Abstract
The red coloration of litchi fruit depends on the accumulation of anthocyanins. The anthocyanins level in litchi fruit varies widely among cultivars, developmental stages and environmental stimuli. Previous studies on various plant species demonstrate that anthocyanin biosynthesis is controlled at the transcriptional level. Here, we describe a litchi R2R3-MYB transcription factor gene, LcMYB1, which demonstrates a similar sequence as other known anthocyanin regulators. The transcription levels of the LcMYB1 and anthocyanin biosynthetic genes were investigated in samples with different anthocyanin levels. The expression of LcMYB1 was strongly associated with tissue anthocyanin content. LcMYB1 transcripts were only detected in anthocyanin-accumulating tissues and were positively correlated with anthocyanin accumulation in the pericarps of 12 genotypes. ABA and sunlight exposure promoted, whereas CPPU and bagging inhibited the expression of LcMYB1 and anthocyanin accumulation in the pericarp. Cis-elements associated with light responsiveness and abscisic acid responsiveness were identified in the promoter region of LcMYB1. Among the 6 structural genes tested, only LcUFGT was highly correlated with LcMYB1. These results suggest that LcMYB1 controls anthocyanin biosynthesis in litchi and LcUFGT might be the structural gene that is targeted and regulated by LcMYB1. Furthermore, the overexpression of LcMYB1 induced anthocyanin accumulation in all tissues in tobacco, confirming the function of LcMYB1 in the regulation of anthocyanin biosynthesis. The upregulation of NtAn1b in response to LcMYB1 overexpression seems to be essential for anthocyanin accumulation in the leaf and pedicel. In the reproductive tissues of transgenic tobacco, however, increased anthocyanin accumulation is independent of tobacco's endogenous MYB and bHLH transcriptional factors, but associated with the upregulation of specific structural genes.
Introduction
The colors of flowers and fruits mainly result from the accumulation of anthocyanins, a group of secondary metabolites that are synthesized via the flavonoid pathway [1]. The structural genes involved in the anthocyanin biosynthetic pathway of plants include chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT). These genes are well characterized in model plants [2], [3] and fruits including grape [4], apple [5], Chinese bayberry [6], and litchi [7]. In addition, numerous studies demonstrate that anthocyanin accumulation is largely regulated at the transcriptional factors which manipulate the expression of structural genes in the anthocyanin biosynthetic pathway [8].
Among various complex transcription factors [including MYB, basic helix-loop-helix (bHLH) and WD40 protein], MYB appears to be the major determinant of anthocyanin accumulation [9], [10]. Subgroup 6 clade MYBs, PAP1 and PAP2, in Arabidopsis and their orthologs in other plant species are known to be the key regulators of anthocyanin biosynthesis. PAP1 and PAP2 overexpression in Arabidopsis induces anthocyanin biosynthesis in the entire plant [11], [12]. Many MYBs have been identified to control the accumulation of anthocyanin in the tissue or organ of various plants, such as GmMYB-G20-1 in soybean flower [13], [14], BoMYB2 in purple cauliflower [15], LhMYB6 and LhMYB12 in Asiatic hybrid lily [16], VvMYBA in grape [17], [18], and MrMYB1 in bayberry [19]. In apple, the red color of the fruit skin and flesh are controlled by two MYB genes, namely MdMYBA and MdMYB10 [20], [21]. Some MYB transcription factors, however, inhibit the accumulation of anthocyanin. Two R3-MYBs were reported to negatively regulate anthocyanin in Arabidopsis [22], [23]. In strawberry, FaMYB1 represses transcription of anthocyanin-related genes during maturation, and the biosynthesis of proanthocyanidins is inhibited in the leaves of FaMYB1 transgenic Lotus corniculatus [24], [25]. These results suggest functional differences between various R2R3-MYB proteins.
