Abstract
In recent studies, we demonstrated that a deletion of hha caused increased secretion of locus of enterocyte encoded adherence proteins and reduced motility of enterohemorrhagic Escherichia coli (EHEC) O157:H7. In addition to the importance of hha in positive regulation of motility, a two-component quorum sensing pathway encoded by the qseBC genes has been shown to activate bacterial motility in response to mammalian stress hormones epinephrine and norepinephrine as well as bacterially produced autoinducer-3. In this study, we compared regulatory contribution and hierarchy of hha, a member of the Hha/YmoA family of nucleoid-associated proteins, to that of qseBC in the expression of EHEC O157:H7 motility. Since norepinephrine affects motility of EHEC O157:H7 through a qseBC-encoded two-component quorum sensing signaling, we also determined whether the hha-mediated regulation of motility is affected by norepinephrine and whether this effect is qseBC dependent. We used single (Δhha or ΔqseC) and double (Δhha ΔqseC) deletion mutants to show that hha exerts a greater positive regulatory effect in comparison to qseBC on the expression of motility by EHEC O157:H7. We also show that Hha is hierarchically superior in transcriptional regulation of motility than QseBC because transcription of qseC was significantly reduced in the hha deletion mutant compared to that in the parental and the hha-complemented mutant strains. These results suggest that hha regulates motility of EHEC O157:H7 directly as well as indirectly by controlling the transcription of qseBC.
Introduction
Enterohemorrhagic (EHEC) Escherichia coli O157:H7 causes a broad spectrum of diarrheal illnesses, including uncomplicated diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome [1]. Cattle are the major reservoir for EHEC O157:H7, which colonizes the terminal portion called the recto-anal junction or RAJ of the large intestine of these animals [2], [3]. The locus of enterocyte effacement (LEE) [4] encodes a type III secretion system for secreting various LEE and non-LEE-encoded proteins [5] that are required for the colonization of cattle intestines and for the formation of characteristic histopathology, termed attaching and effacing lesions [6], on intestinal tissues [7]–[9]. EHEC O157:H7 colonization of cattle intestines leads to increased fecal shedding of these bacteria, a major risk factor in the contamination of beef and other bovine food products [10].
Motility is essential for pathogenicity of many bacterial pathogens [11], [12], and most EHEC O157:H7 strains associated with large disease outbreaks in humans have been shown to express flagella and tend to be motile [13], [14]. However, the role of flagellar motility in EHEC O157:H7 colonization of bovine intestines and human infections had remained ambiguous. Bovine experimental infection studies have demonstrated that flagella are dispensable for the EHEC O157:H7 colonization in these animals [15]. That motility and/or flagella might not be required for human virulence is suggested by the increased isolation of sorbitol-fermenting non-motile EHEC O157:NM strains from HUS patients in Germany that adhered at significantly higher levels to human colonic epithelial cells and expressed increased amounts of curli [16]. However, in a recent study we have demonstrated that a hha deletion mutant of EHEC O157:H7 expressing LEE at very high levels but showing reduced motility due to the reduced expression of the flagellar gene fliC failed to establish increased colonization of cattle intestines compared to the wild-type strain [17]. Despite unsettled role of motility in EHEC O157:H7 colonization of bovine intestines, there is increasing evidence that flagella might promote adherence of EHEC O157:H7 to the target sites in the large intestine. For example, it has recently been shown that a fliC mutant of EHEC O157:H7 adhered poorly to the cultured primary rectal epithelial cells compared to the fliC complemented mutant strain [18]. In addition, these and other studies have also demonstrated that the flagellar expression is temporally regulated as EHEC O157:H7 bacterial cells show abundant flagella on their cell surfaces during the early stages of adherence to the epithelial cells but the flagellar expression is reduced during the formation of attaching and effacing lesions on these cells [13], [18]. Many enteropathogenic E. coli (EPEC) serotypes have also been shown to require flagella for adherence and formation of microcolonies on HeLa or HEp-2 cells [19], but unlike H7 flagella, purified flagella of EPEC serotypes failed to adhere to the rectal epithelial cells [18], implying a specificity of H7 flagella for the rectal epithelial cells.
Transcriptional regulation of LEE and flagellar genes conferring phenotypes of intimate adherence and motility, respectively, is highly complex. Several transcriptional regulators control the expression of these two important sets of genes in response to complex networks of environmental and physiological cues [20], [21]. For example, the expression of LEE, which consists of five major operons named LEE1 – LEE5, is positively regulated by the master regulator Ler encoded by the ler gene of LEE1 [22]. Transcriptional factors, such as IHF, QseA, GrlA, LrhA, and Pch activate ler transcription [23]–[27] while others, such as H-NS, Hha, Hfq, and SdiA, repress transcription of ler [25], [28]–[33]. Transcriptional regulation of flagellar genes is governed by the master regulator FlhDC complex [34], whose expression is positively modulated by H-NS, Hha, CsrA, OmpR, cAMP-CAP, and QseBC [14], [35]–[38] and negatively by SdiA, GrlA, LrhA, and HdfR [13], [29], [39], [40].
In previous reports, we have demonstrated that a hemolysin modulating protein Hha exerts a negative effect on LEE expression by repressing the transcription of ler and a positive effect on flagellar gene expression by activating flhDC transcription [32], [35]. Similarly, the QseBC encoded quorum sensing system, like Hha, has been shown to exert positive regulatory effects on flagellar gene expression by the activation of flhDC through direct interactions of QseB with the flhDC promoter [14]. In the QseBC system, QseC is a transmembrane histidine sensor kinase that autophosphorylates and transfers the phosphoryl moiety to its cognate response regulator QseB, which interacts with the regulatory sequences of its target genes, such as flhDC, to affect their expression [14], [41]. QseBC also enhances LEE expression through a quorum sensing regulator QseA, which is activated indirectly by QseBC through the activation of quorum sensing qseEF genes in response to bacterially synthesized autoinducers and mammalian stress hormones epinephrine or norepinephrine [42].
