Abstract
Many fungal cell adhesion proteins form functional amyloid patches on the surface of adhering cells. The Candida albicans Agglutinin-like sequence (Als) adhesins are exemplars for this phenomenon, and have amyloid forming sequences that are conserved between family members. The Als5p amyloid sequence mediates amyloid fibril formation and is critical for cell adhesion and biofilm formation, and is also present in the related adhesins Als1p and Als3p. We have developed a fluorescent peptide probe containing the conserved Als amyloid-forming sequence. This peptide bound specifically to yeast expressing Als5p, but not to cells lacking the adhesin. The probe bound to both yeast and hyphal forms of C. albicans. Δals1/Δals3 single and double deletion strains exhibited reduced fluorescence, indicating that probe binding required expression of these proteins. Additionally, the Als peptide specifically stained fungal cells in abscesses in autopsy sections. Counterstaining with calcofluor white showed colocalization with the amyloid peptide. In addition, fungi in autopsy sections derived from the gastrointestinal tract showed colocalization of the amyloid-specific dye thioflavin T and the fluorescent peptide. Collectively, our data demonstrate that we can exploit amyloid sequence specificity for detection of functional amyloids in situ.
Introduction
Amyloids are structured insoluble aggregates consisting of many molecules of the same protein. These protein arrays are often associated with disease states such as AL amyloidosis and Alzheimer’s diseases. Nevertheless, there are proteins whose amyloids are a normal part of an organism’s biology [1], [2]. Such functional amyloids exist in many organisms; from prokaryotes to eukaryotes [3]–[6]. They are integral to many biological functions such as cell-cell contacts, biofilm formation, scaffolding and substrate adhesion [6]–[9]. In fact, microbial amyloids are not only important for biofilm formation but also to modulate host-microbe interactions [7], [8], [10]. Many microbes utilize functional amyloids to attach to and colonize the host; which may allow for host infection or commensalism [6], [7], [10].
Candida albicans is a fungal member of the human microbiome [11]–[13]. This yeast colonizes the skin, oral, gastrointestinal and urogenital tracts of healthy individuals and can cause nosocomial infections in immunocompromised patients [11], [14], [15]. Candida infections can be superficial or systemic, including, respectively oral thrush and often-fatal candidemias [15]. Additionally, C. albicans forms mixed fungal-bacterial biofilms with increased resistance to antifungals and antibacterials [16]–[19]. The ability of this fungus to aggregate, adhere to host cells, form biofilms, and even modulate the host response is attributed to amyloid-forming adhesion proteins belonging to the Als adhesin family [7], [20]–[24].
Cell adhesion is critical to both the commensal and infectious states of C. albicans [25]–[28]. Among the adhesins, the Agglutinin-like sequence (Als) family includes cell wall-linked glycoproteins encoded at 8 loci [29], [30]. Each member of the Als family contain N-terminal Ig-like invasin domains (Ig) which define substrate specificity and a variable number of 36-amino acid tandem repeats (TR domain) that contribute to substrate binding [31]–[34]. There is a glycosylated serine-threonine–rich domain and a GPI anchor that covalently links the protein to the cell [35]. Located between the Ig and TR domains is the T domain, which contains the amyloid-forming sequence [36], [37]. This sequence mediates aggregation of Als proteins into cell surface clusters termed nanodomains composed of functional amyloids [38], [39].
Functional amyloids formed by the C. albicans adhesin Als5p have been extensively studied using a Saccharomyces cerevisiae expression system [7], [23], [36]–[40]. Soluble Als5p forms amyloid fibers that show characteristic binding of the amyloid binding dyes thioflavin-T, thioflavin-S and Congo red [36], [37]. Amyloid-like patches formed on the cell surface are critical for the functional properties of Als5p, including cell aggregation, biofilm formation, and host immunomodulation [7], [38], [39]. Data from these studies are proving to be a predictive model for studying functional amyloids formed on the surface of C. albicans in situ [38], [39], [41]. Recently, amyloids have been demonstrated on fungi in abscesses in the human intestinal tract. Moreover these amyloids bind to human serum amyloid P component, a soluble pattern recognition receptor with anti-inflammatory properties [42]. Thus assessing the role of these amyloids in infections is important for our understanding of fungal disease.
We have recently demonstrated the presence of surface amyloids on fungi in invasive disease in humans, but the nature of these amyloids is unknown. They might be of host or fungal origin, or both, and their composition is unknown. Therefore, we have established a method to label an amyloid sequence and have demonstrated amyloid composed of Als proteins in infected tissue.
