Abstract
Tacrolimus is a widely used immunosuppressive drug for preventing the rejection of solid organ transplants. The efficacy of tacrolimus shows considerable variability, which might be related to genetic variation among recipients. We conducted a retrospective study of 240 Chinese renal transplant recipients receiving tacrolimus as immunosuppressive drug. The retrospective data of all patients were collected for 40 days after transplantation. Seventeen SNPs of CYP3A5, CYP3A4, COMT, IL-10 and POR were identified by the SNaPshot assay. Tacrolimus blood concentrations were obtained on days 1–3, days 6–8 and days 12–14 after transplantation, as well as during the period of the predefined therapeutic concentration range. Kruskal–Wallis test was used to examine the effect of genetic variation on the tacrolimus concentration/dose ratio (C 0/D) at different time points. Chi-square test was used to compare the proportions of patients who achieved the target C 0 range in the different genotypic groups at weeks 1, 2, 3 and 4 after transplantation. After correction for multiple testing, there was a significant association of C 0/D with CYP3A5*3, CYP3A4*1G and CYP3A4 rs4646437 T>C at different time points after transplantation. The proportion of patients in the IL-10 rs1800871-TT group who achieved the target C 0 range was greater (p = 0.004) compared to the IL-10 rs1800871-CT and IL-10 rs1800871-CC groups at week 3 after transplantation. CYP3A5*3, CYP3A4 *1G, CYP3A4 rs4646437 T>C and IL-10 rs1800871 C>T might be potential polymorphisms affecting the interindividual variability in tacrolimus metabolism among Chinese renal transplant recipients.
Introduction
Tacrolimus is an effective immunosuppressive drug widely used in solid organ transplantation to prevent rejection [1]. It is characterized by a narrow therapeutic range and large inter- and intraindividual variability in its pharmacokinetics [2]. Therefore, daily drug monitoring and dosage adjustment of tacrolimus are widely used so that the concentrations of the drug can be adjusted to achieve the target trough blood concentration (C 0) range [3]. In current clinical practice, it can take several weeks to reach the target C 0 range and transplant recipients experience significant risk of graft rejection or toxicity during this period; so, it is very important to achieve a stable maintenance dose as quickly as possible [4]. However, dose requirement and the length of time required to reach the target C 0 range show significant interindividual and interethnic variability. Full understanding of this mechanism is highly desirable for the patients to improve the therapeutic efficacy and reduce the side effects.
Tacrolimus is metabolized mainly by biotransformation enzymes cytochrome P450 (CYP) 3A4 and 3A5 [3], [5]. The single nucleotide polymorphism (SNP) 6986A>G in intron 3 of the CYP3A5 gene, referred to as CYP3A5*3, results in a splicing defect and the absence of protein activity, unlike the A nucleotide with normal protein activity, referred to as CYP3A5*1. Patients carrying at least one CYP3A5*1 allele are named CYP3A5 expressers and those with CYP3A5*3/*3 genotype are named CYP3A5 nonexpressers [6]. It has been shown that CYP3A5 expressers require a higher maintenance tacrolimus dose and longer time to achieve the target tacrolimus C 0 compared to CYP3A5 nonexpressers among organ transplant recipients [7]–[12]. Moreover, a study revealed that the CYP3A5 rs28365085 T>C might have functional consequence on CYP3A5 activity [13]. Besides the SNPs of CYP3A5 gene, the functional variants of CYP3A4 gene may also influence tacrolimus pharmacokinetics. Wang et al. reported that CYP3A4 *22 (rs35599367, intron 6 C>T) markedly affects CYP3A4 mRNA level and could serve as a biomarker for predicting response to CYP3A4-metabolized drugs [14]. The CYP3A4 rs33972239 delT locates in exon 13 of CYP3A4 gene. So it is a susceptible variant affecting the enzyme activity. He et al. reported that CYP3A4*1G (rs2242480, 20230 C>T) allele can increase the activity of the CYP3A4 enzyme [15]. In addition, schirmer et al. reported that CYP3A4 rs4646437 T>C can affect the hepatic CYP3A4 protein expression levels [16]. Cytochrome P450 oxidoreductase (POR) is required for drug metabolism by all microsomal cytochrome P450 enzymes. Zhang et al. reported that SNPs in the POR gene influence the rates of P450-mediated drug metabolism in patients [17]. Other studies reported that POR rs1057868 C>T and POR rs2868177 A>G are associated with CYP3A activity [17], [18]. These SNPs associated with the CYP3A function might influence tacrolimus pharmacokinetics. In a multicenter study, Jacobson et al. reported that rs2239393 A>G and rs4646312 T>C of catechol-O-methyltransferase (COMT) gene are associated with variation of tacrolimus C 0 /D [19]. This information suggests that the genetic polymorphisms of COMT gene may also affect tacrolimus metabolism. Interleukin-10 (IL-10) can regulate CYP3A enzyme activity. It is reported that the administration of IL-10 down-regulated CYP3A activity by 12% in healthy subjects [20]. Thus, the CYP3A-dependent tacrolimus metabolism may be influenced by IL-10 gene polymorphisms. In addition to the genetic mechanism, clinical factors associated with tacrolimus pharmacokinetics have been reported [21].
Although several factors have been confirmed to impact on tacrolimus pharmacokinetics, some factors with the potential to influence tacrolimus metabolism need to be investigated, especially in different ethnic groups. The aim of this retrospective study was to evaluate the influence of CYP3A4, CYP3A5, COMT, IL-10 and POR SNPs on C 0/D and the length of time required to reach the target C 0 range during the early phase after transplantation in a group of Chinese renal transplant recipients.
Materials and Methods
Study Design and Patient Population
The study protocol was approved by the Ethical Committee of Nanfang Hospital, an affiliate of the Southern Medical University, China. Written informed consent was obtained from all recipients before their participation in the study. The retrospective study population, from the Nanfang Hospital in Guangzhou, consisted of the renal transplant recipients who received tacrolimus as immunosuppressant between January 2007 and August 2012. Patients with conditions that could affect tacrolimus pharmacokinetics and pharmacodynamics were excluded. Exclusion criteria were hepatitis B (58 patients), hepatitis C (6), cancer (5), systemic lupus erythematosus (SLE) with long-term hormone therapy (4), liver and renal transplantation (7), second renal transplantation (10), acute rejection (5), <18 years old (2). Finally, a total of 240 patients were eligible for the retrospective study. Demographic characteristics, laboratory test results and drug administration history were extracted from electronic medical records. The retrospective data of all patients were collected for 40 days after transplantation.
