SUMMARY
Abnormal activation of Wnt/β-catenin-mediated transcription is associated with a variety of human cancers. Here we report that LATS2 inhibited oncogenic Wnt/β-catenin-mediated transcription by disrupting the β-catenin/BCL9 interaction. LATS2 directly interacted with β-catenin and to be present on Wnt target gene promoters. Mechanistically, LATS2 inhibited the interaction between BCL9 and β-catenin and subsequent recruitment of BCL9, independent of LATS2 kinase activity. LATS2 was down-regulated and inversely correlated with the levels of Wnt target genes in human colorectal cancers. Moreover, nocodazole, an anti-microtubule drug, potently induced LATS2 to suppress tumor growth in vivo by targeting β-catenin/BCL9. Our results suggest that LATS2 is not only a key tumor suppressor in human cancer, but may also be an important target for anti-cancer therapy.
INTRODUCTION
The Hippo signaling pathway plays a critical role in oncogeneis by regulating cell proliferation, epithelial-mesenchymal transition and apoptosis (Zhao et al., 2010; Wu et al., 2003; Harvey, et al., 2003; Zhao, et al., 2008). Mammalian LATS (Large Tumor Suppressor) 1 and 2, the homologs of Wts in Drosophila, are serine/threonine kinases and key components of the Hippo signaling pathway (Dong, et al., 2007; Xu et al., 1995; Justice et al., 1995; Yabuta et al., 2000). In canonical Hippo signaling, LATS 1 and 2 phosphorylate YAP and promote YAP cytoplasmic retention and degradation, resulting in inhibition of cell proliferation and oncogenesis (Huang et al., 2005; Zhao et al., 2008). While most studies have focused on canonical Hippo signaling, some studies suggest that individual components of the Hippo signaling pathway can regulate cell survival, oncogenesis, and cytokinesis independent of the Hippo-YAP cascade (Zhao et al., 2010). For example, LATS1 was found to regulate cytokinesis by inhibiting LIMK1 (Yang et al., 2004); and LATS2 bound to Mdm2 to activate p53, serving as a novel checkpoint for the maintenance of proper chromosome number (Aylon et al., 2006). Moreover, genetic studies showed that Lats1 and Lats2 may have unique functions in mouse development and oncogenesis. While Lats1-deficient mouse developed ovarian tumors and soft-tissue sarcomas (St John et al., 1999), the deletion of Lats2 in mice resulted in embryonic lethality (McPherson et al., 2004). Interestingly, Lats2-deficient mouse embryo fibroblasts (MEF) lost contact inhibition and displayed centrosome amplification and genomic instability (McPherson et al., 2004).
The Wnt/β-catenin signaling pathway plays critical roles in development, stem cell self-renewal and oncogenesis (MacDonald et al., 2009). The constitutive activation of Wnt/β-catenin signaling has been found to be associated with a variety of human cancers such as colorectal cancer, prostate cancer, and squamous cell carcinoma (SCC) (Morin et al., 1997; Korinek et al., 1997; You et al., 2002; Chen et al., 2001; Li and Wang, 2008). Wnt/β-catenin signaling promotes cell proliferation, survival and invasive growth through β-catenin/Tcf-mediated transcription (Morin et al., 1997; Korinek et al., 1997). In the absence of Wnt/β-catenin signaling, Wnt target genes are silenced by Tcf/Lef family proteins and co-repressors of Groucho/TLE1 and histone deacetylase 1 (HDAC1). Wnt signaling leads to an accumulation of cytosolic and nuclear β-catenin. Nuclear β-catenin stimulates gene transcription by recruiting chromatin remodeling complexes and co-activators (MacDonald et al., 2009). Several transcription complexes or co-activators, including BCL9 and Pygopus (PYGO), polymerase-associated factor 1 (PAF1), and SET1 (trithorax), have been identified to be recruited by β-catenin (Parker et al., 2002; Mosimann et al., 2006; Kramps et al., 2002). Importantly, BCL9 is highly expressed in human tumor tissues, and the β-catenin/BCL9 complex has been identified as an important target for cancer therapy (Takada et al., 2012). β-catenin-mediated transcription is tightly regulated by various signaling molecules and pathways. Since there is a crosstalk between Hippo and Wnt/β-catenin signaling (Zecca and Struhl, 2010; Varelas et al., 2010), we initially set out to explore how the Hippo signaling pathway regulates Wnt/β-catenin-mediated transcription. Unexpectedly, we found that LATS2 was capable of directly inhibiting β-catenin/BCL9-mediated transcription independent of the canonical Hippo-YAP signaling cascade and its associated kinase activity. LATS2 down regulation was found to be associated with the development and metastasis of human colorectal cancer and poor prognosis. Intriguingly, we found that nocodazole, a chemotherapeutic drug that inhibits the polymerization of microtubule, potently inhibited β-catenin/BCL9-mediated transcription and tumor growth by inducing LATS2. Our results suggest that LATS2 may suppress oncogenesis by targeting β-catenin/BCL9 in addition to phosphorylating YAP.
RESULTS
LATS2 inhibits Wnt/β-catenin-mediated transcription
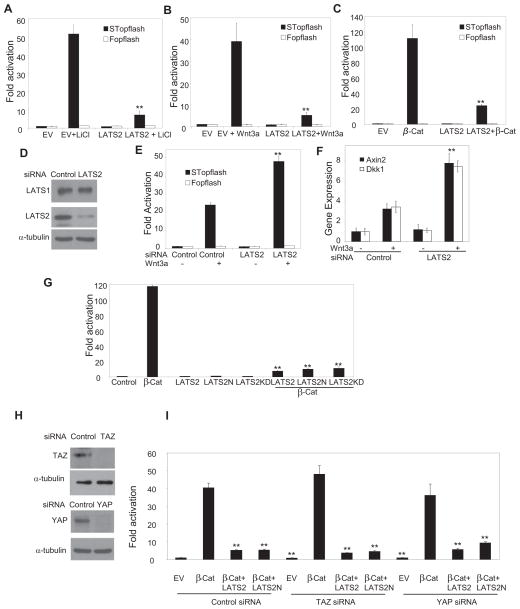
To explore whether LATS2 modulated Wnt/β-catenin-mediated transcription, we utilized a SuperTopflash (STopflash) reporter assay that measures β-catenin/Tcf-mediated transcription (Li and Wang, 2008). Over-expression of LATS2 significantly inhibited β-catenin/Tcf-mediated transcription in HEK293T cells induced by LiCl, an inhibitor of GSK-3β (Fig. 1A). Similarly, over-expression of LATS2 also inhibited β-catenin/Tcf-mediated transcription induced by Wnt-3a (Fig. 1B). Canonical Wnt ligands bind to Frizzled receptors and LRP5/6 receptors to stabilize cytosolic β-catenin, which is involved in multiple signaling cascades. The increased β-catenin translocates to the nucleus to activate gene transcription (MacDonald et al., 2009; Townsley et al., 2004). Recently, Varelas et al (2010) showed that Hippo signaling inhibited Wnt/β-catenin signaling by promoting an interaction between TAZ and DVL2, which is the upstream of β-catenin. To determine the Wnt signaling step that is inhibited by LATS2, we examined whether LATS2 could directly inhibit STopflash reporter activity induced by over-expression of β-catenin-S4A (β-Cat) which carries four alanine substitutions in the GSK3β recognition site. Unexpectedly, over-expression of LATS2 also significantly inhibited β-Cat-induced transcription, suggesting that LATS2 could act on or downstream of β-catenin (Fig. 1C). In contrast, over-expression of LATS1 did not inhibit β-Cat-induced transcription (Fig. S1A and B). Moreover, over-expression of LATS2 did not affect NF-κB or AP-1 reporter activities (Fig. S1C). To test whether endogenous LATS2 regulated β-catenin/Tcf-mediated transcription, we utilized siRNA to knock down LATS2. Western blot analysis confirmed that LATS2, but not LATS1, was efficiently depleted (Fig. 1D). The knockdown of LATS2 significantly enhanced β-catenin/Tcf-mediated transcription induced by Wnt-3a (Fig. 1E). To confirm the specificity of LATS2 siRNA, additional siRNA targeting different LATS2 sequence showed similar effects (Fig. S1D and E). The restoration of LATS2 expression abolished enhanced β-catenin/Tcf-mediated transcription induced by LATS2 siRNA (Fig. S1F). Moreover, we found that the knockdown of LATS2 also enhanced the expression of AXIN2 and DKK1, two well-known Wnt target genes, induced by Wnt-3a (Fig. 1F). To determine whether LATS2 kinase activity was required for the inhibition of Wnt/β-catenin signaling, we examined whether the kinase-dead LATS2 mutant (LATS2-KD) could inhibit β-catenin/Tcf-mediated transcription. Unexpectedly, over-expression of LATS2-KD also potently inhibited β-Cat-induced transcription. To further confirm our results, we deleted the entire kinase domain (amino acid 626 to 1042) of LATS2 and found that the N-terminal fragment (amino acid 1–625) of LATS2 (LATS2N) also strongly inhibited β-Cat-induced transcription (Fig. 1G). Since TAZ had been found to inhibit Wnt/β-catenin signaling (Varelas et al., 2010), we also examined whether the inhibition of β-catenin-mediated transcription by LATS2 was dependent on TAZ. However, the knockdown of TAZ did not affect LATS2-mediated inhibition of β-Cat-induced transcription (Fig. 1H and I). Similarly, the knockdown of YAP did not also modulate LATS2 inhibition (Fig. 1H and I). Moreover, we found that over-expression of YAP or TAZ did not affect inhibit β-catenin-mediated transcription (Fig. S1G and H). Taken together, our results suggest that LATS2 is capable of inhibiting Wnt/β-catenin-mediated transcription independent of canonical Hippo-YAP signaling.
