, saline (S) and poly(I:C) (P) are as in Figure 1. poly(I:C)+B. fragilis (P+BF): n=9/group
(B) Colon expression of tight junction components relative to β-actin. Data for each gene are normalized saline controls. Data for S and P are as in Figure 1. Asterisks directly above bars indicate significance compared to saline control (normalized to 1, as denoted by the black line), whereas asterisks at the top of the graph denote statistical significance between P and P+BF groups. n=8/group
(C) Immunofluorescence staining for claudin 8. Representative images for n=5.
(D) Colon protein levels of claudin 8 (left) and claudin 15 (right). Representative signals are depicted below. Data are normalized to signal intensity in saline controls. n=3/group
(E) Colon expression of IL-6 relative to β-actin. Data are normalized to saline controls. Data for S and P are as in Figure 1. P+BF: n=3/group
(F) Colon protein levels of cytokines and chemokines relative to total protein content. Data are normalized to saline controls. Data for S and P are as in Figure 1. n=10/group
See also Figure S1.
B. fragilis treatment also restores MIA-associated increases in colon IL-6 mRNA and protein levels (Figures 3E and 3F). Levels of other cytokines are altered in both colons and small intestines of MIA offspring (Figures 1D and S1C), but these are not affected by B. fragilis treatment, revealing specificity for IL-6. We further find that recombinant IL-6 treatment can modulate colon levels of both CLDN 8 and 15 in vivo and in in vitro colon organ cultures (data not shown), suggesting that B. fragilis-mediated restoration of colonic IL-6 levels could underlie its effects on gut permeability. Collectively, these findings demonstrate that B. fragilis treatment of MIA offspring improves defects in GI barrier integrity, and corrects alterations in tight junction and cytokine expression.
B. fragilis Treatment Restores Specific Microbiota Changes in MIA Offspring
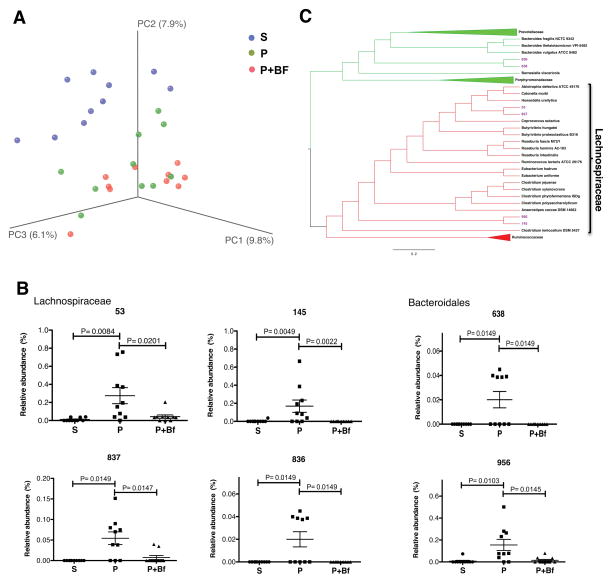
The finding that B. fragilis ameliorates GI defects in MIA offspring prompted us to examine its effects on the intestinal microbiota. No significant differences are observed following B. fragilis treatment of MIA offspring by PCoA (ANOSIM R=0.0060 p=0.4470; Table 3), in microbiota richness (PD: p=0.2980, Observed Species: p=0.5440) and evenness (Gini: p=0.6110, Simpson Evenness: p=0.5600; Figures 4A, S2A and S2B), or in relative abundance at the class level (Figure S2C). However, evaluation of key OTUs that discriminate adult MIA offspring from controls reveals that B. fragilis treatment significantly alters levels of 35 OTUs (Table S2). Specifically, MIA offspring treated with B. fragilis display significant restoration in the relative abundance of 6 out of the 67 OTUs found to discriminate MIA from control offspring (Figure 4B and Table S2), which are taxonomically assigned as unclassified Bacteroidia and Clostridia of the family Lachnospiraceae (Figure 4B and Table S2). Notably, these alterations occur in the absence of persistent colonization of B. fragilis, which remains undetectable in fecal and cecal samples isolated from treated MIA offspring (Figures S2D and S2E). Phylogenetic reconstruction of the OTUs that are altered by MIA and restored by B. fragilis treatment reveals that the Bacteroidia OTUs cluster together into a monophyletic group and the Lachnospiraceae OTUs cluster into 2monophyletic groups (Figure 4C). This result suggests that, although treatment of MIA offspring with B. fragilis may not lead to persistent colonization, this probiotic corrects the relative abundance of specific groups of related microbes of the Lachnospiraceae family as well as unclassified Bacteriodales. Altogether, we demonstrate that treatment of MIA offspring with B. fragilis ameliorates MIA-associated dysbiosis of the commensal microbiota.
Figure 4. B. fragilis Treatment Alters the Intestinal Microbiota and Corrects Species-Level Abnormalities in MIA Offspring.
(A) Unweighted UniFrac-based 3D PCoA plot based on all OTUs. Data for saline (S) and poly(I:C) (P) are as in Figure 2.
(B) Relative abundance of key OTUs of the family Lachnospiraceae (top) and order Bacteroidales (bottom) that are significantly altered by MIA and restored by B. fragilis treatment.
(C) Phylogenetic tree based on nearest-neighbor analysis of 16S rRNA gene sequences for key OTUs presented in panel B. Red clades indicate OTUs of the family Lachnospiraceae and green clades indicate OTUs of the order Bacteriodales. Purple taxa indicate OTUs that are significantly elevated in P and corrected by B. fragilis(BF) treatment.
