Abstract
Cauliflower mosaic virus (CaMV) is a plant pararetrovirus with a double-stranded DNA genome. It is the type member of the genus Caulimovirus in the family Caulimoviridae. CaMV is transmitted by sap inoculation and in nature by aphids in a semi-persistent manner. To investigate the patterns and timescale of CaMV migration and evolution, we sequenced and analyzed the genomes of 67 isolates of CaMV collected mostly in Greece, Iran, Turkey, and Japan together with nine published sequences. We identified the open-reading frames (ORFs) in the genomes and inferred their phylogeny. After removing recombinant sequences, we estimated the substitution rates, divergence times, and phylogeographic patterns of the virus populations. We found that recombination has been a common feature of CaMV evolution, and that ORFs I–V have a different evolutionary history from ORF VI. The ORFs have evolved at rates between 1.71 and 5.81×10−4 substitutions/site/year, similar to those of viruses with RNA or ssDNA genomes. We found four geographically confined lineages. CaMV probably spread from a single population to other parts of the world around 400–500 years ago, and is now widely distributed among Eurasian countries. Our results revealed evidence of frequent gene flow between populations in Turkey and those of its neighboring countries, with similar patterns observed for Japan and the USA. Our study represents the first report on the spatial and temporal spread of a plant pararetrovirus.
Introduction
Studies of the population genetics of plant viruses are important for understanding the evolution of virus-host interactions [1]–[3], because plant viruses sometimes adapt rapidly to new or resistant hosts [4]–[6]. Most evolutionary studies of plant viruses have focused on those with single-stranded RNA (ssRNA) genomes [3], [7]–[9], partly because many plant viruses have such genomes. Another reason for this focus is that they have error-prone RNA polymerases, and therefore evolve at a measurable rate which complicates the creation of resistant plant cultivars. Populations of plant viruses with single-stranded DNA (ssDNA) genomes have also been studied, including those of begomoviruses and mastreviruses in the family Geminiviridae, which also evolve at a measurable rate, are emergent viruses and damage many crops worldwide [10]–[15]. These reports showed that virus populations have been shaped by selection, founder effects, and recombination. On the other hand, there has been little work on the population genetics of plant viruses with double-stranded DNA (dsDNA) genomes.
Cauliflower mosaic virus (CaMV) has a dsDNA genome and is the type species of the genus Caulimovirus in the family Caulimoviridae [16]. Although it infects plants, CaMV is grouped with the hepadnaviruses of animals as a pararetrovirus because it has icosahedral virions and because its replication strategy involves an RNA intermediate [16]. CaMV is transmitted by sap inoculation, and in nature by aphids such as Brevicoryne brassicae, Myzus persicae, and at least 25 other species in a semi-persistent manner. CaMV reduces the yield and quality of brassica crops worldwide. In nature, its host range seems to be limited to plants of the family Brassicaceae, but some isolates are able to infect plants of the family Solanaceae experimentally [17].
The genome of CaMV is a circular dsDNA molecule of about 8000 nt with three short single-stranded regions: two in one strand, one in the other [18]. It has seven open reading frames (ORFs) and large and small intergenic regions [16]. Located between ORF VI and ORF I, the large intergenic region contains the pregenomic RNA 35S promoter, the RNA polyadenylation signal, and the minus-strand primer-binding site. The small intergenic region, containing the 19S promoter, is located between ORFs V and VI. The genome encodes six viral gene products that have been detected in planta. Protein P1 is the cell-to-cell movement protein, P2 is the aphid transmission factor, P3 is the virion-associated aphid transmission factor, P4 is the coat protein precursor, P5 is the polyprotein precursor of proteinase, reverse transcriptase, and ribonuclease, and P6 is the translation transactivator/viral silencing suppressor and also the major protein of the inclusion body matrix [19]. No protein encoded by ORF VII has been detected in planta and the function of this ORF is still unknown [20].
The possibility of controlling a pathogen is improved if we know when, where, and how it first became established in the host population of interest, namely its ‘centre of emergence’. This is analogous to the ‘centre of diversity’ of crop species [21]. Brassicas are major vegetable crops for human and animal consumption worldwide; most cultivars originated in South-West Eurasian countries [22]–[26]. Many plant viruses infect these crops, with Turnip mosaic virus (TuMV), Cucumber mosaic virus (CMV), and CaMV being particularly well known. We assessed the population structure of TuMV in a number of previous studies [9], [27]–[29].
Only nine full nucleotide sequences of CaMV genomes have been reported so far [30], providing insufficient data to characterize the population structure of the virus. Here, we report the genomic sequences of CaMV isolates from brassica hosts in the Eurasian region. We analyzed our 69 new sequences in combination with nine published sequences to estimate the phylogeny, the evolutionary timescale, and the degree of divergence between populations in different countries. Our analyses provide insights into the spatial and temporal evolution of several CaMV populations.
Materials and Methods
Virus isolates and host tests
We surveyed the brassica crop-producing areas of Greece, Iran, Turkey, and Japan during the growing seasons of 2001–2010. All collected samples were tested by direct double-antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) [31] using the antiserum to CaMV (BIOREBA, Switzerland). Some of the Japanese isolates were gifts from NIAS Genebank, Japan, whereas the remaining isolates were collected from private gardens and fields, with permission from owners. No specific permissions were required for the locations/activities. Our field studies did not involve endangered or protected species. Details of the CaMV isolates, their place of origin, original or common (English) host plant, year of isolation, and host type are shown in Table S1 in File S1.
All of the isolates were sap-inoculated to the Japanese Brassica rapa cv. Hakatasuwari plants and serially cloned through single lesions at least three times using chlorotic local lesions that appeared approximately 10 days after the inoculation. The biological cloning step is important because CaMV was often in mixed infections with TuMV and/or CMV, and some plants contained a mixture of CaMV strains (data not shown). Hence, there is a possibility that artificial recombination events will be detected in the sequence data. Purified CaMV isolates were propagated in Brassica rapa cv. Hakatasuwari plants. Plants infected systemically with each of the CaMV isolates were homogenized in 0.01 M potassium phosphate buffer (pH 7.0) and mechanically inoculated onto young Japanese cultivars of B. rapa cv. Hakatasuwari, and Raphanus sativus cvs. Akimasari-2go, Taibyo-sobutori and Everest, B. oleracea var. capitata cvs. Shinsei, Ryosan 2go and Soushu, B. oleracea var. botrytis cv. Snow queen, B. oleracea var. italica cv. Challenger, B. oleracea var. Gongylodes cv. Grand duke, B. napus cv. Otsubu, B. pekinensis cv. Nozaki 1-go, and B. campestris var. Narinosa cv. Tatsuai. Inoculated plants were kept for at least four weeks in a glasshouse at Saga University at 25°C.
Viral DNA and sequence data
Viral DNAs were extracted from CaMV-infected B. rapa leaves using DNeasy Plant Mini Kit (Qiagen K.K., Japan). The DNAs were amplified using high-fidelity Platinum Pfx DNA polymerase (Invitrogen, Japan). The PCR products were separated by electrophoresis in agarose gels and purified using a QIAquick Gel Extraction kit (Qiagen K.K., Japan). Sequences from each isolate were determined using at least three overlapping independent PCR products to cover the complete genome. The sequences of the PCR products or cloned fragments of adjacent regions of the genome overlapped by at least 300 nt to ensure that they were from the same genome and were not from different components of a genome mixture. Each PCR product was sequenced by primer walking in both directions using a BigDye Terminator v3.1 Cycle Sequencing Ready Reaction kit (Life Technologies, Japan) and an Applied Biosystems Genetic Analyzer DNA model 310. Ambiguous nucleotides in any sequence were checked in sequences obtained from at least three to five other independent plasmids, which were cloned into EcoR V site of plasmid pZErO-2. Sequence data were assembled using BioEdit v5.0.9 [32].
The genomic sequences of the 76 isolates were used for a range of evolutionary analyses. The genomic sequence of ID1 isolate of Horseradish latent virus (HRLV, Accession code NC_018858) was used as an outgroup because BLAST searches showed that it was most closely and consistently related to those of CaMV. We aligned all genes via the corresponding amino acid sequences using CLUSTAL X2 [33] with TRANSALIGN (kindly supplied by Georg Weiller, Australian National University). ORF I to ORF V sequences were then reassembled to form concatenated ORF I–V sequences of 5,106 nt. We discarded overlapping sequences between ORF III and ORF IV (9 nt) and between ORF IV and ORF V (23 nt). The aligned ORF VI sequences were 1,554 nt in length.