Although both Hha and QseBC affect flagellar gene expression by increasing the transcription of the master regulator FlhDC [14], [35], [43], neither the relative contribution nor the hierarchy of these two genetic systems in the regulation of flhDC and bacterial motility is fully understood. In this study, we used single gene deletion mutants, lacking either the hha (Δhha) or the qseC (ΔqseC) gene, to compare the magnitude of the regulatory effects of Hha to that of the well-characterized positive effects of QseBC on bacterial motility [14] in the presence or absence of norepinephrine. We selected the qseC deletion mutant for this comparison since the qseC mutant has previously been shown to exhibit reduced motility due to its inability to respond to norepinephrine [14], [44]. In addition, we used a double deletion mutant, lacking hha and qseC (Δhha ΔqseC), to confirm both the magnitude of contribution and hierarchy of Hha relative to QseBC in the complex regulatory network controlling motility of EHEC O157:H7.
Materials and Methods
Bacterial strains, culture media, and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. A stx2 deletion mutant strain (NADC 6431) [32] derived from a streptomycin-resistant mutant of enterohemorrhagic Escherichia coli (EHEC) O157:H7 strain 86–24 [45], [46] was used as a parent strain for constructing the isogenic hha (NADC 6491), qseC (NADC 6488), and hha qseC (NADC 6528) deletion mutants (Table 1). Bacterial strains were cultivated in Luria-Bertani broth (LB) (Sigma-Aldrich, St. Louis, MO) or LB agar supplemented with antibiotics (Sigma-Aldrich) as needed (streptomycin, 100 mg liter−1; kanamycin 50 mg liter−1; and carbenicillin 100 mg liter−1).
Table 1. Bacterial strains and plasmidsa .
| Strain or plasmid | Genotype and description | Source or reference |
| E. coli strains | ||
| NADC 5570 | stx2+ and streptomycin-resistant E. coli O157:H7 | [45], [46] |
| strain 86-24 | ||
| NADC 6431 | Δstx2 derivative of NADC 5570 | [32] |
| NADC 6491 | Δhha mutant strain of NADC 6431 | [32] |
| NADC 6488 | ΔqseC mutant strain of NADC 6431 | This study |
| NADC 6528 | ΔqseC mutant strain of NADC 6491 | This study |
| TOP 10 | F- mcrA Δ (mrr-hsdRMS-mcrBC) Φ80lacZΔM15 | Life Technologies |
| ΔlacX74 recA1 araD139 Δ (ara-leu)7697 galU | ||
| galK rpsL (StrR) endA1 nupG | ||
| Plasmids | ||
| pSMART-LC | Cloning vector | Lucigen |
| pCRXL and pCR2.1 | Cloning vectors | Life Technologies |
| pAM450 | Plasmid with a temperature-sensitive origin of | [32], [46] |
| replication | ||
| pSM400 | pCRXL containing a 3.0 kb US-DS fragment for | This study |
| deleting the qseC gene | ||
| pSM488 | qseC deletion plasmid constructed by cloning a | This study |
| 3.0 kb US-DS fragment of pSM400 at the XbaI | ||
| site of pAM450 | ||
| pSM639 | pSMART-LC containing 3.51 kb fragment for | This study |
| complementing the qseC deletion mutant | ||
| pSM197R | pCR2.1 carrying a 630 bp DNA fragment | [32], [56] |
| encoding hha | ||
Detailed descriptions of the construction of bacterial strains and plasmids listed in this table are provided under material and methods section.
Recombinant DNA procedures
For constructing an in-frame qseC deletion mutant of EHEC O157:H7 strain NADC 6431, a 1.5 kb DNA fragment upstream (US) and a 1.5 kb DNA fragment downstream (DS) of the qseC ORF of this strain were isolated by PCR using primer pairs qseC-USF/qseC-USR and qseC-DSF/qseC-DSR, respectively (Table 2). The XbaI restriction sites were built into primers qseC-USF and qseC-DSR and BglII restriction sites were built into primers qseC-USR and qseC-DSF. The US and DS fragments were ligated together in the 5′US-DS3′ order at the BglII site located at the 3′ and 5′ ends of the US and DS fragments, respectively. The 3.0 kb US-DS fragment was cloned into pCRXL and electroporated into E. coli TOP10 bacterial cells according to the manufacturer's instructions (Life Technologies, Grand Island, NY) to generate a recombinant plasmid pSM400 (Table 1). The pSM400 plasmid DNA was digested with XbaI, digested DNA subjected to standard agarose gel electrophoresis, and 3.0 kb US-DS XbaI fragment was extracted from an agarose gel using a Gel Extraction Kit according to the manufacturer's instructions (Qiagen, Valencia, CA). The 3.0 kb US-DS XbaI fragment was cloned at the XbaI site of a plasmid (pAM450) temperature-sensitive for its replication [32], [46]. The recombinant of pAM450 carrying the 3.0 kb XbaI fragment (pSM488) (Table 1) was electroporated into the parent strain (NADC 6431) for deleting the qseC gene by using a previously described method [32]. The resulting qseC deletion mutant was named NADC 6488 (Table 1). The hha mutant (NADC 6491) described in a previous study [32] was used as a host strain for the plasmid pSM488 that allowed deletion of the gene qseC to generate a hha qseC double deletion mutant (NADC 6528) (Table 1) by using the same procedure as was used for deleting the qseC gene. The deletion of the qseC gene in the parental and the hha mutant strains was confirmed by PCR using the qseC deletion primers listed in Table 2. A 3.51 kb DNA fragment containing the qseB and qseC genes was isolated from EHEC O157:H7 strain 5570 by PCR using the primers qseBC-USF and qseBC-DSR and cloned into a low-copy vector pSMART-LC (Lucigen Corporation, Middleton, WI). The recombinant qseBC-pSMART-LC (pSM639) was used for in trans complementation of the qseC and hha qseC deletion mutants. We used plasmid pSM197R (pCR2.1 carrying hha), whose construction has been described in a previous report [32], for in trans complementation of the hha and hha qseC deletion mutants for hha. Empty vectors pSMART-LC and pCR2.1 were electroporated into the parent strain NADC 6431 to construct strains NADC 6431/pSMART-LC and NADC 6431/pCR2.1 that were used as controls (Table 1).
