Abstract
A better understand the ecology of microbes and their role in the global ecosystem could be achieved if traditional ecological theories can be applied to microbes. In ecology organisms are defined as specialists or generalists according to the breadth of their niche. Spatial distribution is often used as a proxy measure of niche breadth; generalists have broad niches and a wide spatial distribution and specialists a narrow niche and spatial distribution. Previous studies suggest that microbial distribution patterns are contrary to this idea; a microbial generalist genus (Desulfobulbus) has a limited spatial distribution while a specialist genus (Methanosaeta) has a cosmopolitan distribution. Therefore, we hypothesise that this counter-intuitive distribution within generalist and specialist microbial genera is a common microbial characteristic. Using molecular fingerprinting the distribution of four microbial genera, two generalists, Desulfobulbus and the methanogenic archaea Methanosarcina, and two specialists, Methanosaeta and the sulfate-reducing bacteria Desulfobacter were analysed in sediment samples from along a UK estuary. Detected genotypes of both generalist genera showed a distinct spatial distribution, significantly correlated with geographic distance between sites. Genotypes of both specialist genera showed no significant differential spatial distribution. These data support the hypothesis that the spatial distribution of specialist and generalist microbes does not match that seen with specialist and generalist large organisms. It may be that generalist microbes, while having a wider potential niche, are constrained, possibly by intrageneric competition, to exploit only a small part of that potential niche while specialists, with far fewer constraints to their niche, are more capable of filling their potential niche more effectively, perhaps by avoiding intrageneric competition. We suggest that these counter-intuitive distribution patterns may be a common feature of microbes in general and represent a distinct microbial principle in ecology, which is a real challenge if we are to develop a truly inclusive ecology.
Introduction
In order to better understand the ecology of microbes and their role in the global ecosystem, it is essential to determine whether ecological ideas and theories that have been derived from studies on plants and animals are also applicable to microbes [1]–[4]. However, it may be that, due to differences in scale and physiologies between micro- and macroorganisms, there will be principles of ecology that are difficult to reconcile between the two. Developing an inclusive ecology represents a substantial challenge as for many years a primary assumption in microbial ecology was that while all microbes could be found in all environments, community structures were shaped by local environmental factors, exemplified in Baas-Becking's statement “Everything is everywhere, but the environment selects” [5]. In contrast traditional ecology, where endemism and biogeography are the norm, can be exemplified by the ecological truism that “all species are always absent from almost everywhere” [6]. However, microbial communities have been shown to have distinct spatial distribution patterns [7]–[11], yet it is unclear what processes structure microbial communities or whether ecological ideas, such as biogeography [12], [13], are readily applicable to microbes.
The direct application of ecological theory to microbes is difficult because of the complexity of defining microbial taxonomic groups and the extent of microbial genetic and phenotypic diversity [14], [15]. These problems particularly confound the whole bacterial assemblage analyses that are commonly used in microbial ecology. Such broad-brush studies have no equivalent in traditional ecology where studies are usually constrained within tight taxonomic boundaries. Ecologists avoid the problems common to studies in microbial ecology as they use these model species/genera to represent larger groups [16], [17]. Such an approach has rarely been applied to microorganisms primarily because of the difficulty in defining homogeneous species. This issue can be avoided by studying model microbial genera if the selected genera are phylogenetically and functionally homogeneous [18]. Therefore, to study the environmental distribution of microbes we selected model genera that fitted these specific criteria and could also be defined as either specialists or generalists.
A specialist or generalist organism is most succinctly defined as having either a narrow or wide potential niche respectively. However, defining the actual extent of an organism's niche, the n-dimensional space that affects an organism's growth and survival [19], is clearly extremely challenging, if not impossible. Therefore, within traditional ecology spatial distribution has become an accepted proxy measure of niche breadth for many taxa, so it is assumed that a specialist organism with a narrow niche will have a limited spatial distribution and a generalist a wide spatial distribution [20]–[22]. In microbial ecology such assumptions, which underpin many ideas in ecology, have never been properly tested. There are microbes that utilise a vast array of carbon sources for energy and growth and are also capable of respiring more than one type of compound, thus are metabolically flexible with multiple electron donors and acceptors and so can confidently be defined as generalists with wide potential niches. Conversely, there are microbes that have very specific and limited metabolic needs and are clearly highly specialised with narrow potential niches. Thus, metabolic flexibility represents a fundamentally important aspect of a microbe's ecological function and, we propose, represents a reasonable proxy for niche breadth. Therefore, if the spatial distribution of metabolically generalist and specialist microbial model genera is a good proxy for niche breadth this would be a strong indicator that some rules that govern the distribution of large organisms such as plants and animals can also be applied to microbes.
