Abstract
To date, our understanding of the role of X-Box Binding Protein 1 (XBP1) in metabolic processes was limited to its ability to up-regulate ER folding capacity and thereby, to increase insulin sensitivity. Here, we demonstrate that XBP1s interacts with Forkhead box O1 (FoxO1) transcription factor and directs it to proteasome-mediated degradation. Our results provide the first evidence that, in addition to its regulatory effects on the ER system and insulin sensitivity, XBP1s can independently regulate glucose homeostasis through its interaction with FoxO1. Indeed, a DNA binding defective mutant of XBP1s, which does not have the ability to increase ER folding capacity, is still capable of reducing blood glucose levels and increasing glucose tolerance in the severely obese and diabetic ob/ob mice. XBP1-mediated degradation of FoxO1 might lead to development of new therapeutic approaches for treatment of type 2 diabetes.
INTRODUCTION
Obesity and its associated complications constitute serious public health threats in the 21st century1. Obesity is the leading cause of insulin resistance and type 2 diabetes2–5. Thus, understanding the molecular links between obesity and insulin resistance is of critical importance in developing effective treatments for both insulin resistance and for type 2 diabetes.
Endoplasmic Reticulum (ER) is a sophisticated luminal network for the synthesis, maturation, folding and transport of the secretory and membrane proteins6,7. Conditions that lead to perturbations in ER homoestasis can create a condition defined as ER stress and lead to activation of a complex signaling network called the unfolded protein response (UPR)8–10. The UPR cascade is primarily initiated by type-I transmembrane kinases, PKR-like endoplasmic reticulum kinase (PERK), inositol requiring enzyme-1 (IRE1) and a type-II transmembrane protein, activating transcription factor-6 (ATF6)6–10.
IRE1 possesses kinase as well as endoribonuclease activity11,12. The endoribonuclease domain of IRE1 cleaves the mRNA of a transcription factor called X-Box Binding Protein-1 (XBP1), and initiates the removal of a 26bp intron from the full length XBP1 mRNA. The resulting combination of two open reading frames creates a translational frame shift, which leads to the translation of a higher molecular weight protein, called the spliced form of XBP1 (XBP1s)13–15. XBP1s is a highly active transcription factor and functions as one of the master regulators of ER folding capacity16,8–10,17.
In recent years, we and others have shown that increased ER stress in obesity plays a key role in the development of pathologies such as insulin resistance and type 2 diabetes18–20. We have identified XBP1 as a major regulator of ER folding capacity in obesity and demonstrated that haploinsufficiency of XBP1 in mice was sufficient to create severe insulin resistance and diabetes20. More recently we have also established a causal link between XBP1, ER stress and development of leptin resistance in obesity21.
Forkhead transcription factors of the FoxO subfamily have emerged as a shared component among pathways regulating diverse cellular functions, such as differentiation, metabolism, proliferation, and survival22,23,24. In particular, among the various members of the FoxO subfamily, Forkhead box O1 (FoxO1) acts as a cardinal regulator of whole body energy homeostasis including hepatic glucose output, adipocyte and muscle differentiation, and feeding behavior in the brain22,23. FoxO1, in coordination with PPARγ co-activator 1α (PGC-1α), increases the gene expression of phosphoenolpyruvate carboxykinase (Pck) and glucose-6 phosphatase (G6pc) and regulates gluconeogenesis25,26. Although this mechanism is beneficial during fasting states for preventing hypoglycemia in healthy people, it is one of the main pathologies that lead to the development of fasting as well as postprandial hyperglycemia in insulin-resistant conditions. Activation of insulin receptor signaling, ultimately resulting in phosphorylation and activation of Akt, leads to phosphorylation of FoxO1 on three different residues and is thought to be one of the main mechanisms that is responsible for exclusion of FoxO1 from the nucleus22,23. In addition, other mechanisms such as acetylation27–29, methylation30 and degradation31–33 have been shown to alter FoxO1 activity22,23.
In our current work we identified a novel mechanism and demonstrated that XBP1s directly interacts with FoxO1 to promote its degradation by the 26S proteasome system and investigated the details and consequences of this interaction.
RESULTS
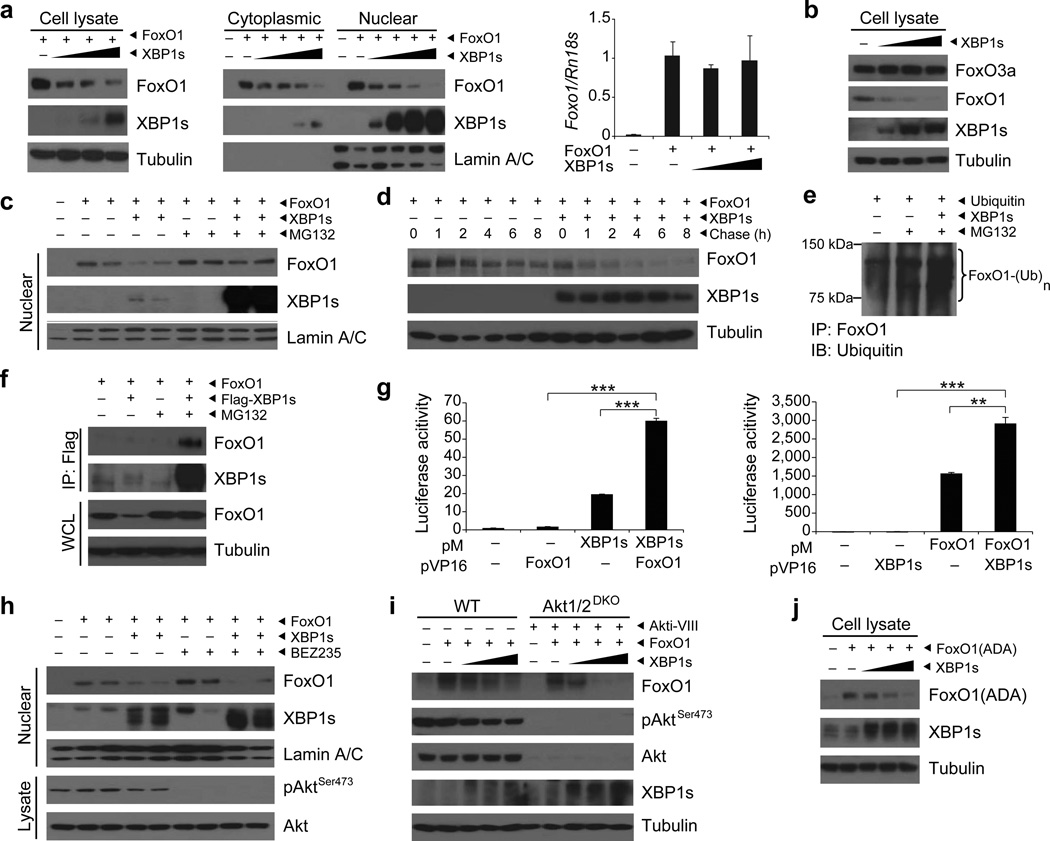
Considering the central roles of FoxO1 and XBP1s in regulating metabolic homeostasis, as well as the ability of both molecules to interact with other transcription factors, we sought to investigate whether XBP1s directly affects FoxO1. To test this possibility, we co-infected mouse embryonic fibroblasts (MEFs) with a constant dose of FoxO1-encoding adenovirus (Ad-FoxO1), along with increasing doses of XBP1s-expressing adenovirus (Ad-XBP1s) and analyzed FoxO1 and XBP1s protein levels in total cell lysates and in cytoplasmic and nuclear fractions.
When XBP1s expression was increased, FoxO1 protein levels declined dramatically in total cell lysates as well as in cytoplasmic and nuclear fractions (Fig. 1a). We next assessed the levels of Foxo1 mRNA in the presence or absence of XBP1s. Our results showed that Foxo1 mRNA levels were unaltered when FoxO1 was co-expressed with XBP1s (Fig. 1a). Furthermore, we investigated whether XBP1s can also affect other members of FoxO protein family such as FoxO3a. However, we did not observe any alterations in the FoxO3a levels after XBP1s expression (Fig. 1b).
Figure 1.