B. fragilis Treatment Corrects ASD-Related Behavioral Abnormalities
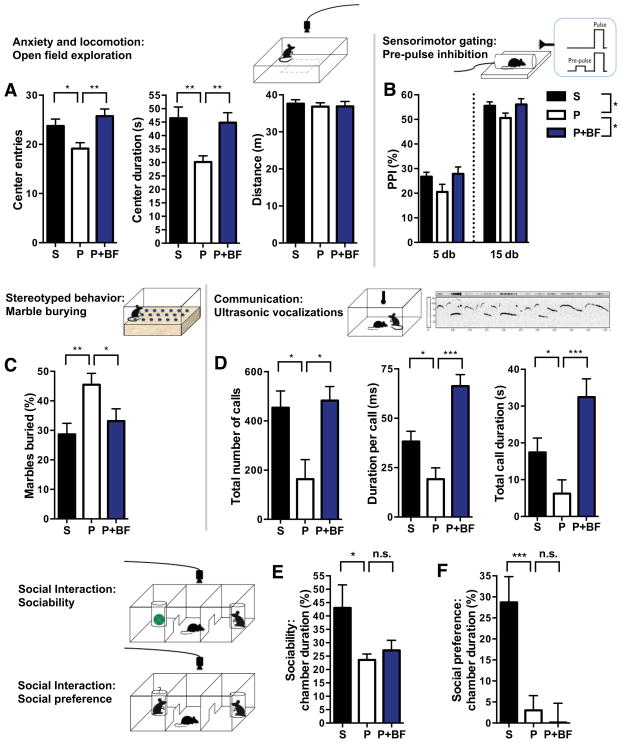
Studies suggest that GI issues in children with ASD can contribute to the development, persistence, and/or severity of symptoms (Buie et al., 2010; Coury et al., 2012). To explore the potential impact of GI dysfunction on core ASD behavioral abnormalities, we tested whether B. fragilis treatment impacts autism-related behaviors. We replicated previous findings that adult MIA offspring display cardinal behavioral features of ASD (Malkova et al., 2012). Open field exploration involves mapping an animal’s movement in an open arena to measure locomotion and anxiety (Bourin et al., 2007). MIA offspring display decreased entries and time spent in the center of the arena, which is indicative of anxiety-like behavior (Figure 5A; compare saline (S) to poly (I:C) (P)). The pre-pulse inhibition (PPI) task measures the ability of an animal to inhibit its startle in response to an acoustic tone (“pulse”) when it is preceded by a lower-intensity stimulus (“pre-pulse”). Deficiencies in PPI are a measure of impaired sensorimotor gating, and are observed in several neurodevelopmental disorders, including autism (Perry et al., 2007). MIA offspring exhibit decreased PPI in response to 5 or 15 db pre-pulses (Figure 5B). The marble burying test measures the propensity of mice to engage repetitively in a natural digging behavior that is not confounded by anxiety (Thomas et al., 2009). MIA offspring display increased stereotyped marble burying compared to controls (Figure 5C). Ultrasonic vocalizations are used to measure communication by mice, wherein calls of varying types and motifs are produced indifferent social paradigms (Grimsley et al., 2011). MIA offspring exhibit ASD-related deficits in communication, as indicated by reduced number and duration of ultrasonic vocalizations produced in response to a social encounter (Figure 5D). Finally, the three-chamber social test is used to measure ASD-related impairments in social interaction (Silverman et al., 2010). MIA offspring exhibit deficits in both sociability, or preference to interact with a novel mouse over a novel object, and social preference (social novelty), or preference to interact with an unfamiliar versus a familiar mouse (Figure 5E and 5F). Altogether, these behavioral assays measure the cardinal diagnostic symptoms of ASD, in addition to ASD-associated anxiety and deficient sensorimotor gating, and confirm that the MIA model reflects behavioral features of autism.
Figure 5. B. fragilis Treatment Ameliorates Autism-Related Behavioral Abnormalities in MIA Offspring.
(A) Anxiety-like and locomotor behavior in the open field exploration assay. n=35–75/group
(B) Sensorimotor gating in the pre-pulse inhibition (PPI) assay. n=35–75/group
(C) Repetitive marble burying assay. n=16–45/group
(D) Ultrasonic vocalizations produced by adult male mice during social encounter. n=10/group
S=saline+vehicle, P=poly(I:C)+vehicle, P+BF=poly(I:C)+B. fragilis. Data were collected simultaneously for poly(I:C)+B. fragilisΔPSA and poly(I:C)+B. thetaiotaomicron treatment groups(See also Figures S3 and S4).
Remarkably, oral treatment with B. fragilis ameliorates many of these ASD-related behaviors. B. fragilis-treated MIA offspring do not exhibit anxiety-like behavior in the open field (Figure 5A; compare poly(I:C) (P) to poly(I:C)+B. fragilis (P+BF)), as shown by restoration in the number of center entries and duration of time spent in the center of the arena. B. fragilis improves sensorimotor gating in MIA offspring, as indicated by increased combined PPI in response to 5 and 15 db pre-pulses (Figure 5B), with no significant effect on the intensity of startle to the acoustic stimulus (data not shown). B. fragilis-treated mice also exhibit decreased levels of stereotyped marble burying and restored communicative behavior(Figure 5C and 5D). Interestingly, B. fragilis raises the duration per call by MIA offspring to levels exceeding those observed in saline controls (Figure 5D), suggesting that despite normalization of the propensity to communicate (number of calls), there is a qualitative difference in the types of calls generated with enrichment of longer syllables.