We tested the idea that the extent of spatial distribution along an estuarine gradient is related to metabolic flexibility in two model microbial genera that are both distributed along the full length of the Colne estuary, Essex, UK [23], [24]. Estuaries are natural environmental gradients that have been used to show spatial and evolutionary differentiation in macro-organisms [17], [25] and are hotspots of biogeochemical cycling and microbial activity [26]. The model genera were the sulfate-reducing bacteria Desulfobulbus, a generalist genus that can respire both sulfur and nitrogen oxyanions, use fermentation for growth and metabolise a range of carbon sources [27], [28] and the methanogenic archaeal genus Methanosaeta, a metabolic specialist that uses acetate as both electron acceptor and donor and as its sole carbon source [29]. Genotypes (at approximately the species level) of the generalist Desulfobulbus genus were spatially restricted to distinct regions of the estuarine salinity gradient in a manner similar to that classically described for estuarine macrofauna [17] while genotypes of the specialist Methanosaeta showed no such differential distribution and were monotonically distributed along the estuary [8], [30]–[32].
These data challenge assumptions from traditional ecology that spatial distribution is a proxy measure of niche breadth leading us to propose the hypothesis that, in contrast to traditional ecological ideas, the breadth of the spatial distribution of specialist and generalist microbes is inversely related to the breadth of their potential niches. However, this hypothesis is derived from data from only two genera that are from different Domains and have very different metabolisms and so we cannot exclude the possibility that the differences seen in their distributions are due to their phylogenetic or metabolic differences and not differences in their metabolic flexibility. Therefore, to test the hypothesis above and to determine whether metabolic flexibility in general does influence the spatial distribution of microbes we expanded this study to include the most generalist methanogenic archaeal genus, Methanosarcina, and the highly specialised sulfate-reducing bacterial genus, Desulfobacter. Methanosarcina is the only methanogen genus able to perform all three methanogenic respiratory pathways [33] while Desulfobacter are SRB that respire sulfur oxyanions only, primarily whilst oxidising acetate [34]. Analysis of these additional genera produces a dataset that includes a generalist and specialist genus from both the bacterial and the archaeal Domains and that are methanogens and sulphate-reducers and so will address whether the specific metabolism or phylogeny of these four model genera or indeed their metabolic flexibility is a driver of the counter-intuitive differential distribution of Methanosaeta and Desulfobulbus detected previously.
Results
16S rRNA gene fragments, amplified by genus-specific PCR (Table S1 in File S1), were used to analyse the distribution of all four genera in sediment along the full length of the Colne estuary, Essex, UK (Figure S1) using the molecular fingerprinting method Denaturing Gradient Gel Electrophoresis (DGGE; Supplementary information). Profiles of representative bands are shown in Figure 1 (for complete profiles see Figure S3). Genotype distribution patterns from metabolic generalist (Methanosarcina and Desulfobulbus) and specialist (Methanosaeta and Desulfobacter) model genera were highly dissimilar. Both generalist models showed a restricted spatial distribution, in which particular genotypes were only found in certain regions of the estuary, while both specialist models showed no such restricted distribution, with all bands detected all along the estuary.
Figure 1. Corrected mean band intensities of representative bands from replicated DNA-DGGE analyses from the 11 sites (from the marine site 1 to the freshwater site 11) along the Colne estuary, Essex, UK for: A. Desulfobulbus, B. Methanosarcina, C. Methanosaeta and D. Desulfobacter.
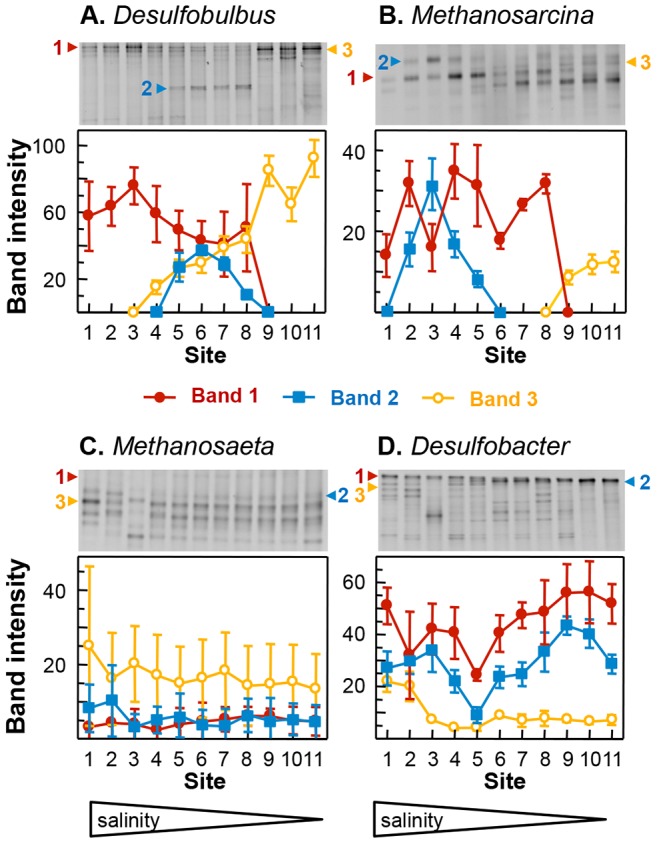
Numbered arrows on each gel image indicate band numbers. [Band 1 (line with solid circle) Band 2 (line with open square) Band 3(line with open circle)]. Error bars represent the standard error of the mean (SEM, n = 3). A complete profile is given in Figure S3.