Fig. 1. LATS2 inhibits Wnt/β-catenin-mediated transcription independent of Hippo-YAP signaling.
(A) LATS2 inhibited STopflash reporter activities induced by LiCl in 293T cells. Experiments were performed at least three times. Values are mean ± s.d. for triplicate samples from a representative experiment. **P< 0.01, Student’s t-test.
(B) LATS2 inhibited STopflash reporter activities induced by Wnt-3a in 293T cells. Values are mean ± s.d. for triplicate samples from a representative experiment. **P < 0.01, Student’s t-test.
(C) LATS2 inhibited STopflash reporter activities induced by β-Cat in 293T cells. β-Cat, β-catenin. Values are mean ± s.d. for triplicate samples from a representative experiment. **P< 0.01, Student’s t-test.
(D) LATS2, but not LATS1, was knocked down by siRNA in 293T cells.
(E) LATS2 knockdown enhanced STopflash reporter activities induced by Wnt-3a. Values are mean ± s.d. for triplicate samples from a representative experiment. **P < 0.01, Student’s t-test.
(F) LATS2 knockdown enhanced the expression of AXIN-2 and DKK1 induced by Wnt-3a. Values are mean ± s.d. for triplicate samples from a representative experiment. **P < 0.01, Student’s t-test.
(G) LATS2 inhibited β-catenin-mediated transcription independent of its kinase activity. Values are mean ± s.d. for triplicate samples from a representative experiment. **P < 0.01, Student’s t-test.
(H) SiRNA knocked down TAZ and YAP.
(I) TAZ or YAP knockdown did not affect LATS2 inhibition of β-catenin-mediated transcription. Values are mean ± s.d. for triplicate samples from a representative experiment. **P< 0.01, Student’s t-test. See also Fig. S1.
LATS2 directly interacts with β-catenin
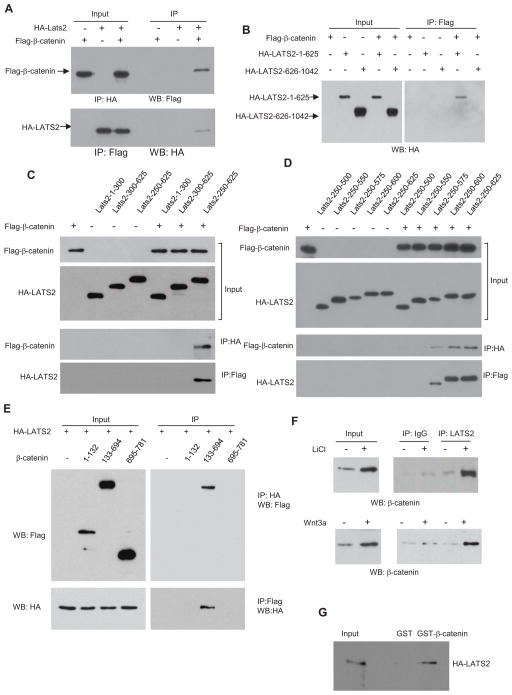
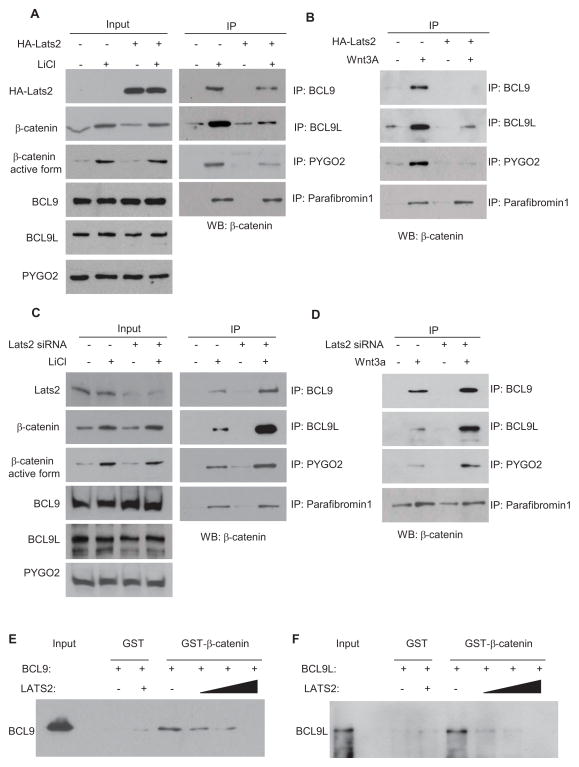
Since LATS2 potently inhibited β-catenin-mediated transcription, we first performed co-immunoprecipitation (Co-IP) to examine whether LATS2 interacted with β-catenin. Flagβ-catenin and HA-LATS2 were expressed either individually or in combination in HK293T cells. The nuclear extracts were prepared and immunoprecipitated with anti-HA antibodies. Flag-β-catenin could be detected in the immunoprecipitates with anti-HA antibodies or vice versa (Fig. 2A). However, the interaction between LATS2 and β-catenin was very weaker or could be detected in the cytoplasm (Fig. S2A). Since the kinase domain deletion mutant LATS2N retained inhibition of β-catenin-mediated transcription, we examined whether LATS2N could bind to β-catenin. Co-IP revealed that HA-LATS2N, but not the C-terminal domain (amino acid 626 to 1042) of LATS2 (HA-LATS2C), interacted with β-catenin (Fig. 2B). Further mapping revealed that amino acids 250–575 of LATS2 were sufficient for interaction with β-catenin (Fig. 2C and D). Interestingly, unlike their kinase domains, the sequence similarity between LATS1 and LATS2 in that region is extremely low (Yabuta et al., 2000). β-catenin contains the C-terminal transactivation domain and multiple arm repeats which play critical roles in interacting with and recruiting other transcription components (MacDonald et al., 2009). We found that the LATS2 did not interact with the transactivation domain of β-catenin (data not shown). To further map the amino acid region of β-catenin which was required for interaction, we generated three fragments of β-catenin and found that β-catenin-133–694 containing arm repeats was able to pull down LATS2 (Fig. 2E) or vice versa. The D162 and D164 residues of β-catenin are critical for the interaction between β-catenin and BCL9. However, the mutation of these two sites did not affect β-catenin interaction with LATS2 (Fig. S2B). To further determine whether endogenous LATS2 and β-catenin interact, we treated 293T cells with LiCl and isolated the nuclear extracts from these cells. As shown in Fig. 2F, while anti-LATS2 antibodies negligibly pulled down β-catenin without stimulation, a significant amount of β-catenin was IP-ed by anti-LATS2 antibodies, as compared with IgG, upon LiCl stimulation. Similarly, we found that Wnt-3a also induced the complex formation between β-catenin and LATS2. To further confirm our results and determine whether β-catenin directly interacted with LATS2, we generated the recombinant glutathione S-transferase (GST)-β-catenin fusion protein and in vitro translated HA-LATS2 proteins. Our GST pull-down assay revealed that β-catenin could directly bind to LATS2 in vitro (Fig. 2G). Taken together, our results suggest that LATS2 may inhibit β-catenin-mediated transcription by directly interacting with β-catenin.