Results
A fluorescent labeled peptide containing the amyloid sequence from Als5p binds to Als5p-expressing yeast cells
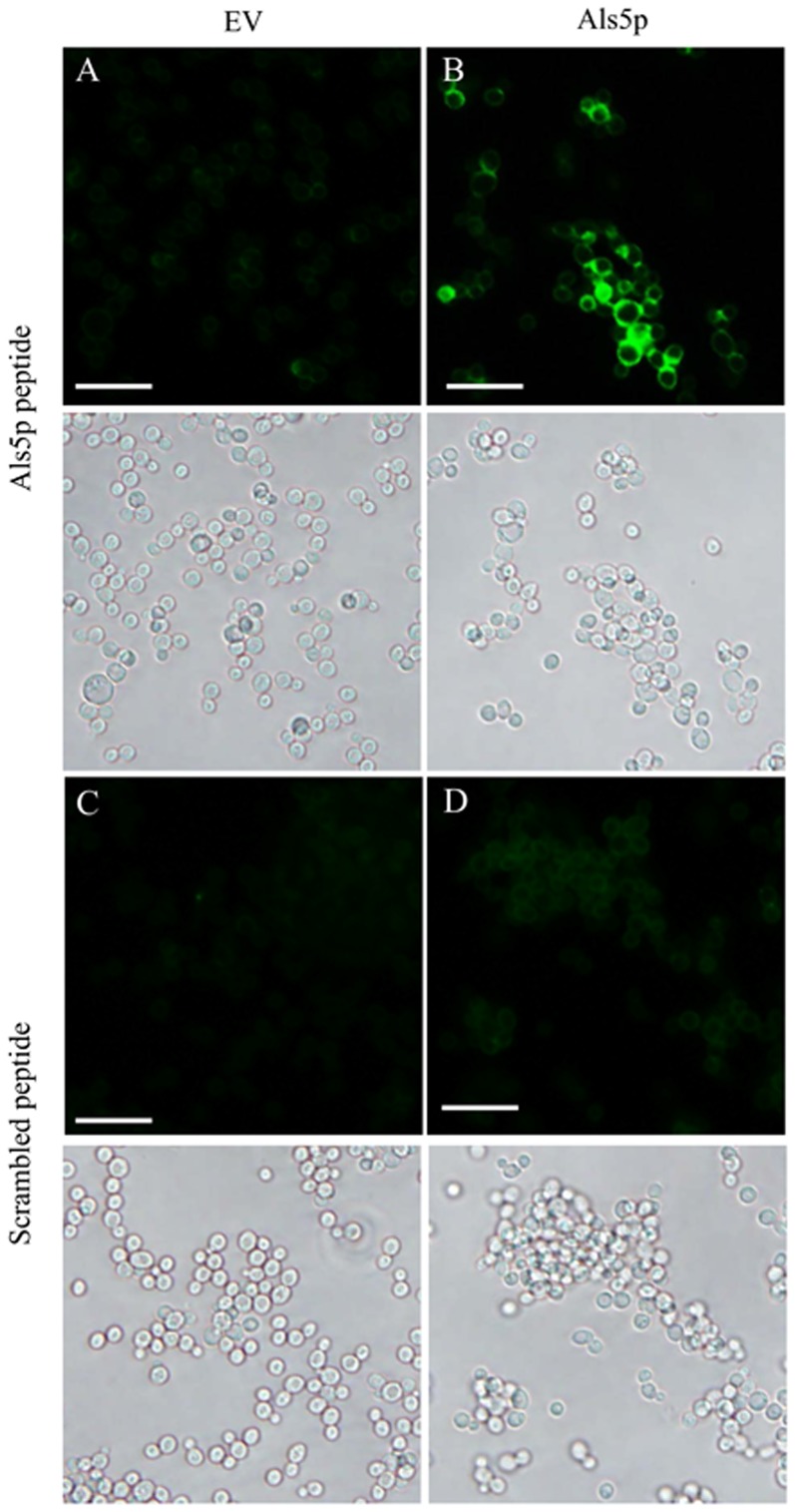
Previously we have shown that a peptide containing the amyloid sequence common to Als1p, Als3p, and Als5p (SNGIVIVATTRTV) can form amyloids, and that exogenous peptide can reverse the loss of aggregation in S. cerevisiae expressing an amyloid deficient mutation Als5pV326N [36], [39]. We proposed that this reestablishment of aggregation occurs by peptide binding to the mutant protein and serving as a template for amyloid formation [36], [40], [43]. To determine the ability of this protein sequence to bind S. cerevisiae-expressing Als5p, we labeled a peptide containing the amyloid forming sequence of Als5p with fluorescein. The labeled amyloid-forming peptide bound specifically to a strain of S. cerevisiae expressing Als5p and not to cells harboring the empty vector (Figure 1A and 1B). A scrambled peptide (VITGNTNIRTSVA), containing the same amino acid composition, stained cells poorly (Figure 1C and 1D).
Figure 1. Als5p expressing S. cerevisiae cells stained with fluorescent peptides.
S. cerevisiae harboring an empty vector (A,C) or expressing Als5p (B, D) were stained with amyloid peptide (200 µg/ml; A, B) or a scrambled-sequence peptide of identical composition (200 µg/ml; C, D). Lower micrographs are bright field images. All scale bars are 30 µm.