Recombination analyses
We investigated recombination in the genomic sequences using RDP [34], GENECONV [35], BOOTSCAN [36], MAXCHI [37], CHIMAERA [38], and SISCAN [39], all implemented in RDP4 [40]. We also analyzed the data using the original PHYLPRO [41], SISCAN version 2 [39]. These analyses were done using default settings for the different detection programs and a Bonferroni-corrected P-value cut-off of 0.01, and overlapping 100- and 50-nt slices. These analyses also assessed which non-recombinant sequences contained regions that were most closely related to those of the recombinant sequences, indicating the lineages that most likely provided those regions of the recombinant genomes. For simplicity, we refer to these as the ‘parental isolates’ of recombinants. To examine the impact of gaps introduced when aligning the CaMV sequences to the outgroup, we checked for evidence of recombination after aligning the CaMV with the outgroup excluded. Finally, the aligned sequences were analyzed for recombination using the Recombination Analysis Tool (RAT) [42]. This analysis compared the percentage of nucleotide similarities using a sliding window of 30 nt, allowing detection of breakpoints among sequences.
We included the recombinant genomes in our analyses of individual ORFs when there was no evidence of within-ORF recombination, but discarded recombinant genomes for our phylogenetic estimates of rates and timescales. Moreover, we discarded 192 nt and 93 nt of the 5′ and 3′ ends, respectively, from the aligned ORF VI 1,554 nt. Specifically, we discarded both of the ends from the major recombination sites that were found, and used 1,269 nt for the subsequent ORF VI analyses.
Estimation of substitution rates and divergence times
The phylogenetic relationships of the sequences and of their constituent ORFs were estimated using the Neighbor-Net method in SPLITSTREE v4.11.3 [43], and using maximum likelihood in PhyML v3 [44]. For the maximum-likelihood (ML) analysis, we used the general time-reversible (GTR) model of nucleotide substitution, with rate variation among sites modelled using a gamma distribution and a proportion of invariable sites (GTR+Γ4+I). This model was selected in R [45] using the Bayesian information criterion, which has been shown to perform well in a variety of scenarios [46]. Branch support was evaluated by bootstrap analysis based on 1000 pseudoreplicates. The maximum-likelihood trees were compared using PATRISTIC [47].
We performed Bayesian phylogenetic analyses in BEAST v1.7.5 [48] to estimate the evolutionary rate and timescale of CaMV. The sampling times of the sequences were used as calibrations for the molecular clock. We used Bayes factors to select the best-fitting molecular-clock model and coalescent prior for the tree topology and node times. We compared strict and relaxed (uncorrelated exponential and uncorrelated lognormal) molecular clocks [49] and compared five demographic models (constant population size, expansion growth, exponential growth, logistic growth, and the Bayesian skyline plot). We also tested for clocklike evolution using a regression of root-to-tip distances on viral sampling times in the software Path-O-Gen v1.3 (http://tree.bio.ed.ac.uk/software/pathogen).
Posterior distributions of parameters, including the tree, were estimated by Markov Chain Monte Carlo (MCMC) sampling. Samples were drawn every 104 MCMC steps over a total of 108 steps, with the first 10% of samples discarded as burn-in. Acceptable sampling from the posterior and convergence to the stationary distribution were checked using the diagnostic software Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer/).
To estimate substitution rates and divergence times from heterochronous sequence data, the sampling times need to have a sufficient spread in relation to the substitution rate [50]. We investigated the temporal structure in our data sets by comparing our rate estimates with those from ten date-randomized replicates. A data set was considered to have sufficient temporal structure when the mean rate estimate from the original data set was not contained in any of the 95% credibility intervals of the rates estimated from the date-randomized replicates. This follows the approach taken in previous studies [51], [52].
The spatial population dynamics of CaMV through time were inferred in BEAST using a diffusion model with discrete location states [48]. This approach uses an explicit model that describes the migration of CaMV lineages throughout their evolutionary history. The most important pairwise diffusions can be identified using Bayes factors [53]. Using SPREAD [54] and Google Earth (http://www.google.com/earth), we produced a graphical animation of the estimated spatio-temporal movements of CaMV lineages.
Demographic analyses
DnaSP v5.0 [55] was used to estimate haplotype and nucleotide diversities. Haplotype diversity refers to the frequency and number of haplotypes in the population. Nucleotide diversity estimates the average pairwise differences among sequences. Nonsynomymous (dN) and synonymous (dS) substitution (dN/dS) ratios were calculated for seven ORFs using the Pamilo-Bianchi-Li (PBL) method in MEGA v5 [56]. The program Structure v2.3.4 [57] was used to test for evidence of genetic structure among subpopulations and to identify individuals that were admixed or had migrated among populations. To select the number of clusters that best represented population structure, we performed analyses with 1 to 10 subpopulations (K = 1 to 10), sampling from 106 Markov chain steps after a burn-in of 105 steps. We identified the maximum delta-K value to determine the best-supported number of subdivisions in the populations [58].
Results
Biological characteristics of the CaMV isolates
A total of more than 1000 samples collected during the 2001–2010 growing seasons in Greece, Iran, Japan, and Turkey were tested by DAS-ELISA. About 70 plants of B. napus (oilseed rape), B. oleracea (cabbage), B. oleracea var. italica (broccoli), B. oleracea var. botrytis (cauliflower), R. sativus (radish) and other brassicas were found to be infected with CaMV. The viruses were found in commercial fields as well as in home gardens.
Brassica and Raphanus plants were systemically infected by most isolates. Although they had very minor differences in pathogenicity in Brassica and Raphanus plants, we concluded that most isolates were of a similar pathotype. In contrast, three (JPNN, JPNS1, and JPNS2) of the ten isolates collected in Japan showed very faint symptoms in both Brassica and Raphanus plants, and we call these attenuated isolates.
Genome sequences
The complete genomes of 67 CaMV isolates were sequenced in the present study. The genomes of Eurasian isolates determined in the present study were 7984–8063 nt in length, with ORF lengths of 978–984 nt (ORF I), 459–480 nt (ORF II), 390 nt (ORF III), 1458–1512 nt (ORF IV), 2025–2040 nt (ORF V), 1560–1575 nt (ORF VI), and 285–291 nt (ORF VII). Furthermore, the large intergenic regions located between ORFs VI and VII were 704–784 nt in length, whereas the small intergenic regions located between ORFs V and VI were 103–104 nt in length. All of the motifs reported for different caulimovirus-encoded proteins were found. The new genomic sequences determined in this study are available in DDBJ/EMBL/GenBank databases with accession codes AB863136–AB863202.
Patristic distance plots
We made pairwise comparisons of the maximum-likelihood trees of the individual ORFs using PATRISTIC. All pairwise plots of the distances in the trees inferred from the ORFs I, II, III, and IV gave similar patterns. This is illustrated by the plot of ORF I against ORF III distances (Figure 1A), in which the two sets of distances had a linear correlation coefficient of 0.516 (p<0.001). The plots of the ORF V distances against those of ORF I to IV showed that ORF V might have two slightly different but overlapping populations of distances (data not shown). By contrast, plots of the ORF VI distances against those of ORFs I–V, either individually (Figure 1B) or concatenated (Figure 1C), showed that there were two completely distinct lineages of ORF VI, and these were distinct from those in ORF V. Furthermore, plots of the Group A and Group B ORF VI distances against those of the concatenated ORFs I–V (Figure 1D and E) showed that the two sublineages were distinct. The patristic distances of the ORF VII tree gave much more complex patterns when plotted against those of the other ORFs. However, because ORF VII is much shorter than the other ORFs, it is possible that this apparent complexity is an artefact of sampling. Overall, the PATRISTIC plots supported concatenation of ORFs I–V for subsequent evolutionary analyses. We analyzed ORF VI separately and omitted ORF VII from our analyses.
Figure 1. Multidimensional scaling of tree-to-tree patristic distances.