Table 2. Primers used for PCR and QRT-PCR.
| Primer | Nucleotide sequencea | Locationb |
| E. coli Primers c | ||
| qseC-USF | GATTCTAGAGGCTTTGGTTAACAGGAGAAAG | 3975818–3975839 |
| qseC-USR | GAAAGATCTAGATCTGGTACGAATAAAAT | 3977194–3977173 |
| CACTACCG | ||
| qseC-DSF | GCGAGATCTAGATCTAGGGTAAGACTTTTG | 3978576–3978598 |
| CTAAATTC | ||
| qseC-DSR | GACTCTAGACACGACTATTCAACCGCATAG | 3980066–3980046 |
| qseBC-F | GATTCTAGAGGCTTTGGTTAACAGGAGAAAG | 3975818–3975839 |
| qseBC-R | GATCTCTAGAGAGTACGGATTTCTGCGATTAC | 3979328–3979297 |
| qseC F (qseC deletion | CTGGCGAACCCTTAACACTG | 3977001–3977020 |
| confirmation primer) | ||
| qseC R (qseC deletion | TCGTTCAGTTGACCATTGGAG | 3978750–3978729 |
| confirmation primer) | ||
| qseC F (QRT-PCR) | CTGGCGAACCCTTAACACTG | 3977507–3977526 |
| qseC R (QRT-PCR) | TCCAGACAAAACGCCATTGA | 3977626–3977607 |
| qseC p (QRT-PCR) | FAM/TGGCGATAACGGAGAAGATATTCC/TAM | 3977529–3977552 |
| rpoA F (QRT-PCR) | GGCTTGACGATTTCGACATC | 4242887–4242906 |
| rpoA R (QRT-PCR) | GGTGAGAGTTCAGGGCAAAG | 4242997–4242978 |
| rpoA p (QRT-PCR) | TGAAGTTATTCTTACCTTGAATAAATC | 4242976–4242942 |
Nucleotide sequences of primers used in this study were selected from the published genome sequence of E. coli O157:H7 strain EDL933 [57] with the accession number AE005174.2.
Location refers to the position of primer sequence in the genome of EDL933 [57].
Subscripts F, R, and P denotes forward primer, reverse primer, and TaqMan probe; (QRT-PCR) denotes quantitative reverse transcriptase-based PCR.
Quantitative reverse transcription PCR (QRT-PCR)
Bacterial strains were grown for five h (A600 of 1.1 to 1.2) in low-glucose DMEM (Life Technologies). One ml aliquots of these bacterial cultures were mixed with two ml of RNA-Protect solution and the total RNA was isolated by using the RNeasy Kit according to the manufacturer's instructions (Qiagen). RNA was subjected to DNase treatment by using the Ambion TURBO DNA-free Kit to remove DNA contamination according to the manufacturer's instructions (Life Technologies). QRT-PCR was performed by adding 25 ng of DNase-treated RNA, 0.75 µM each of antisense and sense primers, 0.25 µM of a TaqMan probe (labeled at the 5′ end with FAM reporter and at 3′ end with TAMRA quencher) (Integrated DNA Technologies, Coralville, IA), to a QRT-PCR Master Mix (Agilent Technologies, Santa Clara, CA). Primers and labeled probes used for QRT-PCR are listed in Table 2. QRT-PCR was carried out in Mx3005P qPCR System (Agilent) by using the following parameters: 50°C for 30 min for cDNA synthesis; 95°C for 10 min; 35 cycles of 95°C for 30 sec, 55°C for 60 sec, and 72°C for 30 sec for cDNA amplification and detection of amplification products. The fold-change in the gene expression was plotted by adjusting the expression of a target gene to one for the parent (calibrator) strain and by normalizing to the expression of the house-keeping gene rpoA.
Determination of bacterial motility
Bacterial strains were grown in LB broth containing-100 µg ml−1 of carbenicillin at 37°C with shaking (175 rpm) for 18–24 h. These cultures were standardized to OD600 of 4.00 and 2 µl from each culture was spotted on soft-agar motility plates (DMEM containing 0.3% Noble agar, 100 µg ml−1 carbenicillin and with or without 50 µM norepinephrine). The diameters of motility halos produced around the point of inoculations were measured after incubating motility plates for 18–24 h at 37°C.
Statistical analyses
A two sample, nonparametric Mann-Whitney test was used to determine the significance of the differences in motilities and expression of selected genes in the mutant and the complemented mutants relative to the parental strain. The differences were considered significant at p<0.05.