Immunosuppressant Regimens and Tacrolimus Measurement
All patients were treated with a combination of immunosuppressants consisting of tacrolimus, mycophenolate mofetil and steroids. The first oral administration of tacrolimus was given approximately 12 h after the transplantation. The initial dosage was calculated according to the weight of the patient (0.10 mg/kg body weight, twice a day) and subsequently adjusted according to the trough blood concentration (C 0), which was measured by the Microparticle Enzyme ImmunoAssay on an IMx analyzer (Abbott Laboratories, Chicago, IL). Patients' C 0 were measured every other day after transplantation during hospitalization and twice a week after they were discharged from the hospital. The predefined C 0 range was 10–12 ng/ml, and the stable maintenance tacrolimus dose was the dosage at which the target C 0 range was achieved for more than 2 consecutive days and following C 0 values were within the range 9–14 ng/ml. This dosage did not change and was considered to be the stable maintenance tacrolimus dose. The length of time required to reach the target C 0 range was the period from transplantation to the time when patients achieved the stable maintenance tacrolimus dose D. C 0 concentration was dose-corrected (C 0/D) using the corresponding 24 h dose on a mg/kg basis. C 0/D on days 1–3, 6–8 and 12–14 after transplantation, as well as the period of the predefined tacrolimus therapeutic range were selected as the representative ratio parameters of the early phase after transplantation. The corresponding laboratory parameters including hemoglobin, hematocrit, albumin, alanine aminotransferase, aspartate aminotransferase, total bilirubin and unconjugated bilirubin were obtained. The relationships between representative ratio parameters and the genetic variants were analyzed in this study.
SNP Genotyping and Linkage Disequilibrium Measurement
Human DNA was extracted from leukocytes in peripheral blood using the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China). The SNPs of the CYP3A5, CYP3A4, COMT, IL-10 and POR genes meeting the following two criteria were selected for our study. (1) It has been reported that the SNPs might affect the corresponding gene activity, or the SNPs are located in the coding region of the gene. (2) The minor allele frequency (MAF) is >5% in the CHB population (data from HapMap). Finally, the CYP3A5 rs776746 A>G (CYP3A5*3 allele), CYP3A5 rs28365085T>C, CYP3A4 rs2242480 C>T (CYP3A4*1G allele), CYP3A4 rs35599367 C>T (CYP3A4*22 allele), CYP3A4 rs4646437 T>C, CYP3A4 rs33972239 delT, POR rs1057868 C>T, POR rs2868177 A>G, COMT rs4646312 T>C, COMT rs2239393 A>G, COMT rs737865 T>C, COMT rs6267 G>T, COMT rs4680 G>A, COMT rs165599 G>A, IL-10 rs1800871 C>T, IL-10 rs1800872 C>A and IL-10 rs1800896 A>G were analyzed in this study. The genotypes of the 17 SNPs were determined by the SNaPshot assay using the Applied Biosystems Multiplex Kit (Life Technologies Corporation, Shanghai, China) [22].All SNPs of 240 patients tested in this study were successfully genotyped and passed quality control. Haplotypes were inferred by a Bayesian statistical method with the PHASE 2.1 software (Stephens and Donnelly 2003). Reconstructed haplotypes were inserted into the Haploview v. 4.2 program to find r 2.
Statistical Analysis
The dose-adjusted tacrolimus trough concentration (C 0/D) is the ratio of the measured tacrolimus trough concentration C 0 divided by the corresponding daily tacrolimus dose D expressed as mg/kg body weight. All values are expressed as mean ± SD. Sample size and statistical power were evaluated using one-way analysis of variance model (unequal n's) based on nQuery advisor version 7.0 (Statistical Solutions, Cork, Ireland). To account for multiple testing, the Bonferroni correction was applied. P values for SNPs less than 0.05/N (N = number of SNPs to be analyzed) were considered as significant. All SNPs identified were tested for deviations from Hardy–Weinberg disequilibrium with the use of a χ2 test. The following analyses were used to evaluate the impact of each SNP on C 0/D and the length of time required to reach the target C 0 range. C 0/D among the three genotypes of these SNPs was compared using the Kruskal–Wallis test. C 0/D between the two genotypes of these SNPs was compared using the Mann–Whitney test. SNPs that were associated significantly with C 0/D were examined for association with the length of time required to reach the target C 0 range. The proportion of patients who achieved the target C 0 range among the different genotypic groups at different time points was analyzed with the χ2 test. All statistical analyses were performed using the SPSS software package (version 13.0, SPSS Inc., Chicago, IL).
Results
Patient characteristics and genotype frequencies
A total of 240 renal transplant recipients were included in this retrospective study. Of these, 183 finally achieved the target C 0 range through drug monitoring and dosage adjustment. The other 57 patients who hardly achieved the target C 0 range would undergo further therapy. Of the 17 SNPs, 14 (except CYP3A5 rs28365085 T>C, CYP3A4*22 and CYP3A4 rs33972239 delT) were identified in the renal transplant recipients. Finally, the 14 SNPs were analyzed in this study. The allele frequencies of the 14 SNPs in 240 patients were in accordance with Hardy–Weinberg equilibrium, and the same results were found in 183 patients with the stable condition. The demographics, clinical characteristics and genotype frequencies of the patients on days 1–3, 6–8 and 12–14 after transplantation, as well as during the period of the predefined tacrolimus therapeutic range are given in Tables 1 and 2.
Table 1. Demographics, clinical characteristics of the Chinese renal transplant recipients.
| Days 1 to 3 | Days 6 to 8 | Days 12 to 14 | Stable condition | |
| n = 240 | n = 240 | n = 240 | n = 183 | |
| Age (years), (mean ± SD) | 41.03±12.22 | 41.03±12.22 | 41.03±12.22 | 41.77±12.14 |
| Gender (male/female) | 161/79 | 161/79 | 161/79 | 124/59 |
| Body weight (kg) (mean ± SD) | 57.94±10.12 | 57.94±10.12 | 57.94±10.12 | 58.25±10.18 |
| Hematocrit (%) | 0.353±0.0559 | 0.339±0.0577 | 0.303±0.0496 | 0.309±0.0413 |
| Hemoglobin (g/L) | 115.2±19.8 | 112.8±18.8 | 100.8±16.4 | 101.2±14.0 |
| Albumin (g/L) | 35.3±4.5 | 35.9±4.9 | 37.0±5.3 | 38.6±4.5 |
| Alanine aminotransferase (ALT), (U/L) | 18.0±14.0 | 27.9±35.6 | 40.6±45.4 | 29.7±31.2 |
| Aspartate aminotransferase(AST), (U/L) | 19.3±11.6 | 22.2±16.4 | 22.5±14.0 | 17.7±9.0 |
| Total bilirubin (TBIL), (μmol/L) | 10.22±4.43 | 11.19±4.57 | 9.21±3.70 | 8.99±3.31 |
| Unconjugated bilirubin(IBIL), (μmol/L) | 7.12±3.15 | 7.80±3.34 | 6.24±2.69 | 6.08±2.55 |
| Tacrolimus dose (mg/day) | 6.64±1.56 | 7.02±2.16 | 7.88±2.97 | 8.30±3.15 |
| Tacrolimus concentration (ng/ml) | 10.8±5.4 | 10.3±4.0 | 10.1±3.5 | 11.0±1.3 |
| Concentration/Dose Ratio (ng/ml)/(mg/kg) | 95.5±49.1 | 95.7±57.0 | 86.3±52.7 | 90.8±47.1 |
Table 2. Frequencies of allelic variants in the Chinese renal transplant recipients.