Fig. 2. LATS2 interacts with β-catenin.
(A) LATS2 and β-catenin interacted when overexpressed in 293T cells.
(B) The kinase domain of LATS2 was not required for the interaction between β-catenin and LATS.
(C and D) The amino acids 250–575 of LATS2 were required for interaction with β-catenin.
(E) β-catenin-133–694 containing arm repeats interacted with LATS2. 293T cells were co-transfected with HA-LATS2 and a variety of β-catenin deletion mutants.
(F) Endogenous β-catenin interacted with LATS2 upon LiCl or Wnt-3a stimulation.
(G) GST-β-catenin fusion protein pulled down HA-LATS2 protein in vitro. See also Fig. S2.
LATS2 is down-regulated and inversely correlated with β-catenin-mediated transcription in human colorectal cancers
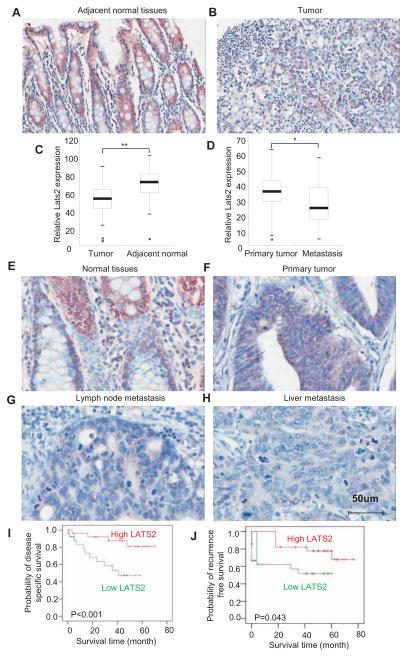
To examine whether LATS2 played an inhibitory role in human colorectal cancer development, we strictly compared LATS2 expression in human colorectal cancer tissues with matched adjacent normal colorectal tissues using human Colon Cancer Screen-13 tissue microarray consisting of 50 samples. The intensity of immunostaining was qualitatively measured using Image-Pro Plus 6.0 image analysis software. Immunostaining revealed that LATS2 expression was significantly decreased in human colorectal cancer tissues compared in matched adjacent normal colorectal tissues (Fig. 3A–C). To examine whether LATS2 expression was inversely correlated with human colorectal progression, we stained LATS2 in human primary colorectal tumors and metastatic colorectal tumors grown in the liver or lymph node. LATS2 was nearly undetectable in human colorectal cancer metastasis from liver and lymph node compared with normal and primary colorectal cancer tissues (Fig. 3D–H and Fig. S3).
Fig. 3. LATS2 expression is inversely associated with human colorectal cancer development and prognosis.
(A and B) LATS2 expression was decreased in human colorectal tumor tissues compared with adjacent normal tissues. TMA was stained with anti-LATS2 polyclonal antibodies.
(C) Quantitative measurement of LATS2 in human colorectal tumor tissues and adjacent normal tissues. The intensities of immunostaining were quantitatively measured using Image-Pro Plus 6.0 image analysis software. P < 0.01. n = 50.
(D) Quantitative measurement of LATS2 in human primary colorectal tumors and metastatic tumors from the lymph node and liver. P < 0.05. n = 30.
(E–H) LATS expression was lost in human metastatic colorectal tumors from the lymph node or liver. TMA consisting of primary tumors and metastatic tumors from lymph node and liver was stained with anti-LATS2 polyclonal antibodies.
(I) LATS2 expression was inversely associated with the survival times of patients with colorectal cancer. The published colon cancer dataset (GSE17537) containing patient follow-up information was used. Kaplan-Meier survival plot was performed for disease-specific survival and the log-rank test was applied to test the survival differences using SPSS 17.0 software. P < 0.001
(J) LATS2 expression was inversely associated with the recurrence-free survival times of patients with colorectal cancer. P < 0.05.
See also Table S1–4 and Fig. S3.
To further determine whether LATS2 is an important tumor suppressor gene in human colorectal cancer, we examined whether LATS2 downregulation was associated with patient prognosis using the colon cancer dataset (GSE17537) in Gene Expression Omnibus (GEO). Patients were dichotomized into two groups as above or below median for LATS2 expression value. Kaplan-Meier survival plot revealed that LATS2 expression was inversely associated with the survival times of patients with colorectal cancers using the log-rank test (P < 0.001; Fig. 3I). Interestingly, patients with high LATS2 expression also had longer recurrence-free survival times than those with low LATS2 expression (P < 0.043; Fig.3J).
To determine LATS2 as a negative regulator of Wnt/β-catenin-mediated transcription in human cancers, we immunostained the expression of LATS2, β-catenin, AXIN2 and MMP7 in human colorectal cancer tissue microarray consisting of 150 samples. Pearson correlation analysis showed that LATS2 was found to be inversely associated with tumor grade (Table S1). Using β-catenin as a control variable, LATS2 was also inversely associated with tumor grade (Table S2). Consistent with other studies, we found that over 85% of TMA samples were positively stained for cytoplasmic and nuclear β-catenin. Pearson correlation analysis showed that β-catenin expression was correlated with the levels of AXIN2 (R2 = 0.383, P < 0.001) and MMP7 (R2 = 0.333, P < 0.001) (Table S2). To determine whether LATS2 negatively regulated the expression of AXIN2 and MMP7 in vivo, partial correlation analysis was performed using β-catenin as a control variable. We found that LATS2 expression was inversely correlated with the levels of AXIN2 (R2 = −0.311; P < 0.01) and MMP7 (R2 = −0.259; P < 0.01) (Table S4). Taken together, our results suggest that LATS2 is a negative regulator of oncogenic β-catenin-mediated transcription in human colorectal cancers.