Expression pattern analysis and DNA-protein interaction assays indicate that different MYBs activate different anthocyanin biosynthetic structural genes. PAL, CHS, and DFR are activated in pap1-D plants by linking an enhancer sequence to PAP [11]. In grape berry, MybA gene regulates anthocyanin biosynthesis via UFGT expression [17]. MdMYB1 cDNA increases luciferase enzyme activity when cobombarded with constructs containing the MdDFR and MdUFGT promoters [26]. This suggests that MdMYB1 can activate the expression of these two anthocyanin pathway genes in apple. Similarly, the activity of Arabidopsis DFR promoter was significantly increased when MdMYB10 was cotransformed with an apple bHLH [21]. The coexpression of GbMYB2 and GbMYC1 activates the GbDFR and GbANS promoters in tobacco leaf protoplasts [27]. Pr-D overexpression in cauliflower specifically activates a bHLH transcription factor and a subset of anthocyanin structural genes that encode BoF3′H, BoDFR, and BoANS [28].
It is well known that both intrinsic and extrinsic factors affect the biosynthesis of anthocyanins. Anthocyanin accumulation is influenced by various environment conditions, such as light, temperature, and phytohormones. In apple [26], pear [29], peach [30], and bayberry [6], anthocyanin biosynthesis is enhanced by sunlight. Generally, shading fruit by bagging with a dark material inhibits the biosynthesis of anthocyanins, and rapid anthocyanin accumulation is noticeable after bag removal. High temperature inhibits, while low temperature enhances anthocyanin accumulation in apple fruit [31], [32]. ABA promotes anthocyanin synthesis during grape development and grape cell culture [33]–[35]. Jasmonate (JA) regulates anthocyanin accumulation in Arabidopsis by degrading JA ZIM-domain (JAZ) proteins, preventing the interactions between JAZ proteins and bHLH and MYB transcription factors [36].
Litchi (Litchi chinensis Sonn.) is an economically important evergreen fruit crop in the Sapindaceae family. The fruit consist of drupes with a white translucent edible aril that is surrounded by the pericarp. The red color on the pericarp of litchi results from the accumulation of anthocyanins [37]. Color differences between litchis are mainly due to variations in the anthocyanin concentration in the pericarp. The biosynthesis of anthocyanins in the pericarp of litchi demonstrates cultivar, developmental, and environmental differences. And one structural gene, LcUFGT, has been shown to have major role in these differences [7]. However, this mechanism has not been characterized at the level of transcriptional regulation.
In this study, an R2R3-MYB gene, LcMYB1, was isolated from the pericarp of ‘NMC’, a red litchi cultivar. Its expression patterns in different tissues, pericarp developmental stages, color genotypes, and ABA and light stimuli were assessed, and the correlation between anthocyanin accumulation and the expression of other anthocyanin biosynthetic genes was investigated. Furthermore, the functional activity of this transcription factor in driving anthocyanin accumulation was evaluated using two heterologous transgenic assays. In these studies, the behavior of LcMYB1 suggests that it is responsible for red pigmentation in litchi fruit. ABA and sunlight exposure enhance anthocyanin accumulation, mainly by upregulating LcMYB1 expression. The overexpression of LcMYB1 in transformed tobacco lines indicates that the efficient induction of anthocyanin biosynthesis by LcMYB1 in vegetative tissues depends on the coexpression of the bHLH protein, NtAn1b.