| Genotypes (n,%) | Days 1 to14 | Stable condition |
| N = 240 | N = 183 | |
| CYP3A5 (*1/*1, *1/*3, *3/*3) | 21/103/116 | 17/81/85 |
| (8.75%,42.92%,48.33%) | (9.29%,44.26%,46.45%) | |
| CYP3A5 rs28365085 T>C (T/T, T/C, C/C) | 240/0/0 | 183/0/0 |
| (100%,0%,0%) | (100%,0%,0%) | |
| CYP3A4 (*1/*1, *1/*1G, *1G/*1G) | 131/90/19 | 98/73/12 |
| (54.58%,37.50%,7.92%) | (53.55%, 39.89%, 6.56%) | |
| CYP3A4 rs4646437 T>C (T/T, T/C, C/C) | 10/80/150 | 6/63/114 |
| (4.17%,33.33%,62.50%) | (3.28%,34.42%,62.30%) | |
| CYP3A4 (*1/*1,*1/*22, *22/*22) | 240/0/0 | 183/0/0 |
| (100%,0%,0%) | (100%,0%,0%) | |
| CYP3A4 rs33972239 delT (−/−, −/T, T/T) | 240/0/0 | 183/0/0 |
| (100%,0%,0%) | (100%,0%,0%) | |
| POR rs1057868 C>T (C/C, C/T, T/T) | 101/107/32 | 67/90/26 |
| (42.08%,44.58%,13.34%) | (36.61%,49.18%,14.21%) | |
| POR rs2868177 A>G (A/A, A/G, G/G) | 84/104/52 | 65/85/33 |
| (35.00%, 43.33%,21.67%) | (35.52%,46.45%,18.03%) | |
| COMT rs4646312 T>C (T/T, T/C, C/C) | 115/98/27 | 92/73/18 |
| (47.92%, 40.83%, 11.25%) | (50.27%,39.89%,9.84%) | |
| COMT rs2239393 A>G (A/A, A/G, G/G) | 116/96/28 | 93/71/19 |
| (48.33%, 40.00%, 11.67%) | (50.82%,38.80%,10.38%) | |
| COMT rs737865 T>C (T/T, T/C, C/C) | 126/92/22 | 99/70/14 |
| (52.50%,38.33%, 9.17%) | (54.10%,38.25%,7.65%) | |
| COMT rs6267 G>T (G/G, G/T, T/T) | 213/26/1 | 163/19/1 |
| (88.75%,10.83%, 0.42%) | (89.07%,10.38%,0.55%) | |
| COMT rs4680 G>A (G/G, G/A, A/A) | 133/86/21 | 97/69/17 |
| (55.42%, 35.83%, 8.75%) | (53.01%,37.70%,9.29%) | |
| COMT rs165599 G>A (G/G, G/A, A/A) | 54/138/48 | 38/106/39 |
| (22.50%, 57.50%, 20.00%) | (20.77%,57.92%,21.31%) | |
| IL-10 rs1800871 C>T (C/C, C/T, T/T) | 15/111/114 | 8/84/91 |
| (6.25%, 46.25%, 47.50%) | (4.37%,45.90%,49.73%) | |
| IL-10 rs1800896 A>G (A/A, A/G, G/G) | 217/23/0 | 168/15/0 |
| (90.42%, 9.58%, 0%) | (91.80%,8.20%,0%) | |
| IL-10 rs1800872 C>A (C/C, C/A, A/A) | 15/112/113 | 8/85/90 |
| (6.25%, 46.67%, 47.08%) | (4.37%,46.45%,49.18%) |
Single genetic polymorphism analysis for association with tacrolimus C0/D
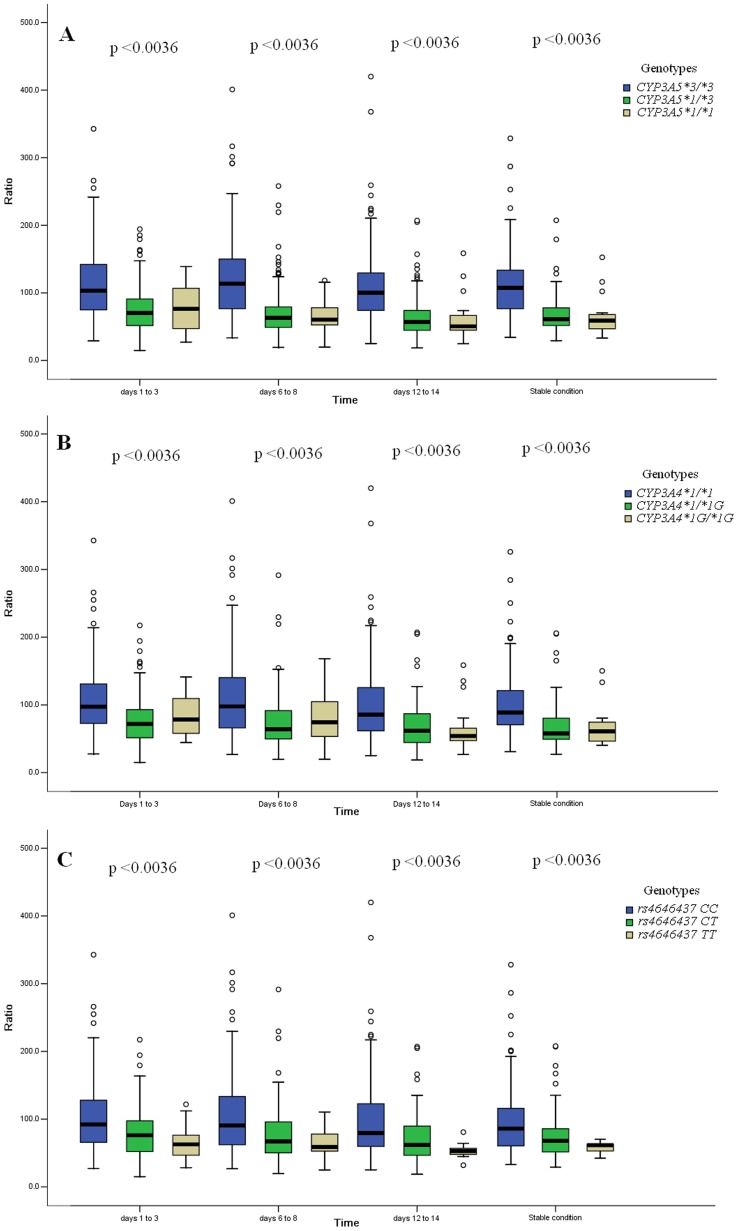
We examined the association of the 14 genotypic variants with tacrolimus C 0/D at different time points after transplantation. The level of significance has been adjusted according to the Bonferroni correction (p bonf <0.0036). Of the 14 variants, CYP3A5*3, CYP3A4*1G and CYP3A4 rs4646437 T>C presented a significant association with tacrolimus C 0/D at different time points after transplantation (Tables 3 and 4; Figure 1). Tacrolimus C 0/D of the patients with CYP3A5 *3/*3 was highest among the different genotypic groups of CYP3A5*3 (Figure 1A). C 0/D of the patients with CYP3A4 *1/*1was highest among the different genotypic groups of CYP3A4*1G (Figure 1B). C 0/D of the patients with CYP3A4 rs4646437-CC was highest among the different genotypic groups of CYP3A4 rs4646437 T>C (Figure 1C). Moreover, the IL-10 rs1800871 C>T and IL-10 rs1800872 C>A presented a marginal association (p<0.05) with C 0/D at the time point when the patients achieved the maintenance dose (Table 4). However, impact of IL-10 rs1800871 C>T and IL-10 rs1800872 C>A on C 0/D was not statistically significant after applying Bonferroni correction. None of the other 9 variants demonstrated a significant association with C 0/D at any time point. In addition, the minimum sample sizes needed for 80% power for analysis of CYP3A5*3, CYP3A4*1G and CYP3A4 rs4646437 T>C were estimated, and the sample size (240 patients) is enough to assure the statistical power and conclusion (Table S1).