Results
Hha affects motility by a greater magnitude than QseBC
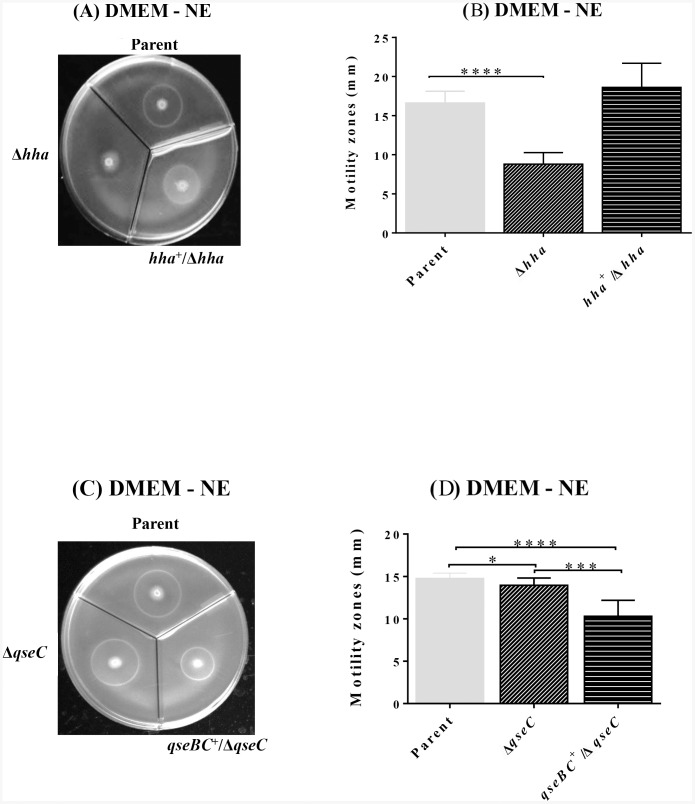
Since both Hha and QseBC enhance motility of EHEC O157:H7 by the transcriptional activation of flhDC [14], [35], we first determined the magnitude of the effect of Hha on the expression of motility by comparing the motility of a hha deletion mutant to that of a qseC deletion mutant, which according to the published reports shows significantly reduced motility in comparison to the parent strain [14], [44]. In the present study, we observed that the motility of the hha deletion mutant (8.8 mm±0.47; p = <0.0001) was highly reduced compared to the parental (16.6 mm±0.48) (Figs. 1A and 1B) and the qseC deletion mutant strains (14.0 mm±0.27; p = 0.0177) (Figs. 1C and 1D). The complementation of the hha deletion mutant with a hha-carrying plasmid (pSM197R) restored motility of the mutant strain (18.6 mm±0.98) similar to the parental level (16.6 mm±0.48) as there was no significant (p = 0.1065) difference in the motilities of these two strains (Figs 1A and 1B). On the other hand, the magnitude of motility (14.0 mm±0.3) of the qseC deletion mutant (NADC 6488) was only slightly but significantly lower (p = 0.0177) than the motility (14.7 mm±0.02) of the parental strain (NADC 6431) (Figs. 1C and 1D). In addition, in trans complementation of the qseC deletion mutant with a plasmid-cloned copy of qseBC (pSM639) resulted in a significant reduction in motility (10.3 mm±0.6; p = <0.0001) compared to both the qseC mutant (14.0 mm±0.3) and the parental strain (14.7 mm±0.2) (Figs. 1C and 1D). This negative effect of qseBC-mediated complementation could presumably result from the increased expression of QseBC causing abnormal stoichiometry of non-phosphorylated to phosphorylated QseB that in turn reduces motility of the complemented qseC mutant in the absence of NE.
Figure 1. Relative effects of the hha and qseC deletions on bacterial motility.
Three biological and a total of nine technical replicates of the overnight bacterial cultures of each strain were standardized to equivalent optical densities (A 600 nm = 4.00) and spotted in 2 µl aliquots on the motility agar plates containing or lacking norepinephrine (NE). Figures 1A and 1B show the motility zones produced around the point of bacterial inoculations and plot of their corresponding diameters (mm), respectively, for hha (Δhha), hha-complemented (hha +/Δhha) mutants, and the parent strain. Figures 1C and 1D show the motility zones produced around the point of bacterial inoculations and plot of their corresponding diameters (mm), respectively, for qseC (ΔqseC), qseC-complemented (qseBC +/ΔqseC) mutants, and the parent strain. Statistical significance of the differences in the motilities of the mutant and the complemented mutant relative to the parent strain is indicated by * (p = 0.0177); *** (p = <0.003); **** (p = <0.0001).
Norepinephrine enhances motility irrespective of the presence or absence of hha or qseC
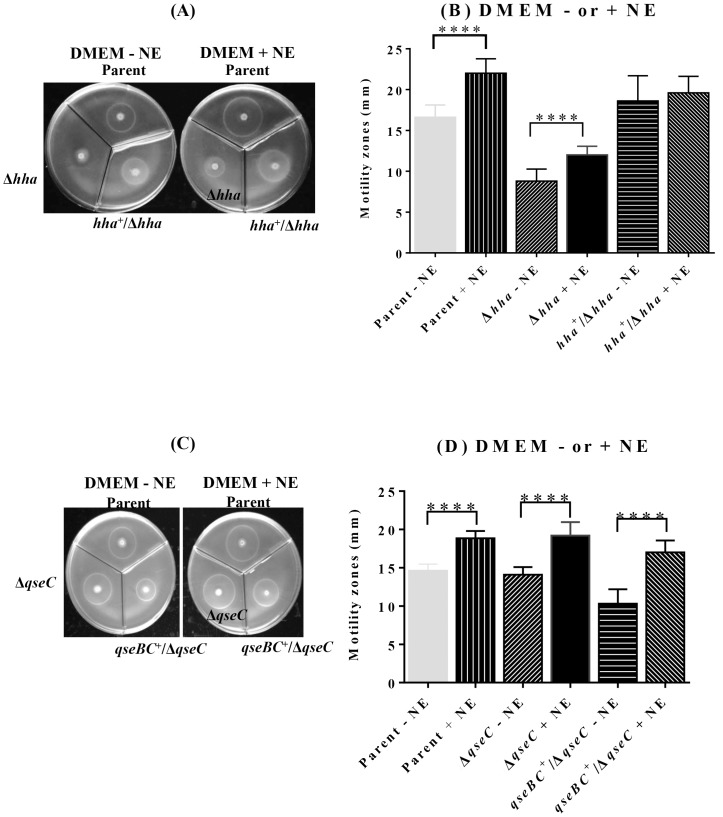
Catecholamines, such as norepinephrine (NE) and epinephrine (E), have been shown to enhance EHEC O157:H7 motility by activation of the qseBC-encoded quorum sensing signaling system, and the mutants lacking qseC are unable to sense NE and therefore show reduced motility [47], [48]. The NE dependency of the QseBC-mediated positive regulation of motility is reflected in the absolute requirement of the phosphorylated QseB, the transcriptional factor responsible for the up-regulation of flhDC encoding FlhDC, the master positive regulator of motility [14], [44]. However, there is no evidence to date demonstrating any direct or indirect role of NE or QseBC in the Hha-mediated positive regulation of motility in EHEC O157:H7. A comparison of the motility of the hha mutant grown with or without NE revealed that although the motility of the hha mutant increased from 8.8 mm±0.47 without NE to 12.0 mm±0.33 (p = <0.0001) with NE, but it was still lowest in magnitude compared to the motility of the parental strain (16.6 mm±0.48 without NE to 22.0±0.56 with NE; p = <0.0001) and the hha mutant complemented with the hha-carrying plasmid pSM197R (18.6 mm±0.98 without NE to 19.6±0.64 with NE; p = 0.251) (Figs. 2A and 2B). As shown in Figs. 2C and 2D, motility of the parental strain increased from 14.6 mm±0.28 in the absence of NE to 18.8 mm±0.3 (p = <0.0001) in the presence of NE. Similarly, motility of the qseC mutant increased from 14.10 mm±0.31 without NE to 19.0 mm±0.56 with NE (p = <0.0001) and motility of the qseC-complemented mutant also increased from 10.0 mm±0.6 in the absence of NE to 17.0 mm±0.5 (p = <0.0001) in response to NE (Figs. 2C and 2D).