LATS2 inhibits BCL9/BCL9L recruitment by β-catenin
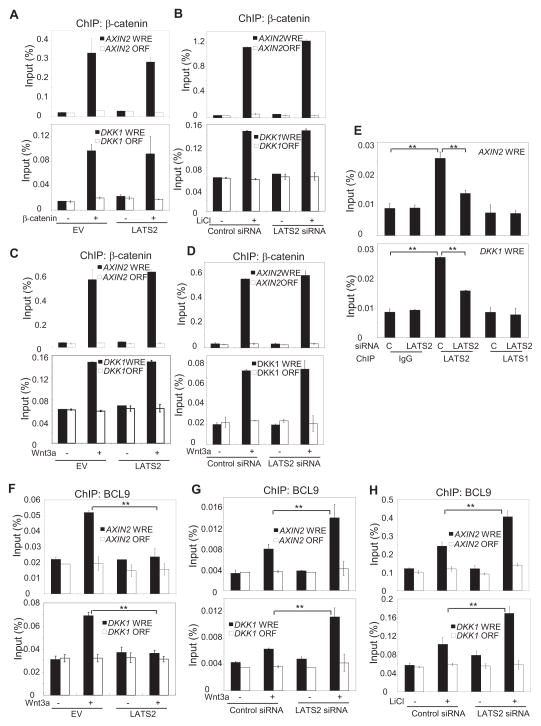
To explore how LATS2 inhibited β-catenin-mediated transcription, we performed chromatin immunoprecipitation (ChIP) assays to determine whether LATS2 would affect the recruitment of β-catenin to the Wnt-regulated enhancer (WRE). As a strict negative control, a region located in the open reading frame (ORF) was also examined. We chose AXIN2 and DKK1 for our ChIP assays because their promoters are tightly regulated by Wnt/β-catenin. As shown in Fig. 4A, over-expression of LAST2 did not inhibit the level of β-catenin on the AXIN2 and DKK1 promoters by over-expression of β-Cat. Conversely, the knockdown of LATS2 did not affect β-catenin binding to the AXIN2 and DKK1 promoters induced by LiCl (Fig. 4B). Similarly, the over-expression or knockdown of LATS2 did not modulate β-catenin binding to Axin-2 and DKK1 promoters induced by Wnt-3a (Fig. 4C and D). Interestingly, ChIP assays revealed that LATS2, but not LATS1 was present on the AXIN2 and DKK1 promoters. The knock-down of LATS2, but not LATS1, reduced its binding on the AXIN2 and DKK1 promoters (Fig. 4E). To activate transcription, β-catenin recruits an array of transcription co-activators, including BCL9 and PYGO, p300/CBP and the PAF1 complex, to the Wnt target gene promoter (Parker et al., 2002; Mosimann et al., 2006; Kramps et al., 2002; Townsley et al., 2004; Gu et al., 2009). Thus, we explored whether LATS2 might interfere with β-catenin-mediated recruitment of transcription co-activators to the Wnt target gene promoter. Over-expression or knock-down of LATS2 did not affect the recruitment of CBP/p300 to the AXIN2 and DKK1 promoters induced by Wnt-3a (data not shown). Importantly, over-expression of LATS2 inhibited recruitment of BCL9 to the AXIN2 and DKK1 promoters induced by Wnt-3a (Fig. 4F); the knock-down of LATS2 enhanced BCL9 binding to the AXIN2 and DKK1 promoters upon Wnt-3a or LiCl stimulation (Fig. 4G and H).
Fig. 4. LATS2 inhibits β-catenin-mediated recruitment of BCL9 to the Wnt target gene promoter.
(A) 293T cells were co-transfected with β-catenin and LATS2 as indicated. The ChIP-enriched DNAs were quantitatively measured using Real-time PCR with AXIN-2- and DKK1-specific primers. A region located in the open reading frame (ORF) was utilized as a negative control.
(B) Cells were transfected with LATS2 or control siRNA for 48 hr and then treated with LiCl for 2 hr. ChIP assays were performed as described in (A).
(C) Cells were transfected with LATS2 for 24 hr and then treated with Wnt-3a. ChIP assays were performed as described in (A).
(D) Cells were transfected with LATS2 or control siRNA and then treated with Wnt-3a. ChIP assays were performed as described in (A).
(E) Cells were transfected with LATS2 or control siRNA and ChIPed with anti-LATS2, anti-LATS1 or IgG. Values are mean ± s.d. for triplicate samples from a representative experiment. **P< 0.01, Student’s t-test.
(F) Cells were transfected with LATS2 or vector control and then treated with Wnt-3a. ChIP assay were performed using anti-BCL9 antibodies. Values are mean ± s.d. for triplicate samples from a representative experiment. **P< 0.01, Student’s t-test.
(G and H) Cells were transfected with LATS2 or control siRNA for 48 hr and then treated with Wnt-3a or LiCl. ChIP assays were performed using anti-BCL9 antibodies. Values are mean ± s.d. for triplicate samples from a representative experiment. **P < 0.01, Student’s t-test.
To further confirm LATS2 inhibited BCL9 recruitment by β-catenin, we also performed co-IP experiments to determine whether LATS2 interfered with the interaction between β-catenin and BCL9. As shown in Fig. 5A, upon LiCl stimulation, significant amount of β-catenin was pulled down by anti-BCL9 or anti-PYGO2 antibodies. BCL9L, a homolog of BCL9, was also found to interact with β-catenin to promote Wnt target gene transcription. We found that LiCl could also induce the interaction between β-catenin and BCL9L. However, the β-catenin levels pulled down by anti-BCL9, anti-BLC9L, or anti-PYGO2 antibodies were reduced when LATS2 was over-expressed. Similarly, LAT2 also inhibited the interaction between the active form of β-catenin, BCL9, BCL9L and PYGO2 as determined using anti-active form of β-catenin. Over-expression of LATS2 inhibited the interaction between β-catenin, BCL9, BCL9L and PYGO2 induced by Wnt-3a (Fig. 5B). In contrast, the knockdown of LATS2 enhanced β-catenin-BCL9/BCL9L complex formation upon LiCl or Wnt-3a stimulation (Fig. 5C and D). As a control, over-expression or knockdown of LATS2 did not affect the interaction between β-catenin and parafibromin 1 (Fig. 5A–D). Finally, we performed in vitro binding assays by using human GST-β-catenin fusion proteins and in vitro-translated full-length LATS2, BCL9 and BCL9L. While GST-β-catenin, but not GST, formed a complex with BCL9, the addition of LATS2 disrupted the BCL9 and β-catenin complex in a dose-dependent fashion (Fig. 5E). Similarly, LATS2 also interfered with the BCL9L and β-catenin complex in a dose-dependent fashion (Fig. 5F). We found that BCL9 and BCL9L were weakly expressed in HT29 cells compared with other colorectal cell lines, including SW620, HCT116 and SW480 (Fig. S4A). Consistent with our findings, over-expression of LATS2N could not inhibit STopflash reporter activities in HT29 cells (Fig. S4B). Of note, it was probably due to the induction of apoptosis so that we were able to overexpress the full-length LATS2 in HT29 cells. In contrast, over-expression of LATS2N or LATS2 significantly inhibited STopflash reporter activities in SW620 and SW480 and HCT116 cells (Fig. S4C and D). LATS2 and LATS2N were unable to inhibit β-catenin-mediated transcription in HCT116 cells and 293T cells in which BCL9 and BCL9L were depleted (Fig. S4E and F). Moreover, over-expression of LATS2 and LATS2N inhibited STopflash reporter activities in both HCT116 p53+/+ and HCT116 p53−/− cells, indicating that their inhibition is independent of p53 (Fig. S4G). LATS2N also significantly inhibited the expression of Wnt target genes in SW480 cells, but not in HT29 cells (Fig. S4H–J).
Fig. 5. LATS2 inhibits the interaction between β-catenin and BCL9.