The Als5p amyloid-forming peptide binds to C. albicans in both yeast and hyphal forms
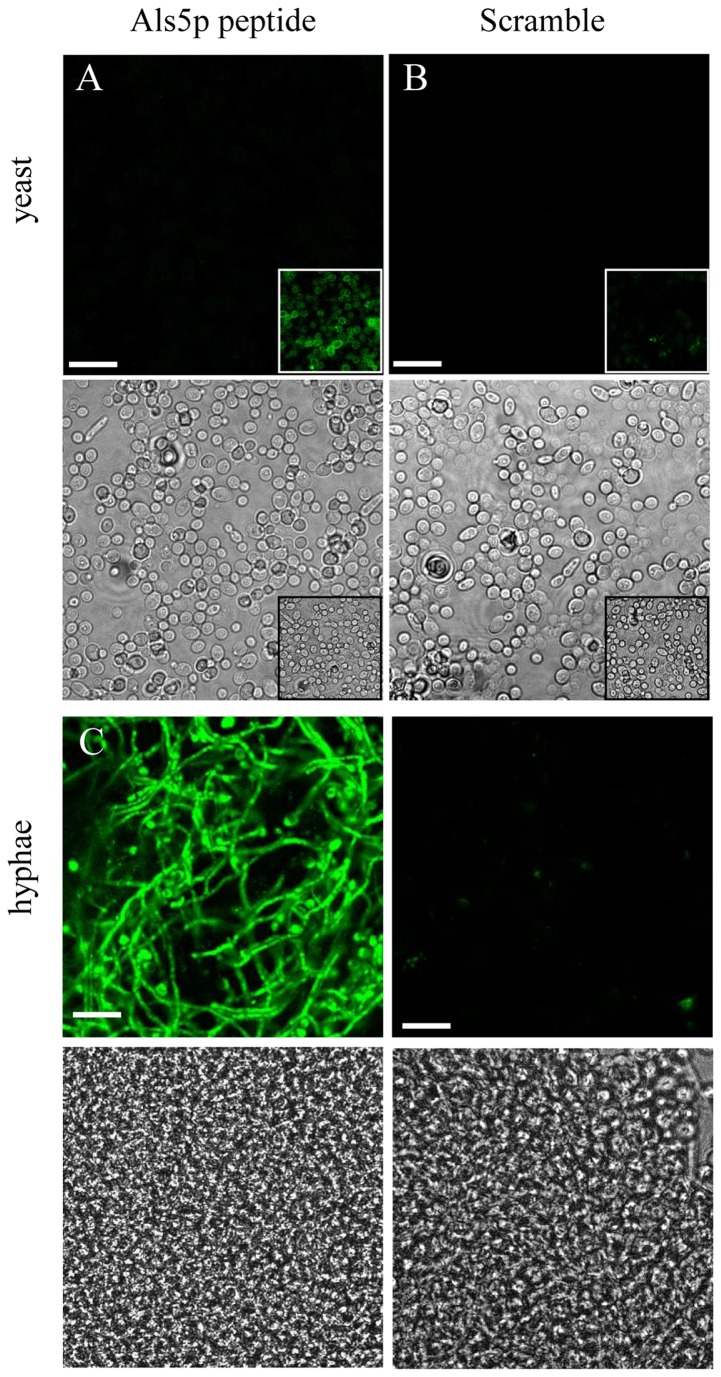
Als1p and Als5p are constitutively expressed at low levels in both yeast and hyphal forms of C. albicans [25]. However, the hyphal form of the fungus expresses high levels of Als3p, and in some conditions Als1p as well [21], [44]. Since Als1p, Als3p, and Als5p contain identical amyloid forming sequences, we hypothesized that the Als5p peptide would therefore bind to both hyphae and yeast forms [36]. Fluorescence microscopy revealed a higher level of fluorescence in hyphae versus yeast (Figure 2A and B). This binding was sequence-specific in both cases, because much less cellular fluorescence was seen when the cells were treated under similar conditions with the scrambled peptide (Figure 2B).
Figure 2. Confocal micrographs of C. albicans yeast and hyphae binding Als5p peptide.
C. albicans cultured in spider medium, with 20 µg/ml (A, C) amyloid or (B, D) scrambled sequence peptide. Fields were imaged containing (A, B) yeast or (C, D) hyphae at a gain of 6.5. Inset images in A and B were acquired at a higher gain of 7.0.
C. albicans als1/als1 als3/als3 deletion strains show reduced binding of the Als amyloid-forming peptide
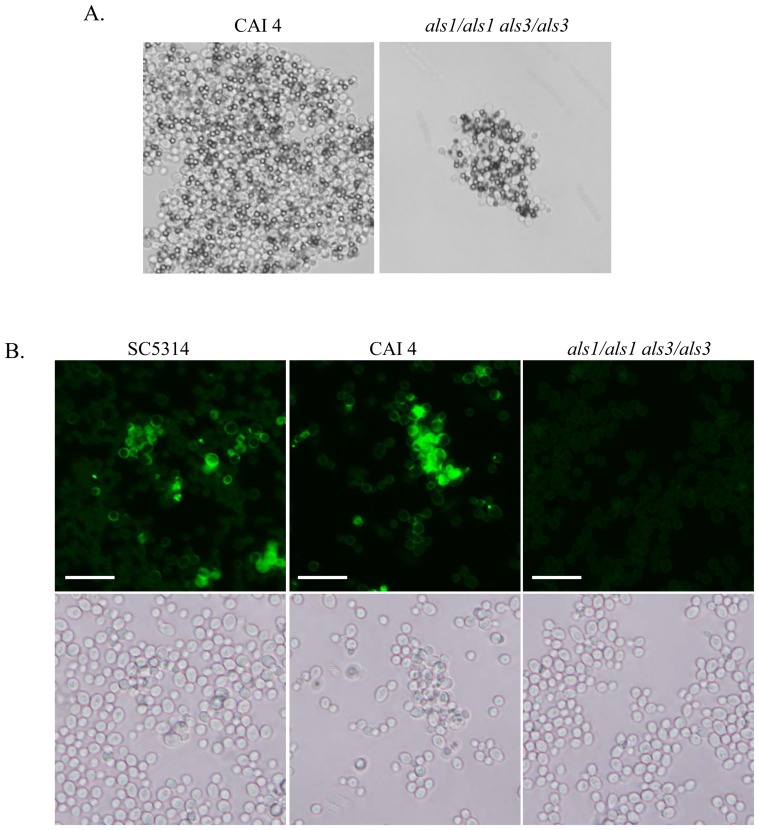
Als1p and Als3p are major adhesins displayed on yeast phase and hyphae, respectively [21], [44]; therefore, we hypothesized that the amyloid-forming peptide binds not only to the amyloid region of Als5p but also to the homologous regions of these other adhesins. We tested this hypothesis with an als1/als1 als3/als3 deletion strain. To confirm that the als1/als1 als3/als3 cells display decreased adhesins we tested for an aggregation defect. The deletion strain aggregated, although less strongly than the parental strain CAI4 (Figure 3A). The labeled peptide stained yeast forms of C. albicans CAI4 and SC5314 more brightly than the yeast of the als1/als1 als3/als3 mutant (Figure 3B).