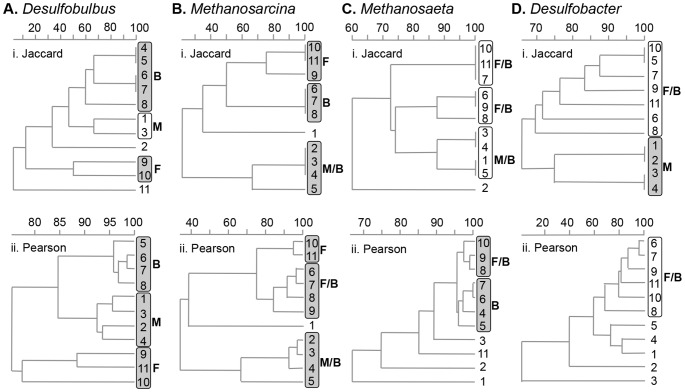
Cluster analysis of both DGGE band presence/absence (Jaccard) and total DGGE profile (Pearson) indicated geographically coherent clustering for the generalist models, Desulfobulbus and Methanosarcina (Figure 2A and B). The specialist models, Methanosaeta and Desulfobacter, showed inconsistent and geographically incoherent clustering with generally high overall similarities between sites (>59%) across the two methods (Figure 2C and D). Unconstrained Canonical Correspondence Analysis (CCA) of the DGGE fingerprints showed some clustering according to site for all genera (Figure S4). However, very low eigenvalues (scales on the CCA; 0.015–0.048) in the CCA of the specialist genera (Figure S4C and D) indicated a poor correlation between genotypic dissimilarity matrixes and environmental variables while eigenvalues (0.181–0.680) were higher for the generalist models (Figure S4A and B). Mantel and partial Mantel tests revealed a significant correlation (p<0.05) between the geographic distance between sites and distribution of the generalist models (Table S2A and B in File S1) whether based on Band Intensity, Jaccard or Pearson analyses. Very limited significant correlations were found for Desulfobacter and Methanosaeta (Table S2C and D in File S1), with no clear coherent correlations across all three methods of analysis.
Figure 2. Cluster analyses of the DNA-DGGE profiles: i. Jaccard and ii. Pearson analyses of A. Desulfobulbus; B. Methanosarcina; C. Methanosaeta; D. Desulfobacter.
Marine (M), Marine/Brackish (M/B), Brackish (B), Freshwater/Brackish (F/B) and Freshwater (F) clusters are circled. Shaded clusters are geographically continuous, unshaded clusters are not.
Analysis of RNA (in this case 16S rRNA) provides an indication that the organisms detected are not only present but also active in an environment. Banding patterns in the RNA-DGGE analysis of both Desulfobulbus and Methanosaeta (Figure S5) were very similar to those seen in the DNA-DGGE (Figure 1 and Figure S3). As with DNA-DGGE Desulfobulbus had a restricted pattern of spatial distribution, while Methanosaeta genotypes were constantly detected all along the estuary [30]. Cluster analyses showed spatially coherent clustering for Desulfobulbus clusters (Fig 3A) and a weak but inconsistent clustering for Methanosaeta (Fig 3B). Unconstrained CCAs showed strong geographic clustering for Desulfobulbus (Eigen values 0.381–0.628; Figure S6A) and no clustering for Methanosaeta (Eigen values 0.075–0.138; Figure S6B). Mantel and partial Mantel tests (Table S3 in File S1) revealed a clearly significant correlation between geographic distance and the Desulfobulbus distribution pattern (p<0.05). While all Mantel tests were significant for Methanosaeta (p<0.05) no coherent correlation could be seen with partial Mantel tests. Importantly, DNA- and RNA-based Desulfobulbus distribution patterns were very similar, as observed previously for Methanosaeta [30]. This implies that detected genotypes (from analysis of DNA) are active (from analysis of RNA), and thus DNA based DGGE fingerprints represent a satisfactory representation of metabolically active populations. Pyrosequence analysis of functional genes from Desulfobulbus (dsrA) and Methanosaeta (mcrA) along the Colne estuary also shows a clear difference in distribution patterns between these two model genera, supporting the conclusions above [32].
Figure 3. Cluster analyses of the RNA-DGGE profiles: i. Jaccard and ii. Pearson analyses of A. Desulfobulbus; B. Methanosarcina.