Table 3. Comparison of the tacrolimus C 0/D in the different groups classified by genetic variant genotypes.
| Genotype | n = 240 | (days1 to 3)C0/D | p | (days6 to 8)C0/D | p | (days 12 to 14) C0/D | p |
| CYP3A5*1/*1 | 21 | 78.4±35.1 | 68.5±27.1 | 60.7±32.7 | |||
| CYP3A5*1/*3 | 103 | 76.9±37.1 | 2.93×10−9 | 72.9±41.2 | 3.87×10−13 | 65.2±33.7 | 2.79×10−14 |
| CYP3A5*3/*3 | 116 | 115.0±53.2 | 120.9±62.4 | 109.6±59.3 | |||
| CYP3A4*1/*1 | 131 | 107.7±53.2 | 111.2±62.2 | 100.5±59.7 | |||
| CYP3A4*1/*1G | 90 | 79.8±40.4 | 7.07×10−5 | 76.6±45.0 | 9.58×10−7 | 69.9±36.6 | 1.64×10−6 |
| CYP3A4*1G/*1G | 19 | 85.6±32.1 | 79.5±37.3 | 65.3±35.8 | |||
| CYP3A4 rs4646437 TT | 10 | 68.7±29.2 | 66.8±25.6 | 53.8±12.8 | |||
| CYP3A4 rs4646437 TC | 80 | 81.9±40.0 | 4.32×10−4 | 80.8±47.6 | 7.41×10−4 | 71.5±39.1 | 3.48×10−5 |
| CYP3A4 rs4646437 CC | 150 | 104.5±52.3 | 105.6±60.9 | 96.3±57.8 | |||
| POR rs1057868 CC | 101 | 96.2±45.1 | 95.4±53.6 | 84.7±52.9 | |||
| POR rs1057868 CT | 107 | 93.9±52.7 | 0.714 | 99.0±61.4 | 0.422 | 90.1±54.7 | 0.444 |
| POR rs1057868 TT | 32 | 98.3±50.2 | 85.8±52.7 | 78.3±44.9 | |||
| POR rs2868177 AA | 84 | 94.0±55.6 | 94.6±62.8 | 83.9±54.9 | |||
| POR rs2868177 AG | 104 | 93.8±43.8 | 0.471 | 93.2±54.1 | 0.411 | 83.4±40.7 | 0.471 |
| POR rs2868177 GG | 52 | 101.1±48.7 | 102.6±53.6 | 95.7±68.2 | |||
| COMT rs4646312 TT | 115 | 95.0±48.5 | 97.5±55.0 | 87.5±55.3 | |||
| COMT rs4646312 TC | 98 | 97.3±53.0 | 0.994 | 93.562.9 | 0.469 | 84.1±52.5 | 0.620 |
| COMT rs4646312 CC | 27 | 90.9±36.6 | 96.1±43.1 | 88.5±42.3 | |||
| COMT rs2239393 AA | 116 | 95.7±48.6 | 97.6±54.9 | 87.8±55.1 | |||
| COMT rs2239393 AG | 96 | 96.7±53.2 | 0.966 | 93.4±63.4 | 0.413 | 83.9±53.0 | 0.577 |
| COMT rs2239393 GG | 28 | 90.5±36.0 | 96.0±42.3 | 88.0±41.6 | |||
| COMT rs737865 TT | 126 | 94.7±47.3 | 95.4±56.2 | 86.0±57.0 | |||
| COMT rs737865 TC | 92 | 95.4±53.1 | 0.749 | 93.1±60.8 | 0.153 | 85.4±48.1 | 0.632 |
| COMT rs737865 CC | 22 | 100.0±43.1 | 108.1±45.2 | 91.5±47.3 | |||
| COMT rs6267 GG | 213 | 94.7±49.1 | 96.1±59.1 | 85.7±49.4 | |||
| COMT rs6267 GT | 26 | 102.2±50.7 | 0.775 | 93.8±37.3 | 0.646 | 91.2±76.5 | 0.972 |
| COMT rs6267 TT | 1 | 84.5 | 60.7 | 74.4 | |||
| COMT rs4680 GG | 133 | 98.9±52.6 | 96.4±58.7 | 87.2±56.5 | |||
| COMT rs4680 GA | 86 | 90.2±42.7 | 0.594 | 94.8±54.0 | 0.936 | 84.2±47.1 | 0.962 |
| COMT rs4680 AA | 21 | 95.9±50.8 | 95.3±61.0 | 88.4±51.7 | |||
| COMT rs165599 GG | 54 | 105.0±50.8 | 103.2±58.0 | 87.7±43.0 | |||
| COMT rs165599 GA | 138 | 90.3±45.4 | 0.117 | 91.4±58.7 | 0.144 | 82.6±56.7 | 0.089 |
| COMT rs165599 AA | 48 | 99.7±55.9 | 99.6±50.8 | 95.3±50.4 | |||
| IL-10 rs1800871 CC | 15 | 88.1±40.1 | 85.4±54.7 | 75.7±40.2 | |||
| IL-10 rs1800871 CT | 111 | 92.1±47.8 | 0.330 | 91.1±58.5 | 0.104 | 82.6±57.2 | 0.129 |
| IL-10 rs1800871 TT | 114 | 99.7±51.4 | 101.5±55.8 | 91.2±49.4 | |||
| IL-10 rs1800896 AA | 217 | 96.3±48.9 | 0.222 | 96.7±54.7 | 0.114 | 86.6±50.6 | 0.290 |
| IL-10 rs1800896 AG | 23 | 88.1±51.5 | 86.7±76.6 | 83.2±70.6 | |||
| IL-10 rs1800872 CC | 15 | 88.0±40.1 | 82.6±55.5 | 74.0±40.9 | |||
| IL-10 rs1800872 CA | 112 | 92.1±47.8 | 0.352 | 91.6±58.0 | 0.086 | 83.4±56.8 | 0.174 |
| IL-10 rs1800872 AA | 113 | 99.8±51.4 | 101.6±56.1 | 90.8±49.7 |
Table 4. Comparison of the tacrolimus C 0/D in the different genotypic groups on the time achieving target blood tacrolimus concentrations.