(A and B) LATS2 inhibited the interaction between β-catenin and BCL9, BCL9L or PYGO2 induced by LiCl and Wnt3a.
(C and D) LATS2 knockdown enhanced the interaction between β-catenin and BCL9, BCL9L or PYGO2 induced by LiCl and Wnt3a.
(E) LATS2 inhibited the direct interaction between β-catenin and BCL9 in vitro.
(F) LATS2 inhibited the direct interaction between β-catenin and BCL9L in vitro.
See also Fig. S4.
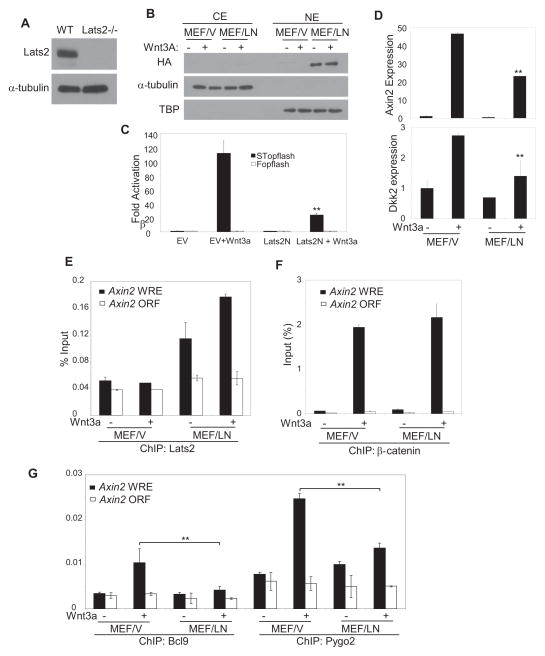
The knock-out of Lats2 in mice results in embryonic lethality (McPherson et al., 2004). To further confirm the essential role of LATS2 in the inhibition of β-catenin-mediated transcription, we utilized Lats2−/−MEF (Fig. 6A). We found that the basal levels of DKK2 were increased Lats2−/−MEFs compared with Lats2+/+MEFs (data not shown). To rule out the non-specific effects of cell immortalization, we attempted to reintroduce Lats2 back in Lats2−/− MEFs. Unfortunately, we were unable to obtain stable clones probably due to growth suppression mediated by over-expression of Lats2 through Hippo signaling. However, we were able to stably express LATS2N in Lats2−/−MEFs. Interestingly, Western blot analysis revealed that most LATS2N protein was detected in nuclear fractions in Lats2−/−MEFs expressing LATS2N (Lats2−/−MEF/LATS2N) (Fig. 6B). STopflash report assays showed that LATS2N significantly inhibited β-catenin/Tcf-mediated transcription induced by Wnt-3a (Fig. 6C). Importantly, the expressions of Axin2 and Dkk1 induced by Wnt-3a were significantly inhibited in Lats2−/−MEF/LATS2N cells as compared to control Lats2−/−MEF cells transduced with empty vector (Lats2−/−MEF/V) (Fig. 6D). ChIP assays using anti-LATS2 antibodies revealed that LATS2N was detected in the Axin2 promoter (Fig. 6E). While LATS2N did not impair β-catenin binding to the Axin2 promoter induced by Wnt-3a, the recruitment of BCL9 and PYGO2 to the Axin2 promoters induced by Wnt-3a was significantly reduced in Lats2−/−MEF/LATS2N as compared with Lats2−/−MEF/V (Fig. 6G). Moreover, we found that ectopic expression of LATS2N also inhibited human colorectal cell tumor growth in vivo by inhibiting the β-catenin/BCL9 interaction (data not shown).
Figure 6. LATS2 kinase domain deletion mutant inhibits Wnt/β-catenin-mediated transcription Lats2−/−MEFs.
(A) Confirmation of LATS2 deletion in LATS2−/− MEFs by Western blot.
(B) Lats2−/−MEFs were stably expressed with HA-LATS2N. Nuclear proteins were isolated from Lats2−/−MEFs expressing HA-LATS2N (MEF/LN) or control vector (MEF/V) and probed with anti-HA antibodies.
(C) LATS2N inhibited STopflash reporter activities induced by Wnt3a. Values are mean ± s.d. for triplicate samples from a representative experiment. **P < 0.01, Student’s t-test.
(D) LATS2N inhibited the expression of Axin2 and Dkk1 induced by Wnt3a. Values are mean ± s.d. for triplicate samples from a representative experiment. **P< 0.01, Student’s t-test.
(E and F) Both Lats2−/−MEF/V and Lats2−/−MEF/LATS2N Cells were treated with Wnt-3a and ChIP-ed with anti-LATS2 or anti-β-catenin. The ChIP-enriched DNA was measured using Real-time PCR with the Axin-2-specific primers.
(G) LATS2N inhibited the recruitment of BCL9 and PYGO2 to the Axin promoter induced by Wnt3a. Values are mean ± s.d. for triplicate samples from a representative experiment. **P< 0.01, Student’s t-test.
LATS2 is required for nocodazole-mediated inhibition of tumor growth by targeting β-catenin/BCL9
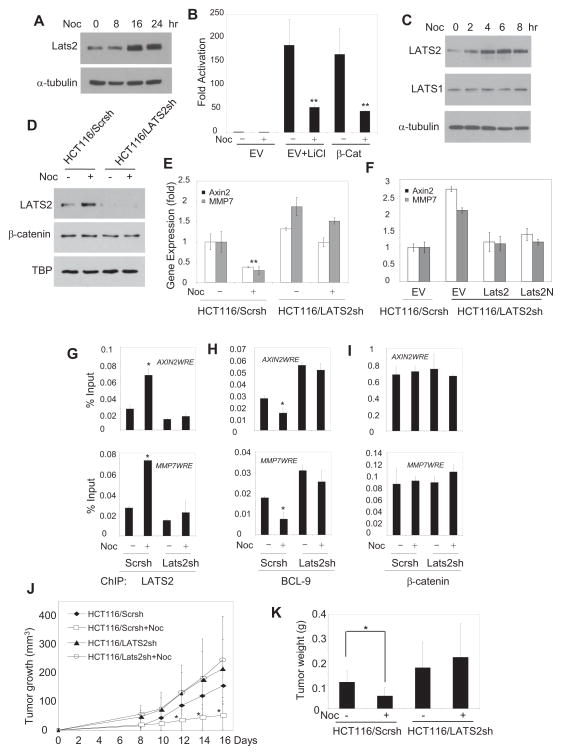
Nocodazole is a chemotherapeutic drug that exerts its effect in tumor cells by inhibiting the polymerization of microtubules. Elegant studies by Aylon et al (2006) showed that nocodazole potently induced LATS2 expression and inhibited cell growth. Based on all the above-mentioned findings, we hypothesized that nocodazole could exert its anti-tumor effect by inhibiting Wnt/β-catenin signaling via the induction of LATS2 in addition to targeting microtubules. As shown in Fig. 7A, nocodazole potently induced LATS2 expression in HEK293T cells. Consistently, nocodazole significantly inhibited β-catenin-mediated transcription induced by LiCl or β-Cat overexpression (Fig. 7B). We also found that nocodazole induced LATS2, but not LATS1, in a time-dependent manner in HCT116 cells (Fig. 7C) and SW480 cells (Fig. S5A). To test whether LATS2 was required for nocodazole-mediated inhibition of β-catenin-mediated transcription, we utilized shRNA to knock-down LATS2. Western blot confirmed that shRNA efficiently knocked down nuclear LATS2 proteins in HCT116 cells while there were no effects on nuclear β-catenin (Fig. 7D). While nocodazole significantly suppressed the expression of AXIN2 and MMP7 in HCT116 cells expressing scramble shRNA (HCT116/Scrsh), it could not affect the expression of AXIN2 and MMP7 in HCT116 cells expressing LATS2 shRNA (HCT116/LATS2sh) (Fig. 7E). The restoration of LATS2 or LATS2N in HCT116/LATS2sh cells suppressed the expression of AXIN2 and MMP7, confirming the specificity of LATS2 shRNA (Fig. 7F). Moreover, ChIP assays found that nocodazole induced LATS2 binding to the promoter of AXIN2 or MMP7 in HCT116/Scrsh cells, but not in HCT116/LATS2sh cells (Fig. 7G). Consistently, we found that the occupancy of BCL9 on the promoter of AXIN2 or MMP7 was reduced in HCT116/Scrch cells, but not in HCT116/LATS2sh cells, upon nocodazole treatment (Fig. 7H). In contrast, nocodazole did not interfere with β-catenin presence on the promoter of AXIN2 or MMP7 (Fig. 7I). The constitutive activation of Wnt/β-catenin plays a critical role in human colorectal tumor growth. Since nocodazole inhibited Wnt/β-catenin by inducing LATS2, we further examined whether LATS2 was required for nocodazole-mediated anti-tumor growth in vivo. Both HCT116/Scrsh and HCT116/LATS2sh cells were inoculated into nude mice and subsequently treated with nocodazole. Without nocodazole treatment, we found that the tumors generated from HCT116/LATS2sh cells in nude mice grew faster than those from HCT116/Scrsh cells. While nocodazole significantly suppressed tumor growth of HCT116/Scrsh cells implanted in nude mice, it was unable to inhibit tumor growth of HCT116/LATS2sh cells (Fig. 7J and K), implicating that nocodazole could target oncogenic β-catenin/BCL9 transcription in addition to other anti-tumor effects.