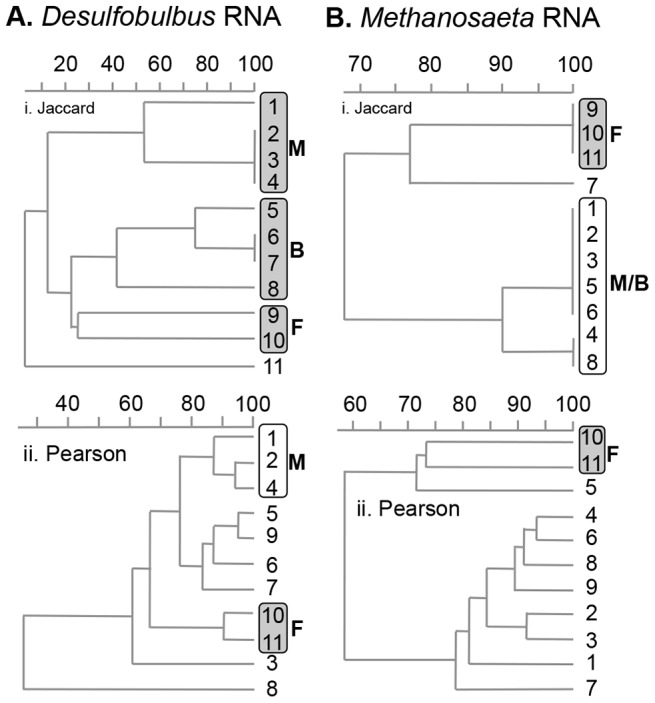
Marine (M), Marine/Brackish (M/B), Brackish (B), Freshwater/Brackish (F/B) and Freshwater (F) clusters are circled. Shaded clusters are geographically continuous, unshaded clusters are not.
These data show that metabolic flexibility does appear to directly affect the distribution of microbial genera; metabolic generalists (Desulfobulbus and Methanosarcina) have a specialist spatial distribution strongly correlated with environmental variables and metabolic specialists (Methanosaeta and Desulfobacter) have a generalist spatial distribution along an estuarine gradient that is not correlated to environmental factors.
Discussion
Here we show that within microbial genera metabolic flexibility appears to have a profound effect on the spatial distribution patterns of those genera, with members of metabolic generalist genera showing a narrower spatial distribution than metabolic specialist genera. These seemingly counter-intuitive distribution patterns suggest that microbes are distributed in ways that differ from most plants and animals and that these differences would have to be accounted for in a truly inclusive ecology. The comparison described here, using two specialist and two generalist genera from across both archaeal and bacterial domains and both the terminal oxidation processes of sulphate reduction and methanogenesis, suggests that neither phylogeny nor metabolism are major driving forces in the detected distribution of these four genera.
The distribution of the generalist methanogen genus Methanosarcina was assessed for the first time, to our knowledge, indicating a differential distribution pattern with genotypes restricted to marine, marine-brackish, brackish-freshwater and freshwater regions of the estuary. Methanosarcina strains have been isolated from various freshwater and marine environments and isolates are able to use at least two of the three methanogenic pathways (acetoclastic, hydrogenotrophic and methylotrophic methanogenesis), but with important differences in their substrate preference [33]. In an estuary, acetate concentrations are higher in freshwater regions [35] and decrease substantially due to consumption by SRB where sulfate is freely available, which is generally in all but the most freshwater sediments [36]. Methanosarcina are known to use one-carbon compounds, such as methylamines, that are more abundant in marine environments [37]. Thus, both the salinity gradient and the variable availability of different substrates (such as acetate and methylamines) along the estuarine gradient could be strong drivers of Methanosarcina's differential spatial distribution, in a similar way to the distribution of Desulfobulbus along the estuary [8], [31]. Thus, the distribution patterns of the generalist model genera are similar to those observed for estuarine macroorganisms, such as Gammarus [17], [25] and comparable to classic spatial distributions seen in other systems [16].
Desulfobacter distribution was also assessed for the first time, to our knowledge. Some bands appear to fluctuate along the estuary, but no correlation was found with the environmental variables, although some weak site-related clustering was observed (Figure 2D). Thus, the only supportable conclusion is that all genotypes were present all along the estuary. Such an undifferentiated distribution pattern, also seen with Methanosaeta, may be a general property of metabolically specialist microbes. Thus, these data support the proposed hypothesis that, in contrast to traditional ecological ideas, the breadth of the spatial distribution of specialist and generalist microbes is inversely related to the breadth of their potential niche and indicate that the pattern of spatial distribution of a microbe is directly linked to its metabolic flexibility.
Whilst definitions of specialisation are varied and often context dependent [38] most ecologists define taxa according to the breadth of their niche; specialists have narrow niches and generalists have broad niches with spatial distribution often used as a proxy measure of niche breadth (e.g. [20], [21], [39]). Thus, in many cases organisms are defined as specialists or generalists based on their existing population structure but not directly on their intrinsic biological properties. Such a distribution-based definition of specialist/generalist has never been used to describe microbial populations and in this study the metabolic specialists would be classified as ecological generalists and vice versa. From an ecological perspective it is counter-intuitive that metabolic versatility should limit the spatial distribution of a species and metabolic restriction result in a broader spatial distribution [39] yet, for these four model genera in this estuary, such contrasting patterns of spatial distribution are unequivocal.