XBP1s binds to FoxO1 and promotes its degradation. (a) XBP1s and FoxO1 protein levels in MEFs expressing FoxO1 and increasing levels of XBP1s. Foxo1 mRNA levels were analyzed by qPCR. (b) Endogenous FoxO3a and FoxO1 protein levels in MEFs expressing increasing levels of XBP1s. (c) Nuclear FoxO1 and XBP1s protein levels in MEFs treated with DMSO or MG132. (d) Pulse-chase analysis of FoxO1 stability in MEFs overexpressing XBP1s. (e) Ubiquitinylated FoxO1 levels in MEFs expressing ubiquitin and XBP1s after DMSO or MG132 treatment. (f) Immunoblotting of FoxO1 and XBP1s in XBP1s–immunoprecipitates from MG132-treated MEFs, expressing FoxO1 and XBP1s. Total FoxO1 protein level were determined in whole cell lysates (WCL). (g) Mammalian two-hybrid assay with pM-XBP1s and pVP16-FoxO1 or pM-FoxO1 and pVP16-XBP1s. (h) Nuclear FoxO1 and XBP1s protein levels in MEFs treated with 1 µM BEZ235 for 4 h. Phospho-AktSer473 and total Akt levels were determined in cell lysates. (i) FoxO1, pAktSer473, total Akt and XBP1s levels in WT and Akt1/2 double knockout (Akt1/2-DKO) MEFs treated with DMSO or 20 µM Akt inhibitor for 30 min. (j) Phosphorylation resistant mutant FoxO1(ADA) protein levels in MEFs expressing FoxO1(ADA) and increasing levels of XBP1s. Each experiment was independently reproduced three times. Error bars are ± s.e.m.; P values were determined by Student’s t-test (** P < 0.01, *** P < 0.001).
Next, to test whether the decrease in FoxO1 protein levels associated with the presence of XBP1s was due to enhanced degradation of FoxO1, we co-expressed FoxO1 and XBP1s in MEFs and subsequently exposed the cells to DMSO or to the 26S proteasome inhibitor MG132 (10 µM) for 2 h. MG132 treatment blocked XBP1s-induced degradation of FoxO1 (Fig. 1c), indicating that XBP1s enhances FoxO1 degradation. To document this more directly, we performed a pulse-chase experiment for FoxO1 at the presence or absence of XBP1s. Degradation rate of FoxO1 was markedly increased in the presence of XBP1s (Fig. 1d). We also documented that expression of XBP1s markedly increased the ubiquitination of FoxO1 (Fig. 1e).
These observations prompted us to test whether XBP1s binds directly to FoxO1. For this purpose we co-expressed FoxO1 and a Flag-tagged XBP1s (Flag-XBP1s) in MEFs and also treated one group of cell with MG132 (20 µM) for 2 h to block the degradation of FoxO1. We failed to observe any interaction under normal conditions, with no inhibition of proteasome activity (Fig. 1f); this may have been due to a rapid turnover of the XBP1s-FoxO1 complex and low FoxO1 levels in the whole cell lysates (WCL) from which XBP1s was immunoprecipitated (Fig. 1f). However, when MG132 was utilized to inhibit the 26S proteasome system, XBP1s immunoprecipitation also pulled down FoxO1, demonstrating that these two proteins are, in fact, able to interact directly (Fig. 1f). To further support our claim that XBP1s directly interacts with FoxO1, we performed mammalian two-hybrid assays for XBP1s and FoxO1 (Supplementary Fig. 1a,b). Transfection of the CHO cells with both pM-XBP1s and pVP16-FoxO1 yielded a significantly (P < 0.001) higher luciferase signal when compared to that obtained with use of pM-XBP1s alone (Fig. 1g). Furthermore, with use of the reverse strategy (cloning XBP1s to pVP16 vector, and FoxO1 to pM vector), the mammalian two-hybrid assay similarly created a significantly (P < 0.0012) higher luciferase signal than that generated by the pM-FoxO1 alone (Fig. 1g). Collectively, our results indicate that XBP1s directly binds to FoxO1, and promotes its degradation.
To rule out the possibility that a low level basal Akt activity persists even under the conditions of starvation with 0.5% FBS containing media, but is not detectable via immunoblotting, we applied BEZ235 compound, which is a highly effective dual PI3K/mTOR inhibitor, and evaluated whether this further inhibition of Akt would block the effect of XBP1s on FoxO1. Treatment of MEF cells with BEZ235 (1 µM, 4 hours) after the starvation period slightly increased nuclear levels of FoxO1 when the cells were infected with Ad-FoxO1 alone (Fig. 1h), most likely due to inhibition of remnant basal Akt activity. However, when the cells were co-infected with Ad-XBP1s and Ad-FoxO1, and then exposed to BEZ235 compound, nuclear levels of FoxO1 remained reduced (Fig. 1h), indicating that the action of XBP1s on FoxO1 is Akt-independent. In a control experiment we pre-treated cells with BEZ235 (1 µM) and then stimulated with insulin (500 nM) for 15 minutes. BEZ235, indeed, completely blocked insulin-stimulated Akt activity (Supplementary Fig. 1c). These results provided support that effect of XBP1s on FoxO1 is Akt independent. However, to fully validate this observation, we first used Akt1/2 double knockout cells. To exclude the possibility that Akt3 might play a role in mediating the XBP1s-mediated FoxO1 degradation, we infected the cells with increasing doses of XBP1s and transfected with a constant dose of FoxO1. Subsequently, cells were treated with a specific Akt inhibitor (Akti-VIII, 20µM). Overexpression of XBP1s in the Akt1/2 double knockout cells, which were further treated with Akt inhibitor, again led to a dramatic reduction in the FoxO1 protein levels (Fig. 1i). These results suggest that XBP1s does not probably need FoxO1 phosphorylation to induce its degradation. However, to fully prove the hypothesis that XBP1s-induced FoxO1 degradation is completely independent of FoxO1 phosphorylation, we investigated whether XBP1s can still lead to degradation of phosphorylation-resistant FoxO1 (FoxO1-ADA). FoxO1-ADA is mutated on three residues (T24A/S253D/S316A) and is resistant to effects of Akt34. XBP1s expression still led to degradation of FoxO1 (Fig. 1j), documenting that XBP1s does not require FoxO1 phosphorylation to induce its degradation.
We previously demonstrated that reduction of ER stress and increased insulin sensitivity can only be achieved by high levels of XBP1s expression20. Recently, depletion of FoxO1 was shown to result in increased glucose tolerance and improved metabolic homeostasis, even in the liver specific IRS1/2 double knockout mice35. Given these findings, we reasoned that if the interaction between XBP1s and FoxO1 plays a dominant role in the regulation of glucose homeostasis, and if the effects of XBP1s on FoxO1 that have been demonstrated in cell culture systems (Fig. 1) hold true also in vivo, then XBP1s expression, at a level that is insufficient to modulate ER stress and insulin sensitivity, may still lead to improved glucose homeostasis. We explored this idea with a series of experiments in which we injected three increasing doses of Ad-XBP1s and Ad-LacZ into the tail vein of obese, insulin resistant and diabetic ob/ob mice.
First, nine-week-old ob/ob mice were injected with 1 × 107 PFU g−1 of Ad-XBP1s or Ad-LacZ. This dose of Ad-XBP1s only slightly increased XBP1s protein levels in the liver (Supplementary Fig. 2a), but did not significantly alter the expression of XBP1s-target genes (such as Dnajb9, Pdia3, and Hspa5) (Supplementary Fig. 2b). Blood glucose levels on post-injection days three and five were not significantly reduced in the Ad-XBP1s-injected group compared with the control mice (Supplementary Fig. 2c). Circulating insulin levels on post-injection day 9 were unaffected (Supplementary Fig. 2d), however, a glucose tolerance test (GTT) performed on post-injection day 5 documented a significant improvement in glucose tolerance (Supplementary Fig. 2e). Also, an insulin tolerance test (ITT) on day 7 demonstrated that insulin-stimulated disposal of glucose was not different between Ad-LacZ- and Ad-XBP1s-injected groups (Supplementary Fig. 2f). To investigate whether this dose of Ad-XBP1s can effectively increase insulin action, we infused either saline or insulin (0.25 IU kg−1) through the portal vein into the livers of ob/ob mice on post-injection day nine. No differences in insulin-stimulated IR- and IRS1-tyrosine and AktSer473 phosphorylations were detected (Supplementary Fig. 2g). However, levels of FoxO1 protein in total liver lysates and also in the nuclear fractions were significantly reduced in the Ad-XBP1s-injected animals, but no changes were noted in the abundance of FoxO1 mRNA (Supplementary Fig. 2h, i).