Fig. 7. Nocodazole induces LATS2 to blockβ-catenin/BCL9-mediated transcription and inhibits tumor growth in vivo.
(A) Nocodazole induced LATS2 in HEK293 T cells by Western blot.
(B) Nocodazole significantly suppressed β-catenin/Tcf-mediated transcription induced by LiCl or β-Cat overexpression. P < 0.01.
(C) Nocodazole induced LATS2, but not LATS1, in a time-dependent manner in HCT116 cells.
(D) The knock-down of nocodazole-induced LATS2 by shRNA. Nuclear proteins from HCT116 cells were isolated and probed with anti-LATS2 antibodies.
(E) Nocodazole suppressed the expression of AXIN2 and MMP7 dependent of LATS2. Both HCT116/Scrsh and HCT116/LATS2sh cells were treated with nocodazle and the expression of AXIN2 and MMP7 was determined by Real-time RT-PCR.P < 0.01.
(F) The restoration of LATS2 or LATS2N suppressed the expression of AXIN2 and MMP7 in HCT116/LATS2sh cells.
(G) Nocodazole induced LATS2 binding the promoter of AXIN2 or MMP7 by ChIP assays. Both HCT116/Scrsh and HCT116/LATS2sh cells were treated with nocodazole, and the promoter of AXIN2 or MMP7 was ChIP-ed with anti-LATS2 antibodies. P < 0.05.
(H) Nocodazole inhibited the occupancy of BCL-9 on the promoter of AXIN2 or MMP7 via the induction of LATS2. Both HCT116/Scrch and HCT116/LATS2sh cells were treated with nocodazole and the promoter of AXIN2 or BCL9 was ChIP-ed with anti-BCL-9 antibodies. P < 0.05.
(I) Nocodazole did not affect β-catenin binding to the promoter of AXIN2 or MMP7.
(J) LATS2 was required for nocodazole-mediated inhibition of tumor growth in nude mice. N = 7–8. P < 0.05.
(K) Comparisons of actual tumor weights at the end of nocodazole treatment. At the end of experiments, tumors from nude mice were dissected and weighted. * P < 0.05.
See also Fig. S5.
DISCUSSION
Given the critical roles of Wnt/β-catenin in development, stem cell function and oncogenesis, Wnt/β-catenin signaling is regulated by variety of signaling pathways and molecules (MacDonald et al., 2009). LATS2 as tumor suppressor has been found to be mutated or down-regulated in several human cancers (Takahashi et al., 2005; Lee, et al., 2009; Li et al., 2011). However, the molecular mechanism by which LATS2 suppresses oncogenesis is not fully understood. Recent studies suggest that LATS2 can inhibit oncogenesis by phosphorylating the transcription factors YAP and TAZ in the canonical Hippo signaling pathway. Our results reveal that LATS2 also is a nuclear repressor of β-catenin-mediated transcription by disrupting interaction between β-catenin and BCL9/PYGO2. Intriguingly, the anti-microtubule compound nocodazole can inhibit tumor growth by targeting β-catenin/BCL9 via the induction of LATS2, providing novel insights into nocodazole-mediated anti-tumor effects. Our results suggest that LATS2 may be an important target for anti-cancer therapies.
Wnt/β-catenin signaling is abnormally activated in a variety of human cancers. β-catenin/TCF-mediated transcription plays important roles in tumor development and progression. There are several negative regulators that have been found to inhibit β-catenin/TCF-mediated transcription. For example, the nemo-like kinase (NLK) has been found to inhibit β-catenin/TCF binding to DNA by phosphorylating LEF1 and TCF4 (Ishitani et al., 1999). Here we identified LATS2 as a new nuclear repressor which targeted the interaction between β-catenin and the co-activator BCL-9. Intriguingly, unlike NLK, LATS2 kinase activity was not required for the inhibition of β-catenin/TCF-mediated transcription. Biochemical and ChIP assays revealed that LATS2 inhibited the recruitment of BCL9/PYGO2, but not β-catenin to the Wnt target gene promoters. BCL9 is the co-activator of β-catenin that has been found to bind to the N-terminal arm-repeats while most of the transcription co-activators interact with the C-terminal domain of β-catenin (Kramps et al., 2002; Fiedler et al., 2008). It is possible that LATS2 might interfere with BCL-9 recruitment by interacting with β-catenin. Supporting this notion, our in vitro binding assay demonstrated that LATS2 could disrupt the BCL-9 and β-catenin complex.
Canonical Hippo signaling has been found to inhibit Wnt/β-catenin signaling through TAZ which interacts with DVL2 (Varelas et al., 2010). Both TAZ and YAP are phosphorylated by LATS1 and LATS2. However, we found that knock-down of TAZ did not affect LATS2 inhibition of β-catenin-mediated transcription. Importantly, we found that LATS2 directly interacted with β-catenin and occupied on the Wnt target gene promoters. Our results suggest that nuclear LATS2 may play a direct role in the suppression of oncogenesis. While recent studies focus on the phosphorylation of TAZ and YAP by LATS1 and LATS2 in the cytoplasm, earlier studies have shown that LATS2 was strongly detected in the nucleus (Yabuta et al., 2000). LATS2 has been found to inhibit androgen-regulated gene expression by interacting with the androgen receptor in the nucleus (Powzaniuk et al., 2004). Moreover, oncogenic H-Ras activation has been found to induce LATS2 accumulation in the nucleus. The inhibition of LATS2 expression overcomes a p53-dependent stress checkpoint and apoptosis and promotes H-Ras-mediated transformation (Aylon et al., 2009; Aylon et al., 2010). While the underlying mechanisms which control nuclear accumulation of LATS2 are unknown, it is possible that a variety of oncogenic signaling pathways may affect the subcellular localization of LATS2. In fact, we found that LATS2 can be detected in both the cytoplasm and nucleus of various tumor cell lines (data not shown). Although the nuclear localization signal (NLS) is not identified in LATS2, interestingly, the kinase domain deletion mutant LATS2N was predominantly detected in the nucleus, suggesting that NLS may be present at the N-terminal of LATS2. Alternatively, it is possible that the kinase domain of LATS2 may promote the cytoplasmic localization of LATS2. Consistent with the nuclear function of LATS2, our in vivo tumor growth model demonstrated that the LATS2N could potently suppress tumor growth by inhibiting β-catenin-dependent tumor growth.
