Abstract
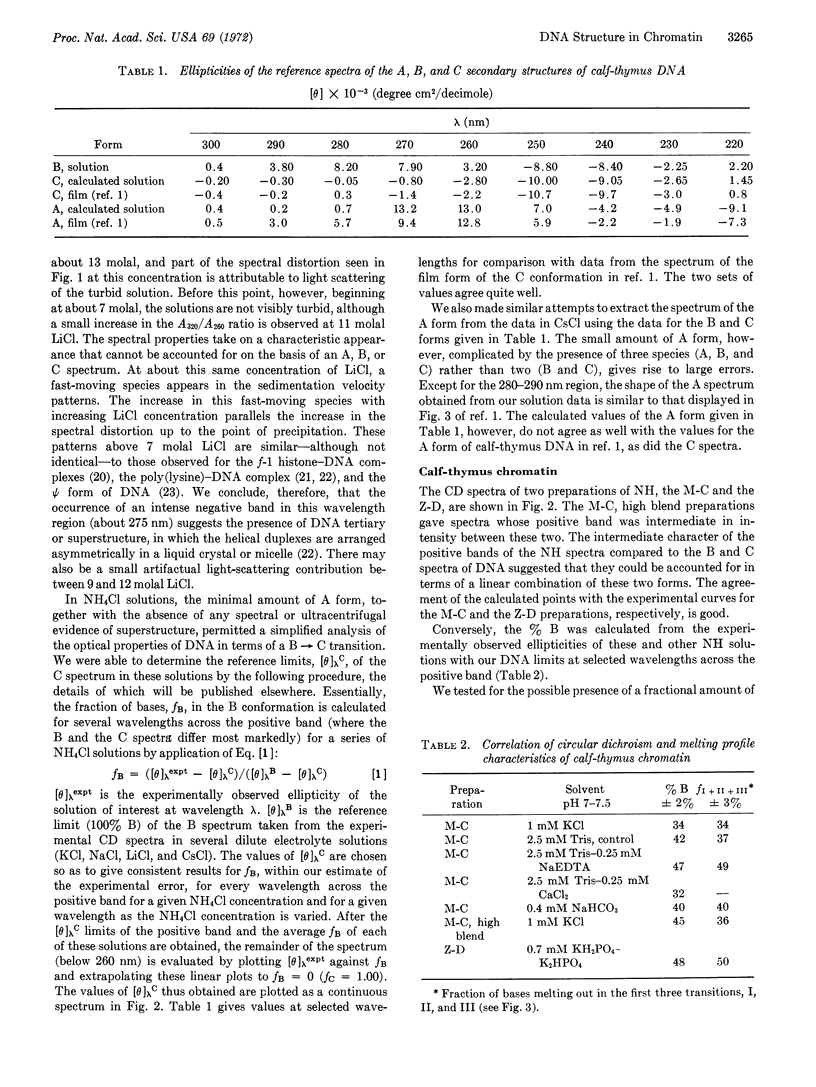
We have established empirical limits for the circular dichroism spectra appropriate for aqueous solutions of the B, C, and A forms of calf-thymus DNA and have analyzed DNA conformations in biological structures. The circular dichroism spectrum above 250 nm of purified calf-thymus chromatin can be satisfactorily accounted for as a linear combination of contributions of the B and C reference spectra without invoking higher-order structures such as supercoils. The amount of A contribution, if any, is below the limit of detection (≤4%). The fraction of bases in the B conformation depends on the method of isolation of nucleohistone, and ranges from 30-50%. The B content of a given preparation is increased by addition of a chelating agent and decreased by addition of divalent ions. More radical increases ensue upon protein removal. Nuclease treatment results in a dramatic decrease in B content. The fraction of bases melting out in the lower transitions of the complex melting profile of a given chromatin preparation corresponds to its B content. We propose a model for chromatin structure in which part of the DNA duplex is exposed or accessible to the solvent and is in the B conformation. The remainder of the base pairs and ribophosphate backbone are protected from interaction with the solvent by efficient histone coverage and are in the C conformation. Divalent ions modulate the distribution of bases between these two conformations.