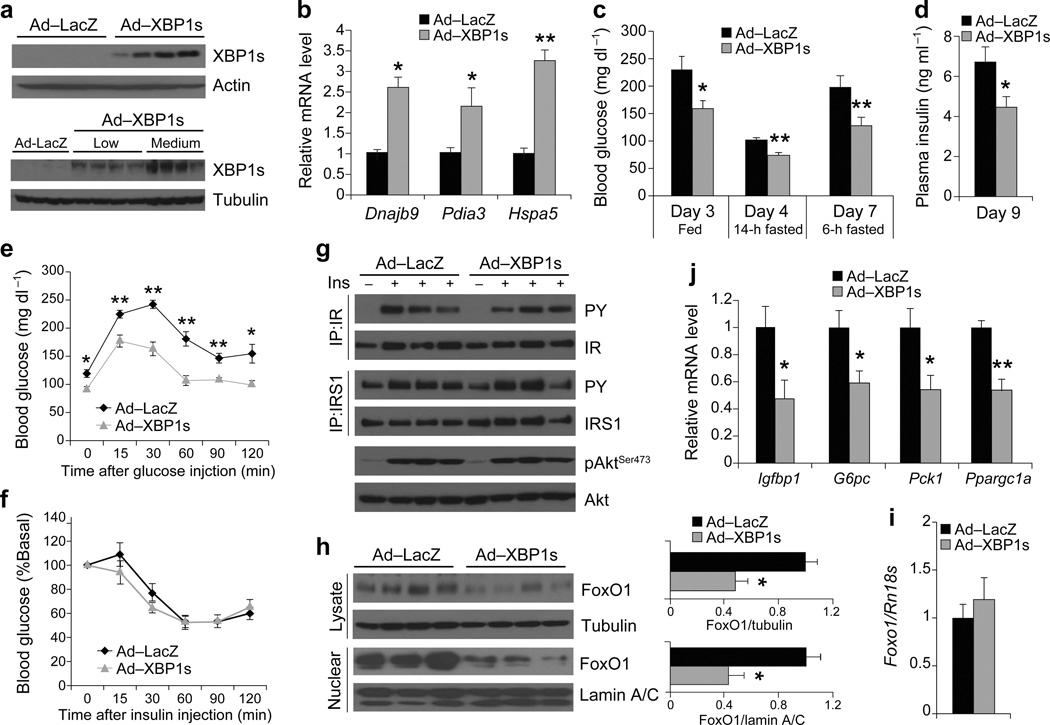
We next injected a higher dose (4 × 107 PFU g−1) of Ad-LacZ and Ad-XBP1s through the tail vein of 7–week old male ob/ob mice. This dose of Ad-XBP1s significantly increased XBP1s levels compared to the previous lower dose (Fig. 2a), and a corresponding 2–3 fold increase in the mRNA levels for Dnajb9, Pdia3, and Hspa5 (Fig. 2b). Blood glucose levels were significantly reduced in XBP1s-overexpressing mice in different nutritional states (Fig. 2c). Moreover, plasma insulin levels were significantly down-regulated relative to the control group (Fig. 2d) and, as documented by GTT, the glucose disposal rate in the Ad-XBP1s-injected group was significantly enhanced when compared with the Ad-LacZ-injected group (Fig. 2e). However, ITT revealed no alteration in insulin-stimulated glucose clearance from the circulation in the Ad-XBP1s-injected group relative to controls (Fig. 2f).
Figure 2.
Medium-level expression of XBP1s in ob/ob mice improves glucose homeostasis without altering the insulin receptor signaling. Seven-week-old, male, ob/ob mice were injected with Ad-LacZ (n = 6) or Ad-XBP1s (n = 6) (4 × 107 PFU g−1) through tail vein. (a) XBP1s protein levels in the liver of ob/ob mice injected with low or medium dose of adenovirus. (b) Relative mRNA levels of XBP1s target genes, Dnajb9, Pida3 and Hspa5 in the liver of Ad-LacZ- or Ad-XBP1s-injected mice. (c) Blood glucose levels in fed, 6h and 14h fasting on indicated days after injections. (d) Plasma insulin levels on day 9, (e) GTT on day 5, and (f) ITT on day 7 after the injections. (g) IR and IRS1 tyrosine and AktSer473 phosphorylations with or without insulin stimulation in the liver on post-injection day 9. (h) Total and nuclear levels of FoxO1 protein in the liver of Ad-LacZ- or Ad-XBP1s-injected ob/ob mice. Graph depicts total FoxO1/tubulin and nuclear FoxO1/lamin A/C ratios. Relative mRNA levels of (i) Foxo1, (j) Igfbp1, G6pc, Pck1, and Ppargc1a in the liver of adenovirus-injected mice. Experiments were repeated in five independent cohorts. Error bars are ± s.e.m.; P values were determined by Student’s t-test (* P < 0.05, ** P < 0.01, *** P < 0.001).
To analyze IR signaling in the medium-dose XBP1s injected group, we infused either saline or insulin (0.25 IU kg−1) through the portal vein into the liver of the Ad-LacZ and Ad-XBP1s-injected ob/ob mice on post-injection day nine, and analyzed the activation of IR, IRS1 and Akt. No differences were seen between the two groups in terms of insulin-stimulated tyrosine phosphorylation of IR and IRS1, nor in phosphorylation of AktSer473 (Fig. 2g). However, a dramatic decrease was observed in the total and nuclear levels of FoxO1 protein in the Ad-XBP1s-injected group (Fig. 2h), though mRNA levels were unaltered (Fig. 2i). In parallel with these observations, the expression of Igfbp1, G6pc, Pck1 and Ppargc1a was also significantly reduced in the Ad-XBP1s injected group (Fig. 2j). Corresponding to the significant suppression of gluconeogenic gene expression, the pyruvate tolerance test (PTT) revealed that the hyperglycemic response to a pyruvate challenge was significantly lower in the XBP1s overexpressing group (Supplementary Fig. 3a); however, glycogen levels in the livers of both groups were unaltered after XBP1s overexpression (Supplementary Fig. 3b).
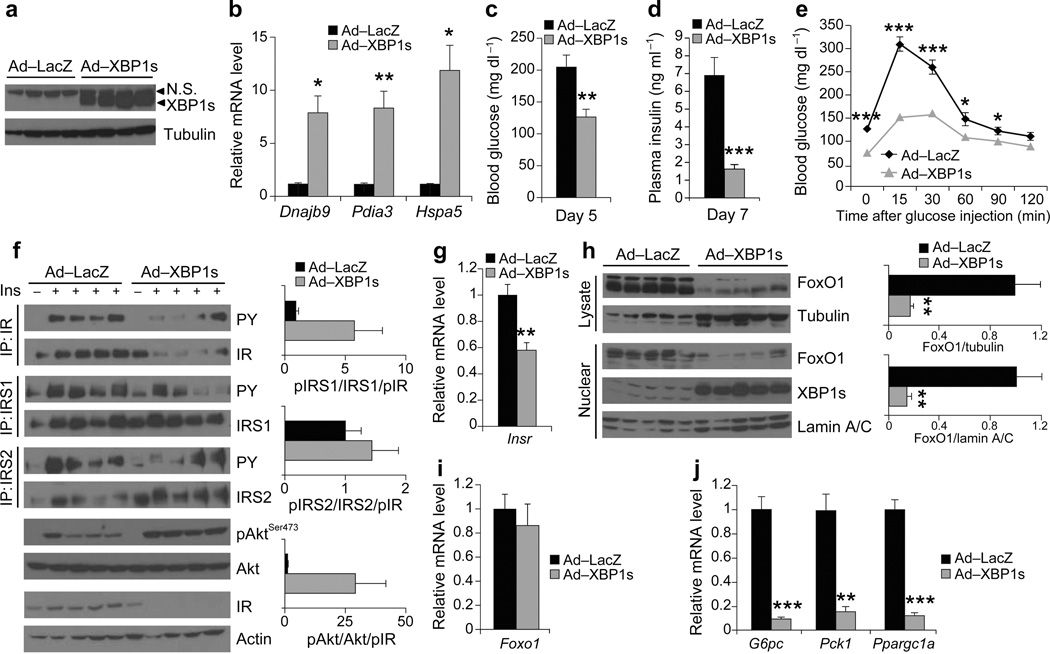
In a third group of mice, we increased the Ad-XBP1s and Ad-LacZ dose to 1.8 × 108 pfu g−1. XBP1s protein levels were further increased in this group compared to the previous group (Fig. 3a), and expression levels of the chaperones were up-regulated from 7- to 12-fold (Fig. 3b). Levels of blood glucose (day 5) and insulin (day 7) were significantly different between the Ad-LacZ and Ad-XBP1s-injected group (Fig. 3c, d), while the GTT demonstrated that the glucose disposal curve was greatly enhanced after administration of glucose in the Ad-XBP1s-injected group, relative to controls (Fig. 3e).