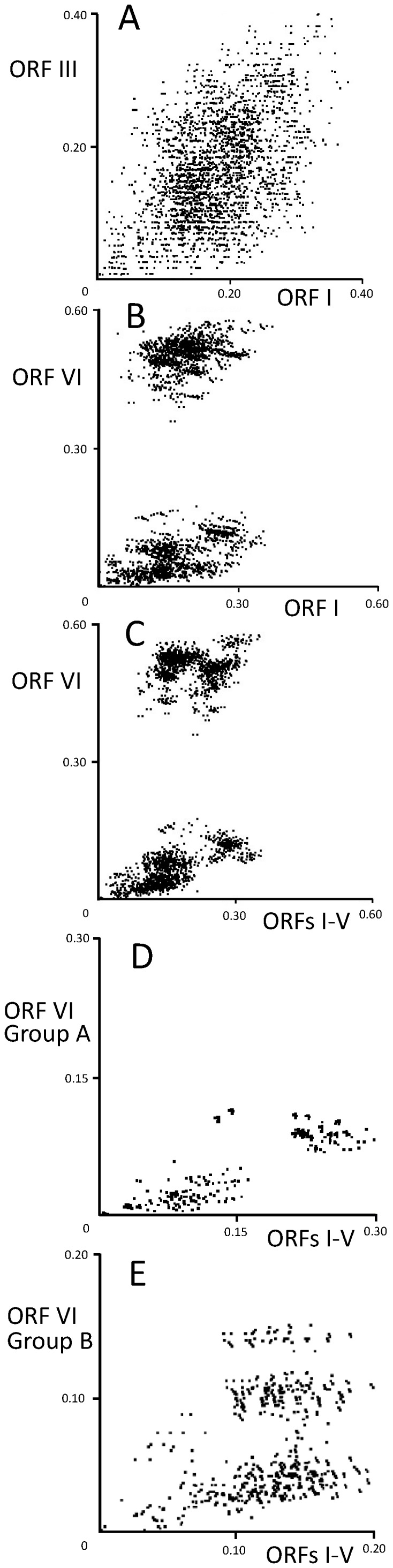
ORF I vs ORF III isolates (A); ORF I vs ORF VI isolates (B); ORFs I–V vs ORF VI isolates (C); ORFs I–V Group A vs ORF VI isolates (D); and ORFs I–V Group B vs ORF VI isolates (E).
Recombination analyses
Clear evidence of non-tree like evolution was indicated by the SplitsTree analyses (Figure S1 in File S1). These indicated that there might be recombinant regions in both ORFs I–V and ORF VI. We analyzed the protein-encoding gene sequences of 67 CaMV isolates and nine published sequences for evidence of recombination. Many clear recombination sites were detected throughout the CaMV genomes (Table 1, Figure S2 in File S1). Sites were found at 5′ and 3′ sequences of ORF VI at nt 5996 in the genomes of isolates from Iran and Japan, and at nt 7348 in Greek isolates. Some recombination sites were found in other Turkish genomes, but many were not statistically significant.
Table 1. Tentative and clear recombination sites in Cauliflower mosaic virus genomes.
| Isolate | Position (nt)a | ORF | Parental isolate | Recombination detection programb | P-valuec | |
| Major | Minor | |||||
| B29 | 3296-3946 | IV–V | TUR50 | Unknown (TUR4) | BSRSoP | 3.81×10−9 |
| 5996-7341 | VI | Unknown (TUR50) | TUR4 | RGBMCSRSo | 2.14×10−31 | |
| BBC | 3259-3946 | IV–V | TUR50 | Unknown (TUR4) | BSRSoP | 6.48×10−10 |
| 4214-5995 (UD) | V | Unknown (TUR263) | TUR50 | RGMCSo | 3.41×10−17 | |
| Cabbage S | 3298-4078 | IV–V | TUR50 | Unknown (TUR4) | GBSRSoP | 2.02×10−9 |
| 6239-74 | VI–VII | TUR285 | CM1841 | RGBMCSRSo | 7.43×10−31 | |
| CM1841 | 3259-4071 | IV–V | TUR50 | Unknown (TUR4) | BSRSoP | 3.80×10−10 |
| 4214-5995 | V–VI | Unknown (TUR263) | TUR50 | RGMC | 3.42×10−15 | |
| CMV-1 | 3259-4031 | IV–V | TUR50 | Unknown (TUR4) | SRSoP | 2.66×10−10 |
| 5887-195 | VI–VII | Unknown (TUR4) | TUR50 | RGBMCSRSo | 7.84×10−34 | |
| CRO180A | 5996-7362 | VI | TUR50 | Unknown (TUR4) | RGBMCSRSoP | 3.10×10−31 |
| D/H | 5957-82 | VI–VII | Unknown (TUR50) | TUR4 | RGBMCSRSoP | 4.21×10−35 |
| GRC83 | 7240-15 | VI–VII | GRC86D | BBC | RGBMCSRSoP | 1.32×10−26 |
| GRC84B | 7240-15 | VI–VII | GRC86D | BBC | RGBMCSRSoP | 1.37×10−24 |
| GRC86B | 4318-7239 | VI | GRC84B | TUR216 | RBMCSRSoP | 3.21×10−10 |
| GRC86D | 7348-615 | VI–VII | TUR94 | Unknown (CM1841) | RGBMCSRSo | 7.28×10−17 |
| GRC87E | 7348-615 | VI–VII | TUR94 | Unknown (CM1841) | RGBMCSRSoP | 3.18×10−13 |
| GRC87G | 7348-615 | VI–VII | TUR94 | Unknown (CM1841) | RGBMCSRSo | 4.21×10−14 |
| GRC91B | 7348-615 | VI–VII | TUR94 | Unknown (CM1841) | RGBMCSRSo | 2.63×10−15 |
| GRC92A | 7348-615 | VI–VII | TUR94 | Unknown (CM1841) | RGBMCSRSo | 7.99×10−15 |
| GRC92C | 7348-615 | VI–VII | TUR94 | Unknown (CM1841) | RGBMCSRSo | 1.83×10−14 |
| GRC92D | 7348-504- | VI–VII | TUR94 | Unknown (CM1841) | RGBMCSRSo | 1.15×10−17 |
| IRN1 | 5969-102 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 2.07×10−35 |
| IRN2 | 5969-102 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSoP | 7.68×10−35 |
| IRN3 | 5969-102 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 2.07×10−35 |
| IRN4 | 5996-195 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 4.08×10−34 |
| IRN5 | 5996-208 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 1.58×10−34 |
| IRN6 | 5944-180 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 4.70×10−34 |
| IRN7 | 5969-76 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSoP | 8.64×10−34 |
| IRN8 | 5962-208 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 4.48×10−35 |
| IRN9 | 5965-64 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 1.53×10−33 |
| IRN10 | 5967-7342 | VI | TUR50 | Unknown (TUR4) | RGBMCSRSo | 3.77×10−34 |
| IRN11 | 5969-42 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSoP | 1.44×10−34 |
| IRN12 | 5965-7342 | VI | TUR50 | Unknown (TUR4) | RGBMCSRSo | 2.59×10−34 |
| IRN13 | 5965-180 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 1.45×10−33 |
| IRN14 | 5952-99 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 2.46×10−33 |
| IRN18 | 5969-212 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSoP | 1.57×10−36 |
| IRN19 | 5965-64 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 1.18×10−35 |
| IRN21 | 5996-180 | VI–VII | TUR50 | Unknown (TUR4) | RGBMCSRSo | 2.93×10−34 |
| JPNHGB340 | 3259-3946 | IV–V | TUR50 | Unknown (TUR4) | BSRSoP | 5.12×10−9 |
| 5996-7341 | VI | Unknown (TUR4) | TUR50 | RGBMCSRSo | 5.81×10−33 | |
| JPNKWB778 | 3265-3946 | IV–V | TUR50 | Unknown (TUR4) | BSRSoP | 3.72×10−9 |
| 5965-7341 | VI | Unknown (TUR4) | TUR50 | RGBMCSRSo | 3.35×10−33 | |
| JPNM | 4214-5964 | V–VI | Unknown (TUR263) | TUR50 | RGMC | 1.26×10−15 |
| JPNN | 5996-7361 | VI | Unknown (TUR4) | TUR50 | RGBMCSRSo | 1.35×10−34 |