Regardless of functional group or phylogeny specific genotypes of generalists are spatially restricted to specific regions of the estuary, whereas specialists are more evenly distributed and seem generally unaffected by gross environmental variables. These data suggest that spatial differentiation in microbes occurs if individuals within a genus harbour sufficient metabolic flexibility to exploit differing conditions along an environmental gradient. Specialists, that by definition would lack the capacity to exploit differing conditions, should exhibit either a highly restricted (distribution is limited by changing environmental factors that preclude growth/survival) or a cosmopolitan distribution along a gradient (environmental factors in the gradient have little or no effect on the distribution of the specialist genera). The later seems to be the case for the specialist genera we have analysed here. This apparent paradox could be explained by the difference in terms of potential and realised niches [21], [38], [39]. We suggest that metabolic specialists are constrained by relatively few factors, usually the availability of one or two specific substrates (in this study, acetate for Methanosaeta and acetate and sulfate for Desulfobacter). As a consequence while metabolic specialists have a narrow potential metabolic niche they are able to occupy much of this niche space. If, as suggested, microbial specialists already occupy a great extent of their potential niche, there is little niche space that could be occupied by diverging strains. With little ecological space available for speciation intrageneric competition may be highly constrained and so species coexist along environmental gradients. For generalists the opposite is true, genotypes are spatially constrained within a narrow realised niche but have a much wider potential niche (see isolate data in Oakley et al. [32]) and so do have niche space that can be exploited by diverging strains. Therefore, we propose that all generalist microbial genotypes could potentially occupy additional niche space but are constrained from doing so by intense intrageneric competition. Such competition would be enhanced if environmental heterogeneity, such as temporal variation in the availability of electron donors and acceptors, was great, as appears to be the case on a on a microscopic level in many environments (e.g. [40]). Therefore, intrageneric competition, in concert with environmental heterogeneity, may effectively produce barriers to dispersal that facilitate localised adaptation and differentiation between strains. Thus, a generalist phenotype results in very different ecological outcomes in microbial communities compared to those seen in traditional ecological studies of plants and animals.
We further suggest that among the four ecological processes proposed by Hanson et al. [41] as drivers of microbial biogeography, speciation through selection by environmental variables is more relevant to generalists. In contrast, it appears that specialists are probably mainly affected by the three other processes: dispersal, mutation and drift; resulting in presumably subtle and small-scale biogeographic patterns. This challenges the primary assumption that mainly abiotic environmental factors must be accounted for to understand the ecology of microbes as there appear to be different ecological rules for different groups.
Most efforts with the aim of including microbes into our existing ecological framework has been based on using broad-brush approached to determine whether microbial communities are similar to plants and animals in their relationships and interactions with the environment. Here we show, using taxonomically focused analyses, that, in contrast to the perceived view in ecology that specialisation can be defined by spatial restriction [22], microbial metabolic specialists have a wider distribution than metabolic generalists. These results illustrate the real challenge we face in integrating microbes within ecological theory and analysis in order to build a truly unified ecology.
Materials and Methods
Ethics statement
No permissions were required to sample the estuarine sites used here as they are freely accessible to the public and not on private land.
Site description and sampling
The sample sites (see Figure S1) and sampling strategy, sediment porewater salinity, Sulfate and acetate measurements (Figure S2) and sample treatment prior to DNA and RNA extraction are described in File S1.
Nucleic acids extraction, 16S rRNA gene PCR and fingerprint analysis of the model genera
DNA and RNA were extracted from all sediment samples using the hydroxyapatite spin-column method [42], [43] and 16S rRNA gene sequences amplified as described in File S1. A new set of primers specific for Methanosarcina 16S rRNA gene (Msc214f and Msc613r; Table S1 in File S1) was designed using the web-based primer design software Primer3 (http://frodo.wi.mit.edu/primer3/) and assessed for specificity and sensitivity with ThermoPhyl ([44]; http://go.warwick.ac.uk/thermophyl). Specificity was checked empirically by amplification of Methanosarcina mazei (DSM 2053) and Methanosarcina acetivorans (DSM 2834) pure culture DNA and using several other archaeal and bacterial negative controls.
Amplified 16S rRNA-gene fragments from both DNA and RNA were analysed using DGGE fingerprinting as described in Information SI. DGGE patterns corresponded extremely well with qPCR and pyrosequence analysis of genotype distribution [32], [45]. In addition RNA-DGGE analysis required only a 2-step PCR but produced almost identical distribution patterns to those from the 3- or 4-step PCRs used in DNA-DGGE analyses, indicating that the additional steps do not introduce artefacts into the analysis. Dissimilarity matrixes were obtained from Pearson correlation and Jaccard analyses of the DGGE gels in GelComparII (Applied Maths, Belgium). Environmental variables were also converted into dissimilarity matrixes and correlations between the two matrices were analysed using Mantel and partial Mantel tests [46]. Canonical Correspondence Analysis (CCA) was used to assess the effect of the environmental gradient on the distribution of organisms as described in File S1.