Figure 3. Aggregation and amyloid peptide staining of C. albicans.
A) Aggregates of wild type (CAI4) and double deletion strains binding with heat denatured BSA-coated magnetic beads. B) The yeast form of C. albicans strains, SC5314, CAI4 and als1/als1 als3/als3 double deletion were probed with 20 µg/ml amyloid peptide. All scale bars are 20 µm.
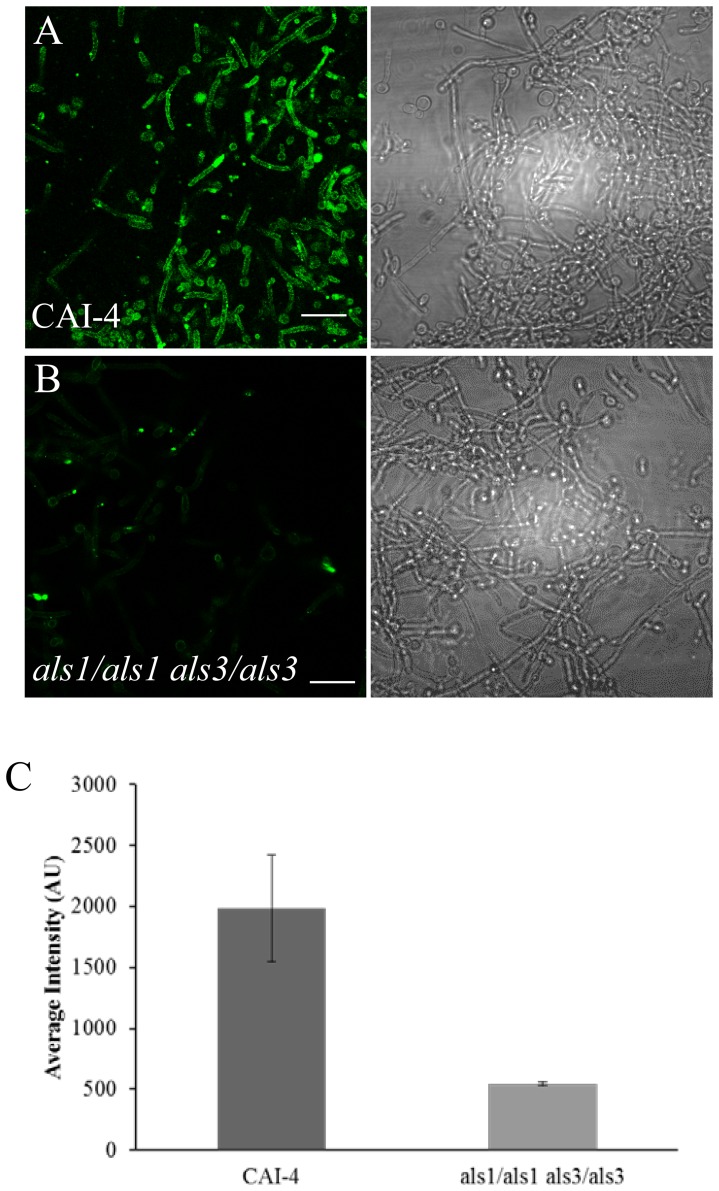
The pattern was also apparent in hyphae induced with spider medium. The hyphal-induced als1/als1 als3/als3 strain consistently exhibited a 75% decrease in fluorescence when compared to the wild-type CAI4 (Figure 4). Single gene deletion strains of als1/als1 and als3/als3 showed variable decreases in labeling (data not shown). Therefore, the majority of the labeling was due to the peptide binding to Als1p or Als3p or both.
Figure 4. Peptide staining of als1/als1 als3/als3 deletion strains.
All strains were induced to form hyphae by incubating overnight in spider media at 37°C. A) CAI4 and B) als1/als1 als3/als3 double deletion strains were probed with 20 µg/ml amyloid peptide with their corresponding brightfield images (right). All scale bars are 40 µm. C) Quantification of the average intensity of images A and B. Error bars represent standard error of the mean (n = 4; see text for details).
Fluorescent peptide binds specifically to fungi in autopsy tissue from candidiasis patients
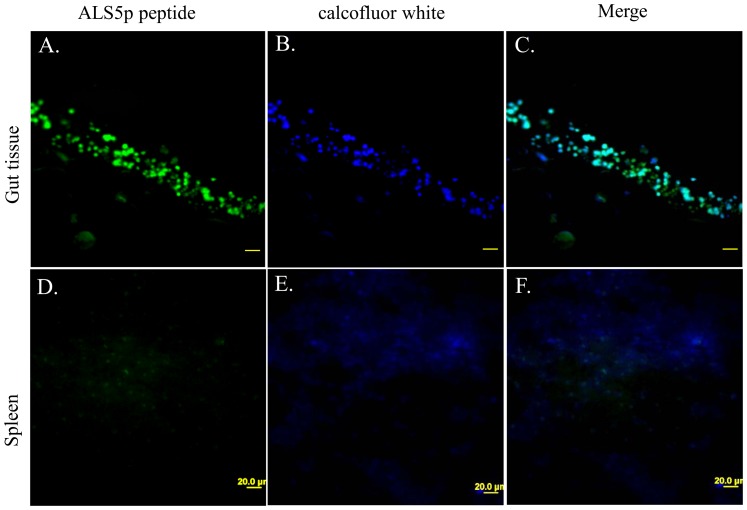
Amyloid binding dyes bind to yeast in autopsy sections of patients with candidiasis, but the composition of the amyloid is not known [8]. We therefore stained with the Als-specific probe, looking for co-localization with the amyloid binding dyes in situ. We stained autopsy gastrointestinal tissue positive for both yeast and filamentous forms of C. albicans (Figure 5). Additionally, we stained autopsy spleen samples with the fungal specific dye calcofluor white and with the amyloid peptide. The amyloid peptide efficiently stained yeast in gastrointestinal autopsy sections, but not in an uninfected spleen (Figure 6A and D). In the stained regions, the amyloid peptide co-localized with calcofluor white (Figure 6C). Negative control spleen samples did not show amyloid peptide staining and only diffuse faint calcofluor white fluorescence (Figure 6D-F).