| JPNS1 | 3259-3946 | IV–V | TUR50 | Unknown (TUR4) | SRSoP | 9.43×10−9 |
| 5996-269 | VI–VII | Unknown (TUR50) | TUR4 | RGBMCSRSo | 1.19×10−36 | |
| JPNS2 | 3259-3946 | IV–V | TUR50 | Unknown (TUR4) | SRSoP | 5.74×10−9 |
| 5996-269 | VI–VII | Unknown (TUR50) | TUR4 | RGBMCSRSo | 1.19×10−36 | |
| JPNUV1 | 4214-5964 | V–VI | Unknown (TUR263) | TUR50 | RGMC | 1.26×10−15 |
| JPNUV26 | 4214-5964 | V–VI | Unknown (TUR263) | TUR50 | RGMC | 1.15×10−16 |
| JPNTKD762 | 3242-3989 | IV–V | TUR50 | Unknown (TUR4) | SRSoP | 2.17×10−8 |
| 5881-210 | VI–VII | Unknown (TUR50) | TUR4 | RGBMCSRSo | 5.12×10−35 | |
| NY8153 | 3296-3946 | IV–V | TUR50 | Unknown (TUR4) | BSRSoP | 3.20×10−11 |
| 2104 (UD) - 5896 (UD) | IV–VI | Unknown (TUR263) | TUR50 | SRSoP | 8.95×10−13 | |
| 5909-164 | VI–VII | Unknown (TUR4) | TUR50 | RGBMCSRSo | 5.70×10−35 | |
| TUR2 | 399-1261 | I–II | TUR249 | TUR59 | RBSR | 2.87×10−5 |
| TUR34 | 4438-5876 | V–VI | Unknown (TUR285) | TUR278 | SRSo | 1.39×10−6 |
| TUR59 | 4511-5948 | V–VI | TUR278 | Unknown (TUR285) | MSRSo | 4.61×10−7 |
| 5996-164 | VI–VII | TUR4 | Unknown (TUR50) | RGBMCSRP | 8.65×10−34 | |
| TUR214 | 1772-2108 | III–IV | TUR2 | TUR12 | BSRSoP | 2.49×10−6 |
| TUR216 | 2832-4937 (UD) | IV–V | TUR249 | Unknown (TUR2) | BSRSo | 2.03×10−16 |
| 5324-7347 | VI | Unknown (TUR306) | GRC92D | M | 1.90×10−5 | |
| TUR220 | 5539-6357 | VI | TUR81 | Unknown (TUR285) | RGB | 5.22×10−5 |
| TUR239 | 34 (UD) -1034 | I | Unknown (TUR4) | TUR244 | RGBSRSo | 3.09×10−10 |
| 1857 (UD) -2799 | V | GRC83 | Unknown (IRN2) | SRSoP | 3.71×10−5 | |
| 4365-5326 (UD) | V–VI | TUR50 | TUR4 | BMSRSoP | 6.57×10−11 | |
| TUR289 | 471 (UD) -2485 | I–IV | TUR84 | Unknown (TUR306) | RGBMCSRSo | 2.00×10−9 |
| TUR306 | 1831-2512 | III–IV | Unknown (TUR94) | TUR84 | BSRSo | 4.52×10−9 |
| W260 | 3259-3946 | IV–V | TUR50 | Unknown (TUR4) | BSRSoP | 2.54×10−9 |
| Xinjing | 627-1661 | I–III | Unknown (IRN19) | IRN21 | RBSoP | 1.96×10−5 |
Recombination sites detected in the CaMV genomes by the recombination detection programs (listed in column 6), from the aligned sequences of the likely recombinant and its ‘parental isolates’. The nucleotide position shows locations of individual genes numbered as in Xinjing genome (AF140604). UD; Undetermined.
Recombinant isolates identified by the recombination detection programs: R (RDP), G (GENECONV), B (BOOTSCAN), M (MAXCHI), C (CHIMAERA) and SR (SISCAN) programs in RDP4, and SO (SISCAN total nucleotide site analysis) in original SISCAN version 2 and P (PHYLPRO) programs. The analyses were done using default settings and a Bonferroni-corrected P-value cut-off of 0.01 in RDP4.
The reported P-value is for the program in bold type and underlined in RDP4 and is the smallest P-value among the isolates calculated for the region in question. P-values smaller than 1.0×10−5 are listed.
Phylogenetic analyses
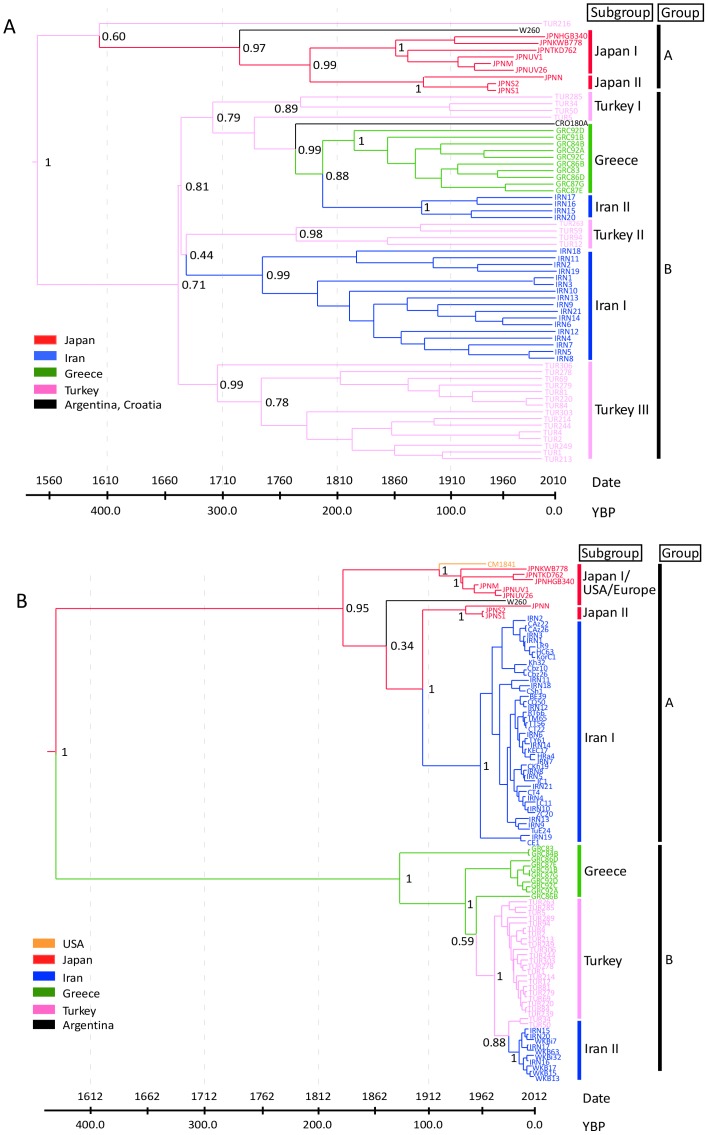
Networks and phylogenetic trees were inferred from concatenated ORFs I–V and from ORF VI. The network inferred from ORFs I–V had short internal branches (Figure S1A in File S1). In contrast, the ORF VI sequences showed two major lineages of CaMV separated by long branches (Figure S1B and Figure S3 in File S1). Each of the subgroups in ORFs I–V and ORF VI contains isolates collected in a geographically confined area.
The major differences between trees from ORFs I–V and from ORF VI are found in the relationships among the subgroups, not the subgroup membership. The maximum-likelihood bootstrapping analysis showed strong support for the various nodes in the ORF VI tree (as in Figure S3 in File S1). In contrast, the tree from ORFs I–V only had strong support at the subgroup level, with the basal nodes having support values below 30%. ORFs I–V and ORF VI yielded maximum-likelihood trees with very different relative branch lengths. In the ORF VI tree, the two basal branches span two-thirds of the mean root-to-tip distance of the tree, compared with only one-tenth in the ORFs I–V tree.
The ORF VI tree partitions most of the sequences into two major groups: Group A consists of Iranian and Japanese/North American/European subgroups, and Group B consists of Greek, Turkish and Iranian subgroups. Although most of the isolates from each country were placed into a single subgroup, those of Iran fell into two. Interestingly, the Iran II isolates that clustered with Turkish isolates in Group B came from the Khorasan Razavi district (see Table S1 in File S1), which is in north-eastern Iran and is not adjacent to Turkey. The topology of Group B showed a geographically hierarchical pattern of evolution, with the Turkish population diverging from the Greek population, and the Iranian population diverging from the Turkish population.