Figure 3.
High-level expression of XBP1s in the liver of ob/ob mice increases insulin sensitivity. Seven-week-old, male, ob/ob mice were injected with Ad-LacZ (n = 6) or Ad-XBP1s (n = 6) (1.8 × 108 PFU g−1) through tail vein. (a) XBP1s protein levels in the liver lysates on day 6 after injection. (b) Dnajb9, Pida3 and Hspa5 mRNA levels in the liver of Ad-LacZ- or Ad-XBP1s-injected mice. (c) Fed blood glucose levels on day 5, (d) plasma insulin levels on day 7, and (e) GTT on day 3 after the adenovirus injections. (f) In vivo insulin receptor signaling in the liver of Ad-LacZ- or Ad-XBP1s-injected ob/ob mice on post injection day 6. Graphs depicts the (phospho/total)/pIR ratios. (g) Relative mRNA levels of Insr in the liver of adenovirus-injected mice on day 6 after injection. (h) Total and nuclear FoxO1 levels in the liver of Ad-LacZ- or Ad-XBP1s-injected ob/ob mice. Graph depicts total FoxO1/tubulin and nuclear FoxO1/lamin A/C ratios. Relative mRNA levels of (i) Foxo1 and (j) G6pc, Pck1, and Ppargc1a in the liver of Ad-LacZ- or Ad-XBP1s-injected ob/ob mice. Experiments were repeated in three independent cohorts. N.S: non-specific. Error bars are ± s.e.m.; P values were determined by Student’s t-test (* P < 0.05, ** P < 0.01, *** P < 0.001).
We also analyzed insulin-induced activation of IR signaling subsequent to infusion of insulin through the portal vein. Interestingly, total IR levels were dramatically reduced in the high dose XBP1s-injected ob/ob livers (Fig. 3f). While pIR/IR ratio was slightly increased (Supplementary Fig. 3c), total amount of tyrosine phosphorylated IR was significantly reduced (P < 0.01, 6.5 fold) (Supplementary Fig. 3c). Insulin-stimulated tyrosine phosphorylation of IRS1 was unaltered but that of IRS2 was significantly reduced (~2.5 fold), while AktSer473 phosphorylation was significantly increased (3.5 fold) (Supplementary Fig. 3c). However, there is an important point to note; the IR signal transduction is driven by the total amount of tyrosine phosphorylated IR. Under high levels of XBP1s expression, this parameter is 6.5 times less in the XBP1s-expressing ob/ob group than the controls. Despite this dramatic reduction in the total signal generated from IR, in the Ad-XBP1s injected mice, insulin-stimulated tyrosine phosphorylations of IRS1 was unaltered, IRS2 was only reduced 2.5 fold and AktSer473 phosphorylation was increased 3.5 times (Fig. 3f, Supplementary Fig. 3c), indicating that IR-IRS and IRS-Akt axis is actually markedly enhanced. Indeed, when the total values of pIRS1/IRS are divided by pIR value, a 5-fold higher activation of IRS1 and a 15-fold enhancement in that for AktSer473 was seen in the Ad-XBP1s group relative to controls. Overall then, we conclude that reflection of each unit of IR tyrosine phosphorylation in the cell is dramatically enhanced in the presence of high levels of XBP1s expression. The decrease in levels of total IR protein and mRNA (Fig. 3f, g) when XBP1s was over-expressed is likely a response to increased insulin sensitivity and reduced blood glucose levels, aimed at preventing hypoglycemia in these mice.
Analysis of protein levels in the total and nuclear fractions derived from the livers of Ad-LacZ and Ad-XBP1s groups showed that FoxO1 protein was further reduced compared to the levels in the previous two XBP1s over-expressing groups: the difference of FoxO1 levels in the total lysates of Ad-XBP1s- versus Ad-LacZ-injected ob/ob mice was 5.9-fold and for the nuclear lysates was 7.1 fold (Fig. 3h). However, FoxO1 mRNA levels were not reduced, indicating that FoxO1 protein is post-transcriptionally regulated by XBP1s (Fig. 3i). And finally, G6pc, Pck1 and Ppargc1a expression was significantly suppressed in the Ad-XBP1s group when compared to controls (Fig. 3j).
The above findings indicated that XBP1s can also regulate glucose homeostasis, independent of its action on the ER folding capacity and on the insulin signaling system; if this is correct, an XBP1s mutant, which is unable to bind to DNA but can still bind to FoxO1 and trigger its degradation, should also reduce blood glucose levels and increase glucose tolerance. To test this possibility, we replaced the DNA binding domain of XBP1s with an artificial nuclear localization signal (Fig. 4a), and verified the ability of this DNA-binding mutant XBP1s (ΔDBD) to bind to FoxO1 and promote its degradation. ΔDBD was able to move to nucleus as XBP1s (Fig. 4a), but was unable to activate endoplasmic reticulum stress element (ERSE) driven luciferase reporter systems and could not initiate the transcriptional program (Fig. 4b). However, ΔDBD retained its capacity to bind (Fig. 4c) and degrade FoxO1 (Fig. 4d) without affecting FoxO1 mRNA levels (Fig. 4e). To explore whether ΔDBD would reduce blood glucose levels in ob/ob mice, and if so, to what extent that is seen with XBP1s, we injected medium dose (4 × 107 PFU g−1) Ad-LacZ, Ad-XBP1s and Ad-ΔDBD adenoviruses via tail vein into ob/ob mice. Measurement of 6-h fasting blood glucose levels on post-injection day three, revealed that ΔDBD reduced blood glucose levels to the same extent as did XBP1s (Fig. 4F). GTT documented that the improvement in glucose tolerance was not significantly different between the XBP1s- and ΔDBD-injected groups (Fig. 4g). FoxO1 protein levels in total lysates were significantly reduced in the XBP1s- and ΔDBD-expressing groups (Fig. 4h) but, similar to our previous results, FoxO1 mRNA levels were unaltered (Data not shown). And while XBP1s significantly increased the mRNA levels of Hspa5, this effect was lost in ΔDBD overexpressing ob/ob livers (Fig. 4i). Furthermore, gene expression of Igfbp1, G6pc and Pck1 was significantly reduced in both XBP1s and ΔDBD overexpressing ob/ob livers (Fig. 4j). These results provide the first evidence that XBP1s, independent of its modulatory effects on ER folding capacity, can regulate glucose homeostasis.
Figure 4.
A DNA binding defective mutant XBP1s (ΔDBD) improves glucose homeostasis in ob/ob mice. (a) Nuclear protein levels of XBP1s and ΔDBD in MEFs infected with the same dose of Ad-XBP1s or Ad-ΔDBD. (b) ERSE-luciferase activity and Hspa5 mRNA levels in MEFs infected with Ad-XBP1s or Ad-ΔDBD. (B) Immunoblotting of FoxO1 and ΔDBD in ΔDBD–immunoprecipitates from MEFs expressing FoxO1 and ΔDBD. Total FoxO1 protein levels were determined in whole cell lysates (WCL). (d) FoxO1 protein levels and (e) Foxo1 mRNA levels in MEFs expressing FoxO1 and increasing levels of ΔDBD. (f–j) Seven-week-old, male, ob/ob mice were injected with Ad-LacZ (n = 6), Ad-XBP1s (n = 6) or Ad-ΔDBD (n = 6) (4 × 107 PFU g−1) through tail vein. (f) 6-h fasted blood glucose levels on day 3 and (g) GTT on day 5 after the adenovirus injections. (h) Phospho-AktSer473, total Akt, FoxO1, XBP1s and ΔDBD protein levels in the liver of Ad-LacZ- or Ad-XBP1s- or Ad-ΔDBD-injected ob/ob mice on day 7 after the injections. Graph depicts total FoxO1/tubulin protein ratio. Relative mRNA levels of (i) Hspa5, (j) Igfbp1, G6pc, and Pck1 in the liver of the liver of Ad-LacZ- or Ad-XBP1s- or Ad-ΔDBD-injected ob/ob mice. Error bars are ± s.e.m.; P values were determined by Student’s t-test (* P < 0.05, ** P < 0.01, *** P < 0.001).