Keywords: circular dichroism, B-C structure
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AMES B. N., DUBIN D. T. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J Biol Chem. 1960 Mar;235:769–775. [PubMed] [Google Scholar]
- Bonner J., Dahmus M. E., Fambrough D., Huang R. C., Marushige K., Tuan D. Y. The Biology of Isolated Chromatin: Chromosomes, biologically active in the test tube, provide a powerful tool for the study of gene action. Science. 1968 Jan 5;159(3810):47–56. doi: 10.1126/science.159.3810.47. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Cohen G., Eisenberg H. Deoxyribonucleate solutions: sedimentation in a density gradient, partial specific volumes, density and refractive index increments, and preferential interactions. Biopolymers. 1968;6(8):1077–1100. doi: 10.1002/bip.1968.360060805. [DOI] [PubMed] [Google Scholar]
- FULLER W., WILKINS M. H., WILSON H. R., HAMILTON L. D. THE MOLECULAR CONFIGURATION OF DEOXYRIBONUCLEIC ACID. IV. X-RAY DIFFRACTION STUDY OF THE A FORM. J Mol Biol. 1965 May;12:60–76. doi: 10.1016/s0022-2836(65)80282-0. [DOI] [PubMed] [Google Scholar]
- Fasman G. D., Schaffhausen B., Goldsmith L., Adler A. Conformational changes associated with f-1 histone-deoxyribonucleic acid complexes. Circular dichroism studies. Biochemistry. 1970 Jul 7;9(14):2814–2822. doi: 10.1021/bi00816a010. [DOI] [PubMed] [Google Scholar]
- HEARST J. E., VINOGRAD J. The net hydration of T-4 bacteriophage deoxyribonuecleic acid and the effect of hydration on buoyant behavior in a density gradient at equilibrium in the ultracentrifuge. Proc Natl Acad Sci U S A. 1961 Jul 15;47:1005–1014. doi: 10.1073/pnas.47.7.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEARST J. E., VINOGRAD J. The net hydration of deoxyribonucleic acid. Proc Natl Acad Sci U S A. 1961 Jun 15;47:825–830. doi: 10.1073/pnas.47.6.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes M., Garrett R. A., Gratzer W. B. Structure of nucleic acid-poly base complexes. Biochemistry. 1970 Oct 27;9(22):4410–4416. doi: 10.1021/bi00824a600. [DOI] [PubMed] [Google Scholar]
- Jordan C. F., Lerman L. S., Venable J. H. Structure and circular dichroism of DNA in concentrated polymer solutions. Nat New Biol. 1972 Mar 22;236(64):67–70. doi: 10.1038/newbio236067a0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Li H. J., Bonner J. Interaction of histone half-molecules with deoxyribonucleic acid. Biochemistry. 1971 Apr 13;10(8):1461–1470. doi: 10.1021/bi00784a030. [DOI] [PubMed] [Google Scholar]
- MARVIN D. A., SPENCER M., WILKINS M. H., HAMILTON L. D. The molecular configuration of deoxyribonucleic acid. III. X-ray diffraction study of the C form of the lithium salt. J Mol Biol. 1961 Oct;3:547–565. doi: 10.1016/s0022-2836(61)80021-1. [DOI] [PubMed] [Google Scholar]
- Maestre M. F., Wang J. C. Circular dichroism of superhelical DNA. Biopolymers. 1971 Jun;10(6):1021–1030. doi: 10.1002/bip.360100608. [DOI] [PubMed] [Google Scholar]
- Maurer H. R., Chalkley G. R. Some properties of a nuclear binding site of estradiol. J Mol Biol. 1967 Aug 14;27(3):431–441. doi: 10.1016/0022-2836(67)90049-6. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H. A super-coil model for nucleohistone. J Mol Biol. 1972 Jul 14;68(1):115–124. doi: 10.1016/0022-2836(72)90267-7. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H., Richards B. M. Super-helical model for nucleohistone. Nature. 1967 Jul 29;215(5100):508–509. doi: 10.1038/215508a0. [DOI] [PubMed] [Google Scholar]
- Permogorov V. I., Debabov V. G., Sladkova I. A., Rebentish B. A. Structure of DNA and histones in the nucleohistone. Biochim Biophys Acta. 1970 Feb 18;199(2):556–558. doi: 10.1016/0005-2787(70)90107-3. [DOI] [PubMed] [Google Scholar]
- Richards B. M., Pardon J. F. The molecular structure of nucleohistone (DNH). Exp Cell Res. 1970 Sep;62(1):184–196. doi: 10.1016/0014-4827(79)90519-6. [DOI] [PubMed] [Google Scholar]
- Shapiro J. T., Leng M., Felsenfeld G. Deoxyribonucleic acid-polylysine complexes. Structure and nucleotide specificity. Biochemistry. 1969 Aug;8(8):3219–3232. doi: 10.1021/bi00836a014. [DOI] [PubMed] [Google Scholar]
- Smart J. E., Bonner J. Selective dissociation of histones from chromatin by sodium deoxycholate. J Mol Biol. 1971 Jun 28;58(3):651–659. doi: 10.1016/0022-2836(71)90030-1. [DOI] [PubMed] [Google Scholar]
- Smart J. E., Bonner J. Studies on the role of histones in the structure of chromatin. J Mol Biol. 1971 Jun 28;58(3):661–674. doi: 10.1016/0022-2836(71)90031-3. [DOI] [PubMed] [Google Scholar]
- Tuan D. Y., Bonner J. Optical absorbance and optical rotatory dispersion studies on calf thymus nucleohistone. J Mol Biol. 1969 Oct 14;45(1):59–76. doi: 10.1016/0022-2836(69)90209-5. [DOI] [PubMed] [Google Scholar]
- Tunis-Schneider M. J., Maestre M. F. Circular dichroism spectra of oriented and unoriented deoxyribonucleic acid films--a preliminary study. J Mol Biol. 1970 Sep 28;52(3):521–541. doi: 10.1016/0022-2836(70)90417-1. [DOI] [PubMed] [Google Scholar]
- Tunis M. J., Hearst J. E. Optical rotatory dispersion of DNA in concentrated salt solutions. Biopolymers. 1968;6(8):1218–1223. doi: 10.1002/bip.1968.360060816. [DOI] [PubMed] [Google Scholar]