Genetic population structure
We compared the haplotype and nucleotide diversities of CaMV populations and subpopulations in each country (data not shown). The haplotype diversity in most groups exceeded 0.95. The nucleotide diversity of ORF VI from the Japanese samples in Group A was greater (0.03849) than those of Iran and USA, whereas greater diversity was found in the Greek samples in Group B (although only a small number of Greek isolates were used for these calculations). Nucleotide diversity was highest in Iran (0.06934). In ORFs I–V, nucleotide diversity was higher in Turkey (0.02776) than in Greece, Iran, or Japan. In estimating these genetic differences, we assumed that the population of each country evolved independently, although the sampling area in each country might influence our estimates.
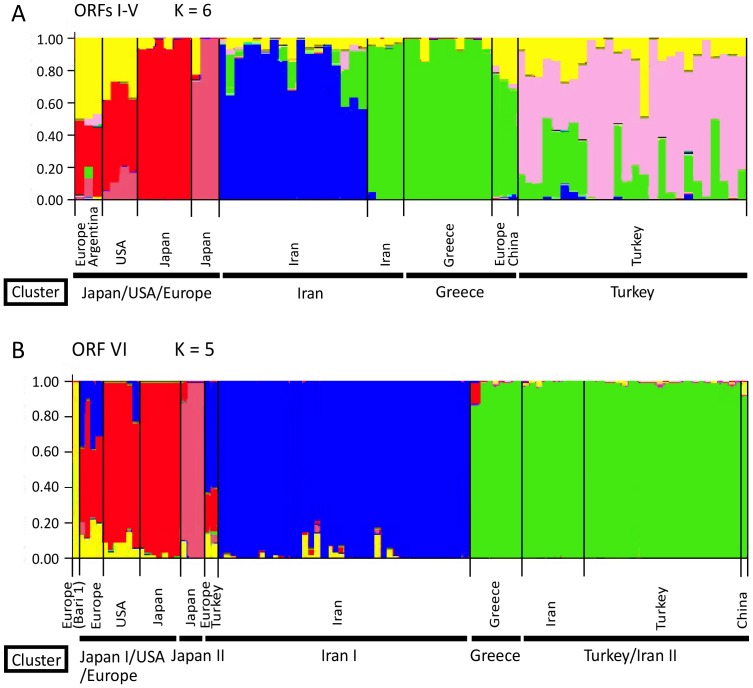
The cluster-based method implemented in Structure was used to identify individuals that were admixed or had migrated among brassica-infecting CaMV populations. Our analysis supported six subpopulations in ORFs I–V (Figure 2A) and five in ORF VI (Figure 2B). Many individuals contain substantial numbers of nucleotide polymorphisms that are apparently characteristic of ORFs I–V subpopulations, that are colour-coded in Figure 2. The Japan/USA/Europe cluster consisted of yellow, red, and dark pink subpopulations, and the Japanese isolates seemed to be divided into two subpopulations. On the other hand, the Iranian cluster consisted of yellow, green, and blue subpopulations, with the last two being dominant. Turkish clusters consisted of yellow, light pink, and green populations, with the light pink subpopulation being predominant. All of the clusters included the yellow subpopulation, and this might be ancestral the ancestral population. Most individual clusters have a predominant subpopulation in ORF VI (Figure 2B). The major subpopulations of Japan, Iran, and Turkey were red/dark pink, blue, and green, respectively. The Bari 1 isolate was part of the yellow subpopulation, which might be the ancestral isolate of the CaMV subpopulation seen in the Neighbor-Net tree (Figure S1B in File S1). Although the proportion of the yellow subpopulation was small in all clusters, the subpopulation was admixed with other individuals in all clusters. Our results suggest that CaMV became geographically segregated, but with frequent spread between regions.
Figure 2. Cluster-based analysis of population subdivision using Structure.
The results are grouped by population of origin for each individual. Each individual is represented by a column. The number of clusters is indicated by the value of K: ORFs I–V, K = 6 (A), ORF VI, K = 5 (B). The colour proportion for each bar represents the posterior probability of assignment of each individual to one of six clusters (A) and one of five clusters (B) of genetic similarity. Clusterings correspond to those shown in Figure S1 in File S1.
Evolutionary rates and timescales
We used a Bayesian phylogenetic method to estimate the evolutionary rates and timescales for the individual genomic regions. Based on the results of our PATRISTIC analyses, we analyzed a concatenated alignment of ORFs I–V and a separate alignment of ORF VI. The best-supported demographic models were exponential growth for ORFs I–V and constant size for ORF VI (Table S2 in File S1). For both data sets, a relaxed-clock model provided a better fit than the strict-clock model (Table 2). To determine whether there was temporal structure in the ORFs I–V and ORF VI data sets, we fitted a linear regression between collection date and the root-to-tip genetic divergence using Path-O-Gen v1.3 (Figure S4 in File S1). For ORFs I–V and ORF VI, we obtained respective R-squared values of −0.201 and 0.160, and respective P-values of 0.104 and 0.119. These results indicate that the relationship between collection date and sampling time is not significant, so the molecular clock hypothesis is rejected for these data sets.
Table 2. Details of the data sets used for estimation of nucleotide substitution rate and time to the most recent common ancestor for Cauliflower mosaic virus.
| Parameter | Open reading frame | |
| I–V | VI | |
| Best-fit substitution model | GTR+I+Γ4 | GTR+I+Γ4 |
| Best-fit molecular clock model | Relaxed Uncorrelated Exponential | Relaxed Uncorrelated Exponential |
| Best-fit population growth model | Exponential growth | Constant size |
| Sequence length (nt) | 5106 | 1269 |
| No. of sequences | 66 | 97 |
| Sampling date range | 1960–2010 | 1960–2012 |
| Chain length (in millions) | 100 | 100 |
| TMRCAa (years) | 491 (86–1270) | 431 (113–886) |
| Substitution rate (nt/site/year) | 1.71×10−4 (1.45×10−5–3.87×10−4) | 5.81×10−4 (2.47×10−4–9.47×10−4) |
| dN/dSb | 0.069 | 0.201 |
| No. of variable sites | 1074 | 448 |
Time to the most recent common ancestor.
Nonsynomymous (dN) and synonymous (dS) substitution (dN/dS) ratios were calculated for seven ORFs using the Pamilo-Bianchi-Li (PBL) method in MEGA v5 [56].
Nonetheless our analyses of date-randomized replicates revealed that the sampling times of ORFs I–V and ORF VI had sufficient temporal structure for calibration of the molecular clock (Figure S5 in File S1). This was indicated by the smaller 95% credibility intervals of the rate estimates from the original data set compared with the date-randomized replicates. In addition, the mean posterior rate estimates from the original data were not contained with the 95% credibility intervals of the rate estimates from the date-randomized replicates. The mean estimated substitution rates were 1.71×10−4 subs/site/year for ORFs I–V and 5.81×10−4 subs/site/year for ORF VI (Table 2). Estimates of the age of the root were 491 years for ORFs I–V and 431 years for ORF VI (Table 2, Figure 3).
Figure 3. Bayesian phylogenetic estimates from ORFs I–V and ORF VI of Cauliflower mosaic virus.
Maximum-clade–credibility trees from BEAST analyses of 66 and 97 isolates of ORFs I–V (A) and ORF VI (B), respectively. Branch colours correspond to the most probable geographic location of their descendent nodes.
Patterns of viral migration
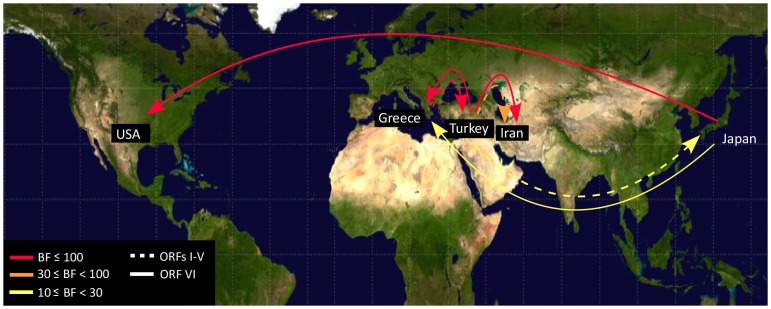
Our Bayesian phylogenetic analysis of the origin and global spread of CaMV showed strong Bayes factor (BF) support from ORFs I–V hat the virus had spread from Turkey to Greece (BF = 205) and to Iran (BF = 61) (Figure 4). There was also some support for spread from Turkey to Japan (BF = 14). The ORF VI data supported spread from Greece to Turkey (BF = 230) and to Iran (BF = 128), and from Japan to USA (BF = 112).