Supporting Information
Map of the Colne estuary, Essex, UK showing the 11 sampling sites along the full extent of the estuary.
(TIF)
Porewater concentrations (mM) of A. chloride (salinity); B. sulphate from the 11 sampled sites along the Colne estuary. Error bars represent the standard error of the mean (SEM, n = 3).
(TIF)
Corrected mean band intensities from replicated DNA-DGGE analyses from the 11 sites (from the marine site 1 to the freshwater site 11) along the Colne estuary, Essex, UK for: A. Desulfobulbus , B. Methanosarcina , C. Methanosaeta and D. Desulfobacter . Band numbers are indicated on the left of gel images. Error bars represent the standard error of the mean (SEM, n = 3).
(TIF)
Unconstrained Canonical Correspondence Analysis tests correlating environmental variables (geographic distance (GD), chloride (Cl) and sulphate (S)) with model genera's genotypic distribution patterns from DNA-DGGE for A. Desulfobulbus , B. Methanosarcina , C. Methanosaeta and D. Desulfobacter . Geographical groupings of samples are circled and labelled.
(TIF)
Corrected mean band intensities from replicated RNA-DGGE analyses from the 11 sites (from the marine site 1 to the freshwater site 11) along the Colne estuary, Essex, UK for: A. Desulfobulbus , B. Methanosaeta . Band numbers are indicated on the left of gel images. Error bars represent the standard error of the mean (SEM, n = 3).
(TIF)
Unconstrained Canonical Correspondence Analysis tests correlating environmental variables (geographic distance (GD), chloride (Cl) and sulphate (S)) with model genera's genotypic distribution patterns from RNA-DGGE for A. Desulfobulbus , B. Methanosaeta . Geographical groupings of samples are circled and labelled.
(TIF)
Contains Table S1–S3 and detailed description of Materials and Methods used and basic results with associated references. Table S1: List of PCR primers and annealing temperatures used in this study. *TD - These PCRs were Touchdown PCRs. Initial annealing temperature is +10°C and reduces by 1°C per cycle for the first 10 cycles of the PCR. Table S2. Mantel tests correlating environmental variables (geographic distance (D), chloride (C) and sulphate (S)) with model genera's DNA-DGGE based genotypic distribution patterns (based on Band intensities, Pearson and Jaccard analyses). Table S3. Mantel tests correlating environmental variables (geographic distance (GD), chloride (C) and sulphate (S)) with model genera's RNA-DGGE based genotypic distribution patterns (based on Band intensities, Pearson and Jaccard analyses).
(DOCX)
Funding Statement
This study was performed as part of an EU Commission funded Marie Curie Excellence Grant for Teams, MicroComXT (MEXT-CT-2005-024112) to KJP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Falkowski PG, Fenchel T, Delong EF (2008) The microbial engines that drive Earth's biogeochemical cycles. Science 320: 1034–1039. [DOI] [PubMed] [Google Scholar]
- 2. Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, et al. (2007) The role of ecological theory in microbial ecology. Nat Rev Microbiol 5: 384–392. [DOI] [PubMed] [Google Scholar]
- 3. Robinson CJ, Bohannan BJM, Young VB (2010) From structure to function: The ecology of host-associated microbial communities. Microbiol Mol Biol Rev 74: 453–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA (2012) The application of ecological theory toward an understanding of the human microbiome. Science 336: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baas-Becking LGM (1934) Geobiologie of inleiding tot de milieukunde (in Dutch) The Hague, the Netherlands: W.P. van Stockum & Zoon.
- 6.Begon M, Harper JL, Townsend CR (1996) Ecology: Individuals, populations and communities. Oxford, UK: Blackwell Science Ltd.