| Genotype | n = 183 | Stable conditions (C 0/D) | p |
| CYP3A5*1/*1 | 17 | 67.3±29.9 | |
| CYP3A5*1/*3 | 81 | 69.5±27.9 | 3.01×10−13 |
| CYP3A5*3/*3 | 85 | 115.7±52.0 | |
| CYP3A4*1/*1 | 98 | 105.0±51.4 | |
| CYP3A4*1/*1G | 73 | 74.3±35.5 | 1.12×10−6 |
| CYP3A4*1G/*1G | 12 | 74.6±34.4 | |
| CYP3A4 rs4646437 TT | 6 | 60.6±9.4 | |
| CYP3A4 rs4646437 TC | 63 | 78.8±38.8 | 1.11×10−3 |
| CYP3A4 rs4646437 CC | 114 | 99.0±50.4 | |
| POR rs1057868 CC | 67 | 92.4±46.6 | |
| POR rs1057868 CT | 90 | 90.9±49.5 | 0.728 |
| POR rs1057868 TT | 26 | 86.1±40.5 | |
| POR rs2868177 AA | 65 | 92.0±50.8 | |
| POR rs2868177 AG | 85 | 86.5±39.4 | 0.563 |
| POR rs2868177 GG | 33 | 99.4±56.9 | |
| COMT rs4646312 TT | 92 | 91.1±48.6 | |
| COMT rs4646312 TC | 73 | 87.3±46.0 | 0.152 |
| COMT rs4646312 CC | 18 | 103.2±43.2 | |
| COMT rs2239393 AA | 93 | 91.4±48.4 | |
| COMT rs2239393 AG | 71 | 87.2±46.5 | 0.2 |
| COMT rs2239393 GG | 19 | 101.1±43.0 | |
| COMT rs737865 TT | 99 | 89.6±49.8 | |
| COMT rs737865 TC | 70 | 90.2±44.1 | 0.328 |
| COMT rs737865 CC | 14 | 101.4±43.0 | |
| COMT rs6267 GG | 163 | 92.3±48.6 | |
| COMT rs6267 GT | 19 | 78.1±31.0 | 0.561 |
| COMT rs6267 TT | 1 | 74.4 | |
| COMT rs4680 GG | 97 | 89.0±45.5 | |
| COMT rs4680 GA | 69 | 94.3±49.9 | 0.749 |
| COMT rs4680 AA | 17 | 86.6±45.9 | |
| COMT rs165599 GG | 38 | 94.3±42.1 | |
| COMT rs165599 GA | 106 | 89.351.3 | 0.363 |
| COMT rs165599 AA | 39 | 91.3±40.1 | |
| IL-10 rs1800871 CC | 8 | 73.5±36.8 | |
| IL-10 rs1800871 CT | 84 | 84.1±46.9 | 0.017 |
| IL-10 rs1800871 TT | 91 | 98.4±47.1 | |
| IL-10 rs1800896 AA | 168 | 90.4±44.5 | 0.710 |
| IL-10 rs1800896 AG | 15 | 95.4±72.0 | |
| IL-10 rs1800872 CC | 8 | 76.5±36.8 | |
| IL-10 rs1800872 CA | 85 | 84.5±46.6 | 0.046 |
| IL-10 rs1800872 AA | 90 | 98.0±47.6 |
Figure 1. Box-and-whisker plot of tacrolimus C 0/D for different genotypic groups.
The boxes represent the median, 25th and 75th percentiles of the data. The circles represent deviant cases. The X-axis gives the times (days 1–3, 6–8 and 12–14, and the period of stable conditions after transplantation). The Y-axis gives the C 0/D. Genetic variants are: A, CYP3A5*3; B, CYP3A4*1G; and C, CYP3A4 rs4646437 T>C. The p values among the genetic groups are given above the box-and-whisker plot.
Difference in the length of time required to reach the target C0 range
According to the above data, CYP3A5*3, CYP3A4*1G, CYP3A4 rs4646437 T>C, IL-10 rs1800871 C>T and IL-10 rs1800872 C>A might be associated with C 0/D. We also evaluated the relationships between the five variants and the length of time required to reach the target C 0 range. The proportion of patients who achieved the target C 0 range was compared for the different genotypic groups at weeks 1, 2, 3 and 4 after transplantation (Table 5). The level of significance has been adjusted according to the Bonferroni correction (p bonf <0.01). The proportion of patients in CYP3A4*1/*1 group who achieved the target C 0 range at week 1 was higher (p = 0.041) compared to the CYP3A4*1/*1G and CYP3A4*1G/*1G groups. However, the significance was lost after Bonferroni correction. The proportion of patients in the IL-10 rs1800871-TT group who achieved the target C 0 range at week 3 was higher (p = 0.004) compared to the IL-10 rs1800871-CT and IL-10 rs1800871-CC groups. There was no significant difference among the other variant groups at any time point.