Figure 4. Patterns of Cauliflower mosaic virus migration jointly estimated across the two ORF regions.
ORFs I–V and ORF VI migrations are shown by solid and dashed lines. Lines connecting discrete regions indicate statistically supported ancestral state changes and their thicknesses denote statistical support. There are five categories of support. In increasing order, line thicknesses indicate 6≤BF<10 (positive support); 10≤BF<30 (strong support); 30≤BF<100 (very strong support); and BF≥100 (decisive support). Migration line was not shown when they were represented by only a single sample.
Discussion
We aimed to understand the migration dynamics and spread of CaMV in their natural hosts by utilizing over 50 years of surveillance data. Our analyses show that the samples from Europe, Japan, Middle East and USA, including the regions where various Brassicaceae were first domesticated, seems to have captured a significant sample of the global genetic diversity of CaMV. The presence of as-yet-uncollected CaMV infecting different non-brassica plant species may have biased our analysis against the detection of heterotopic processes. We recently presented a similar case study for TuMV evolution using wild orchid and brassica isolates [9].
Our comparisons of the ML trees of the individual ORFs using PATRISTIC showed that the ORFs I–V shared similar evolutionary histories, and this was different from that of ORF VI (Figure 1). The ORF I–V proteins are expressed from 35S RNA, whereas ORF VI protein is from 19S RNA. ORF VI protein is the major component of cytoplasmic inclusion bodies and the structures called viroplasms, which are thought to be ‘virion factories’. Additionally, this protein is an essential determinant of host range, affects symptom severity [20], and is known to transactivate the translation of ORFs I–V from the polycistronic 35S protein [20], [59]. Interestingly, attenuated isolates of three Japanese JPNN, JPNS1, and JPNS2 were found in the present study, and the isolates grouped together in the ORF VI tree (Figure S1B in File S1).
Recombination is an important source of genetic variation not only for CaMV [30], [60] but also for many other plant viruses [3], [13], [61]–[63]. We report several phylogenetic patterns that might have resulted from recombination in CaMV and that have not previously been found in the isolates from North America [30]. Additionally, although recombination sites have not been found in the ORF VI region [64], we found that many isolates from Europe, Iran, Japan and USA isolates were recombinants, with sites located at the 5′ and 3′ ends of ORF VI (Table 1, Figure S2 in File S1). Our results suggest that these two sites are recombination hot spots in CaMV. The recombination hot spot at the 5′ end in ORF VI is located in the middle of reported virulence/avirulence [65] and pathogenicity domains [59], [66]. The present geographical distributions of the various CaMV recombinant lineages imply that there have been complex patterns of CaMV movement throughout the world.
Our estimates of the genetic population structure have shown that there has been frequent spread between regions (Figure 2). However, the structure of ORF VI (Figure 2B) showed clear geographical segregation at the primary divergence of the CaMV population, which was not shown by ORFs I–V (Figure 2A). The same divergences were shown by the Neighbor-Net trees of the same data (Figure S1 in File S1). Our Bayesian phylogenetic analysis revealed that ORFs I–V and ORF VI support different local migration patterns for CaMV. For instance, ORFs I–V showed that CaMV migrated from Turkey to Greece and Iran, whereas ORF VI data set showed that the virus from Greece and then spread to Turkey or Iran. This suggests that there was insufficient phylogenetic signal to reveal unequivocally the complex patterns of migration in the CaMV populations in the past. The Neighbour-Net tree (Figure S1B in File S1) was estimated from ORF VI sequences that included one from the Italian Bari1 isolate. The position of this isolate in the ORF VI tree suggests that there might be a third distinct CaMV population that is yet to be sampled and sequenced. The different migration patterns in different regions might reflect characteristics of CaMV transmission and geographical barriers. CaMV is transmitted by aphids in a semi-persistent manner and they are able to only carry the virus for a short time. Mountains, deserts, country-dependent agriculture crops and growing conditions of crops may present obstacles to the spread of aphids, thus limiting the spread of the virus. Physical obstacles have also been reported to be responsible for the strain localization of Rice yellow mottle virus [67] and Tobacco vein banding mosaic virus [68].
CaMV mainly infects brassica crops, including cabbage, broccoli and cauliflower. Non-heading cabbages and kale were probably domesticated before 1000 BC in Eurasia [69], but were not taken to North America and Japan until the 17th and 19th centuries respectively. Broccoli and perhaps cauliflower originated from kale, and first appeared in the east Mediterranean. Broccoli and cauliflower spread from Italy to other European countries around the 16th to 19th centuries, prior to their introduction into North America and Japan in 19th to 20th centuries [70], [71]. Our estimate of the divergences in the tree of ORF VI shows that the primary divergence was around 450 years ago, but the divergences of the subgroup lineages occurred about 100–200 years ago (Figure 3B). Thus our well-supported estimate of the time to the most recent common ancestor of CaMV lineages based on the ORFs VI sequences is consistent with the global trade in broccoli, cauliflower and other brassica species grown as antiscorbutics, from Europe to other parts of the world. This timing also suggests that aphids were not responsible for the primary global spread of CaMV. Further global sampling of CaMV isolates is needed to confirm these results and the discrepancy between the topologies of the ORF I–V and ORF VI trees, nonetheless the age of the ancestor of CaMV fits neatly with the timescale of migration of brassica crops across the world.
We have interpreted our results while assuming that CaMV has evolved in a straightforward manner. We have concluded that the apparent difference in phylogeny between the ORFs I–V and ORF VI genes results from an inadequate phylogenetic signal in ORFs I–V, as shown by the lack of bootstrap support for the basal nodes of trees estimated from those sequences. However it is important to note that the evolution of CaMV, a pararetrovirus, may be unusual. CaMV has an unusually high recombination rate [60], and its populations have very large effective sizes [72]. Another paraeretrovirus, Banana streak virus, exists as both a virus and as endogenous elements integrated within the host genome with, probably, completely different evolutionary rates [73]. It is also noteworthy that the 35S promoter that is widely used in transgenic plant research includes much of ORF VI [74]. Thus, the unexpected should be expected in studies of the molecular phylogenetics of caulimovirids, not only in the gene sequences themselves but also in their behavior in the methods used to analyze them.
In conclusion, our study has shown that (i) recombination is common in CaMV; (ii) ORFs I–V and ORF VI of its genome show different evolutionary patterns; (iii) the ORFs are evolving at a rate in the range of 1.71–5.81×10−4 substitutions/site/year, which is similar to that of RNA and ssDNA viruses; (iv) ORF VI is the most rapidly evolving ORF; (v) there is evidence of at least four geographically confined lineages of CaMV; (vi) CaMV probably spread from a single population to other parts of the world around 400–500 years ago; (vii) CaMV is widely distributed in Eurasian countries; and (viii) there is evidence of frequent spread between Turkey and neighboring countries, and similarly between Japan and the USA. This is the first report on the spatial and temporal spread of a plant pararetrovirus.
Supporting Information
Figures S1–S5 & Tables S1–S2.
Figure S1. Phylogenetic evidence for recombination among Cauliflower mosaic virus from the Europe, Japan, Middle East (Iran and Turkey) and USA. ORFs I–V (A) and ORF VI (B). Neighbor-Net network analysis was performed using SplitsTree4. Horseradish latent virus is used as the outgroup. Formation of a reticular network rather than a single bifurcated tree is suggestive of recombination. The isolates obtained in this study are listed in Table S1 in File S1.
Figure S2. Recombination analysis by RAT plot. Each blue line represents a pairwise sequence comparison. The red curve represents the estimated proportion of recombinants at each position in the alignment. The red vertical lines denote estimated positions of recombination breakpoints, which approximately match the boundaries of the ORF VI region. The estimated nucleotide positions of the recombination sites are shown relative to the 5′ end of the genome, using numbering of the gapped aligned sequences with gaps removed (see Materials and methods). Recombination sites in parentheses are shown relative to the 5′ end of the genome using numbering of the sequence of the Xinjing isolate.
Figure S3. Maximum-likelihood tree estimated from ORF VI of 105 non-recombinant Cauliflower mosaic virus isolates. Nodes are labelled with bootstrap support percentages.
Figure S4. Regression of root-to-tip distance (inferred from Maximum-likelihood trees) against year of isolation for the gene with the smallest number of sequences in each ORF region.