Results
Isolation of LcMYBs from the pericarp of litchi
Using degenerate primers, a 283-base pair (bp) product was amplified using ‘NMC’ cDNA. The full-length cDNA was obtained using 3′ and 5′ RACE. This full-length cDNA, namely LcMYB1, is a 940 bp transcript that encodes a protein of 281 amino acids. When translated into protein, LcMYB1 displays distinct R2 and R3 MYB repeat domains (Fig. 1). In the R3 domain, LcMYB1 shows residues that in accordance with amino acid motif [DE]Lx2[RK]x3Lx6Lx3R predicted to allow interaction with bHLHs, suggesting that it may interact with bHLH proteins [38]. In the variable region, the small motif [K/R]Px3[K/T][F/Y] is conserved [26]. This motif is part of the motif that was previously reported as KPRPR[S/F]F (motif 6) [39]. LcMYB1 demonstrated a high degree of homology with other MYB transcription factors (Fig. 1). The amino acid identity over the R2R3 DNA-binding domain are 86.7% similar to Chinese bayberry MrMYB1, 83.8% similar to Arabidopsis PAP1, 81.9% similar to grape VvMYBA1, and 80.0% similar to apple MdMYB10 respectively.
Figure 1. Protein sequence alignment of the R2R3 DNA-binding domains of LcMYB1 and the other known anthocyanin MYB regulators in other species.
The R2 and R3 domains are underlined. The bHLH-binding motif is boxed in the R3 domain. Motif 6 was previously identified in the C-terminal domain of anthocyanin-related MYBs in Arabidopsis [39]. The accession number of these proteins (or translated products) are as follows in the GenBank database: MrMYB1, GQ340767; MdMYB1, ABK58136; MdMYB10, DQ267896; PyMYB10, ADN26574; GmMYB10, ACM62751.1; PhAN2, AAF66727; LeANT1, AAQ55181; CsRuby, AFB73909; MrMYB1, GQ340767; VlMYBA1-1, BAC07537; VvMYBA1, BAD18977; VvMYBA2, BAD18978; GhMYB10, CAD87010; AtPAP1, AAG42001; AtPAP2, AAG42002; AtMYB113, NP_176811; AtMYB114, NP_176812; AmROSEA1, ABB83826; AmROSEA2, DQ275530; AmVENOSA, DQ275531; and FaMYB1, AF401220.
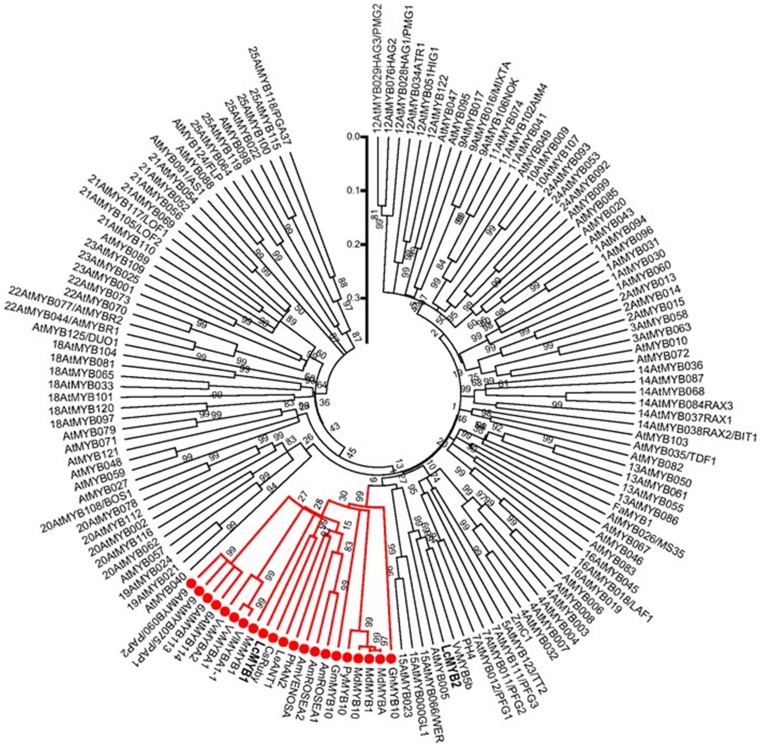
The phylogenetic analysis indicates that LcMYB1 is closely related to the subgroup 6 MYBs in Arabidopsis: the nearest MYB is CsRuby, which shared 87.1% and 49.3% amino acid identity with the R2R3 DNA-binding domain and the entire protein, respectively (Fig. 2). LcMYB1 is clustered with the R2R3 MYB transcription factors that are involved in the regulation of anthocyanin biosynthesis in other plant species. These results suggest that LcMYB1 may demonstrate similar functions as subgroup 6 MYBs in Arabidopsis [12], which play an important role in regulating anthocyanin synthesis.