Although B. fragilis-treated MIA offspring exhibit improved communicative, repetitive, anxiety-like and sensorimotor behavior, they retain deficits in sociability and social preference (Figure 5E). Interestingly, this parallels the inability to improve social behavior by administration of risperidone to ASD individuals (Canitano and Scandurra, 2008) and to CNTNAP2 knockout mice, a genetic mouse model for ASD (Penagarikano et al., 2011). These data suggest that there may be differences in the circuitry or circuit plasticity governing social behavior as compared to the other behaviors, and that B. fragilis treatment may modulate specific circuits during amelioration of ASD-related behavioral defects.
Interestingly, behavioral improvement in response to B. fragilis treatment of MIA offspring is not associated with changes in systemic immunity (Figure S3A–D) and is not dependent on polysaccharide A (PSA), a molecule previously identified to confer immunomodulatory effects by B. fragilis (Figure S3E) (Mazmanian et al., 2008; Ochoa-Reparaz et al., 2010; Round and Mazmanian, 2010). Furthermore, amelioration of behavior is not specific to B. fragilis, as similar treatment with Bacteroides thetaiotaomicron also significantly improves anxiety-like, repetitive and communicative behavior in MIA offspring (Figure S4). This is consistent with our finding that B. fragilis treatment improves ASD-related behavior in the absence of evident colonization of B. fragilis in the GI tract (Figure S2D and S2E), and thus, may be functioning through persistent shifts in the composition of resident microbiota (Figure 4). There is, however, some degree of specificity to bacterial treatment, as administration of Enterococcus faecalis has no effect on anxiety-like and repetitive behavior in MIA offspring (data not shown).
The Serum Metabolome is Modulated by MIA and B. fragilis Treatment
Metabolomic studies have shown that gut microbial products are found in many extra-intestinal tissues, and molecules derived from the microbiota may influence metabolic, immunologic and behavioral phenotypes in mice and humans (Blumberg and Powrie, 2012; Nicholson et al., 2012). Given that MIA offspring display increased gut permeability, tight junction defects and dysbiosis, we hypothesized that gut bacteria may affect the metabolome of mice. We utilized gas chromatography/liquid chromatography with mass spectrometry (GC/LC-MS)-based metabolomic profiling to identify MIA-associated changes in serum metabolites. 322 metabolites were detected in sera from adult mice (Table S5). Interestingly, MIA leads to statistically significant alterations in 8% of all serum metabolites detected (Table S4). Furthermore, postnatal B. fragilis treatment has a significant effect on the serum metabolome, altering 34% of all metabolites detected (Table S5 and Figure S5).
B. fragilis Treatment Corrects Levels of MIA-Induced Serum Metabolites
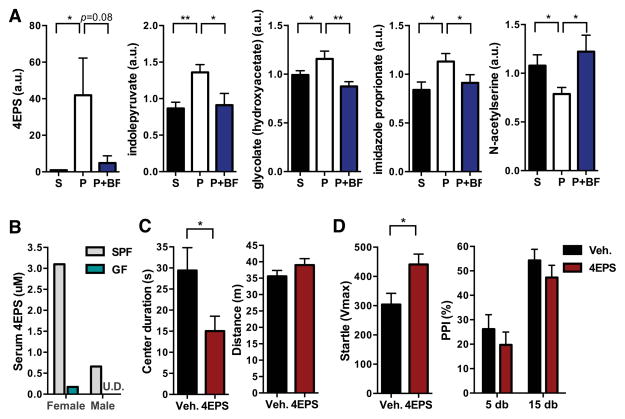
Consistent with the notion that increased intestinal permeability leads to leakage of gut-derived metabolites into the bloodstream, we hypothesized that B. fragilis-mediated improvement of intestinal barrier integrity would restore serum levels of certain metabolites. We therefore focused on serum metabolites that are significantly altered by MIA treatment and restored to control levels by B. fragilis treatment. The most dramatically affected metabolite is 4-ethylphenylsulfate (4EPS), displaying a striking 46-fold increase in serum levels of MIA offspring that is completely restored by B. fragilis treatment (Figure 6A). This metabolite is of particular interest because of the reported production of 4EPS by GI microbes and proposed role for 4EPS in communication by mice (Lafaye et al., 2004). Moreover, we find that compared to conventionally-colonized (SPF; specific pathogen free) mice, germ-free (GF) mice display nearly undetectable levels of serum 4EPS, indicating that serum 4EPS is derived from, or modulated by, the commensal microbiota (Figure 6B). Interestingly, 4EPS is suggested to be a uremic toxin, as is p-cresol (4-methylphenol), a chemically related metabolite reported to be a possible urinary biomarker for autism (Altieri et al., 2011; Persico and Napolioni, 2013). MIA offspring also exhibit elevated levels of serum p-cresol, although the increase does not reach statistical significance (Table S5). The fact that 4EPS shares close structural similarity to the toxic sulfated form of p-cresol (4-methylphenylsulfate; 4MPS) is intriguing as the two metabolites may exhibit functional overlap (Figure S6A).
Figure 6. B. fragilis Treatment Corrects MIA-Mediated Changes in 4-ethylphenylsulfate (4EPS), a Microbe-Dependent Metabolite that Induces Anxiety-like Behavior.
(A) Relative quantification of metabolites detected by GC/LC-MS that are significantly altered by MIA and restored by B. fragilis treatment. n=8/group
(B) Serum concentrations of 4EPS detected by LC-MS. n=1, where each represents pooled sera from 3–5 mice
(C) Anxiety-like and locomotor behavior in the open field exploration assay. n=10/group
(D) Potentiated startle reflex in the pre-pulse inhibition (PPI) assay. n=10/group
S=saline+vehicle, P=poly(I:C)+vehicle, P+BF=poly(I:C)+B. fragilis, SPF=specific pathogen-free (conventionally-colonized), GF=germ-free, Veh.=vehicle (saline), 4EPS=4-ethylphenylsulfate. U.D.=undetectable. See also Figures S5, S6 and S7 and Tables S3 and S4.