Table 5. The impact of the genetic variants on the time to achieve the target blood tacrolimus concentrations.
| Week 1 | Week 2 | Week 3 | Week 4 | |||||
| Stable conditions | Stable conditions | Stable conditions | Stable conditions | |||||
| Yes/No (n) | p | Yes/No (n) | p | Yes/No (n) | p | Yes/No (n) | p | |
| CYP3A5*3/*3 | 9/107 | 0.058 | 39/77 | 0.298 | 72/44 | 0.802 | 79/37 | 0.369 |
| CYP3A5*1/*3 or *1/*1 | 3/121 | 34/90 | 75/49 | 91/33 | ||||
| CYP3A4*1/*1 | 10/121 | 0.041 | 43/88 | 0.296 | 79/52 | 0.855 | 89/42 | 0.280 |
| CYP3A4*1/*1G or 1G/*1G | 2/107 | 29/80 | 67/42 | 81/28 | ||||
| CYP3A4 rs4646437 CC | 9/141 | 0.360 | 49/101 | 0.329 | 95/55 | 0.307 | 106/44 | 0.942 |
| CYP3A4 rs4646437 TC or TT | 3/87 | 24/66 | 51/39 | 64/26 | ||||
| IL-10 rs1800871 TT | 5/109 | 0.679 | 38/76 | 0.351 | 77/37 | 0.004 | 87/27 | 0.076 |
| IL-10 rs1800871 CT or CC | 7/119 | 35/91 | 62/64 | 83/43 | ||||
| IL-10 rs1800872 AA | 5/108 | 0.700 | 37/76 | 0.461 | 75/38 | 0.098 | 86/27 | 0.091 |
| IL-10 rs1800872 CA or CC | 7/120 | 36/91 | 71/56 | 84/43 | ||||
Linkage between CYP3A4 SNPs and CYP3A5*3 in tacrolimus metabolism
The CYP3A4 and CYP3A5 genes are located in 7q21.1. We analyzed the linkage disequilibrium (LD) between the CYP3A4 and CYP3A5 variants. There was a moderate degree of LD between CYP3A4*1/*1G (rs2242480 C>T) and CYP3A5*1/*3 (rs776746 A>G) (r 2 = 0.502) and a low degree of LD between CYP3A4 rs4646437 T>C and CYP3A5*1/*3 (rs776746 A>G) (r 2 = 0.244). We investigated the effect of the CYP3A4*1/*1G and CYP3A4 rs4646437 T>C polymorphisms on the dose-adjusted tacrolimus concentration (C 0/D) among CYP3A5 expressers and nonexpressers (Tables 6 and 7). There was no significant difference in C 0/D between patients with the CYP3A4*1G allele and the *1/*1 genotype. The same results were found between patients with the CYP3A4 rs4646437 T allele and the CYP3A4 rs4646437 CC genotype.
Table 6. Tacrolimus C 0/D in CYP3A4*1/*1G genotypes classified by different CYP3A5 expressers.
| CYP3A5 expresser | p | CYP3A5 nonexpresser | p | |||
| CYP3A4*1/*1 | CYP3A4*1/*1G+ *1G/*1G | CYP3A4*1/*1 | CYP3A4*1/*1G+ *1G/*1G | |||
| N | 25 | 99 | 106 | 10 | ||
| (days1 to 3) C0/D | 75.4±37.4 | 77.6±36.6 | 0.681 | 115.3±53.7 | 112.4±50.0 | 0.875 |
| (days6 to 8) C0/D | 73.1±46.0 | 71.9±37.4 | 0.988 | 121.0±62.3 | 127.9±66.6 | 0.791 |
| (days 12 to 14) C0/D | 58.3±23.8 | 66.0±35.4 | 0.480 | 110.5±61.2 | 100.2±32.1 | 0.890 |
Table 7. Tacrolimus C 0/D in CYP3A4 rs4646437 genotypes classified by different CYP3A5 expressers.
| CYP3A5 expresser | p | CYP3A5 nonexpresser | p | |||
| CYP3A4 rs4646437 CC | CYP3A4 rs4646437 TC + TT | CYP3A4 rs4646437 CC | CYP3A4 rs4646437 TC + TT | |||
| N | 46 | 78 | 104 | 12 | ||
| (days1 to 3) C0/D | 79.0±37.1 | 76.1±36.5 | 0.658 | 115.8±54.2 | 108.4±44.9 | 0.744 |
| (days6 to 8) C0/D | 70.9±38.7 | 73.0±39.5 | 0.668 | 121.7±62.7 | 120.4±62.1 | 0.878 |
| (days 12 to 14) C0/D | 64.1±28.8 | 64.6±36.1 | 0.649 | 110.6±61.7 | 101.0±32.0 | 0.935 |
Discussion
This retrospective study examined the contribution of gene polymorphisms to the dose-adjusted tacrolimus concentration (C 0/D) and the length of time required to reach the target trough blood concentration range (C 0) in Chinese renal transplant recipients. In accord with the results of earlier studies [7]–[11], we found that CYP3A5*3 presented a significant association (p<0.0036) with tacrolimus C 0/D at different time points after transplantation (Figure 1A). This result further validated that the CYP3A5*3 allele was strongly associated with tacrolimus pharmacokinetics. In addition, the CYP3A4 *1G allele and CYP3A4 rs4646437 T>C were associated (p<0.0036) with C 0/D at different time points after transplantation (Figure 1B and 1C). This is the first report of association between CYP3A4 rs4646437 T>C and tacrolimus pharmacokinetics. Because the CYP3A4 and CYP3A5 genes are both located in 7q21.1, the LD between CYP3A4 SNPs and CYP3A5 6986A>G might influence the impact of CYP3A4 SNPs on the tacrolimus C 0/D. Crettol et al. reported that the CYP3A4 rs4646437C>T influenced cyclosporine pharmacokinetics, the rs4646437-T carriers requiring higher cyclosporine dose. They found also that the rs4646437-T allele was in strong LD (r 2 = 0.82) with the CYP3A5*1 allele in Caucasian renal transplant recipients [23]. In this study, there was a moderate degree of LD between CYP3A4*1/*1G (rs2242480 C>T) and CYP3A5*1/*3 (rs776746 A>G) (r 2 = 0.502) and a low degree of LD between CYP3A4 rs4646437 T>C and CYP3A5*1/*3 (rs776746 A>G) (r 2 = 0.244). Miura et al. reported that the CYP3A4*1/*1G might affect interindividual variability in tacrolimus pharmacokinetics among CYP3A5 expressers [24]. Zuo et al. reported that CYP3A4*1G can influence the oral clearance (CL/F) of tacrolimus in CYP3A5 expressers or nonexpressers among Chinese renal transplant recipients [25]. We divided the patients into CYP3A5 expressers and nonexpressers, and examined the impact of CYP3A4 variants on C 0/D in different CYP3A5 expresser groups. There was no significant difference of C 0/D between patients with the CYP3A4*1G allele and the *1/*1 genotype among the different CYP3A5 expresser groups (Table 6). The same result was found between patients with the CYP3A4 rs4646437-T allele and the CYP3A4 rs4646437-CC genotype (Table 7). This results indicated that the LD with CYP3A5*1/*3 might be one reason for the association between the CYP3A4 SNPs and C 0/D although the LD was not strong in our study population. So, the impact of the two SNPs on tacrolimus metabolism needs further investigation. Zhang et al. reported that liver transplantation recipients with donors who had the IL-10 rs1800896-AA genotype had higher C 0/D values compared to donors with the IL-10 rs1800896-AG genotype [20]. They found also that the C 0/D values of liver transplantation recipients with donors who had a low IL-10 production genotype (rs1800871-TT, rs1800872-AA) were higher compared to a high IL-10 production genotype (rs1800871-CC or CT, rs1800872-CC or AC) and they suggested that the expression level of the IL-10 gene could influence C 0/D. In this study, IL-10 gene variants (IL-10 rs1800871 C>T, IL-10 rs1800872 C>A) presented a marginal association (p<0.05) with C 0/D of renal recipients during the period of the predefined tacrolimus therapeutic range. However, the difference was not significant after correction by Bonferroni method. Since the Bonferroni method is very conservative, the effect of IL-10 rs1800871 C>T and IL-10 rs1800872 C>A on tacrolimus needs further investigation. In addition, six susceptible COMT variants and two susceptible POR variants were analyzed; however, none of these variants had a significant association with C 0/D. Moreover, the variants of CYP3A5 rs28365085 C, CYP3A4*22 and CYP3A4 rs33972239 delT were not found in this study, although there are reports that they can affect tacrolimus pharmacokinetics [26]–[28]. This phenomenon revealed that the genetic background of tacrolimus metabolism varies among ethnic groups.