Nocodazole is an anti-tumor agent that inhibits the polymerization of microtubule and disrupts microtubule cytoskeleton. Prolonged treatment with nocodazole induces cell cycle arrest and/or apoptosis. Nocodazole has been found to induce LATS2 to translocate to the nucleus where LATS2 binds to Mdm2 and stabilizes p53, resulting in the activation of p53-mediated transcription (Aylon et al., 2006). By identifying the crosstalk between Wnt/β-catenin and LATS2, we found that nocodazole was a potent inhibitor of Wnt/β-catenin signaling in addition to targeting microtubule cytoskeleton, providing novel insights into the molecular mechanisms of nocodazole-mediated anti-tumor effect. In human colorectal cancer, the tumor suppressor gene adenomatous polyposis coli (APC) that plays an essential role in β-catenin degradation is most commonly mutated. If APC is normally expressed, β-catenin is often found to be mutated. The outcome of APC or β-catenin mutations results in the constitutive activation of β-catenin/Tcf-mediated transcription. As a therapeutic strategy for treating human colorectal cancer, it is important to target β-catenin-mediated transcription instead of the upstream sites of the Wnt/β-catenin signaling pathway. Intriguingly, we found that nocodazole inhibited oncogenic Wnt/β-catenin signaling in a LATS2-dependent manner by disrupting the β-catenin/BCL9 complex. Supporting with this notion, the depletion of LATS2 significantly compromised the inhibition of tumor growth by nocodazole in vivo. Taken together, our results suggest that LATS2 is not only an important tumor suppressor in the development and metastasis of human colorectal cancer, but may also be an important target for anti-cancer therapy.
EXPERIMENTAL PROCEDURES
Plasmids, oligonucleotides, and antibodies
HA-LATS2, HA-LATS2KD and YAP expression vectors were described previously (Zhao et al., 2007). LATS2 deletion mutant constructs were prepared using the pcDNA3.1 (Invitorgen) or retroviral expression vector pQCXIP (Clontech) by standard PCR subcloning. LATS1 expression vector was purchased from Addgene. The plasmids SuperTopflash reporters, pActin-β-galactosidase, CMV-β-galactosidase were used as previously described (Li and Wang, 2008). Primary antibodies were obtained from the following sources: β-catenin (BD); anti-Flag and α-tubulin (Sigma-Aldrich); anti-HA (Covance); anti-TBP, anti-BCL9 and anti-BCL9L (Abcam); anti-PYGO2 and anti-YAP (Santa Cruz); anti-LATS1 and anti-LATS2 (Bethyl Laboratories); and anti-LATS2 antibody for immunostaining (McPherson et al., 2004). RT–PCR primers were purchased from Sigma: human Dkk1 (5′-agcctcttaactccttggca-3′ and 5′-tccatgtcactgggttccta-3′); mouse Axin2 (5′-CTGGCTCCAGAAGATCACAA-3′ and 5′-AGGTGACAACCAGCTCACTG-3′); mouse Dkk1 (5′-GGTGATGCAGGCACTAGAGA-3′ and 5′-AAGCCTGCACAAATGACAAG-3′). The sequences of ChIP primers were; mouse Axin2 WRE (5′-GGCTGCGCTTTGATAAGGTCCT-3′ and 5′-ATCGCGAACGGCTGCTTATTTT-3′); mouse Axin2 negative control (5′-TTACTGGCCAACGTGATGAT-3′ and 5′-GCTGTCAGGCACAGAGAGAG-3′). The siRNA oligonucleotides of LATS2 (On-TargetPlus siRNA) and YAP (On-TargetPlus SMARTpool siRNA) were purchased from Thermo Scientific. The shRNA sequence for LATS2 knockdown is 5′-GCACGCATTTTACGAATTC-3′. All other sequences of RT and ChIP primers were as previously described (Li and Wang, 2008).
Cell culture, transfection and luciferase reporter assay
Human embryonic kidney 293T, HCT116, SW480 and SCC-1 cells were cultured in DMEM containing 10% FBS and antibiotics (streptomycin and penicillin) at 37°C in a 5% CO2/95% air atmosphere. MEF cells were maintained in DMEM supplemented with 15% FBS and NE-AA (none essential amino acids) at 37°C, 5% CO2/95% air atmosphere in humidified incubator. For transient transfections, 5×105 cells were seeded into 12 well plates for 12h and then transfected using Lipofectamine 2000 reagents according to the manufacturer’s protocol (Invitrogen). The total amount of DNA in each individual well was kept constant by adding pcDNA3.1 empty vectors. Luciferase and β-galactosidase activity of total cell lysates were determined using Luc-Screen and GalactoStar kits (Tropix), respectively.
For transfection of siRNA, 293T cells were plated at 40–50% confluence and transfected with various amount of siRNA using Oligofectamine reagents (Invitrogen). For siRNA combined with luciferase reporter experiments, 100 ng of SuperTopflash and 50 ng of CMV–β-galactosidase constructs were transfected using Lipofectamine2000 (Invitrogen) 24 hours after the transfection of siRNA. To knock down LATS2 in HCT116 and SW480 cells, LATS2 shRNA or control shRNA were constructed in pLKO.1 vector (Addgene). To over-express LATS2 or LATS2 deletion mutants in SW480 and MEF cells, LATS2 were subcloned into retroiviral expression vector pQCXIP (Clontech) by standard PCR. To stably knock-down or over-express LATS2, the cells were seeded in 6 well plated and incubated overnight. Next day, respective viral particles were added evenly to the plates. After 24h, the medium was replaced with fresh medium and incubated over night. From next day onwards, the cells were selected using puromycin (2 μg/ml) for at least 3–4 days. The knockdown efficiency was determined immunoblot analysis.
Western blot analysis, Real-time RT-PCR, Co-IP, and GST-Pull down
For each sample 5 ×106 293T cells were lysed in lysis buffer for 10 min on ice. After centrifugation at 10,000g at 4°C, the supernatants were incubated with antibodies at 4 °C for 2 hours and followed by incubation with protein A or protein G–Sepharose (GE Healthcare) for 1 hour. For nuclear co-IP, the nuclear extracts were prepared from 1.5 ×106 of 293T cells, as described previously. Immunoprecipitates were washed three times with PBS and 0.1% NP40 buffer at 4 °C. For each GST-pull down assay, 2μg of GST or GST fusion β-catenin protein were incubated with glutathione beads (GE) at 4 °C for 1 hour and followed by PBS wash. The precipitates beads were then incubated in 1 ml PBS with various amount of in vitro translated BCL9 or LATS2 at 4 °C overnight and followed by washing buffer for 5 times. Proteins bound to the beads were eluted with SDS-loading buffer at 98 °C for 2 min and then subjected to SDS–PAGE. Western blot analysis was performed as described previously (Li and Wang, 2008).
Total RNA from each sample cells was purified using Trizol reagents and cDNA was synthesized with random primers using SuperScript III (Invitrogen). Quantitative RT–PCR analysis was carried out with iQ SYBR Green Supermix (BioRad) on an iCycler iQ real-time PCR detection system (BioRad). GAPDH levels were used as a loading control for real-time PCR.