In addition to 4EPS, MIA offspring display significantly increased levels of serum indolepyruvate, a key molecule of the tryptophan metabolism pathway, which is restored to control levels by B. fragilis treatment (Figure 6A). Indolepyruvate is generated by tryptophan catabolism and, like 4EPS, is believed to be produced by gut microbes (Smith and Macfarlane, 1997) (Figure S6B). Moreover, the elevation in serum indolepyruvate observed in MIA offspring is reminiscent of reported increases inindolyl-3-acryloylglycine (IAG) in human ASD (Bull et al., 2003), which is involved in GI homeostasis and produced by bacterial metabolism (Keszthelyi et al., 2009). MIA offspring also exhibit increased levels of serum serotonin (0.05<p<0.10; Tables S3 and S4), reflecting another alternation in tryptophan metabolism, analogous to the well-established hyperserotonemia endophenotype of autism. MIA also leads to altered serum glycolate, imidazole propionate and N-acetylserine levels (Figure 6A), which are corrected by B. fragilis treatment. How changes in these metabolites may be relevant to ASD or GI dysfunction is currently unknown, but may be an exciting area for future study. These findings demonstrate that specific metabolites are altered in MIA offspring and normalized by B. fragilis treatment, with at least two molecules (4EPS and indolepyruvate) having potential relevance to ASD.
A Serum Metabolite Induces ASD-Related Behavior
MIA-dependent increases of specific metabolites, and their restoration by B. fragilis, suggest that small molecules may play a role in ASD-related behaviors. To test this hypothesis, we examined whether increasing serum 4EPS is sufficient to cause any ASD-related behavioral abnormalities in naïve mice. Mice were treated with 4EPS potassium salt (Figures S7A–C) or vehicle, daily from 3 weeks of age (when MIA offspring display gut permeability) to 6 weeks of age (when behavior testing begins). Remarkably, systemic administration of the single metabolite, 4EPS, to naïve wild-type mice is sufficient to induce anxiety-like behavior similar to that observed in MIA offspring (Figure 6C). Relative to vehicle-treated controls, mice exposed to 4EPS travel comparable distances in the open field but spend less time in the center arena (Figure 6C). Also, in the PPI test, 4EPS-treated mice exhibit increased intensity of startle in response to the unconditioned primary stimulus, but no significant alterations in PPI (Figure 6D), representing anxiety-associated potentiation of the startle reflex (Bourin et al., 2007). Conversely, there are no significant differences between 4EPS-treated versus saline-treated mice in marble burying or USV behavior (Figures S7D and S7E), suggesting that elevating serum 4EPS levels specifically promotes anxiety-like behavior. While not a core diagnostic criterion, anxiety is a common co-morbidity that may contribute to cardinal ASD symptoms. Furthermore, it is possible that complex behaviors may be modulated by combinations of metabolites. In summary, these data reveal that elevated systemic levels of a metabolite regulated by gut microbes causes an ASD-related behavior, suggesting that molecular connections between the gut and the brain maybe associated with autism.
DISCUSSION
ASD is a complex disorder with poorly defined etiologies and no effective or targeted cure. Here we demonstrate that in addition to displaying cardinal behavioral and neuropathological symptoms of ASD, offspring of immune-activated mothers exhibit defective GI integrity, dysbiosis of the commensal microbiota and alterations in serum metabolites that are similar to endophenotypes observed in ASD individuals. Collectively, these findings reveal MIA as a model with face validity for co-morbid GI symptoms and microbiome profiles found in ASD. We find that oral treatment with the human commensal B. fragilis corrects intestinal permeability defects, as well as altered levels of tight junction proteins and cytokines. The ability of B. fragilis to selectively ameliorate MIA-associated increases in colon IL-6 is interesting as this cytokineis required for the development of behavioral deficits in MIA offspring (Smith et al., 2007). Many cytokines including IL-6 regulate tight junction expression and intestinal barrier integrity, and further, a variety of enteric microbes are known to regulate intestinal tight junction and cytokine levels (Turner, 2009). Our study suggests that B. fragilis is able to ameliorate leaky gut by directly targeting tight junction expression, cytokine production and/or microbiome composition. Intriguingly, recent analysis in humans shows that among the Bacteroidaceae family, only a single phylotype most closely related to B. fragilis is selectively depleted in ASD children compared to matched controls, and most dramatically in those subjects with more severe GI issues (Dae-Wook Kang and Rosa Krajmalnik-Brown, personal communication). Thus, the correlation between B. fragilis and improved intestinal health is present in both mice and humans.
Consistent with the role of GI microbes in regulating intestinal permeability and metabolic homeostasis (Nicholson et al., 2012; Wikoff et al., 2009), we show that B. fragilis treatment corrects MIA-associated changes in specific serum metabolites that appear to have a gut origin, suggesting B. fragilis may prevent leakage of harmful molecules from the GI lumen. In a proof-of-concept test of the this hypothesis, we reveal that the microbially-modulated metabolite 4EPS, which is elevated in the circulation by MIA and restored by B. fragilis treatment, is sufficient to induce anxiety-like behavior in naïve mice. These data indicate that metabolomic changes contribute to the onset and/or persistence of autism-related behavioral abnormalities. Notably, we show that commensal microbes are required for the production of serum 4EPS in mice. Several species of Clostridium are believed to be producers of the precursor 4-ethylphenol (Nicholson et al., 2012), consistent with our findings that levels of the Lachnospiraceae family of Clostridia and serum 4EPS are elevated in MIA offspring, and both are corrected by B. fragilis treatment. Moreover, the structural similarity of 4EPS to p-cresol, which also derives from Clostridium species (Persico and Napolioni, 2013), suggests they may be produced through similar biosynthetic pathways (see Figure S6A). Although not all autism-like behaviors are affected by 4EPS alone, our results warrant the examination of several other serum metabolites, perhaps in combination, for their potential to impact the spectrum of autism-related behaviors.