Figure 2. Phylogenetic relationships between Arabidopsis MYB transcription factors and anthocyanin-related MYBs in other species.
LcMYB1 clusters next to CsRudy, within the anthocyanin subgroup 6 MYBs. The subgroup numbers were previously described [12]. The tree was constructed using MEGA 5, neighboring-joining phylogeny testing, and 1,000 bootstrap replicates. The accession numbers for the genes in the other species are provided in Figure 1.
In addition, LcMYB2, which is homologous to LcMYB1, was isolated from our transcriptome sequences data, and full-length cDNA was obtained using 3′and 5′ RACE. However, the expression pattern and phylogenetic analysis demonstrated that LcMYB2 may not be involved in the regulation of anthocyanin biosynthesis (Fig. S1); thus, LcMYB2 was not further analyzed in this paper.
Anthocyanin contents and the expressions of biosynthetic genes in litchi
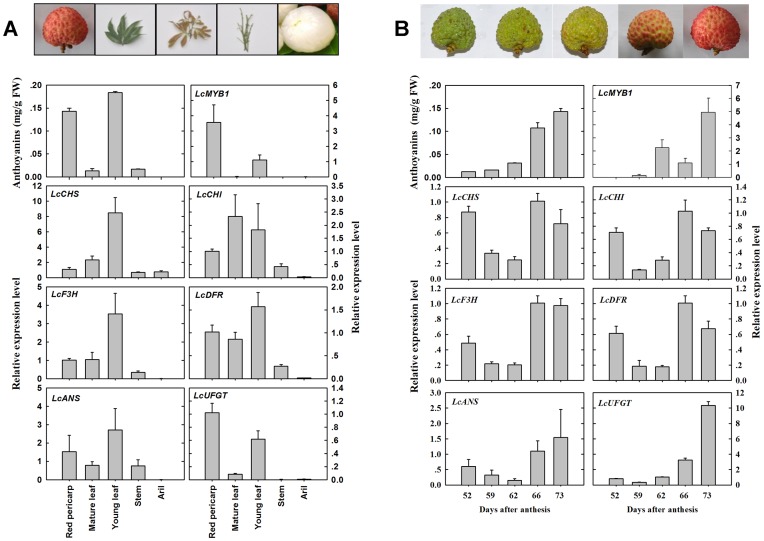
The total anthocyanin content in different tissues of ‘NMC’ was measured using the pH-differential spectrum method. Content varies greatly between different tissues (Fig. 3 A). Red tissues, including mature pericarp and young leaf, contained much higher levels of anthocyanins, while extremely low or no anthocyanins was detected in non-red tissues, including mature leaf, young stem, and aril.
Figure 3. Anthocyanin contents and expressions of biosynthetic genes.
(A) Anthocyanin contents and expression analysis of LcMYB1 and anthocyanin biosynthetic structural genes in different tissues of ‘NMC’ litchi including red pericarp, mature and young leaves, young stem and aril. (B) Anthocyanin contents and expression analysis of LcMYB1 and anthocyanin biosynthetic structural genes in the pericarp of ‘NMC’ litchi during fruit development. The Lcactin gene was used to normalize the expression levels of the genes under identical conditions. The vertical bars represent the standard error of triplicate experiments.