We also showed that glucose-stimulated insulin secretion (GSIS) is not altered after glucose (2 g kg−1) injection (Supplementary Fig. 4), indicating that enhanced GSIS is not a contributing factor in the phenotype of Ad-XBP1s injected ob/ob mice.
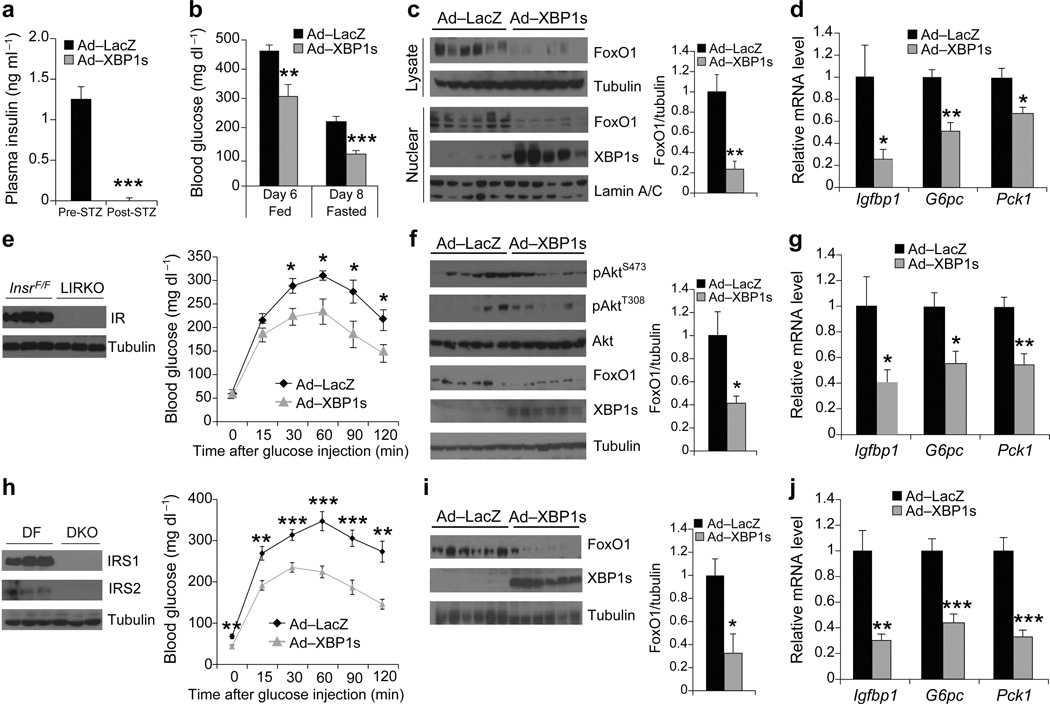
Our data thus indicate that XBP1s can also reduce blood glucose levels independent of its insulin sensitizing effects. This led us to hypothesize that XBP1s also exerted its effects under conditions of insulin deficiency, or of IR or IRS1/2 double deficiency. To test whether XBP1s would exert an effect in the complete absence of insulin, we injected 8-week-old male C57BL/6 mice with streptozotocin (STZ) (200 mg kg−1), to ablate insulin-producing β-cells and create an insulin-deficient condition. Seven days after STZ-injection, blood glucose levels of most mice were over 400 mg dl−1 (Supplementary Fig. 5a) and insulin levels were not detectable (Fig. 5a). On the ninth day after STZ injection, we chose the mice with the highest blood glucose levels (~500 mg kg−1), and injected them through tail vein with either Ad-LacZ or with Ad-XBP1s. Six days after virus injection, we found that XBP1s overexpression had significantly reduced blood glucose levels at fed state in the STZ-injected insulin-deficient (diabetic) mice (Fig. 5b). Furthermore, 8 days after virus injection, blood glucose levels were reduced to euglycemia in the Ad-XBP1s group at 12-hour fasting state (221.0 ± 17.1 vs. 109.2 ± 12.4 mg kg−1, P < 0.001) (Fig. 5b). These results indicate that XBP1s has the ability to reduce blood glucose levels even in the complete absence of insulin. An analysis of protein levels showed that total and nuclear levels of FoxO1 in liver were significantly reduced in the Ad-XBP1s-injected STZ-group when compared with the Ad-LacZ-injected control STZ-group (Fig. 5c). Furthermore, mRNA levels of Igfbp1, G6pc and Pck1 were significantly reduced in the XBP1s overexpressing STZ-injected group (Fig. 5d).
Figure 5.
XBP1s can also improve glucose homeostasis in insulin independent manner. (a–d) Eight-week-old, male, STZ-treated mice were injected with Ad-XBP1s (n = 9) or Ad-LacZ (n = 9) (1.5 × 108 PFU g−1) though tail vein. (a) Plasma insulin levels before and after STZ treatment. (b) Fed and 12-h fasted blood glucose levels on indicated days. (c) Total and nuclear FoxO1 protein levels and (d) Liver Igfbp1, G6pc, and Pck1 mRNA levels 10 days after adenovirus injections. (e–g) Eight-week-old, male, LIRKO mice were injected with Ad-LacZ (n = 8) or Ad-XBP1s (n = 8) (4 × 107 PFU g–1) through tail vein. (e) Liver IR protein levels and GTT on day 4 after adenovirus injections. (f) FoxO1, XBP1s, pAktSer473, pAktThr308 and total Akt levels and (g) Igfbp1, G6pc, and Pck1 mRNA levels in the liver of Ad-XBP1s- or Ad-LacZ-injected LIRKO mice 8 days after adenovirus injection. (h–j) Eight-week-old, male DKO mice were injected with Ad-LacZ (n = 8) or Ad-XBP1s (n = 8) (7.5 × 107 PFU g–1) through tail vein. (h) Liver IRS1 and IRS2 protein levels in Irs1Flox/Flox;Irs2Flox/Flox (DF) and DKO mice. GTT was performed on post injection day 4. (i) FoxO1 and XBP1s protein levels and (j) Igfbp1, G6pc, and Pck1 mRNA levels in the liver of adenovirus-injected DKO mice on day 8 after the injections. Error bars are ± s.e.m.; P values were determined by Student’s t-test. (* P < 0.05, ** P < 0.01, *** P < 0.001).
Next, we over-expressed XBP1s in the livers of liver-specific IR knockout (LIRKO) mice by tail vein injection of Ad-XBP1s or Ad-LacZ. As reported previously36, IR levels were dramatically reduced in the livers of LIRKO (InsrFlox/Flox; Albumin-Cre) mice when compared with levels in the InsrFlox/Flox mice (Fig. 5e). GTT performed on post-injection day 4 showed that XBP1s expression led to significant increases in glucose tolerance (Fig. 5e). Insulin-stimulated glucose disposal was not significantly altered during ITT (Supplementary Fig. 5b). Similar to the results obtained in the ob/ob models, FoxO1 protein levels were significantly reduced (Fig. 5f). Moreover, basal Akt phosphorylations was not altered (Fig. 5f). mRNA levels of Igfbp1, G6pc and Pck1 were all significantly reduced in the XBP1s overexpressing LIRKO mice (Fig. 5g) while chaperone gene expression was significantly upregulated (Supplementary Fig. 5c).
We next repeated the same experiment in liver-specific IRS1/2 double knockout mice (DKO), which lack both IRS1 and IRS2 in the liver (Fig. 5h). As for the results obtained with the LIRKO mice, overexpression of XBP1s in the livers of DLKO mice led to a significant increase in glucose tolerance (Fig. 5h) and reduced FoxO1 protein levels (Fig. 5i). Performance of ITT revealed no differences between the XBP1s and control group (Supplementary Fig. 5d). In parallel with the other XBP1s overexpression models, mRNA levels of Igfbp1, G6pc and Pck1 were significantly reduced (Fig. 5j) and the gene expression of the chaperones was significantly upregulated (Supplementary Fig. 5e).