Remarkably, B. fragilis treatment ameliorates abnormal communicative, stereotyped, sensorimotor and anxiety-like behaviors in MIA offspring, supporting emerging evidence for a gut-brain link in ASD. A role for commensal bacteria in modulating behavioris supported by studies revealing differences between GF and SPF mice in anxiety-like (Heijtz et al., 2011), locomotor and social behavior (Desbonnet et al., 2013). Treatment of SPF animals with commensal microbes can ameliorate depressive (Bravo et al., 2011) and anxiety-like behavior in mice (Bercik et al., 2011), and probiotic treatment has been beneficial in treating psychological distress and chronic fatigue symptoms in humans (Messaoudi et al., 2011; Rao et al., 2009). Our findings provide a novel mechanism by which a human commensal bacterium can improve ASD-related GI deficits and behavioral abnormalities in mice, possibly explaining the rapid increase in ASD prevalence by identifying the microbiome as a critical environmental contributor to disease. We propose the transformative concept that autism is, at least in part, a disease involving the gut that impacts the immune, metabolic and nervous systems, and that microbiome-mediated therapies may be a safe and effective treatment for ASD.
EXPERIMENTAL PROCEDURES
See supplemental information for additional details and references.
Animals and MIA
Pregnant C57BL/6N mice (Charles River; Wilmington, MA) were injected i.p. on E12.5 with saline or 20 mg/kg poly(I:C) according to methods described in ref. (Smith et al., 2007). All animal experiments were approved by the Caltech IACUC.
B. fragilis treatment
Mice were selected at random for treatment with Bacteroides fragilis(ATCC 9343) or vehicle, every other day for 6 days at weaning. 1×1010CFU of freshly grown B. fragilis, or vehicle, in 1.5% sodium bicarbonate was administered in sugar-free applesauce over standard food pellets. The same procedure was used for mutant B. fragilisΔPSA and B. thetaiotaomicron.
Intestinal permeability assay
Mice were fasted for 4 hours before gavage with 0.6 g/kg 4 kDa FITC-dextran (Sigma Aldrich). 4 hours later, serum samples were read for fluorescence intensity at 521 nm using an xFluor4 spectrometer (Tecan). Mice were fed 3% dextran sulfate sodium salt (DSS; MP Biomedicals) in drinking water for 7 days to induce colitis.
Intestinal qRT-PCR, Western blots, and cytokine profiles
Gut tissue was flushed with HBSS and either a) homogenized in Trizol for RNA isolation and reverse transcription according to ref. (Hsiao and Patterson, 2011) or b) homogenized in Tissue Extraction Reagent I (Invitrogen) containing protease inhibitors (Roche) for protein assays. For cytokine profiling, mouse 20-plex cytokine arrays (Invitrogen) were run on the Luminex FLEXMAP 3D platform by the Clinical Immunobiology Correlative Studies Laboratory at the City of Hope (Duarte, CA). Western blots were conducted according to standard methods and probed with rabbit anti-claudin 8 or rabbit anti-claudin 15 (Invitrogen) at 1:100 dilution.
Microbial DNA extraction, 16S rRNA gene amplification and pyrosequencing
Bacterial DNA was extracted from mouse fecal pellets using the MoBioPowerSoil Kit following protocols benchmarked by the NIH Human Microbiome Project. The V3-V5 regions of the 16S rRNA gene were PCR amplified using individually barcoded universal primers containing linker sequences for sequencing using a multiplexed 454-Titanium pyrosequencer.
16S rRNA gene sequence analysis
16S data was processed and its diversity was analyzed using QIIME 1.6 software package (Caporaso et al., 2010b). OTUs were assigned taxonomic classification using the basic BLAST classifier (Altschul et al, 1990). For tree-based alpha- and beta diversity analyses, representative sequences for each OTU were aligned using PyNAST (Caporaso et al., 2010a) and a phylogenetic tree was constructed based on this alignment using Fast Tree (Price et al., 2009). Beta diversity was assessed from unweighted UniFrac, using the Analysis of Similarity (ANOSIM; Fierer et al 2010), Permutational multivariate analysis of variance (PERMANOVA;(Anderson, 2008)), Permutational analysis of multivariate dispersions (PERMDISP;(Anderson et al., 2006)), and Moran’s I.
Identification of differences in specific OTUs
Key OTUs were identified using: (1) Metastats comparison (White et al., 2009) and (2) Random Forests algorithm, under QIIME (Knights et al., 2011) or coupled with Boruta feature selection, in the Genbore emicrobiome tool set (Riehle et al., 2012), and (3) Galaxy platform-based LDA Effect Size analysis (LEfSe; (Segata et al., 2011)). Key OTUs were than aligned using the SINA aligner (http://www.arb-silva.de/aligner/; (Pruesse et al., 2012), compared to the SILVA reference database release 111 (Quast et al., 2013) using Arb (Ludwig et al., 2004) and visualized using Fig Tree (http://tree.bio.ed.ac.uk/software/figtree/). Heat maps of key OTUs were generated by extracting their relative abundance from the OTU table and clustering data by correlation using Cluster 3.0 (de Hoon et al., 2004). Abundance data was visualized using Java TreeView (Saldanha, 2004).