Figure 5. Gomori’s Methenamine Silver stained autopsy section.
. Candida yeast and filamentous forms (hyphae and pseudohyphae) invading human gastrointestinal epithelium at the luminal border [8]. Tissue is a green-blue and the fungi are black.
Figure 6. Autopsy sections stained with Als5p amyloid peptide and calcofluor white.
A-C) Confocal microscopy of gut section stained with A) amyloid peptide (200 µg/ml), B) calcofluor white, C) merged image. D-F) Spleen section stained with D) amyloid peptide (200 µg/ml), E) calcofluor white, F) merged image.
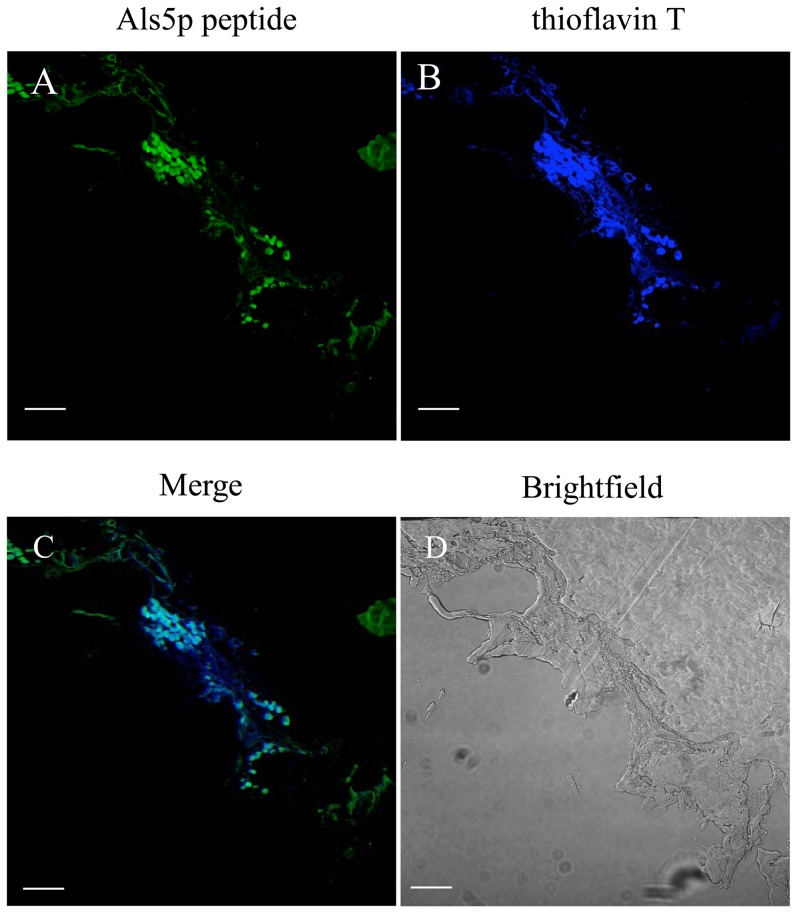
To demonstrate that the peptide was staining the same structures previously observed with amyloid dyes [8], we co-stained autopsy samples from candidiasis patients with thioflavin T and the Als5p amyloid peptide (Figure 7). Yeast and hyphae were positive for both thioflavin T and the amyloid peptide and showed co-localization of the dyes (Figure 7A-C). These results demonstrated that the sequence-specific peptide stained Als amyloids in the tissue.
Figure 7. Autopsy section stained with Als5p amyloid peptide and thioflavin T.
Confocal images of A-C) gut section stained with A) amyloid peptide (200 µg/ml), B) thioflavin T (100 nM), C) merged image, D) brightfield. All scale bars are 40 µm.
Discussion
We have demonstrated that a fluorescent peptide, designed to target the amyloid-forming sequence from the C. albicans adhesin Als5p, bound specifically to S. cerevisiae cells that express Als5p. This peptide also labeled C. albicans yeast and hyphal forms. The peptide showed significantly reduced binding to a strain deleted for the adhesins Als1p and Als3p. Thus, the amyloid peptide bound specifically to proteins that have the same amyloid sequence as Als5p. The peptide co-localized with the fungal-specific dye calcofluor white and with a specific amyloid dye, thioflavin T. These results demonstrate the peptide’s specificity in situ. This peptide is a sensitive probe for fungi in autopsy specimens from patients inflicted with candidiasis. We propose that the approach presented here has potential as a new tool for detection of functional amyloids, and specific peptide designs for disease treatment.