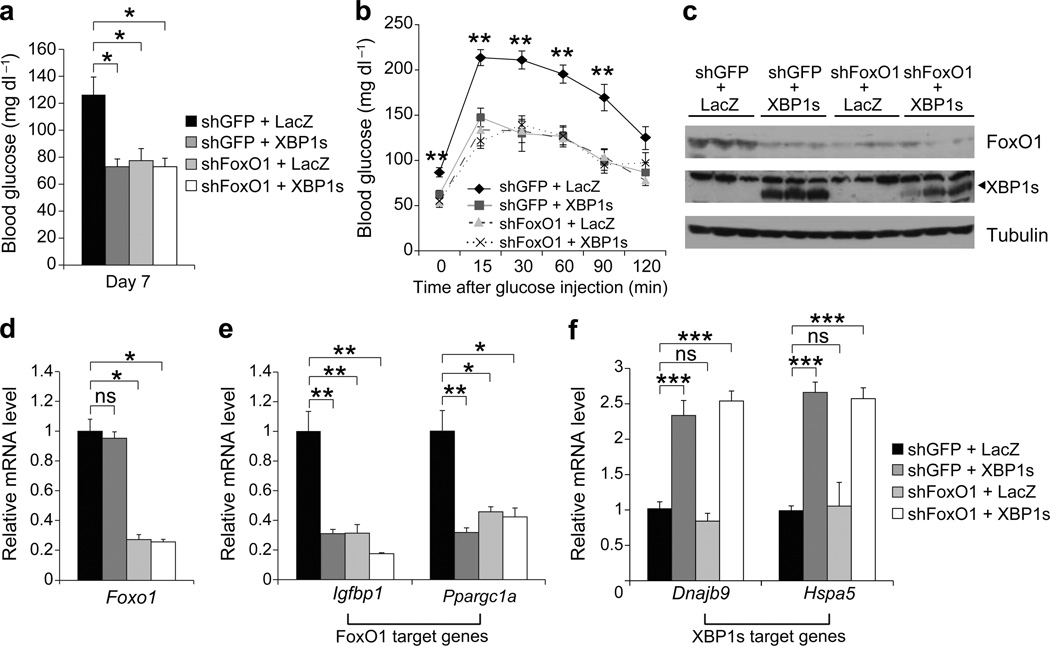
If the regulatory effects of low or medium levels of XBP1s on glucose homeostasis rely mainly on the ability of XBP1s to degrade FoxO1, it would be expected that medium levels of XBP1s should have no further effect on glucose homeostasis when FoxO1 is depleted in the liver. To evaluate this possibility, we overexpressed XBP1s in the liver, while simultaneously depleting FoxO1 with use of an adenovirus that expresses FoxO1-shRNA (Ad-shFoxO1). The first group of ob/ob mice was injected with Ad-LacZ (the control virus for Ad-XBP1s) and Ad-shGFP (control virus for Ad-shFoxO1), the second group with Ad-XBP1s plus Ad-shGFP, the third group with Ad-shFoxO1 and Ad-LacZ, and the fourth group with Ad-XBP1s and Ad-shFoxO1. Analysis of blood glucose levels on post injection day 7 revealed that ob/ob mice in which either XBP1s was overexpressed, FoxO1 was depleted, or XBP1s was overexpressed plus FoxO1 was also depleted, had significantly lower blood glucose levels when compared with the control ob/ob group (Ad-LacZ+Ad-shGFP) (Fig. 6a). However, we saw no difference in the blood glucose levels of these three groups, indicating that XBP1s cannot further reduce blood glucose levels in the absence of FoxO1.
Figure 6.
Enhanced glucose tolerance after medium-level expression of XBP1s is FoxO1-dependent. Seven-week-old, male, ob/ob mice were injected with Ad-shGFP + Ad-LacZ (n = 5), Ad-shGFP + Ad-XBP1s (n = 5), Ad-shFoxO1 + Ad-LacZ (n = 5) or Ad-shFoxO1 + Ad-LacZ (n = 5) through tail vein. (a) Blood glucose levels on day 7, and (b) GTT on day 5 after the adenovirus injections. (c) FoxO1 and XBP1s protein levels in the liver of adenovirus-injected ob/ob mice on day 7 after injections. Relative mRNA levels of (d) Foxo1, (e) Igfbp1 and Ppargc1a, (f) Dnajb9 and Hspa5 on day 7 after adenovirus injections. Error bars are ± s.e.m.; P values were determined by Student’s t-test. (*p<0.05, **p<0.01, ***p<0.001).
Next we performed a GTT, and documented that glucose tolerance was also enhanced to similar levels in the ob/ob mice with overexpression of XBP1s, depletion of FoxO1, as well as in ob/ob mice in which XBP1s was overexpressed plus FoxO1 was depleted (Fig. 6b). Analysis of FoxO1 protein levels showed that FoxO1 protein was similarly reduced significantly in Ad-XBP1s, Ad-shFoxO1, and also in the Ad-XBP1s/Ad-shFoxO1 injected groups (Fig. 6c); however, mRNA levels were only reduced in the FoxO1-depleted and XBP1s-overexpressing/FoxO1 depleted groups (Fig. 6d). We then analyzed expression levels of genes regulated by FoxO1 and XBP1s: as expected, expression of FoxO1 target genes, such as Igfbp1 and Ppargc1a, was significantly down-regulated in XBP1s-overexpressing, FoxO1-depleted as well as in XBP1s-overexpressing/FoxO1-depleted groups, where FoxO1 levels were significantly reduced (Fig. 6e). However, mRNA levels for XBP1s target genes were only up-regulated in the XBP1s-overexpressing groups (Fig. 6f), and not in those with only a depletion of FoxO1. Collectively, these results indicate that the regulatory effects of medium levels of XBP1s are primarily dependent on the ability of XBP1s to induce degradation of FoxO1.
Previous studies have shown that in lean mice with liver-specific deletion of the FoxO1 gene, blood glucose levels were unaltered under fed conditions. However, following 48 hours of starvation, these mice displayed about a 30% decrease in their blood glucose levels, due to an inability to up-regulate gluconeogenesis37. To investigate whether XBP1s would produce a similar phenotype, we injected medium dose (4 × 107 PFU g−1) Ad-LacZ and Ad-XBP1s into the tail vein of wt-lean mice. Overexpression of XBP1s did not alter fed blood glucose levels and did not lead to any differences in GTT and ITT (Supplementary Fig. 6a–c). However, when the mice were taken to starvation for 48 hours on post-injection day four, the XBP1s overexpressing group had significantly lower blood glucose levels (22%, P <0.05) relative to controls after the 48-hour starvation period on day six (Supplementary Fig. 6d).
To investigate whether deficiency of XBP1 in the liver would lead to accumulation of FoxO1 in the liver and thus create glucose intolerance, we used the Xbp1Flox/Flox mouse model, and depleted XBP1 protein by tail vein injection of an adenovirus expressing Cre-Recombinase (Ad-Cre); injection of Ad-LacZ was used as a control. We first injected lean Xbp1Flox/Flox mice (fed on a normal diet) with Ad-Cre and Ad-LacZ at 6 weeks of age; we followed these mice for 5 weeks post injection, but did not detect any alterations in blood glucose levels and in GTT (Supplementary Fig. 6e, f). This was probably due to the fact that the organism can cope with deficiency of XBP1 under normal conditions: in other words, in the absence of ER stress, there is not much need for XBP1s. Indeed, our previous work documented that Xbp1+/− mice do not develop glucose intolerance, nor insulin resistance, when they are fed a normal diet20. Next, the Xbp1Flox/Flox mice were placed on a HFD at three weeks of age; after 4 weeks of HFD feeding, the Xbp1Flox/Flox mice were injected through tail vein with Ad-Cre or Ad-LacZ. To investigate whether Ad-Cre injection completely eliminates XBP1 expression, we administered tunicamycin (3 mg kg−1, intraperitoneal) to the mice on post-injection day 15; and demonstrate that a dramatic increase in the levels of XBP1s protein seen in the control group was blocked by injection of Ad-Cre, indicating that Ad-Cre expression led to almost complete deletion of XBP1 in the livers of Xbp1Flox/Flox mice (Supplementary Fig. 7a).