Behavioral testing
MIA and control offspring were behaviorally tested as in refs. (Hsiao et al., 2012) and (Malkova et al., 2012). Mice were tested beginning at 6 weeks of age for pre-pulse inhibition, open field exploration, marble burying, social interaction and adult ultrasonic vocalizations
Metabolomics screening
Serum samples were extracted and analyzed on GC/MS, LC/MS and LC/MS/MS platforms by Metabolon, Inc. For LC/MS and LC/MS/MS, samples were run on a Waters ACQUITY UPLC and Thermo-Finnigan LTQ mass spectrometer. For GC/MS, samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization. Chemical entities were identified by comparison to metabolomic library entries of purified standards.
4EPS sufficiency experiments
Wildtype mice were injected i.p. with saline or 30 mg/kg 4EPS potassium salt daily from 3 to 6 weeks of age. A dose-response curve was generated by measuring serum 4EPS levels at various times after i.p. injection of 30 mg/kg 4EPS (Figure S7C). Mice were behaviorally tested as described above from 6 to 9 weeks of age.
Statistical Analysis
Statistical analysis was performed using Prism software (Graphpad). Data are plotted in the figures as mean ± SEM. Differences between two treatment groups were assessed using two-tailed, unpaired Student t test with Welch’s correction. Differences among three or more groups were assessed using one-way ANOVA with Bonferroni post hoc test. Two-way repeated measures ANOVA with Bonferroni post hoc test was used for analysis of PPI and CD4+ T-cell stimulation data. Two-way ANOVA with contrasts was used for analysis of the metabolite data. Significant differences are indicated in the figures by *p<0.05, **p<0.01, ***p<0.001. Notable near-significant differences (0.5<p<0.1) are indicated in the figures. Notable non-significant (and non-near significant) differences are indicated in the figures by “n.s.”.
Supplementary Material
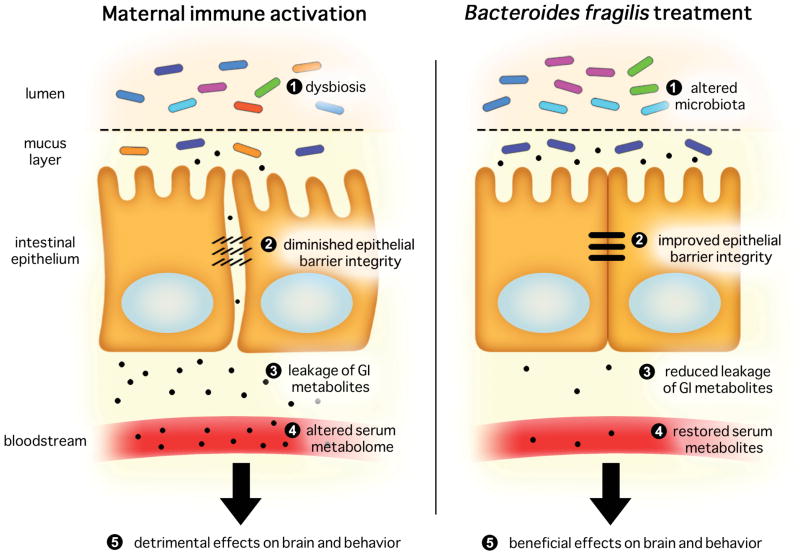
Figure 7. Proposed Microbiota-Gut-Brain Connection in the MIA Model.
Adult offspring of immune-activated mothers exhibit dysbiosis of the gut microbiota, with corresponding increases in intestinal permeability, altered intestinal expression of tight junction components and cytokines, changes in the serum metabolome and abnormal autism-related behaviors. Treatment of MIA offspring with B. fragilis further alters the composition of the gut microbiota, improves gut barrier integrity and restores levels of particular serum metabolites that can cause downstream effects on brain and behavior.
HIGHLIGHTS.
The MIA model recapitulates GI comorbidities associated with autism.
Targeting the microbiota treats specific ASD-like symptoms in the MIA model.
Gut microbes regulate metabolites that alter behavior in animals.
The gut microbiome may contribute to the pathophysiology of ASD.
Acknowledgments
We acknowledge Reyna Sauza, Jaime Rodriguez and Taren Thron for caring for the animals, Dr. Michael Fischbach (UCSF) for advising on pathways of 4EPS and indolepyruvate synthesis, Dr. NadimAjami (Baylor) for providing helpful comments on the manuscript, Greg Donaldson (Caltech) for conducting experiments on microbial viability, Dr. Kym Faull (UCLA) for conducting pilot GC/MS experiments, Dr. Alessio Fasano (Massachusetts General) for conducting pilot microbiota sequencing experiments and Dr. Jerrold Turner (U Chicago) for providing histological analysis of intestinal sections. This work was supported by a Caltech Innovation Fellowship (to EYH), Autism Speaks Weatherstone Fellowship (to EYH), NIH/NRSA Pre-doctoral Fellowship (to EYH), Human Frontiers Science Program Fellowship (to GS), DOD Graduate Fellowship (to JAC), NSF Graduate Research Fellowship (to JAC), Autism Speaks Trailblazer Award (to PHP and SKM), Caltech Innovation grant (to PHP and SKM), Congressionally Directed Medical Research Award (to PHP and SKM), Weston Havens Award (to PHP and SKM), Callie D. McGrath Charitable Foundation awards (to PHP) and NIMH grant MH100556 (to PHP and SKM).