We propose that the specific peptide binding observed exploits the sequence specificity of amyloid formation. Both amyloid nucleation and growth are dependent on sequence similarity [39], [41], [43], [45]. Among C. albicans Als proteins, Als1p, Als3p, and Als5p share amyloid sequences and thus are predicted to bind the same amyloid peptide [36]. We have previously shown that the amyloid peptide can potentiate in vitro amyloid formation, cell aggregation, and biofilm formation [36], [39]. In contrast, a mutated non-amyloid version of the peptide inhibits these activities; therefore, we expected the specific binding of the labeled amyloid-forming peptide. The specificity was supported by several approaches. First only Als-expressing S. cerevisiae bound the peptide, and not a scrambled sequence peptide. Second, the peptide labeled C. albicans in the pattern expected for expression of Als1p, Als3p and Als5p, showing stronger binding to hyphae than to yeast cells in vitro (Figure 2). Collectively, these data indicate that the amyloid sequence-specific peptides can be exploited to design probes to study amyloid forming proteins. This specificity is based on the peptide’s ability to template amyloid formation and to be incorporated in the amyloid structure.
Amyloid dyes, such as thioflavin T and Congo red have been used as pathologists’ gold standard for the detection of amyloids [46], [47]. Although these dyes are inhibitory at high concentrations they can be titrated down to concentrations useful for detection without functional interference [37], [39]. These dyes recognize not only fungal cell wall amyloids, but they also recognize host amyloids. Therefore these dyes would not distinguish the contribution of amyloid from the host or the microbe. The specificity of the Als peptide allows for the resolution of the amyloid source.
Our peptide-based approach allows detection of amyloid without interfering with amyloid function. For instance, this ability could be useful in amyloid-targeted antifungal drug discovery through the ability to mark and quantify amyloid interactions. A similar method has been developed for detecting and quantifying amyloid-beta peptide aggregation by labeling peptide with a quantum nanodot [48]. Presumably a nanodot-coupled Als peptide in a similar assay would lead to discovery of amyloid-targeted antifungal drugs; the signal would decrease as amyloids were inhibited. Thus we could measure the disruption of fungal amyloids in a library screen; thereby indicating to what extent candidate drugs disrupt functional amyloids.
In addition to in vitro assays, our peptide approach coupled with nanodot technology and multiphoton microscopy could be utilized as a real-time detection of the initiation and progression of infection by following amyloid kinetics in animal models of infection. This approach has been utilized to track amyloid initiation and maturation in Alzheimer’s disease mouse models [49].
Thus we have used a novel tool to document the presence of a specific fungal amyloid in infections. This new method confirms and extends our previous report of amyloids in autopsy samples from patients with invasive candidiasis [8]. Specifically, functional Als amyloids are present in situ in infected tissue. We note that the probe successfully labeled an uncharacterized C. albicans strain in these tissues, indicating that this clinical strain contains the highly conserved amyloid forming sequence observed in characterized laboratory strains [36]. In addition, C. albicans amyloids bind to serum amyloid P component, a possible modulator of innate immunity, both in vitro and in situ [8], [42]. Thus the results also support mechanistic studies demonstrating that Als protein amyloid formation is necessary for cell adhesion, fungal aggregation, biofilm formation and binding of serum amyloid P component [8], [37], [39], [40]. The amyloid labeling approach described here may also have more general application for other microbe-derived amyloids [1]–[3], [6].
Materials and Methods
Ethics statement
The tissue used in this study was part of a University of Arizona IRB-approved study where we were granted a waiver/exemption for autopsy material. The University of Arizona permission for autopsy is signed by the next of kin and grants use of tissue for research (unspecified).
Strains and growth conditions
Candida albicans strains CAI4 (CAI4-URA3) and als1/als1 als3/als3 (CJN1348) strains were from Aaron Mitchell (Carnegie Mellon) with the exception of SC5314 which was obtained from ATCC (www.atcc.org) [50]. Als5p (pJL1), Als5pV326N (pJL1V326N) and empty vector (pJL1EV) S. cerevisiae strains were generated as previously described [39].
S. cerevisiae strains were cultured as previously described [39]. Briefly, S. cerevisiae strains were grown at 30°C on CSM-trp plates enriched with 40 mg/L of adenine and 2% galactose. Liquid cultures were grown at 24°C in galactose and adenine enriched CSM-trp with shaking at 170 rpm to an approximate OD600 = 1.0.
C. albicans strains were grown at 30°C on YPD agar plates. For induction of hyphae, C. albicans cells from overnight cultures were washed 3 times with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4) buffer, and diluted to OD600 = 0.5 in Spider medium and incubated for 16 h at 37°C with shaking at 170 rpm [51].
Peptide labeling
Previously described peptides containing the Als5p amyloid sequence and scrambled sequence were synthesized at The Rockefeller University Proteomics Resource Center (New York, New York) [36], [39]. Peptides were labeled, utilizing fluorescein isothiocyanate adsorbed onto celite, according to the manufacturer’s suggested protocol with minor changes (Sigma-Aldrich, St. Louis, MO). More specifically, the lyophilized peptides were reconstituted in 0.1 M carbonate-bicarbonate buffer, pH 9.0 at a concentration of 1 mg/ml and FITC-celite was added to a final concentration of 1.5 mg/ml and incubated at 24°C for 18h. Celite was removed by centrifugation at 2000 rpm for 3 min. Peptides were aliquoted in tubes at a stock concentration of 1 mg/ml and stored at –80°C.
Fungal-peptide hybridization
Yeast cultures were grown as described above. 2×107 yeast cells were washed 3 times with PBS and incubated with 200 or 20 µg/ml of peptide in PBS for 20 min. at room temperature. After incubation cells were washed 3 times with PBS. Hyphal-induced cultures were washed 3 times with PBS and incubated at room temperature with FITC-labeled Als5p amyloid peptide (200 µg/ml or 20 µg/ml) for 30 min. Cultures were washed 3 times with PBS and assessed by microscopy.