The transcription levels of LcMYB1 and the structural genes that encode the enzymes of the anthocyanin pathway were determined in different litchi tissues using qRT-PCR and gene-specific primers (Fig. 3 A). Notable differences were observed. Extremely low or undetectable levels of LcMYB1 and the six tested structural genes were noticed in non-red stem and aril. In non-red mature leaf, the expression levels of LcMYB1 and LcUFGT were also low or undetectable, while the expression levels of other genes including LcCHS, LcCHI, LcF3H, LcDFR and LcANS were detectable. In red pericarp, relatively high transcription levels of LcMYB1, LcDFR, LcANS, and LcUFGT were observed. The expression levels of LcMYB1 and the six structural genes were all high in the anthocynin-rich young leaf.
The developmental patterns of anthocyanin content and the expression levels of LcMYB1 and the six structural genes in the pericarp of ‘NMC’ are shown in Figure 3 B. The rapid accumulation of anthocyanins occurred during the late stage of fruit development. The content of anthocyanins was low (<0.03 mg g−1 FW) in the pericarp before 62 days after full bloom (DAFB), but increase dramatically to 0.11 mg g−1 FW and 0.14 mg g−1 FW at 66 and 73 DAFB, respectively. In parallel with the accumulation of anthocyanins, the expression levels of LcMYB1 and LcUFGT were enhanced as the fruit developed toward full maturity. For the rest of the structural genes, the expression levels were relative high at the first sampling date, decreased afterwards, and increased along with rapid anthocyanin accumulation.
Cultivar differences and the effects of manipulation treatments on LcMYB1 expression
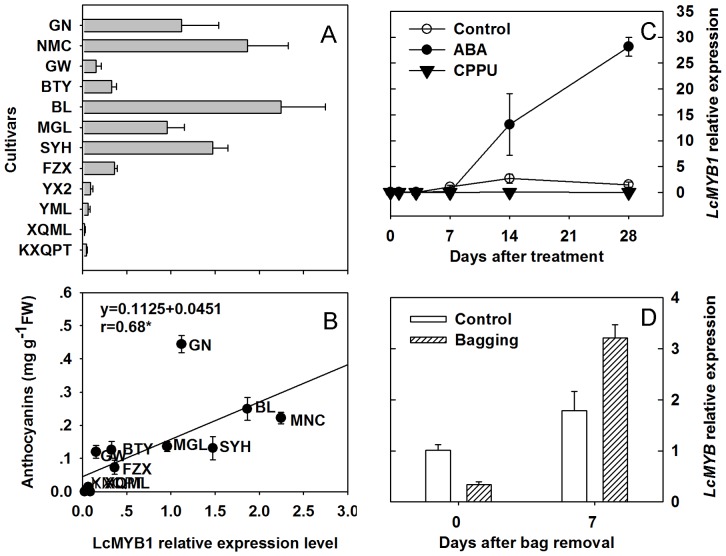
According to our previous study [7], anthocyanin content in the pericarp varies greatly between the 12 tested cultivars. In the present study, the expression of LcMYB1 and its correlation with anthocyanin in the pericarps of these 12 cultivars was assessed. Consistent with pericarp anthocyanin content, the non-red cultivars ‘KXQPT’, ‘XQML’, ‘YML’ and ‘YX2’ displayed extremely low levels of LcMYB1 expression, while noticeable LcMYB1 expression was observed in the rest red cultivars (Fig. 4 A). The expression of LcMYB1 was significantly and positively correlated with anthocyanin content in the pericarps of the tested cultivars (r = 0.68) (Fig. 4 B).
Figure 4. LcMYB1 expression in the pericarp of different cultivars and manipulation treatments.
A, LcMYB1 expression of in the pericarp of twelve cultivars. B, The correlationship between LcMYB1 expression and anthocyanin content among the twelve cultivars. Anthocyanin contents were obtained from Wei et al. [7], Effects of ABA and CPPU on the expression of LcMYB1. D, Effects of bagging and bag removal on LcMYB1 expression.