Depletion of XBP1 for 14 days resulted in a significant increase in 6-hour fasted blood glucose levels (Supplementary Fig. 7b). Circulating insulin levels were significantly up-regulated in the Ad-Cre injected group by 14 days post-injection (Supplementary Fig. 7c). A GTT done on day 17 revealed that Ad-Cre injected mice developed significantly higher levels of glucose intolerance (Supplementary Fig. 7d), and glucose clearance from circulation during ITT (day 24) was significantly reduced in XBP1-depleted group (Supplementary Fig. 7e). On day 30, we infused mice through portal vein with insulin and analyzed the activation of the IR signaling system: insulin-stimulated tyrosine phosphorylation of the IR was significantly reduced (Supplementary Fig. 7f), whereas, no significant changes were seen in IRS1 tyrosine phosphorylation (Supplementary Fig. 7f). Furthermore, we also observed a significant decrease in insulin-stimulated AktSer473 phosphorylation (Supplementary Fig. 7f). To investigate whether we could dissect out the effects of insulin signaling and XBP1s on FoxO1 in this model, we used another group of mice, and undertook studies of insulin receptor signaling at a relatively early time point (day 20); in these animals, IR signaling was again significantly reduced (data not shown). Evaluation of the total and nuclear levels of FoxO1 documented a significant accumulation of this transcription factor in the livers of the Xbp1Flox/Flox mice that were injected with Ad-Cre (Supplementary Fig. 7g). Finally we analyzed the gene expression pattern of gluconeogenic genes and demonstrated that G6pc gene expression was significantly up-regulated in the Ad-Cre injected Xbp1Flox/Flox mice (Supplementary Fig. 7h). Gene expression for Pck1 and Ppargc1a were unaltered (Supplementary Fig. 7h).
DISCUSSION
In recent years we and others have demonstrated that increased ER stress associated with obesity plays a key role in the development of several pathologies such as insulin resistance and type 2 diabetes, and also of leptin resistance in the brain18–21,38. Our findings have revealed that XBP1 is a critical prerequisite for the organism to maintain glucose homeostasis under conditions of high calorie intake and obesity21.
To date, the role of XBP1s in metabolic homeostasis is believed to be restricted to ability of this transcription factor to upregulate ER folding capacity and thus increase insulin sensitivity. Our current results add another dimension to the roles of XBP1s, and provide the first evidence that this protein can also regulate glucose homeostasis independent of its ability to regulate the ER folding capacity and to reduce ER stress. In our current work we have shown that XBP1s directly binds to FoxO1, and promotes its degradation through the 26S proteasome pathway, even when Akt activity is inhibited on three phosphorylation sites, which are required for exclusion of FoxO1 from nucleus, can no longer be phosphorylated. We have recently demonstrated that the migration of XBP1s to nucleus is severely impaired in obesity conditions, and insulin signaling increases XBP1s nuclear translocation39. It is interesting to note that the role of IR signaling on XBP1s activity is not evolved in mice in the same manner when compared with the lower organisms such as C. elegans40. In C. elegans, loss of Daf-2 (IR-homolog), contrary to the mice, leads to an increase in the activity of IR/IRE1/XBP1 axis40.
One of the other surprising findings in the current work is the ability of a DNA-binding defective mutant XBP1s to increase glucose tolerance and reduce blood glucose levels in ob/ob mice when it is overexpressed in liver tissue. XBP1s is thought to mediate its effects by activation of a complex transcriptional program. Our results provide an important piece of evidence that XBP1s, in addition to triggering the transcriptional program, can regulate crucial processes in the cell via its interaction with other proteins.
We also show that XBP1s possesses anti-gluconeogenic activity through degradation of FoxO1. However, XBP1s can suppress gluconeogenic gene expression in the liver, and can improve glucose tolerance without concomitant alterations in insulin sensitivity. For example, experiments performed with low or medium expression of XBP1s in ob/ob mice do not reveal increased insulin sensitivity, but result in significantly reduced blood glucose and greatly enhanced glucose tolerance; however, these events are accompanied by significant reductions of FoxO1 levels in nuclear fractions of the liver. Nevertheless, XBP1s is capable of improving glucose tolerance in LIRKO mice or in liver-specific IRS1/2 double knockout mice. More importantly, XBP1s overexpression in the liver of STZ-treated type 1 diabetic mice leads to a significant decrease in the blood glucose levels in the fed state, and to euglycemia at the fasted state. These results collectively indicate that increasing the activity of XBP1s in the liver may be beneficial not only for insulin resistant-type 2 diabetics, but also for insulin deficient-type 1 diabetics.
On the other hand, high levels of XBP1s expression are associated with greatly increased insulin sensitivity: thus, ligand binding and phosphorylation of the IR in the presence of XBP1s leads to 25-fold greater downstream activation of Akt, compared to that seen in the control group. In parallel, however, IR protein levels are significantly down-regulated when XBP1s is expressed at high levels, in effect preventing the hypoglycemia that might otherwise be induced by the increased insulin sensitivity.
Uncovering the exact molecular mechanisms of XBP1s-induced degradation of FoxO1 will require further investigation. Our current working hypothesis is that by directly interacting with FoxO1, XBP1s drags FoxO1 to the 26S proteasome system. Our preliminary observations show that XBP1s has a very short half-life, which is around 10 minutes (data not shown); however, FoxO1 has a much longer half-life (~2 hours) than XBP1s (Fig. 1). We propose that during the very fast shuttling of XBP1 from cytoplasm to nucleus, and from nucleus to 26S proteasome, interaction of FoxO1 and XBP1s also directs FoxO1 to the 26S proteasome system, thus leading to its degradation. In addition, ER stress was previously linked to development of inflammation. Recently, it was shown that macrophages that are resident in the liver lead to activation of TL4 signaling in obesity and increase FoxO1 accumulation in the liver41. It is also possible that, increased XBP1s expression may reduce the inflammation and lead to a reduction in the activity of macrophages in the liver and also contribute to the exclusion of FoxO1 from the nucleus by utilizing this pathway.
The link between XBP1s and FoxO1 might lead to the development of new therapeutic approaches for the treatment of type 2 diabetes in obesity. Insulin resistance probably developed as a protective mechanism to block further expansion of an organism during conditions in which the animal is consuming high levels of calories, and also to help in extending the life span of the organism. One of the best-known modifications that extend life is the inhibition of insulin receptor signaling23,42,43. This is not observed only in lower organisms, but also in mammalian systems, as exemplified by the report that adipocyte-specific depletion of the insulin receptor44, or brain-specific depletion of IRS2, significantly increases the life span45. Our results indicate that blood glucose levels of insulin-resistant mice can be regulated without increasing insulin sensitivity, by promoting the interaction of XBP1s with FoxO1. Therapies such as the use of peptides, which can mimic the domain of XBP1s that interacts with FoxO1, or alternative approaches such as use of small molecules that induce moderate increases in XBP1s levels, could hold great potential for the treatment of type 2 diabetes without increasing the insulin sensitivity. Moderate activation of XBP1s in obese states could be highly beneficial for the patients without creating the possible side effects of enhanced insulin sensitivity.
Furthermore, our studies also suggest that defective XBP1s action in obesity conditions is one of the mechanisms that contribute to accumulation of FoxO1 in the nucleus46. Finally, XBP1 and FoxO1 play key roles in many disease states, including metabolism, cancer, neurodegeneration, differentiation and immunology22–24. We believe that convergence of the pathways associated with these two proteins will provide unique insights in diverse fields and will help in understanding the molecular mechanisms of important biological processes.
ONLINE METHODS
Pulse–chase assay
We cultured MEFs in 6–well plates at a density of 2 × 105 cells per well and infected cells with adenoviruses. After 16–h incubation with adenovirus, we washed cells with PBS twice and then starved cells in DMEM (cysteine−, methionine−) supplemented with 10% dialyzed FBS, 10 U ml−1 penicillin, and 1 µg ml−1 streptomycin at 37 °C for 30 min. We added 35S–cysteine and 35S–methionine to the starvation medium at a final concentration of 100 µCi ml−1. After 30–min pulse, we washed cells with PBS twice and incubated cells with complete DMEM at 37 °C for 0, 1, 2, 4, 6, or 8 h. We lysed cells with modified L–RIPA buffer (50 mM Tris–HCl, pH 7.5; 2 mM EGTA; 0.3% CHAPS; 100 mM NaF; 10 mM Na4P2O7; 1 mM Na3VO4; 10 µg ml−1 Leupeptin; 10 µg ml−1 Aproptonin; 2 mM PMSF and 20 nM Okadaic acid) and centrifuged cell lysates at 16,100g, 4 °C for 20 min. We incubated cleared cell lysates with FoxO1–specific antibody overnight and then with Protein A–sepharose beads for additional 2 h at 4 °C. Subsequently, we boiled beads in 2 × Laemmli buffer for 5 min and resolved protein samples by SDS–PAGE, transferred proteins onto polyvinylidene fluoride membranes, and exposed blots to films for autoradiography.