- 7. Cermeno P, Falkowski PG (2009) Controls on diatom biogeography in the ocean. Science 325: 1539–1541. [DOI] [PubMed] [Google Scholar]
- 8. Oakley BB, Carbonero F, van der Gast CJ, Hawkins RJ, Purdy KJ (2010) Evolutionary divergence and biogeography of sympatric niche-differentiated bacterial populations. ISME J 4: 488–497. [DOI] [PubMed] [Google Scholar]
- 9. Whitaker RJ, Grogan DW, Taylor JW (2003) Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301: 976–978. [DOI] [PubMed] [Google Scholar]
- 10. Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Nat Acad Sci USA 103: 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaston KJ (2000) Global patterns in biodiversity. Nature 405: 220–227. [DOI] [PubMed] [Google Scholar]
- 13. Whittaker RJ, Willis KJ, Field R (2001) Scale and species richness: towards a general, hierarchical theory of species diversity. JBiogeog 28: 453–470. [Google Scholar]
- 14. Cohan FM (2006) Towards a conceptual and operational union of bacterial systematics, ecology, and evolution. Phil Trans Roy Soc London B 361: 1985–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Curtis TP, Sloan WT (2004) Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr Opin Microbiol 7: 221–226. [DOI] [PubMed] [Google Scholar]
- 16. MacArthur RH (1958) Population ecology of some warblers of Northeastern coniferous forests. Ecology 39: 599–619. [Google Scholar]
- 17. Spooner GM (1947) The distribution of Gammarus species in estuaries. J Mar Biol Assoc 27: 1–52. [Google Scholar]
- 18. Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, et al. (2010) The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol 8: 523–529. [DOI] [PubMed] [Google Scholar]
- 19. Leibold MA (1995) The niche concept revisited - Mechanistic models and community context. Ecology 76: 1371–1382. [Google Scholar]
- 20. Colles A, Liow LH, Prinzing A (2009) Are specialists at risk under environmental change? Neoecological, paleoecological and phylogenetic approaches. Ecol Lett 12: 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Julliard R, Clavel J, Devictor V, Jiguet F, Couvet D (2006) Spatial segregation of specialists and generalists in bird communities. Ecol Lett 9: 1237–1244. [DOI] [PubMed] [Google Scholar]
- 22. Poisot T, Bever JD, Nemri A, Thrall PH, Hochberg ME (2011) A conceptual framework for the evolution of ecological specialisation. Ecol Lett 14: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nedwell DB, Embley TM, Purdy KJ (2004) Sulphate reduction, methanogenesis and phylogenetics of the sulphate reducing bacterial communities along an estuarine gradient. Aquat Microb Ecol 37: 209–217. [Google Scholar]
- 24. Purdy KJ, Munson MA, Nedwell DB, Embley TM (2002) Comparison of the molecular diversity of the methanogenic community at the brackish and marine ends of a UK estuary. FEMS Microbiol Ecol 39: 17–21. [DOI] [PubMed] [Google Scholar]
- 25. Gaston KJ, Spicer JI (2001) The relationship between range size and niche breadth: a test using five species of Gammarus (Amphipoda). Global Ecol Biogeogr 10: 179–188. [Google Scholar]
- 26. Flynn AM (2008) Organic matter and nutrient cycling in a coastal plain estuary: Carbon, nitrogen, and phosphorus distributions, budgets, and fluxes. J Coastal Res 55: 76–94. [Google Scholar]
- 27. Laanbroek HJ, Abee T, Voogd IL (1982) Alcohol conversions by Desulfobulbus propionicus Lindhurst in the presence and absence of sulfate and hydrogen. Arch Microbiol 133: 178–184. [Google Scholar]
- 28.Kuever J, Rainey FA, Widdel F (2005) Desulfobulbus In: Brenner DJ, Krieg NR and Staley JT, editors. Bergey's Manual of Systematic Bacteriology Volume 2: The Proteobacteria; Part C The Alpha-, Beta-, Delta and Epsilonproteobacteria. New York: Springer. pp. 988–992.
- 29.Patel GB (2001) Methanosaeta In: Brenner DJ, Krieg NR and Staley JT, editors. Bergey's Manual of Systematic Bacteriology Volume 1: The Archaea and the Deeply Branching and Phototrophic Bacteria. New York: Springer. pp. 289–294.
- 30. Carbonero F, Oakley BB, Purdy KJ (2012) Genotypic distribution of a specialist model microorganism, Methanosaeta, along an estuarine gradient: Does metabolic restriction limit niche differentiation potential? Microb Ecol 63: 856–864. [DOI] [PubMed] [Google Scholar]
- 31. Hawkins RJ, Purdy KJ (2007) Genotypic distribution of an indigenous model microorganism along an estuarine gradient. FEMS Microbiol Ecol 62: 187–194. [DOI] [PubMed] [Google Scholar]
- 32. Oakley BB, Carbonero F, Dowd SE, Hawkins RH, Purdy KJ (2012) Contrasting patterns of niche partitioning between two anaerobic terminal oxidizers of organic matter. ISME J 6: 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boone DR, Mah RA (2001) Methanosarcina In: Brenner DJ, Krieg NR and Staley JT, editors. Bergey's Manual of Systematic Bacteriology Volume 1: The Archaea and the Deeply Branching and Phototrophic Bacteria. New York: Springer. pp. 269–276.
- 34.Kuever J, Rainey FA, Widdel F (2005) Desulfobacter In: Brenner DJ, Krieg NR and Staley JT, editors. Bergey's Manual of Systematic Bacteriology Volume 2: The Proteobacteria; Part C The Alpha-, Beta-, Delta and Epsilonproteobacteria. New York: Springer. pp. 961–964.
- 35.Silva S (2004) Activity and diversity of sulphate-reducing bacteria, methanogenic archaea in contrasting sediments from the River Colne estuary. Colchester, UK: University of Essex.