Figure 2. Magnitude of the motility in response to NE signaling in the hha and qseC mutants.
The response to norepinephrine (NE) was determined by spotting the overnight bacterial cultures standardized to equivalent OD600 of 4.00 on soft-motility agar plates. The motility zones produced by each strain after an overnight of incubation at 37°C in the absence of NE were compared to those produced by the same strain in the presence of NE. Figures 2A and 2B show the motility zones produced around the point of bacterial inoculations and plot of their corresponding diameters (mm), respectively, for the parent, hha (Δhha), and hha-complemented (hha +/Δhha) mutants strains in the absence or presence of NE. Figures 2C and 2D show motility zones produced around the point of bacterial inoculations and the plot of their corresponding diameters (mm), respectively, for the parent, qseC (ΔqseC), qseC-complemented (qseBC +/ΔqseC) mutant strains in the absence or presence of NE. Statistical significance of the differences in the motilities of the mutant and the complemented mutants relative to the parental strains is indicated by **** (p = <0.0001).
Regulatory effects of Hha on the expression of motility are of greater magnitude than QseBC
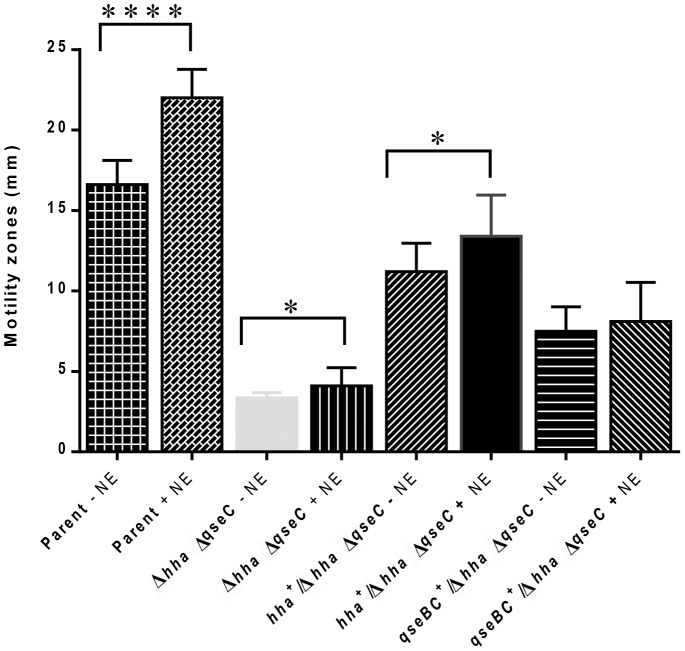
The results using single gene deletions, as described in Figs. 1 and 2, provided the first line of evidence that Hha affects motility by a greater magnitude than QseBC. In order to confirm these results, we examined the motility phenotypes expressed by the hha qseC double deletion mutant (NADC 6528) (Table 1) with or without complementation with hha- or qseBC-recombinant plasmids and in the presence or absence of NE. The motility of the hha qseC mutant was highly and significantly compromised (3.0 mm±0.11 without NE and 4.0 mm±0.36 with NE; p = <0.0001) when compared to the motility phenotypes expressed by the parental strain (17.0 mm±0.48 without and 22.0±0.56 with NE) (Fig. 3), hha (9.0 mm±0.47 without and 12.0 mm±0.33 with NE) (Figs. 2A and 2B) and qseC mutants (14.0 mm±0.31 without and 19.0±0.56) (Figs. 2C and 2D). In trans complementation of the hha qseC double deletion mutant with hha-recombinant plasmid increased the motility of this mutant to 11.0 mm±0.56 without NE and to 13. 4 mm±0.81 with NE (p = 0.0492) (Fig. 3). Overall, we observed about 70% increased motility of hha-complemented hha qseC mutant (11.0 mm±0.56; p = <0.0001) compared to the non-complemented hha qseC mutant (3.30 mm±0.11) in the absence of NE. In the presence of NE, there was also about 70% increase in the motility of hha-complemented hha qseC deletion mutant (13.4 mm±0.81, p = <0.0001) compared to the non-complemented hha qseC mutant (4.0 mm±0.36). On the other hand, complementation with qseBC-carrying plasmid (pSM639) increased motility of the hha qseC mutant by a smaller magnitude than complementation of this mutant with the hha-carrying plasmid (Fig. 3). For example, as shown in Fig. 3, complementation with the qseBC-carrying plasmid restored only 55% (7.5 mm±0.48 without NE) and 49% (8.0 mm±0.77 with NE) of the parental motility to the hha qseC mutant compared to 70% of the parental motility restored to the hha qseC mutant upon complementation with the hha-carrying plasmid in the absence (11.0 mm±0.56; p = <0.0001) or presence (13.4 mm±0.81; p = 0.0001) of NE. Thus, the data presented in Fig. 3 indicates that hha exerts greater control than qseBC in regulating motility of EHEC O157:H7.