Human Tissue-peptide hybridization
Autopsy specimens from patients with histological evidence of invasive candidiasis of the gastrointestinal tract were provided by authors RES and SAK (University of Arizona, Tucson, AZ) in accordance with IRB-approved procedures. Tissues were prepared for staining as previously described [8]. Fixed slides were incubated with 200 µg/ml of peptide for 20 minutes at room temperature. The slides were washed 3X with PBS and stained with 100 nM thioflavin T (in PBS) for 30 minutes. Calcofluor white staining was performed in accordance with the manufacturer’s instructions (Sigma-Aldrich).
Microscopy and quantification of images
Confocal images were acquired using a Nikon Eclipse 90i confocal microscope with 488-nm excitation and 515-nm emission filters for samples probed with the FITC conjugated peptide. Images of samples stained with Thioflavin T or calcofluor white were acquired with 408-nM excitation and 450-nM emission filter settings. Quantified images were analyzed using the Nikon EZ-C1 Gold version 3.90 software. Fluorescent images, representing a single field per sample, were opened and set to view in 1D graph mode with the x-axis set. The highest fluorescent peak expressed in arbitrary units was noted for four y-axis positions in each field. The mean and standard deviation were determined for the four y-axis positions in four fields.
Funding Statement
This work was supported by National Institutes of Health grants R01 GM098616 and SC1 GM083756 to PNL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fowler DM, Koulov AV, Balch WE, Kelly JW (2007) Functional amyloid – from bacteria to humans. Trends Biochem Sci 32(5): 217–224. [DOI] [PubMed] [Google Scholar]
- 2. Maury CPJ (2009) The emerging concept of functional amyloid. J Intern Med 265(3): 329–334. [DOI] [PubMed] [Google Scholar]
- 3. Oli MW, Otoo HN, Crowley PJ, Heim KP, Nascimento MM, et al. (2012) Functional amyloid formation by streptococcus mutans . Microbiology 158(Pt 12): 2903–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Falabella P, Riviello L, Pascale M, Lelio ID, Tettamanti G, et al. (2012) Functional amyloids in insect immune response. Insect Biochem Mol Biol 42(3): 203–211. [DOI] [PubMed] [Google Scholar]
- 5. Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, et al. (2009) Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325(5938): 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwartz K, Boles BR (2013) Microbial amyloids--functions and interactions within the host. Curr Opin Microbiol 16(1): 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bois M, Singh S, Samlalsingh A, Lipke PN, Garcia MC (2013) Does candida albicans Als5p amyloid play a role in commensalism in caenorhabditis elegans? Eukaryot Cell 12(5): 703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gilchrist KB, Garcia MC, Sobonya R, Lipke PN, Klotz SA (2012) New features of invasive candidiasis in humans: Amyloid formation by fungi and deposition of serum amyloid P component by the host. J Infect Dis 206(9): 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR (2012) Diversity, biogenesis and function of microbial amyloids. Trends Microbiol 20(2): 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tükel Ç, Wilson RP, Nishimori JH, Pezeshki M, Chromy BA, et al.. (2009) Responses to amyloids of microbial and host origin are mediated through toll-like receptor 2. Cell Host & [DOI] [PMC free article] [PubMed]
- 11. Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, et al. (2010) Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6(1): e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huffnagle GB, Noverr MC (2013) The emerging world of the fungal microbiome. Trends Microbiol 21: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peleg AY, Hogan DA, Mylonakis E (2010) Medically important bacterial-fungal interactions. Nat Rev Microbiol 8(5): 340–349. [DOI] [PubMed] [Google Scholar]
- 14.Soll DR, Galask R, Schmid J, Hanna C, Mac K, et al. (1991) Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J Clin Microbiol 29(8): 1702–1710. 23 May 2013. [DOI] [PMC free article] [PubMed]
- 15.Gautam H, Kaur R, Goyal R, Bhalla P, Dewan R. (2010) Oral thrush to candidemia: A morbid outcome. Journal of the International Association of Physicians in AIDS Care [DOI] [PubMed]
- 16. Harriott MM, Noverr MC (2010) Ability of Candida albicans mutants to induce staphylococcus aureus vancomycin resistance during polymicrobial biofilm formation. Antimicrob Agents Chemother 54(9): 3746–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klotz SA, Chasin BS, Powell B, Gaur NK, Lipke PN (2007) Polymicrobial bloodstream infections involving Candida species: Analysis of patients and review of the literature. Diagn Microbiol Infect Dis 59(4): 401–406. [DOI] [PubMed] [Google Scholar]
- 18. Rautemaa R, Ramage G (2011) Oral candidosis—clinical challenges of a biofilm disease. Crit Rev Microbiol 37(4): 328–336. [DOI] [PubMed] [Google Scholar]
- 19. Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF (2010) Interaction of Candida albicans cell wall Als3 protein with streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infection and Immunity 78(11): 4644–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aoki W, Kitahara N, Miura N, Morisaka H, Kuroda K, et al. (2012) Profiling of adhesive properties of the agglutinin-like sequence (ALS) protein family, a virulent attribute of Candida albicans . FEMS Immunol Med Microbiol 65(1): 121–124. [DOI] [PubMed] [Google Scholar]
- 21. Liu Y, Filler SG (2011) Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryotic Cell 10(2): 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klotz SA, Gaur NK, De Armond R, Sheppard D, Khardori N, et al. (2007) Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med Mycol 45(4): 363–370. [DOI] [PubMed] [Google Scholar]