Footnotes
Additional Footnotes
EYH, PHP and SKM designed the study, EYH, SWM, SH, JAC and JC performed the experiments and analyzed the data, ERH, TM, GS and JP conducted microbiota sequencing and analysis, SER contributed novel reagents, EYH, SWM, GS, JAC, PHP and SKM wrote the manuscript. All authors discussed the results and commented on the manuscript.
Conflict of interests: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdallah MW, Larsen N, Grove J, Norgaard-Pedersen B, Thorsen P, Mortensen EL, Hougaard DM. Amniotic fluid inflammatory cytokines: Potential markers of immunologic dysfunction in autism spectrum disorders. The world journal of biological psychiatry : the official journal of the World Federation of Societies of Biological Psychiatry. 2013;14:528–538. doi: 10.3109/15622975.2011.639803. [DOI] [PubMed] [Google Scholar]
- Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism--comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11:22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri L, Neri C, Sacco R, Curatolo P, Benvenuto A, Muratori F, Santocchi E, Bravaccio C, Lenti C, Saccani M, et al. Urinary p-cresol is elevated in small children with severe autism spectrum disorder. Biomarkers. 2011;16:252–260. doi: 10.3109/1354750X.2010.548010. [DOI] [PubMed] [Google Scholar]
- Atladottir HO, Pedersen MG, Thorsen P, Mortensen PB, Deleuran B, Eaton WW, Parner ET. Association of family history of autoimmune diseases and autism spectrum disorders. Pediatrics. 2009;124:687–694. doi: 10.1542/peds.2008-2445. [DOI] [PubMed] [Google Scholar]
- Atladottir HO, Thorsen P, Ostergaard L, Schendel DE, Lemcke S, Abdallah M, Parner ET. Maternal infection requiring hospitalization during pregnancy and autism spectrum disorders. J Autism Dev Disord. 2010;40:1423–1430. doi: 10.1007/s10803-010-1006-y. [DOI] [PubMed] [Google Scholar]
- Bauman MD, Iosif AM, Smith SE, Bregere C, Amaral DG, Patterson PH. Activation of the Maternal Immune System During Pregnancy Alters Behavioral Development of Rhesus Monkey Offspring. Biological psychiatry. 2013 doi: 10.1016/j.biopsych.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P, Park AJ, Sinclair D, Khoshdel A, Lu J, Huang X, Deng Y, Blennerhassett PA, Fahnestock M, Moine D, et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil. 2011;23:1132–1139. doi: 10.1111/j.1365-2982.2011.01796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv137. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukthir S, Matoussi N, Belhadj A, Mammou S, Dlala SB, Helayem M, Rocchiccioli F, Bouzaidi S, Abdennebi M. Abnormal intestinal permeability in children with autism. Tunis Med. 2010;88:685–686. [PubMed] [Google Scholar]
- Bourin M, Petit-Demouliere B, Dhonnchadha BN, Hascoet M. Animal models of anxiety in mice. Fundamental & clinical pharmacology. 2007;21:567–574. doi: 10.1111/j.1472-8206.2007.00526.x. [DOI] [PubMed] [Google Scholar]
- Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS, Sourander A, Hinkka-Yli-Salomaki S, McKeague IW, Sundvall J, Surcel HM. Elevated maternal C-reactive protein and autism in a national birth cohort. Mol Psychiatry. 2013 doi: 10.1038/mp.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buie T, Campbell DB, Fuchs GJ, 3rd, Furuta GT, Levy J, Vandewater J, Whitaker AH, Atkins D, Bauman ML, Beaudet AL, et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics. 2010;125(Suppl 1):S1–18. doi: 10.1542/peds.2009-1878C. [DOI] [PubMed] [Google Scholar]
- Canitano R, Scandurra V. Risperidone in the treatment of behavioral disorders associated with autism in children and adolescents. Neuropsychiatr Dis Treat. 2008;4:723–730. doi: 10.2147/ndt.s1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Prevalence of autism spectrum disorders - autism and developmental disabilities monitoring network, 14 sites, United States, 2008. MMWR Surveill Summ. 2012;61:1–19. [PubMed] [Google Scholar]
- Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- Comi AM, Zimmerman AW, Frye VH, Law PA, Peeden JN. Familial clustering of autoimmune disorders and evaluation of medical risk factors in autism. Journal of child neurology. 1999;14:388–394. doi: 10.1177/088307389901400608. [DOI] [PubMed] [Google Scholar]
- Coury DL, Ashwood P, Fasano A, Fuchs G, Geraghty M, Kaul A, Mawe G, Patterson P, Jones NE. Gastrointestinal conditions in children with autism spectrum disorder: developing a research agenda. Pediatrics. 2012;130(Suppl 2):S160–168. doi: 10.1542/peds.2012-0900N. [DOI] [PubMed] [Google Scholar]
- Croen LA, Braunschweig D, Haapanen L, Yoshida CK, Fireman B, Grether JK, Kharrazi M, Hansen RL, Ashwood P, Van de Water J. Maternal mid-pregnancy autoantibodies to fetal brain protein: the early markers for autism study. Biol Psychiatry. 2008;64:583–588. doi: 10.1016/j.biopsych.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- D’Eufemia P, Celli M, Finocchiaro R, Pacifico L, Viozzi L, Zaccagnini M, Cardi E, Giardini O. Abnormal intestinal permeability in children with autism. Acta Paediatr. 1996;85:1076–1079. doi: 10.1111/j.1651-2227.1996.tb14220.x. [DOI] [PubMed] [Google Scholar]
- de Magistris L, Familiari V, Pascotto A, Sapone A, Frolli A, Iardino P, Carteni M, De Rosa M, Francavilla R, Riegler G, et al. Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J Pediatr Gastroenterol Nutr. 2010;51:418–424. doi: 10.1097/MPG.0b013e3181dcc4a5. [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Molecular psychiatry. 2013 doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, Youn E, Summanen PH, Granpeesheh D, Dixon D, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Finegold SM, Downes J, Summanen PH. Microbiology of regressive autism. Anaerobe. 2012;18:260–262. doi: 10.1016/j.anaerobe.2011.12.018. [DOI] [PubMed] [Google Scholar]