Figure 3. Relative contributions of hha and qseC to the regulation of motility.
Relative motility of a hha qseC double deletion mutant was compared to the same mutant complemented with a plasmid-cloned copy of hha or qseC on the soft-agar motility plates lacking or supplemented with norepinephrine (NE). The motility zones produced after an overnight incubation at 37°C are shown as bar graphs. The bars represent mean diameters of the motility zones (mm) ± SEM computed from the three independent bacterial cultures of each strain. Significance of the difference in motility of a strain grown on a soft-agar motility medium lacking or containing NE is shown by asterisks above the brackets. * (p = 0.05); **** (p = <0.0001).
Hha is hierarchically superior to QseBC in regulating motility of EHEC O157:H7
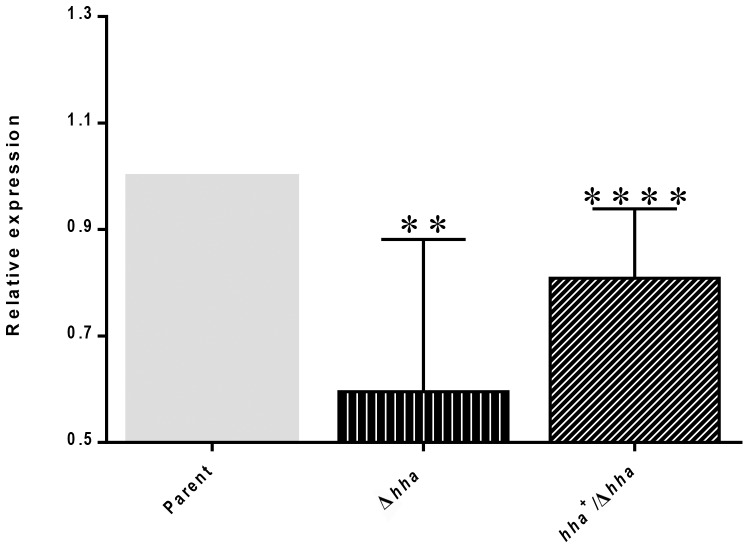
Since complementation with the hha-carrying plasmid (pSM197R) restored highest magnitude of motility to the hha qseC mutant compared to that restored by complementation of this mutant by the qseBC-carrying plasmid, we wanted to determine if the reduced expression of qseBC in the hha mutant strain was responsible for this disparity. QRT-PCR analysis of the total, DNA-free RNA showed 50% reduction in the transcriptional level of qseC in the hha deletion mutant (0.6±0.1; p = 0.0019) compared to the parental strain (1.00) (Fig. 4). The complementation of the hha mutant with the hha-carrying plasmid restored transcriptional levels of qseC to about 80% (0.81±0.04) of that in the parental strain (1.00) (Fig. 4) indicating that hha might be regulating the transcription of the qseBC genes.
Figure 4. Transcriptional activation of qseC by Hha.
The effect of hha on the transcription of qseC was determined by a QRT-PCR assay using a DNA-free RNA prepared from bacterial strains grown in DMEM for 5 h to an A600 of 1.1 to 1.2. The relative expression of qseC in the parent, hha (Δhha), and hha-complemented (hha+/Δhha) mutant strains is shown in the bar graph as means ± SEM of three independent bacterial cultures and nine technical replicates. Significant (p<0.05) differences in qseC transcription between the parent and mutant strains were determined using Mann-Whitney test. ** (p = 0.0019); **** (p = <0.0001).
Discussion
The expression of flagellar motility in E. coli including EHEC O157:H7 requires at least 50 flagellar genes divided into three classes based on the temporal order of their transcription [34]. The flhDC genes of class I flagellar operon encode the master regulator FlhDC, which is essential for the expression of all other flagellar genes [49]. Hha, a nucleoid-associated protein [50], and QseBC, encoding a two-component signal transduction system, are among the multitude of transcriptional systems that activate motility in EHEC O157:H7 by activating flhDC expression [14], [41], [44]. Here we have shown that Hha exerted greater regulatory effect on motility compared to QseBC as motility of the hha deletion mutant was severely compromised compared to that of the parental and the qseC deletion mutant strains.
The use of single gene deletion mutants lacking either hha or qseC showed that Hha is a major activator of motility of EHEC O157:H7 compared to QseBC. By using a hha qseC double deletion mutant we were able to demonstrate that the double mutant suffered the highest reduction in motility confirming that both Hha and QseBC contribute to parental-type motility in EHEC O157:H7. Furthermore, complementation of the hha qseC mutant with a hha- or qseBC-carrying plasmid provided data about the relative contributions of Hha and QseBC to the regulation of motility. The complementation analysis confirmed greater regulatory control of Hha on the expression of motility versus that of QseBC. In addition, complementation with hha not only restored greater proportion of the parental motility to the hha qseC mutant, hha complementation also allowed significant increases in bacterial motility in response to norepinephrine. In contrast, the complementation of the hha qseC mutant with a qseBC-carrying plasmid revealed that the QseBC pathway restored lesser proportion of the parental motility and failed to confer the ability to respond to norepinephrine in the absence of the hha gene. Thus, these data indicated that the cumulative regulatory activities of both Hha and QseBC control the expression of the parental motility phenotype in EHEC O157:H7, albeit Hha plays a greater regulatory role in controlling the motility. Although the QseBC encoded signaling system is reportedly essential for the sensing of bacterially produced anuoinducer-3 (AI-3) and mammalian stress hormones epinephrine and norepinephrine in order to enhance motility [14], [44], [48], both hha and qseC deletion mutants showed significant increases in their motilities in response to norepinephrine. The abilities of the hha and qseC deletion mutants to respond to norepinephrine are similar to those reported for the qseC deletion mutants of Salmonella enterica serovar Typhimurium [51]. Collectively, these findings suggest that other yet unknown genetic systems could allow the hha as well as the qseC mutants to respond to norepinephrine. It is also possible that more than 30 response regulators identified in E. coli could, through a cross-talk with a sensory kinase of a specific two-component system [44], [52], induce the expression of other unknown gene(s) enabling the hha and qseC mutants to respond to norepinephrine and up-regulate their motility.