We examined the relationships between the five SNPs associated with the C 0/D and the length of time required to reach the target C 0 range. Of the five SNPs, IL-10 rs1800871 C>T influenced the proportion of patients who achieved the target C 0 range at weeks 3. MacPhee et al. reported that CYP3A5 nonexpressers achieved the target tacrolimus concentration easily, whereas there was a significant delay for CYP3A5 expressers [12]. In this study, there was no significant difference between the CYP3A5 expressers and CYP3A5 nonexpressers in the proportion of patients who achieved the target C 0 range (Table 5). However, it appeared the genotypic groups with the higher C 0/D, such as the IL-10 rs1800871-TT groups, were able to achieve the target C 0 more easily. According to our data, the proportion of patients in the IL-10 rs1800871-TT group who achieved the target C 0 range was higher (p = 0.004) compared to the IL-10 rs1800871-CT and IL-10 rs1800871-CC groups at week 3. A large proportion of patients achieved the target C 0 range during week 3 after transplantation. So, it appears IL-10 rs1800871 C>T was very important for the ease with which patients were able to achieve the target C 0 range.
Owing to the strict inclusion and exclusion criteria, 97 patients with disease states that might affect tacrolimus pharmacokinetics were excluded. The exclusion of patients with some disease states is necessary because those diseases might affect tacrolimus metabolism and, thus, the results of the study. In addition, we selected days 1–3, 6–8 and 12–14 and the period of the predefined tacrolimus therapeutic range for analysis of the association between genetic polymorphisms and C 0/D. Several time points were selected for the analysis, which was necessary because analysis of one genetic polymorphism at a single time point could produce an unreliable result.
There are several limitations to our study. The number of patients in several genotypic groups was small when the patients were divided into different groups according to genotype, which could influence the study results because of insufficient statistical power. Moreover, we can't confirm that CYP3A4*1G allele and CYP3A4 rs4646437 T>C have independent effect on tacrolimus C 0/D. The mechanism by which IL-10 affects the length of time required to reach the target C 0 range is also unclear and further investigations are needed.
In clinical practice, the immunosuppressive effect of tacrolimus is not equivalent to tacrolimus C 0. However, tacrolimus C 0 is an important parameter to evaluate the immune status of transplant recipients. The latest insight into the genetic mechanism underlying tacrolimus metabolism has proved useful for tacrolimus individualization of organ transplantation patients. Some recent studies have individualized the dosage of tacrolimus on the basis of the CYP3A5 genotype and obtained effective results [29], [30]. In this study, we found a significant association between tacrolimus C 0/D and genotypes CYP3A5*3, CYP3A4*1G and CYP3A4 rs4646437 T>C in Chinese renal transplant recipients. We observed increased proportions of patients with IL-10 rs1800871-TT genotypes who achieved the target C 0 range. Therefore, genotyping of these genetic polymorphisms could potentially benefit Chinese renal transplant recipients by reducing the risk and the length of time needed to reach the target C 0 range, and the results could be useful for the tacrolimus individualization of other organ transplant recipients.
Supporting Information
Sample size and statistical power evaluation based on the different genetic variants.
(DOC)
Acknowledgments
We thank all the patients who participated in this study. We thank all of our colleagues past and present who contributed to this project.
Funding Statement
This work was funded by a programme grant from the National Natural Science Fund of China to Liang Li (NSFC, No. 81101328), the Pearl River Young Talents of Science and Technology in Guangzhou to Liang Li (No. 2013J2200050), the Natural Science Fund of Guangdong to Liang Li (No. S2012040007745), the Research Fund for the President of Nanfang Hospital to Chuan-Jiang Li. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ekberg H, Tedesco-Silva H, Demirbas A, Vítko S, Nashan B, et al. (2007) Reduced exposure to calcineurin inhibitors in renal transplantation. N Engl J Med 357: 2562–2575. [DOI] [PubMed] [Google Scholar]
- 2. Kuypers DRJ, Claes K, Evenepoel P, Maes B, Vanrenterghem Y (2004) Clinical efficacy and toxicity profile of tacrolimus and mycophenolic acid in relation to combined long-term pharmacokinetics in de novo renal allograft recipients. Clin Pharmacol Ther 75: 434–447. [DOI] [PubMed] [Google Scholar]
- 3. Masuda S, Inui K (2006) An up-date review on individualized dosage adjustment of calcineurin inhibitors in organ transplant patients. Pharmacol Ther 112: 184. [DOI] [PubMed] [Google Scholar]
- 4. Wallemacq P, Armstrong VW, Brunet M, Haufroid V, Holt DW, et al. (2009) Opportunities to optimize tacrolimus therapy in solid organ transplantation: report of the European consensus conference. Ther Drug Monit 31: 139–152. [DOI] [PubMed] [Google Scholar]
- 5. de Jonge H, de Loor H, Verbeke K, Vanrenterghem Y, Kuypers DR (2012) In vivo CYP3A4 activity, CYP3A5 genotype, and hematocrit predict tacrolimus dose requirements and clearance in renal transplant patients. Clin Pharmacol Ther 92: 366–375. [DOI] [PubMed] [Google Scholar]
- 6. Xie HG, Wood AJ, Kim RB, Stein CM, Wilkinson GR (2004) Genetic variability in CYP3A5 and its possible consequences. Pharmacogenomics 5: 243–272. [DOI] [PubMed] [Google Scholar]
- 7. Roy JN, Barama A, Poirier C, Vinet B, Roger M (2006) CYP3A4, CYP3A5, and MDR-1 genetic influences on tacrolimus pharmacokinetics in renal transplant recipients. Pharmacogenet Genomics 16: 659–665. [DOI] [PubMed] [Google Scholar]