- Finegold SM, Molitoris D, Song Y, Liu C, Vaisanen ML, Bolte E, McTeague M, Sandler R, Wexler H, Marlowe EM, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002;35:S6–S16. doi: 10.1086/341914. [DOI] [PubMed] [Google Scholar]
- Gondalia SV, Palombo EA, Knowles SR, Cox SB, Meyer D, Austin DW. Molecular characterisation of gastrointestinal microbiota of children with autism (with and without gastrointestinal dysfunction) and their neurotypical siblings. Autism Res. 2012;5:419–427. doi: 10.1002/aur.1253. [DOI] [PubMed] [Google Scholar]
- Gorrindo P, Williams KC, Lee EB, Walker LS, McGrew SG, Levitt P. Gastrointestinal dysfunction in autism: parental report, clinical evaluation, and associated factors. Autism research : official journal of the International Society for Autism Research. 2012;5:101–108. doi: 10.1002/aur.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley JM, Monaghan JJ, Wenstrup JJ. Development of social vocalizations in mice. PloS one. 2011;6:e17460. doi: 10.1371/journal.pone.0017460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijtz RD, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim SH, Voigt RG, Katusic SK, Weaver AL, Barbaresi WJ. Incidence of gastrointestinal symptoms in children with autism: a population-based study. Pediatrics. 2009;124:680–686. doi: 10.1542/peds.2008-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, Krajmalnik-Brown R. Reduced Incidence of and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS One. 2013;8:e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keszthelyi D, Troost FJ, Masclee AA. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol Motil. 2009;21:1239–1249. doi: 10.1111/j.1365-2982.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- Kohane IS, McMurry A, Weber G, MacFadden D, Rappaport L, Kunkel L, Bickel J, Wattanasin N, Spence S, Murphy S, et al. The co-morbidity burden of children and young adults with autism spectrum disorders. PLoS One. 2012;7:e33224. doi: 10.1371/journal.pone.0033224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaye A, Junot C, Ramounet-Le Gall B, Fritsch P, Ezan E, Tabet JC. Profiling of sulfoconjugates in urine by using precursor ion and neutral loss scans in tandem mass spectrometry. Application to the investigation of heavy metal toxicity in rats. J Mass Spectrom. 2004;39:655–664. doi: 10.1002/jms.635. [DOI] [PubMed] [Google Scholar]
- Malkova NV, Yu CZ, Hsiao EY, Moore MJ, Patterson PH. Maternal immune activation yields offspring displaying mouse versions of the three core symptoms of autism. Brain Behav Immun. 2012;26:607–616. doi: 10.1016/j.bbi.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, Bisson JF, Rougeot C, Pichelin M, Cazaubiel M, et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. The British journal of nutrition. 2011;105:755–764. doi: 10.1017/S0007114510004319. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- Onore C, Careaga M, Ashwood P. The role of immune dysfunction in the pathophysiology of autism. Brain Behav Immun. 2012;26:383–392. doi: 10.1016/j.bbi.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parracho HM, Bingham MO, Gibson GR, McCartney AL. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. Journal of medical microbiology. 2005;54:987–991. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry W, Minassian A, Lopez B, Maron L, Lincoln A. Sensorimotor gating deficits in adults with autism. Biological psychiatry. 2007;61:482–486. doi: 10.1016/j.biopsych.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Persico AM, Napolioni V. Urinary p-cresol in autism spectrum disorder. Neurotoxicology and teratology. 2013;36:82–90. doi: 10.1016/j.ntt.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Rao AV, Bested AC, Beaulne TM, Katzman MA, Iorio C, Berardi JM, Logan AC. A randomized, double-blind, placebo-controlled pilot study of a probiotic in emotional symptoms of chronic fatigue syndrome. Gut Pathog. 2009;1:6. doi: 10.1186/1757-4749-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Smith SE, Malkova N, Tse D, Su Y, Patterson PH. Activation of the maternal immune system alters cerebellar development in the offspring. Brain Behav Immun. 2009;23:116–123. doi: 10.1016/j.bbi.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nature reviews Neuroscience. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, Macfarlane GT. Formation of Phenolic and Indolic Compounds by Anaerobic Bacteria in the Human Large Intestine. Microb Ecol. 1997;33:180–188. doi: 10.1007/s002489900020. [DOI] [PubMed] [Google Scholar]
- Smith SE, Li J, Garbett K, Mirnics K, Patterson PH. Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci. 2007;27:10695–10702. doi: 10.1523/JNEUROSCI.2178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol. 2004;70:6459–6465. doi: 10.1128/AEM.70.11.6459-6465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Burant A, Bui N, Graham D, Yuva-Paylor LA, Paylor R. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology. 2009;204:361–373. doi: 10.1007/s00213-009-1466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, Guyonnet D, Legrain-Raspaud S, Trotin B, Naliboff B, et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144:1394–1401. e1394. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BL, Hornig M, Buie T, Bauman ML, Cho Paik M, Wick I, Bennett A, Jabado O, Hirschberg DL, Lipkin WI. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6:e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BL, Hornig M, Parekh T, Lipkin WI. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio. 2012:3. doi: 10.1128/mBio.00261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.