Hha is a global regulator that specifically controls the expression of horizontally acquired genes, many of which are present as pathogenicity islands in E. coli and other members of Enterobacteriaceae [32], [35], [50], [53], [54]. The Hha, a nucleoid-associated protein, reportedly interacts with H-NS and other proteins of the Hha/YmoA family to control gene expression through structuring of DNA and modulating its topology in response to osmolarity and temperature [50], [55]. However, we and others have shown that Hha also interacts directly with the promoter elements of transcriptional regulators that in turn control the expression of virulence genes, flagellar motility, and curli fimbriae in EHEC O157:H7 and Salmonella enterica serovar Typhimurium [32], [35], [53]. Since complementation with a qseBC-carrying plasmid restored the parental level motility to the qseC but not to the hha qseC double deletion mutant suggested that hha might directly control the expression of the qseBC genes. Assessment of the expression of the qseC gene in the hha deletion mutant confirmed that qseC transcription was repressed in the hha mutant and complementation with a hha-carrying plasmid enhanced qseC expression.
In conclusion, we have shown that the expression of the parental motility phenotype of EHEC O157:H7 is collectively and positively controlled by both the hha and qseBC genes, and increased motility in response to norepinephrine occurs independently of hha and qseBC. In addition, we also determined that hha not only plays a greater regulatory role than qseBC in the expression of flagellar motility but it also regulates transcription of the qseBC genes making hha hierarchically superior over qseBC in controlling flagellar motility in EHEC O157:H7.
Acknowledgments
We thank Lindsay Andersen, Matt Inbody, and Kaitlin Johnson for technical assistance in this study. We also thank John Lippolis and Thad Stanton for the critical reviews of this manuscript.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Funding Statement
USDA-ARS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Karmali MA (2004) Infection by Shiga toxin-producing Escherichia coli: an overview. Mol Biotechnol 26: 117–122. [DOI] [PubMed] [Google Scholar]
- 2. Dean-Nystrom EA, Stoffregen WC, Bosworth BT, Moon HW, Pohlenz JF (2008) Early attachment sites for Shiga-toxigenic Escherichia coli O157:H7 in experimentally inoculated weaned calves. Appl Environ Microbiol 74: 6378–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naylor SW, Low JC, Besser TE, Mahajan A, Gunn GJ, et al. (2003) Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun 71: 1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elliott SJ, Yu J, Kaper JB (1999) The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect Immun 67: 4260–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jarvis KG, Kaper JB (1996) Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect Immun 64: 4826–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moon HW, Whipp SC, Argenzio RA, Levine MM, Giannella RA (1983) Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun 41: 1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dean-Nystrom EA, Bosworth BT, Cray WC Jr, Moon HW (1997) Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect Immun 65: 1842–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nart P, Naylor SW, Huntley JF, McKendrick IJ, Gally DL, et al. (2008) Responses of cattle to gastrointestinal colonization by Escherichia coli O157:H7. Infect Immun 76: 5366–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naylor SW, Roe AJ, Nart P, Spears K, Smith DG, et al. (2005) Escherichia coli O157: H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiology 151: 2773–2781. [DOI] [PubMed] [Google Scholar]
- 10. Elder RO, Keen JE, Siragusa GR, Barkocy-Gallagher GA, Koohmaraie M, et al. (2000) Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc Natl Acad Sci U S A 97: 2999–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duan Q, Zhou M, Zhu L, Zhu G (2013) Flagella and bacterial pathogenicity. J Basic Microbiol 53: 1–8. [DOI] [PubMed] [Google Scholar]
- 12. Wright KJ, Seed PC, Hultgren SJ (2005) Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun 73: 7657–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iyoda S, Koizumi N, Satou H, Lu Y, Saitoh T, et al. (2006) The GrlR-GrlA regulatory system coordinately controls the expression of flagellar and LEE-encoded type III protein secretion systems in enterohemorrhagic Escherichia coli . J Bacteriol 188: 5682–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sperandio V, Torres AG, Kaper JB (2002) Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli . Mol Microbiol 43: 809–821. [DOI] [PubMed] [Google Scholar]
- 15. Dobbin HS, Hovde CJ, Williams CJ, Minnich SA (2006) The Escherichia coli O157 flagellar regulatory gene flhC and not the flagellin gene fliC impacts colonization of cattle. Infect Immun 74: 2894–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosser T, Dransfield T, Allison L, Hanson M, Holden N, et al. (2008) Pathogenic potential of emergent sorbitol-fermenting Escherichia coli O157:NM. Infect Immun 76: 5598–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sharma VK, Sacco RE, Kunkle RA, Bearson SM, Palmquist DE (2012) Correlating levels of type III secretion and secreted proteins with fecal shedding of Escherichia coli O157:H7 in cattle. Infect Immun 80: 1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mahajan A, Currie CG, Mackie S, Tree J, McAteer S, et al. (2009) An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157: H7 with bovine intestinal epithelium. Cell Microbiol 11: 121–137. [DOI] [PubMed] [Google Scholar]
- 19. Giron JA, Torres AG, Freer E, Kaper JB (2002) The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol Microbiol 44: 361–379. [DOI] [PubMed] [Google Scholar]
- 20. Mellies JL, Barron AM, Carmona AM (2007) Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun 75: 4199–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tree JJ, Roe AJ, Flockhart A, McAteer SP, Xu X, et al. (2011) Transcriptional regulators of the GAD acid stress island are carried by effector protein-encoding prophages and indirectly control type III secretion in enterohemorrhagic Escherichia coli O157:H7. Mol Microbiol 80: 1349–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elliott SJ, Sperandio V, Giron JA, Shin S, Mellies JL, et al. (2000) The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli . Infect Immun 68: 6115–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Friedberg D, Umanski T, Fang Y, Rosenshine I (1999) Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli . Mol Microbiol 34: 941–952. [DOI] [PubMed] [Google Scholar]