Figure S5. Estimates of nucleotide substitution rates. Mean estimates and 95% credibility intervals are shown. These were estimated from 66 ORFs I–V and 97 ORF VI (see text). In each set of estimates, the first is based on the original data, whereas the remaining ten values are from date-randomized replicates. The 95% credibility intervals of the estimates from the date-randomized replicates do not overlap with the mean posterior estimate from the original data set. In addition, the lower tails of the credibility intervals are long and tend towards zero. These features suggest that there is sufficient temporal structure in the original data sets for rate estimation.
Table S1. Cauliflower mosaic virus isolates analyzed in this study.
Table S2. Detailed results from BEAST analyses of Cauliflower mosaic virus.
(PDF)
Acknowledgments
Isolates analyzed in the present study were officially imported to Japan with permission from the Japanese Plant Protection Station, Ministry of Agriculture, Forestry and Fisheries Japan. We thank A. Golnaraghi, (Saga University, Islam Azad University) and Y. Nagano (Analytical Research Center for Experimental Sciences, Saga University) for their careful technical assistance.
Funding Statement
This work was in part funded by Saga University, and supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 18405022 and 24405026. This work was in part supported by JSPS Postdoctoral Fellowships for Foreign Researchers Grant Number 21•09320 to KO and S. Farzadfar (Saga University). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gibbs AJ, Ohshima K, Phillips MJ, Gibbs MJ (2008) The prehistory of potyviruses: their initial radiation was during the dawn of agriculture. PLoS One 3: e2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sacristán S, García-Arenal F (2008) The evolution of virulence and pathogenicity in plant. Mol Plant Pathoz [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gibbs AJ, Ohshima K (2010) Potyviruses and the digital revolution. Annu Rev Phytopathol 48: 205–223. [DOI] [PubMed] [Google Scholar]
- 4. García-Arenal F, Frail A, Malpica JM (2001) Variability and genetic structure of plant virus populations. Annu Rev Phytopathol 39: 157–186. [DOI] [PubMed] [Google Scholar]
- 5. Ohshima K, Akaishi S, Kajiyama H, Koga R, Gibbs AJ (2010) Evolutionary trajectory of turnip mosaic virus populations adapting to a new host. J Gen Virol 91: 788–801. [DOI] [PubMed] [Google Scholar]
- 6. Gibbs AJ, Fargette D, García-Arenal F, Gibbs MJ (2010) Time-the emerging dimension of plant virus studies. J Gen Virol 91: 13–22. [DOI] [PubMed] [Google Scholar]
- 7. Roossinck MJ (1997) Mechanisms of plant virus evolution. Annu Rev Phytopathol 35: 191–209. [DOI] [PubMed] [Google Scholar]
- 8. García-Arenal F, Frail A, Malpica JM (2003) Variation and evolution of plant virus populations. Int Microbiol 6: 225–232. [DOI] [PubMed] [Google Scholar]
- 9. Nguyen HD, Tomitaka Y, Ho SYW, Duchêne S, Vetten H-J, et al. (2013) Turnip mosaic potyvirus probably first spread to Eurasian brassica crops from wild orchids about 1000 years ago. PLoS One 8: e55336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duffy S, Holmes EC (2008) Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus Tomato yellow leaf curl virus . J Virol 82: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harkins G, Delport W, Duffy S, Wood N, Monjane A, et al. (2009) Experimental evidence indicating that mastreviruses probably did not co-diverge with their hosts. Virol J 6: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lefeuvre P, Martin DP, Harkins G, Lemey P, Gray AJA, et al. (2010) The spread of tomato yellow leaf curl virus from the Middle East to the world. PLoS Pathog 6: e1001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Monjane AL, Harkins GW, Martin DP, Lemey P, Lefeuvre P, et al. (2011) Reconstructing the history of maize streak virus strain a dispersal to reveal diversification hot spots and its origin in southern Africa. J Virol 85: 9623–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rocha CS, Castillo-Urquiza GP, Lima ATM, Silva FN, Xavier CAD, et al. (2013) Brazilian begomovirus populations are highly recombinant, rapidly evolving, and segregated based on geographical location. J Virol 87: 5784–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodelo-Urrego M, Pagán I, González-Jara P, Betancourt M, Moreno-Letelier A, et al. (2013) Landscape heterogeneity shapes host-parasite interactions and results in apparent plant-virus codivergence. Mol Ecol 22: 2325–2340. [DOI] [PubMed] [Google Scholar]
- 16.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (2012) Virus Taxonomy: Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier/Academic Press. pp. 432–433. [Google Scholar]
- 17.Shepherd RJ (1981) CMI/AAB descriptions of plant viruses, no. 243. Commonwealth Mycological Institute, Kew, Surrey, United Kingdom.
- 18. Franck A, Guilley H, Jonard G, Richards K, Hirth L (1980) Nucleotide sequence of Cauliflower mosaic virus DNA. Cell 21: 285–294. [DOI] [PubMed] [Google Scholar]
- 19. Shockey MW, Gardner CO Jr, Melcher U, Essenberg RC (1980) Polypeptides associated with inclusion bodies from leaves of turnip infected with cauliflower mosaic virus. Virology 105: 575–581. [DOI] [PubMed] [Google Scholar]
- 20. Haas M, Bureau M, Geldreich A, Yot P, Keller M (2002) Cauliflower mosaic virus: still in the news. Mol Plant Pathol 3: 419–429. [DOI] [PubMed] [Google Scholar]
- 21.Harlan JR (1998) Distributions of agricultural origins: A global perspective. In: Damania AB, Valkoun J, Willcox G, Qualset CO, editors. Origins of Agriculture and Crop Domestication. ICARDA, Aleppo, Syria. pp. 1–2. [Google Scholar]
- 22.Crisp P (1995) Radish, Raphanus sativus (Cruciferae). In: Smartt J, Simmonds NW, editors. Evolution of Crop Plants. 2nd ed. UK, Harlow: Longman scientific & technical. pp. 86–89. [Google Scholar]
- 23.Hemingway JS (1995) Mustards, Brassica spp. and Sinapis alba (Cruciferae). In: Smartt J, Simmonds NW, editors. Evolution of Crop Plants. 2nd ed. UK, Harlow: Longman scientific & technical. pp. 82–86. [Google Scholar]
- 24.Hodgkin T (1995) Cabbages, kales, etc. Brassica oleracea (Cruciferae). In: Smartt J, Simmonds NW, editors. Evolution of Crop Plants. 2nd ed. UK, Harlow: Longman scientific & technical. pp. 76–82. [Google Scholar]
- 25.MacNaughton IH (1995) Turnip and relatives, Brassica campestris (Cruciferae). In: Smartt J, Simmonds NW, editors. Evolution of Crop Plants. 2nd ed. UK, Harlow: Longman scientific & technical. pp. 62–68. [Google Scholar]
- 26.MacNaughton IH (1995) Swedes and rapes, Brassica napus (Cruciferae). In: Smartt J, Simmonds NW, editors. Evolution of Crop Plants. 2nd ed. UK, Harlow: Longman scientific & technical. pp. 68–75. [Google Scholar]
- 27. Ohshima K, Yamaguchi Y, Hirota R, Hamamoto T, Tomimura K, et al. (2002) Molecular evolution of Turnip mosaic virus: evidence of host adaptation, genetic recombination and geographical spread. J Gen Virol 83: 1511–1521. [DOI] [PubMed] [Google Scholar]
- 28. Tomimura K, Špak J, Katis N, Jenner CE, Walsh JA, et al. (2004) Comparisons of the genetic structure of populations of Turnip mosaic virus in West and East Eurasia. Virology 330: 408–423. [DOI] [PubMed] [Google Scholar]
- 29. Tomitaka Y, Ohshima K (2006) A phylogeographical study of the Turnip mosaic virus population in East Asia reveals an ‘emergent’ lineage in Japan. Mol Ecol 15: 4437–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chenault KD, Melcher U (1994) Phylogenetic relationships reveal recombination among isolates of cauliflower mosaic virus. J Mol Evol 1994: 496–505. [DOI] [PubMed] [Google Scholar]
- 31. Clark MF, Adams AN (1977) Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol 34: 475–483. [DOI] [PubMed] [Google Scholar]
- 32. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 33. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 34. Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16: 562–563. [DOI] [PubMed] [Google Scholar]
- 35.Sawyer SA (1999) GENECONV: A computer package for the statistical detection of gene conversion. Distributed by the author. Department of Mathematics, Washington University in St. Louis. Sawyer website. Available: http://www.math.wustl.edu/~sawyer. Accessed 2013 Dec 6.