Dithiobis (succinimidyl propionate) crosslinking and immunoprecipitation
We infected MEFs with adenovirus for 24 h. Following an overnight starvation, we treated cells with DMSO or 10 µM MG132 for 2 h. We washed MEFs with ice-cold PBS twice to completely remove residual medium and incubated cells with 4 ml of PBS containing 1 mg ml−1 Dithiobis (succinimidyl propionate) at room temperature for 10 min. We stopped the crosslinking reaction by adding 400 µl of 1 M Tris–HCl (pH 8.5) and by incubating at room temperature for 1 min. Then, we washed cells with PBS and lysed with 800 µl of L–RIPA buffer. We centrifuged cell lysates at 16,100g, 4 °C for 20 min and collected supernatants. We incubated supernatants with Flag–specific antibody overnight and then with Protein G–sepharose beads for additional 2 h at 4 °C. We washed beads with L–RIPA buffer supplemented with 150 mM NaCl and boiled beads in 2 × Laemmli buffer for 5 min. Subsequently samples were subjected to Western blotting with FoxO1–specific antibody.
Mammalian two–hybrid assay
We amplified mouse cDNAs encoding XBP1s and FoxO1 using PCR with the following primers:
XBP1s–Fw5’-ATCGTCGACTAATGGTGGTGGTGGCAGCGGCGCCGA-3’
XBP1s–Rv5’-TAGTCTAGATTAGACACTAATCAGCTGGGGGAA-3’
FoxO1–Fw5’-ATCGTCGACTAATGGCCGAAGCGCCCCAGGTGGT-3’
FoxO1–Rv5’-TAGTCTAGATTAGCCTGACACCCAGCTGTGTG-3’
We digested PCR products with restriction enzymes and cloned them into pM or pVP16 vectors. We screened for positive colonies by restriction enzyme digestion and confirmed by sequencing. We seeded CHO cells in 12–well plates at a density of 1 × 105 cells per well and transfected cells with 45 ng of pG5–Luc (Firefly luciferase), 5 ng of pRL–CMV (Renilla luciferase), 230 ng of pM constructs and 230 ng of pVP16 constructs. We lysed cells with 500 µl of lysis buffer (Promega) 24 h after transfection and used 20 µl of cell lysates for luciferase activity measurement with Dual–Luciferase Reporter Assay System (Promega) following the manufacturer’s instruction.
Construction of a DNA binding defective mutant of XBP1s
We replaced XBP1s DNA binding domain (DBD, amino acids 61–90) with an artificial nuclear localization sequence encoding PKKKRKV using PCR. We used pENTR3C–XBP1s as a template and the following oligonucleotides as primers.
Fw: 5’-CCTAAGAAGAAGCGTAAGGTCCTGGAGCAGCAAGTGGTGGATTTG-3’
Rv: 5’-GACCTTACGCTTCTTCTTAGGCAGGTGCGTGAGCCGCTGCCGCTT-3’
PCR conditions were: 94 °C for 3 min; 25 cycles of 94 °C for 30 sec, 55°C for 30 sec, and 72 °C for 3 min; 72°C for 7 min. We phosphorylated PCR products at 5’–terminus with T4 polynucleotide kinase at 37°C for 30 min and then ligated DNA with T4 DNA Ligase at 16°C for 24 h. We identified the positive colonies by restriction enzymes digestion and confirmed by sequencing. We sub-cloned the construct pENTR3C–XBP1(ΔDBD) into pAd vector by homologous recombination to produce an adenovirus vector expressing XBP1(ΔDBD).
Analysis of in vivo insulin receptor signaling
We fasted mice for 6 h (8 am –2 pm) prior to experiments. Following anesthetization of mice with xylazine–ketamine, we infused insulin (0.5 IU kg−1) or saline into mice through the portal vein. Five minutes after infusion, liver tissues were excised, flash frozen in liquid nitrogen, and stored in −80 °C until processing.
Statistical Analysis
We presented data as means ± standard error of the mean (s.e.m.). We used two–tailed Student's t tests to determine P values for statistical significance.
Additional methods. Detailed methodology is described in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Ozcan laboratory for their help during the execution of the experiments. We thank L. Glimcher (Harvard School of Public Health) for providing us with the Xbp1Flox/Flox mouse strain. We are grateful to D. Accili (Columbia University) for providing us with various FoxO1 adenoviruses and to R. Kahn (Joslin Diabetes Center) for his permission to use LIRKO mice in our studies. We thank J. Majzoub (Harvard Medical School) for providing us with mammalian two-hybrid system and R. King (Harvard Medical School) for providing us with an ubiquitin-expressing plasmid. This study was supported by junior faculty start-up funds provided to U.O. by Children’s Hospital Boston, an RO1 grant (R01DK081009) provided to U.O. by US National Institute of Health, and the Timothy Murphy funds provided to Division of Endocrinology, Children’s Hospital Boston.
Footnotes
Authors do not have any conflict of interest.
AUTHOR CONTRIBUTIONS
Y.Z. came up with the hypothesis, designed and performed the experiments, analyzed the data and wrote the manuscript. J.L., C.S., S.W.P, J.C. and J.L. performed the experiments. C.M.R. and S.J.F performed experiments. M.F.W. provided DKO mice. S.B.B. provided LIRKO mice and performed experiments. U.O. came up with the hypothesis, designed and performed the experiments, analyzed the data and wrote the manuscript.
REFERENCES
- 1.James PT, Rigby N, Leach R. The obesity epidemic, metabolic syndrome and future prevention strategies. Eur J Cardiovasc Prev Rehabil. 2004;11:3–8. doi: 10.1097/01.hjr.0000114707.27531.48. [DOI] [PubMed] [Google Scholar]
- 2.Kitamura Y, Accili D. New insights into the integrated physiology of insulin action. Rev Endocr Metab Disord. 2004;5:143–149. doi: 10.1023/B:REMD.0000021436.91347.93. [DOI] [PubMed] [Google Scholar]
- 3.de Luca C, Olefsky JM. Stressed out about obesity and insulin resistance. Nat Med. 2006;12:41–42. doi: 10.1038/nm0106-41. discussion 42. [DOI] [PubMed] [Google Scholar]
- 4.Biddinger SB, Kahn CR. From mice to men: insights into the insulin resistance syndromes. Annu Rev Physiol. 2006;68:123–158. doi: 10.1146/annurev.physiol.68.040104.124723. [DOI] [PubMed] [Google Scholar]
- 5.Saltiel AR. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell. 2001;104:517–529. doi: 10.1016/s0092-8674(01)00239-2. [DOI] [PubMed] [Google Scholar]
- 6.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 7.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 8.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 9.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 10.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 12.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 14.Lee K, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 16.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozcan U, et al. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozcan U, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozcan U, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 21.Ozcan L, et al. Endoplasmic reticulum stress plays a central role in development of leptin resistance. Cell Metab. 2009;9:35–51. doi: 10.1016/j.cmet.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- 23.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 24.Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death. Nat Immunol. 2009;10:1057–1063. doi: 10.1038/ni.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puigserver P, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto M, Accili D. The tangled path to glucose production. Nat Med. 2006;12:33–34. doi: 10.1038/nm0106-33. discussion 34. [DOI] [PubMed] [Google Scholar]
- 27.Daitoku H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motta MC, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 29.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 30.Yamagata K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003;100:11285–11290. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aoki M, Jiang H, Vogt PK. Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc Natl Acad Sci U S A. 2004;101:13613–13617. doi: 10.1073/pnas.0405454101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakae J, Kitamura T, Silver DL, Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest. 2001;108:1359–1367. doi: 10.1172/JCI12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong XC, et al. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 2008;8:65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michael MD, et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- 37.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Ozawa K, et al. The endoplasmic reticulum chaperone improves insulin resistance in type 2 diabetes. Diabetes. 2005;54:657–663. doi: 10.2337/diabetes.54.3.657. [DOI] [PubMed] [Google Scholar]
- 39.Park SW, et al. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat Med. 2010;16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henis-Korenblit S, et al. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci U S A. 2010;107:9730–9735. doi: 10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan W, et al. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 2010 doi: 10.1038/emboj.2010.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 43.Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat Rev Mol Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- 44.Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 45.Taguchi A, Wartschow LM, White MF. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317:369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 46.Altomonte J, et al. Inhibition of Foxo1 function is associated with improved fasting glycemia in diabetic mice. Am J Physiol Endocrinol Metab. 2003;285:E718–E728. doi: 10.1152/ajpendo.00156.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.