- 24. Honda N, Iyoda S, Yamamoto S, Terajima J, Watanabe H (2009) LrhA positively controls the expression of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli by differential regulation of their master regulators PchA and PchB. Mol Microbiol 74: 1393–1341. [DOI] [PubMed] [Google Scholar]
- 25. Laaberki MH, Janabi N, Oswald E, Repoila F (2006) Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157:H7: interplay between Ler, GrlA, HNS and RpoS. Int J Med Microbiol 296: 197–210. [DOI] [PubMed] [Google Scholar]
- 26. Porter ME, Mitchell P, Free A, Smith DG, Gally DL (2005) The LEE1 promoters from both enteropathogenic and enterohemorrhagic Escherichia coli can be activated by PerC-like proteins from either organism. J Bacteriol 187: 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sperandio V, Li CC, Kaper JB (2002) Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli . Infect Immun 70: 3085–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansen AM, Kaper JB (2009) Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli . Mol Microbiol 73: 446–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanamaru K, Tatsuno I, Tobe T, Sasakawa C (2000) SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 38: 805–816. [DOI] [PubMed] [Google Scholar]
- 30. Kendall MM, Gruber CC, Rasko DA, Hughes DT, Sperandio V (2011) Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86–24. J Bacteriol 193: 6843–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharma VK, Bearson SM (2013) Evaluation of the impact of quorum sensing transcriptional regulator SdiA on long-term persistence and fecal shedding of Escherichia coli O157:H7 in weaned calves. Microb Pathog 57: 21–26. [DOI] [PubMed] [Google Scholar]
- 32. Sharma VK, Zuerner RL (2004) Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 186: 7290–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sperandio V (2010) SdiA sensing of acyl-homoserine lactones by enterohemorrhagic E. coli (EHEC) serotype O157:H7 in the bovine rumen. Gut Microbes 1: 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pruss BM, Matsumura P (1997) Cell cycle regulation of flagellar genes. J Bacteriol 179: 5602–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma VK, Bearson BL (2013) Hha controls Escherichia coli O157:H7 biofilm formation by differential regulation of global transcriptional regulators FlhDC and CsgD. Appl Environ Microbiol 79: 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shin S, Park C (1995) Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol 177: 4696–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, et al. (1999) Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181: 7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, et al. (2001) Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli . Mol Microbiol 40: 245–256. [DOI] [PubMed] [Google Scholar]
- 39. Ko M, Park C (2000) H-NS-Dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Bacteriol 182: 4670–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sharma VK, Bearson SM, Bearson BL (2010) Evaluation of the effects of sdiA, a luxR homologue, on adherence and motility of Escherichia coli O157: H7. Microbiology 156: 1303–1312. [DOI] [PubMed] [Google Scholar]
- 41. Clarke MB, Sperandio V (2005) Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli . Mol Microbiol 57: 1734–1749. [DOI] [PubMed] [Google Scholar]
- 42. Reading NC, Torres AG, Kendall MM, Hughes DT, Yamamoto K, et al. (2007) A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J Bacteriol 189: 2468–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clarke MB, Sperandio V (2005) Transcriptional autoregulation by quorum sensing Escherichia coli regulators B and C (QseBC) in enterohaemorrhagic E. coli (EHEC). Mol Microbiol 58: 441–455. [DOI] [PubMed] [Google Scholar]
- 44. Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V (2009) The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC). PLoS Pathog 5: e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, et al. (1988) Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med 109: 705–712. [DOI] [PubMed] [Google Scholar]
- 46. McKee ML, Melton-Celsa AR, Moxley RA, Francis DH, O'Brien AD (1995) Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun 63: 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bansal T, Englert D, Lee J, Hegde M, Wood TK, et al. (2007) Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun 75: 4597–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V (2006) The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103: 10420–10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu X, Matsumura P (1994) The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol 176: 7345–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Madrid C, Balsalobre C, Garcia J, Juarez A (2007) The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol Microbiol 63: 7–14. [DOI] [PubMed] [Google Scholar]
- 51. Bearson BL, Bearson SM (2008) The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog 44: 271–278. [DOI] [PubMed] [Google Scholar]
- 52. Mizuno T (1997) Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli . DNA Res 4: 161–168. [DOI] [PubMed] [Google Scholar]
- 53. Fahlen TF, Wilson RL, Boddicker JD, Jones BD (2001) Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J Bacteriol 183: 6620–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mourino M, Madrid C, Balsalobre C, Prenafeta A, Munoa F, et al. (1996) The Hha protein as a modulator of expression of virulence factors in Escherichia coli . Infect Immun 64: 2881–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ueda T, Takahashi H, Uyar E, Ishikawa S, Ogasawara N, et al. (2013) Functions of the Hha and YdgT proteins in transcriptional silencing by the nucleoid proteins, H-NS and StpA, in Escherichia coli . DNA Res 20: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sharma VK, Carlson SA, Casey TA (2005) Hyperadherence of an hha mutant of Escherichia coli O157:H7 is correlated with enhanced expression of LEE-encoded adherence genes. FEMS Microbiol Lett 243: 189–196. [DOI] [PubMed] [Google Scholar]
- 57. Perna NT, Plunkett G 3rd, Burland V, Mau B, Glasner JD, et al. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409: 529–533. [DOI] [PubMed] [Google Scholar]