- 36. Salminen MO, Carr JK, Burke DS, McCutchan FE (1995) Identification of breakpoints in intergenotypic recombinants of HIV type 1 by Bootscanning. AIDS Res Hum Retroviruses 11: 1423–1425. [DOI] [PubMed] [Google Scholar]
- 37. Maynard Smith J (1992) Analyzing the mosaic structure of genes. J Mol Evol 34: 126–129. [DOI] [PubMed] [Google Scholar]
- 38. Posada D, Crandall KA (2001) Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA 98: 13757–13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16: 573–582. [DOI] [PubMed] [Google Scholar]
- 40. Martin DP, Lemey P, Lott M, Moulton V, Posada D, et al. (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26: 2462–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weiller GF (1998) Phylogenetic profiles: a graphical method for detecting genetic recombinations in homologous sequences. Mol Biol Evol 15: 326–335. [DOI] [PubMed] [Google Scholar]
- 42. Etherington GJ, Dicks J, Roberts IN (2005) Recombination Analysis Tool (RAT): a program for the high-throughput detection of recombination. Bioinformatics 21: 278–281. [DOI] [PubMed] [Google Scholar]
- 43. Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267. [DOI] [PubMed] [Google Scholar]
- 44. Guindon S, Gascuel O (2003) A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 45. Schliep KP (2011) Phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luo A, Qiao H, Zhang Y, Shi W, Ho SY, et al. (2010) Performance of criteria for selecting evolutionary models in phylogenetics: a comprehensive study based on simulated datasets. BMC Evol Biol 10: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fourment M, Gibbs MJ (2006) PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol Biol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lemey P, Rambaut A, Drummond AJ, Suchard MA (2009) Bayesian phylogeography finds its roots. PLoS Comput Biol 5: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drummond AJ, Pybus OG, Rambaut A, Forsberg R, Rodrigo AG (2003) Measurably evolving populations. Trends Ecol Evol 18: 481–488. [Google Scholar]
- 51. Ramsden C, Holmes EC, Charleston MA (2009) Hantavirus evolution in relation to its rodent and insectivore hosts: no evidence for codivergence. Mol Biol Evol 26: 143–153. [DOI] [PubMed] [Google Scholar]
- 52. Firth C, Kitchen A, Shapiro B, Suchard MA, Holmes EC, et al. (2010) Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol Biol Evol 27: 2038–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suchard MA, Weiss RE, Sinsheimer JS (2001) Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol 18: 1001–1013. [DOI] [PubMed] [Google Scholar]
- 54. Bielejec F, Rambaut A, Suchard MA, Lemey P (2011) SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27: 2910–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 56. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 59. Hohn T (2013) Plant pararetroviruses: interactions of cauliflower mosaic virus with plants and insects. Curr. Opin. Virol 3: 1–10. [DOI] [PubMed] [Google Scholar]
- 60. Froissart R, Roze D, Uzest M, Galibert L, Blanc S, et al. (2005) Recombination every day: abundant recombination in a virus during a single multi-cellular host infection. PLoS Biol 3: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265: 218–225. [DOI] [PubMed] [Google Scholar]
- 62. Lefeuvre P, Martin DP, Hoareau M, Naze F, Delatte H, et al. (2007) Begomovirus “melting pot” in the south-west Indian Ocean islands: molecular diversity and evolution through recombination. J Gen Virol 88: 3458–3468. [DOI] [PubMed] [Google Scholar]
- 63. Ohshima K, Tomitaka Y, Wood JT, Minematsu Y, Kajiyama H, et al. (2007) Patterns of recombination in turnip mosaic virus genomic sequences indicate hotspots of recombination. J Gen Virol 88: 298–315. [DOI] [PubMed] [Google Scholar]
- 64. Farzadfar S, Pourrahim R (2013) Biological and molecular variation of Iranian Cauliflower mosaic virus (CaMV) isolates. Virus Genes 47: 347–356. [DOI] [PubMed] [Google Scholar]
- 65. Kobayashi K, Hohn T (2004) The avirulence domain of Cauliflower mosaic virus transactivator/viroplasmin is a determinant of viral virulence in susceptible hosts. Mol Plant-Microbe Interact 17: 475–483. [DOI] [PubMed] [Google Scholar]
- 66. Hapiak M, Li Y, Agama K, Swade S, Okenka G, et al. (2008) Cauliflower mosaic virus gene VI product N-terminus contains regions involved in resistance-breakage, self-association and interactions with movement protein. Virus Res 138: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Traore O, Sorho F, Pinel A, Abubakar Z, Banwo O, et al. (2005) Processes of diversification and dispersion of Rice yellow mottle virus inferred from large-scale and high-resolution phylogeographical studies. Mol Ecol 14: 2097–2110. [DOI] [PubMed] [Google Scholar]
- 68. Zhang C-L, Gao R, Wang J, Zhang G-M, Li X-D, et al. (2011) Molecular variability of Tobacco vein banding mosaic virus populations. Virus Res 158: 188–198. [DOI] [PubMed] [Google Scholar]
- 69. Katz SH, Weaver WW (2003) Encyclopedia of food and culture 2. Scribner 284. [Google Scholar]
- 70. Buck PA (1956) Origin and taxonomy of broccoli. Econ Bot 10: 250–253. [Google Scholar]
- 71. Gray AR (1982) Taxonomy and evolution of broccoli (Brassica oleracea var. italica). Econ Bot 36: 397–410. [Google Scholar]
- 72. Monsion B, Froissant R, Michalakis Y, Blanc S (2008) Large bottleneck size in Cauliflower mosaic virus populations during host plan colonization. PLoS Pathog 4: e1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gayral P, Blondin L, Guidolin O, Carreel F, Hippolyte I, et al. (2010) Evolution of Endogenous Sequences of Banana Streak Virus: What Can We Learn from Banana (Musa sp.) Evolution? J Virol 84: 7346–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Podevin N, du Jardin P (2012) Possible consequences of the overlap between the CaMV 35S promoter regions in plant transformation vectors used and the viral gene VI in transgenic plants. GM Crops and Food: Biotechnology in Agriculture and the Food Chain 3: 296–300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S5 & Tables S1–S2.
Figure S1. Phylogenetic evidence for recombination among Cauliflower mosaic virus from the Europe, Japan, Middle East (Iran and Turkey) and USA. ORFs I–V (A) and ORF VI (B). Neighbor-Net network analysis was performed using SplitsTree4. Horseradish latent virus is used as the outgroup. Formation of a reticular network rather than a single bifurcated tree is suggestive of recombination. The isolates obtained in this study are listed in Table S1 in File S1.
Figure S2. Recombination analysis by RAT plot. Each blue line represents a pairwise sequence comparison. The red curve represents the estimated proportion of recombinants at each position in the alignment. The red vertical lines denote estimated positions of recombination breakpoints, which approximately match the boundaries of the ORF VI region. The estimated nucleotide positions of the recombination sites are shown relative to the 5′ end of the genome, using numbering of the gapped aligned sequences with gaps removed (see Materials and methods). Recombination sites in parentheses are shown relative to the 5′ end of the genome using numbering of the sequence of the Xinjing isolate.
Figure S3. Maximum-likelihood tree estimated from ORF VI of 105 non-recombinant Cauliflower mosaic virus isolates. Nodes are labelled with bootstrap support percentages.
Figure S4. Regression of root-to-tip distance (inferred from Maximum-likelihood trees) against year of isolation for the gene with the smallest number of sequences in each ORF region.
Figure S5. Estimates of nucleotide substitution rates. Mean estimates and 95% credibility intervals are shown. These were estimated from 66 ORFs I–V and 97 ORF VI (see text). In each set of estimates, the first is based on the original data, whereas the remaining ten values are from date-randomized replicates. The 95% credibility intervals of the estimates from the date-randomized replicates do not overlap with the mean posterior estimate from the original data set. In addition, the lower tails of the credibility intervals are long and tend towards zero. These features suggest that there is sufficient temporal structure in the original data sets for rate estimation.
Table S1. Cauliflower mosaic virus isolates analyzed in this study.
Table S2. Detailed results from BEAST analyses of Cauliflower mosaic virus.
(PDF)