- 36. Purdy KJ, Munson MA, Cresswell-Maynard T, Nedwell DB, Embley TM (2003) Use of 16S rRNA-targeted oligonucleotide probes to investigate function and phylogeny of sulphate-reducing bacteria and methanogenic archaea in a UK estuary. FEMS Microbiol Ecol 44: 361–371. [DOI] [PubMed] [Google Scholar]
- 37. Fitzsimons MF, Kahni-Danon B, Dawitt M (2001) Distributions and adsorption of the methylamines in the inter-tidal sediments of an East Anglian Estuary. Environ Exp Bot 46: 225–236. [Google Scholar]
- 38. Devictor V, Clavel J, Julliard R, Lavergne S, Mouillot D, et al. (2010) Defining and measuring ecological specialization. J Appl Ecol 47: 15–25. [Google Scholar]
- 39. Kassen R (2002) The experimental evolution of specialists, generalists, and the maintenance of diversity. J Evolution Biol 15: 173–190. [Google Scholar]
- 40. Jørgensen BB (1977) Bacterial sulphate reduction within reduced microniches of oxidised marine sediments. Mar Biol 41: 7–17. [Google Scholar]
- 41. Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH (2012) Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10: 497–506. [DOI] [PubMed] [Google Scholar]
- 42. Purdy KJ (2005) Nucleic acid recovery from complex environmental samples. Meth Enzymol 397: 271–292. [DOI] [PubMed] [Google Scholar]
- 43. Purdy KJ, Embley TM, Takii S, Nedwell DB (1996) Rapid extraction of DNA and rRNA from sediments by a novel hydroxyapatite spin-column method. Appl Environ Microbiol 62: 3905–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oakley BB, Dowd SE, Purdy KJ (2011) ThermoPhyl: A software tool for designing thermodynamically and phylogenetically optimized quantitative-PCR assays. FEMS Microbiol Ecol 77: 17–27. [DOI] [PubMed] [Google Scholar]
- 45. Purdy KJ, Hurd PJ, Moya-Laraño J, Trimmer M, Oakley BB, et al. (2011) Systems biology for ecology: From molecules to ecosystems. Adv Ecolog Res 43: 88–149. [Google Scholar]
- 46. Mantel N, Fleiss JL (1980) Minimum expected cell-size requirements for the Mantel-Haenszel one-degree-of-freedom chi-square test and a related rapid procedure. Amer J Epidemiol 112: 129–134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Map of the Colne estuary, Essex, UK showing the 11 sampling sites along the full extent of the estuary.
(TIF)
Porewater concentrations (mM) of A. chloride (salinity); B. sulphate from the 11 sampled sites along the Colne estuary. Error bars represent the standard error of the mean (SEM, n = 3).
(TIF)
Corrected mean band intensities from replicated DNA-DGGE analyses from the 11 sites (from the marine site 1 to the freshwater site 11) along the Colne estuary, Essex, UK for: A. Desulfobulbus , B. Methanosarcina , C. Methanosaeta and D. Desulfobacter . Band numbers are indicated on the left of gel images. Error bars represent the standard error of the mean (SEM, n = 3).
(TIF)
Unconstrained Canonical Correspondence Analysis tests correlating environmental variables (geographic distance (GD), chloride (Cl) and sulphate (S)) with model genera's genotypic distribution patterns from DNA-DGGE for A. Desulfobulbus , B. Methanosarcina , C. Methanosaeta and D. Desulfobacter . Geographical groupings of samples are circled and labelled.
(TIF)
Corrected mean band intensities from replicated RNA-DGGE analyses from the 11 sites (from the marine site 1 to the freshwater site 11) along the Colne estuary, Essex, UK for: A. Desulfobulbus , B. Methanosaeta . Band numbers are indicated on the left of gel images. Error bars represent the standard error of the mean (SEM, n = 3).
(TIF)
Unconstrained Canonical Correspondence Analysis tests correlating environmental variables (geographic distance (GD), chloride (Cl) and sulphate (S)) with model genera's genotypic distribution patterns from RNA-DGGE for A. Desulfobulbus , B. Methanosaeta . Geographical groupings of samples are circled and labelled.
(TIF)
Contains Table S1–S3 and detailed description of Materials and Methods used and basic results with associated references. Table S1: List of PCR primers and annealing temperatures used in this study. *TD - These PCRs were Touchdown PCRs. Initial annealing temperature is +10°C and reduces by 1°C per cycle for the first 10 cycles of the PCR. Table S2. Mantel tests correlating environmental variables (geographic distance (D), chloride (C) and sulphate (S)) with model genera's DNA-DGGE based genotypic distribution patterns (based on Band intensities, Pearson and Jaccard analyses). Table S3. Mantel tests correlating environmental variables (geographic distance (GD), chloride (C) and sulphate (S)) with model genera's RNA-DGGE based genotypic distribution patterns (based on Band intensities, Pearson and Jaccard analyses).
(DOCX)