- 8. Haufroid V, Wallemacq P, VanKerckhove V, Elens L, De Meyer M, et al. (2006) CYP3A5 and ABCB1 polymorphisms and tacrolimus pharmacokinetics in renal transplant candidates: guidelines from an experimental study. Am J Transplant 6: 2706–2713. [DOI] [PubMed] [Google Scholar]
- 9. Goto M, Masuda S, Kiuchi T, Ogura Y, Oike F, et al. (2004) CYP3A5*1-carrying graft liver reduces the concentration/oral dose ratio of tacrolimus in recipients of living-donor liver transplantation. Pharmacogenetics 14: 471–478. [DOI] [PubMed] [Google Scholar]
- 10. Zheng H, Webber S, Zeevi A, Schuetz E, Zhang J, et al. (2003) Tacrolimus dosing in pediatric heart transplant patients is related to CYP3A5 and MDR1 gene polymorphisms. Am J Transplant 3: 477–483. [DOI] [PubMed] [Google Scholar]
- 11. Zheng H, Zeevi A, Schuetz E, Lamba J, McCurry K, et al. (2004) Tacrolimus dosing in adult lung transplant patients is related to cytochrome P4503A5 gene polymorphism. J Clin Pharmacol 44: 135–140. [DOI] [PubMed] [Google Scholar]
- 12. MacPhee IA, Fredericks S, Tai T, Syrris P, Carter ND, et al. (2004) The influence of pharmacogenetics on the time to achieve target tacrolimus concentrations after kidney transplantation. Am J Transplant 4: 914–919. [DOI] [PubMed] [Google Scholar]
- 13. Dai Z, Papp AC, Wang D, Hampel H, Sadee W (2008) Genotyping panel for assessing response to cancer chemotherapy. BMC Med Genomics 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W (2011) Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 11: 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He BX, Shi L, Qiu J, Tao L, Li R, et al. (2011) A functional polymorphism in the CYP3A4 gene is associated with increased risk of coronary heart disease in the Chinese Han population. Basic Clin Pharmacol Toxicol 108: 208–213. [DOI] [PubMed] [Google Scholar]
- 16. Schirmer M, Rosenberger A, Klein K, Kulle B, Toliat MR, et al. (2007) Sex-dependent genetic markers of CYP3A4 expression and activity in human liver microsomes. Pharmacogenomics 8: 443–453. [DOI] [PubMed] [Google Scholar]
- 17. Zhang X, Li L, Ding X, Kaminsky LS (2011) Identification of cytochrome P450 oxidoreductase gene variants that are significantly associated with the interindividual variations in warfarin maintenance dose. Drug Metab Dispos 39: 1433–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Jonge H, Metalidis C, Naesens M, Lambrechts D, Kuypers DR (2011) The P450 oxidoreductase *28 SNP is associated with low initial tacrolimus exposure and increased dose requirements in CYP3A5-expressing renal recipients. Pharmacogenomics 12: 1281–1291. [DOI] [PubMed] [Google Scholar]
- 19. Jacobson PA, Oetting WS, Brearley AM, Leduc R, Guan W, et al. (2011) Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation 91: 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X, Wang Z, Fan J, Liu G, Peng Z (2011) Impact of interleukin-10 gene polymorphisms on tacrolimus dosing requirements in Chinese liver transplant patients during the early posttransplantation period. Eur J Clin Pharmacol 67: 803–813. [DOI] [PubMed] [Google Scholar]
- 21. Li L, Li CJ, Zheng L, Zhang YJ, Jiang HX, et al. (2011) Tacrolimus dosing in Chinese renal transplant recipients: a population-based pharmacogenetics study. Eur J Clin Pharmacol 67: 787–795. [DOI] [PubMed] [Google Scholar]
- 22. Li L, Li CJ, Zhang YJ, Zheng L, Jiang HX, et al. (2011) Simultaneous detection of CYP3A5 and MDR1 polymorphisms based on the SNaPshot assay. Clin Biochem 44: 418–422. [DOI] [PubMed] [Google Scholar]
- 23. Crettol S, Venetz JP, Fontana M, Aubert JD, Pascual M, et al. (2008) CYP3A7, CYP3A5, CYP3A4, and ABCB1 genetic polymorphisms, cyclosporine concentration, and dose requirement in transplant recipients. Ther Drug Monit 30: 689–699. [DOI] [PubMed] [Google Scholar]
- 24. Miura M, Satoh S, Kagaya H, Saito M, Numakura K, et al. (2011) Impact of the CYP3A4*1G polymorphism and its combination with CYP3A5 genotypes on tacrolimus pharmacokinetics in renal transplant patients. Pharmacogenomics 12: 977–984. [DOI] [PubMed] [Google Scholar]
- 25. Zuo XC, Ng CM, Barrett JS, Luo AJ, Zhang BK, et al. (2013) Effects of CYP3A4 and CYP3A5 polymorphisms on tacrolimus pharmacokinetics in Chinese adult renal transplant recipients: a population pharmacokinetic analysis. Pharmacogenet Genomics 23: 251–261. [DOI] [PubMed] [Google Scholar]
- 26. Elens L, Bouamar R, Hesselink DA, Haufroid V, van der Heiden IP, et al. (2011) A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin Chem 57: 1574–1583. [DOI] [PubMed] [Google Scholar]
- 27. Elens L, Bouamar R, Hesselink DA, Haufroid V, van Gelder T, et al. (2012) The new CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with an increased risk of delayed graft function and worse renal function in cyclosporine-treated kidney transplant patients. Pharmacogenet Genomics 22: 373–380. [DOI] [PubMed] [Google Scholar]
- 28. Elens L, Hesselink DA, van Schaik RH, van Gelder T (2013) The CYP3A4*22 allele affects the predictive value of a pharmacogenetic algorithm predicting tacrolimus predose concentrations. Br J Clin Pharmacol 75: 1545–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thervet E, Loriot MA, Barbier S, Buchler M, Ficheux M, et al. (2010) Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin Pharmacol Ther 87: 721–726. [DOI] [PubMed] [Google Scholar]
- 30. Chen SY, Li JL, Meng FH, Wang XD, Liu T, et al. (2013) Individualization of tacrolimus dosage basing on cytochrome P450 3A5 polymorphism – a prospective, randomized, controlled study. Clin Transplant 27: E272–E281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample size and statistical power evaluation based on the different genetic variants.
(DOC)