ChIP assays
ChIP assays were performed using a ChIP assay kit (Upstate Biotechnology) according to the manufacturer’s protocol. Cells were incubated with a dimethyl 3,3′-dithiobispropionimidate-HCl (DTBP; Pierce) solution(5 mmol) for 30 min on ice before formaldehyde treatment. For each ChIP reaction, 2 ×106 cells were used. All resulting precipitated DNA samples were quantified by real-time PCR. Data are expressed as the percentage of input DNA. The primer sequences used for real-time PCR were as previously described (Li and Wang, 2008).
In vivo tumor growth and immunostaining
In vivo tumor growth in nude mice was performed as described previously (Li and Wang, 2008). For nocodazle treatment, nude mice (7–8 mice per group) were inoculated with HCT116/Scrsh or HCT116/LATS2sh cells for 6 days and then treated with nocodazole (10 mg/kg) every 2 days for 10 days.
TMAs were purchased from the Lifespan Bioscience. Colon Cancer Screen 12 TMA contains 140 colon adenocarcinoma cores with stage/grade and 10 normal colon cores. Colon Cancer Screen 13 TMA contains 50 colon adenocarcinoma specimens with matched adjacent normal tissues. Colon Cancer Screen 88 TMA contains 30 cases of adenocarcinoma, 30 metastasis, 9 adjacent normal tissues. For immunostaining, antigen retrieval was performed by pressure cooking in a Decloaking chamber (Biocare Medical) in citrate buffer (2.1 g/L citric acid, pH 6.0) at 120 °C for 20 min. TMA was stained with monoclonal antibodies against β-catenin (BD Biosciences), polyclonal antibodies against LATS2, polyclonal antibodies against AXIN2 and polyclonal antibodies against MMP7 at 4 °C overnight. We then incubated sections with horseradish perioxidase-labeled polymer for 60 min, detected the immunocomplexes with AEC+ chromogen (Dako EnVision System) and counterstained with hematoxylin. The intensity of immunostaining was measured by Image-Pro Plus 6.0 image analysis software (Media Cybernetics). The intensity of each image was calculated by normalizing the average integrated OD (IOD) with the total selected area of interest (AOI). Correlation between the expressions of different genes was calculated using SPSS software using SPSS 17.0 software (SPSS Inc., Chicago, Illinois, USA). For patient survival analysis, a colon cancer dataset (GSE17537) containing patient follow-up information was used (Smith et al., 2010). Patients were dichotomized into two groups as above or below median for LATS2 expression value. Kaplan-Meier survival plot was performed for both disease specific and recurrence free survival and the log-rank test was applied to test the survival differences and calculate the p value using SPSS 17.0 software.
Supplementary Material
Highlights.
LATS2 is a nuclear repressor of Wnt/β-catenin-mediated transcription
LATS2 inhibits oncogenic β-catenin signaling by disrupting β-catenin/BCL9 complex
LATS2 expression is inversely correlated with colorectal cancer development
Nocodazole, an anti-microtubule drug, can inhibit β-catenin/BCL9 by inducing LATS2
Acknowledgments
This work was supported by NICDR grants DE015964, DE17684, and DE13848 and NCI grant CA132134 to C.-Y. W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Yabuta N, Besserglick H, Buganim Y, Rotter V, Nojima H, Oren M. Silencing of the Lats2 tumor suppressor overrides a p53-dependent oncogenic stress checkpoint and enables mutant H-Ras-driven cell transformation. Oncogene. 2009;28:4469–4479. doi: 10.1038/onc.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Ofir-Rosenfeld Y, Yabuta N, Lapi E, Nojima H, Lu X, Oren M. The Lats2 tumor suppressor augments p53-mediated apoptosis by promoting the nuclear proapoptotic function of ASPP1. Genes Dev. 2010;24:2420–2429. doi: 10.1101/gad.1954410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Guttridge DC, You Z, Zhang Z, Fribley A, Mayo MW, Kitajewski J, Wang CY. Wnt-1 signaling inhibits apoptosis by activating beta-catenin/T cell factor-mediated transcription. J Cell Biol. 2001;152:87–96. doi: 10.1083/jcb.152.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M, Sánchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Müller J, Evans P, Bienz M. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell. 2008;30:507–518. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, et al. Pygo2 expands mammary progenitor cells by facilitating histone H3 K4 methylation. J Cell Biol. 2009;185:811–826. doi: 10.1083/jcb.200810133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Ishitani T, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- Jimenez-Velasco A, et al. Downregulation of the large tumor suppressor 2 (LATS2/KPM) gene is associated with poor prognosis in acute lymphoblastic leukemia. Leukemia. 2005;19:2347–2350. doi: 10.1038/sj.leu.2403974. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC −/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Kramps T, et al. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109:47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- Lee KH, Goan YG, Hsiao M, Lee CH, Jian SH, Lin JT, Chen YL, Lu PJ. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315:2529–2538. doi: 10.1016/j.yexcr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Li J, Wang CY. TBL1-TBLR1 and beta-catenin recruit each other to Wnt target-gene promoter for transcription activation and oncogenesis. Nat Cell Biol. 2008;10:160–169. doi: 10.1038/ncb1684. [DOI] [PubMed] [Google Scholar]
- Li W, Wang L, Katoh H, Liu R, Zheng P, Liu Y. Identification of a tumor suppressor relay between the FOXP3 and the Hippo pathways in breast and prostate cancers. Cancer Res. 2011;71:2162–2171. doi: 10.1158/0008-5472.CAN-10-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev, Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson JP, et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004;23:3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Parker DS, Jemison J, Cadigan KM. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development. 2002;129:2565–2576. doi: 10.1242/dev.129.11.2565. [DOI] [PubMed] [Google Scholar]
- Powzaniuk M, et al. The LATS2/KPM tumor suppressor is a negative regulator of the androgen receptor. Mol Endocrinol. 2004;18:2011–2023. doi: 10.1210/me.2004-0065. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG, Parlow AF, McGrath J, Xu T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- Takada K, Zhu D, Bird GH, Sukhdeo K, Zhao J-J, Mani M, Lemieux M, Carrasco DE, Ryan J, Horst D, Fulciniti M, Munshi NC, Xu W, Kung AL, Shivdasani RA, Walensky LD, Carrasco DR. Targeted disruption of the BCL9/β-catenin complex inhibits oncogenic Wnt signaling. Sci Transl Med. 2012;4:148ra117. doi: 10.1126/scitranslmed.3003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, et al. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin, Cancer Res. 2005;11:1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Cliffem A, Bienz M. Pygopus and Legless target Armadillo/beta-catenin to the nucleus to enable its transcriptional co-activator function. Nat Cell Biol. 2004;6:626–633. doi: 10.1038/ncb1141. [DOI] [PubMed] [Google Scholar]
- Varelas X, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yabuta N, et al. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics. 2000;63:263–270. doi: 10.1006/geno.1999.6065. [DOI] [PubMed] [Google Scholar]
- Yang X, Hao Y, Li DM, Stewart R, Insogna KL, Xu T. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat Cell Biol. 2004;6:609–617. doi: 10.1038/ncb1140. [DOI] [PubMed] [Google Scholar]
- You Z, et al. Wnt signaling promotes oncogenic transformation by inhibiting c-Myc-induced apoptosis. J Cell Biol. 2002;157:429–440. doi: 10.1083/jcb.200201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Guan, Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Struhl G. A feed-forward circuit linking wingless, fat-dachsous signaling, and the warts-hippo pathway to Drosophila wing growth. PLoS Biol. 2010;8:e1000386. doi: 10.1371/journal.pbio.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.