- 23. Rauceo JM, Gaur NK, Lee KG, Edwards JE, Klotz SA, et al. (2004) Global cell surface conformational shift mediated by a Candida albicans adhesin. Infect Immun 72(9): 4948–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao X, Oh SH, Hoyer LL (2007) Deletion of ALS5, ALS6 or ALS7 increases adhesion of Candida albicans to human vascular endothelial and buccal epithelial cells. Med Mycol 45(5): 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Green CB, Marretta SM, Cheng G, Faddoul FF, Ehrhart EJ, et al. (2006) RT-PCR analysis of Candida albicans ALS gene expression in a hyposalivatory rat model of oral candidiasis and in HIV-positive human patients. Med Mycol 44(2): 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murciano C, Moyes DL, Runglall M, Tobouti P, Islam A, et al. (2012) Evaluation of the role of Candida albicans agglutinin-like sequence (Als) proteins in human oral epithelial cell interactions. PLoS One 7(3): e33362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng G, Wozniak K, Wallig MA, Fidel PL, Jr TrupinSR, et al. (2005) Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect Immun 73(3): 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ganguly S, Mitchell AP (2011) Mucosal biofilms of Candida albicans . Curr Opin Microbiol 14(4): 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sheppard DC, Yeaman MR, Welch WH, Phan QT, Fu Y, et al. (2004) Functional and structural diversity in the Als protein family of Candida albicans . J Biol Chem 279(29): 30480–30489. [DOI] [PubMed] [Google Scholar]
- 30. Hoyer LL, Green CB, Oh SH, Zhao X (2008) Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med Mycol 46(1): 1–15 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salgado PS, Yan R, Taylor JD, Burchell L, Jones R, et al. (2011) Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans . Proc Natl Acad Sci U S A 108(38): 15775–15779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frank AT, Ramsook CB, Otoo HN, Tan C, Soybelman G, et al. (2010) Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot Cell 9(3): 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rauceo JM, De Armond R, Otoo H, Kahn PC, Klotz SA, et al. (2006) Threonine-rich repeats increase fibronectin binding in the Candida albicans adhesin Als5p. Eukaryot Cell 5(10): 1664–1673 10.1128/EC.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoyer LL, Hecht JE (2001) The ALS5 gene of Candida albicans and analysis of the Als5p N-terminal domain. Yeast 18(1): 49–60. [DOI] [PubMed] [Google Scholar]
- 35. Ahmad MF, Yadav B, Kumar P, Puri A, Mazumder M, et al. (2012) The GPI anchor signal sequence dictates the folding and functionality of the Als5 adhesin from Candida albicans . PLoS One 7(4): e35305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Otoo HN, Lee KG, Qiu W, Lipke PN (2008) Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot Cell 7(5): 776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, et al. (2010) Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell 9(3): 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alsteens D, Garcia MC, Lipke PN, Dufrene YF (2010) Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc Natl Acad Sci U S A 107(48): 20744–20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrene YF, et al. (2011) A role for amyloid in cell aggregation and biofilm formation. PLoS One 6(3): e17632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, et al. (2012) Strengthening relationships: Amyloids create adhesion nanodomains in yeasts. Trends Microbiol 20(2): 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alsteens D, Ramsook CB, Lipke PN, Dufrene YF (2012) Unzipping a functional microbial amyloid. ACS Nano 6(9): 7703–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pepys MB (2012) Invasive candidiasis: New insights presaging new therapeutic approaches? J Infect Dis 206(9): 1339–1341. [DOI] [PubMed] [Google Scholar]
- 43. Dumoulin M, Canet D, Last AM, Pardon E, Archer DB, et al. (2005) Reduced global cooperativity is a common feature underlying the amyloidogenicity of pathogenic lysozyme mutations. J Mol Biol 346(3): 773–788. [DOI] [PubMed] [Google Scholar]
- 44. Fu Y, Ibrahim AS, Sheppard DC, Chen Y, French SW, et al. (2002) Candida albicans Als1p: An adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol Microbiol 44(1): 61–72. [DOI] [PubMed] [Google Scholar]
- 45. Morris KL, Rodger A, Hicks MR, Debulpaep M, Schymkowitz J, et al. (2013) Exploring the sequence-structure relationship for amyloid peptides. Biochem J 450(2): 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Styren,Scot D, Hamilton RL, Styren GC, Klunk WE (2000) X-34, A fluorescent derivative of congo red: A novel histochemical stain for Alzheimer's disease pathology. Journal of Histochemistry & Cytochemistry 48(9): 1223–1232. [DOI] [PubMed] [Google Scholar]
- 47. Maezawa I, Hong H, Liu R, Wu C, Cheng RH, et al. (2008) Congo red and thioflavin-T analogs detect A? oligomers. J Neurochem 104(2): 457–468. [DOI] [PubMed] [Google Scholar]
- 48. Tokuraku K, Marquardt M, Ikezu T (2009) Real-time imaging and quantification of amyloid-beta peptide aggregates by novel quantum-dot nanoprobes. PLoS One 4(12): e8492 10.1371/journal.pone.0008492; 10.1371/journal.pone.0008492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dong J, Revilla-Sanchez R, Moss S, Haydon PG (2010) Multiphoton in vivo imaging of amyloid in animal models of Alzheimer's disease. Neuropharmacology 59(4–5): 268–275 10.1016/j.neuropharm.2010.04.007; 10.1016/j.neuropharm.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, et al. (2008) Complementary adhesin function in C. albicans biofilm formation. Curr Biol 18(14): 1017–1024 10.1016/j.cub.2008.06.034; 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chauhan N, Kruppa MD (2009) Standard growth media and common techniques for use with Candida albicans . Methods Mol Biol 499: 197–201 10.1007/978-1-60327-151-6_18; 10.1007/978-1-60327-151-6_18. [DOI] [PubMed] [Google Scholar]