The expression levels of LcMYB1 in response to the ABA and CPPU treatments are shown in Figure 4 C. ABA enhanced while CPPU inhibited anthocyanin accumulation in ‘Feizixiao’ [7]. The expression patterns of LcMYB1 were consistent with anthocyanin accumulation across different treatments. LcMYB1 expression was detected in all of the red pericarps, but not in any of the non-red pericarps. Expression was undetectable before 14 days after treatment (DAT), after which there was a notable expression in the control. In CPPU-treated pericarp, the expression of LcMYB1was hardly detectable throughout the entire experiment period, while a steep increase in LcMYB1 expression was observed 7 days after ABA application.
Bagging and bag removal were used to study the effects of illumination on LcMYB1 expression (Fig. 4 D). In the pericarp of the control fruit, which were grown under normal light conditions, LcMYB1 transcription was approximately 3-fold higher than in the bagged fruit. When the dark-grown litchi fruit were re-exposed to sunlight, LcMYB1 transcription increased by approximately 2-fold within 7 days after bag removal. The effects of bagging and bag removal on the expressions of LcMYB1 in the pericarp generally paralleled the effects on anthocyanin accumulation [7]. This result is similar to the MdMYB1 transcription patterns observed in response to illumination [26].
LcMYB1 function in tobacco
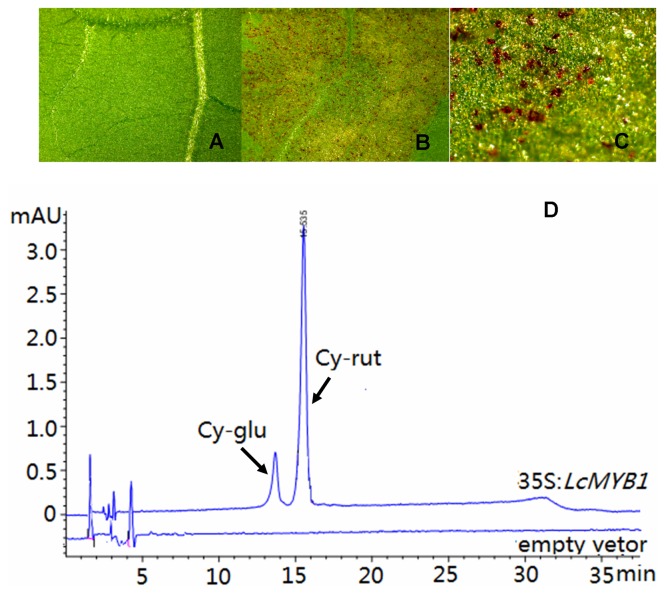
The transient expression levels of anthocyanin-synthesizing transcription factors to prove their function in tobacco leaf are frequently utilized [6], [21], [40]. Pigmentation is evident at the infiltration points on tobacco leaf at 5 days after infiltration with LcMYB1 cDNA (Fig. 5 A–C). To confirm that anthocyanins were synthesized in tobacco after LcMYB1 transformation, leaf tissue sample were extracted and the anthocyanins were analyzed using HPLC. No observable anthocyanin peaks were observed in the extracts of the tobacco leaves that were transformed with the Agrobacterium- carrying empty vector, while two major peaks were observed in the extracts of the tobacco leaves that transformed with the Agrobacterium-carrying 35S:LcMYB1 vector (Fig. 5 D). These two peaks represent cyanidin-3-glucoside and cyanidin-3-rutinoside respectively, which is consistent with the anthocyanin composition observed in the red pericarp of litchi.
Figure 5. Color development in Nicotiana tabacum leaves following transient transformation.
Microscopic images showing anthocyanin accumulation in tobacco leaf infiltrated with: A) an empty vector (1× magnification) or B–C) 35S:LcMYB1 (8× magnification). D) Anthocyanin HPLC profiles of 35S:LcMYB1 extracts from tobacco leaf (top line) and empty vector (bottom line). Peaks identified at 520 nm: cy-glu, cyanidin-3-glucoside; cy-rut, cyanidin-3-rutinoside.
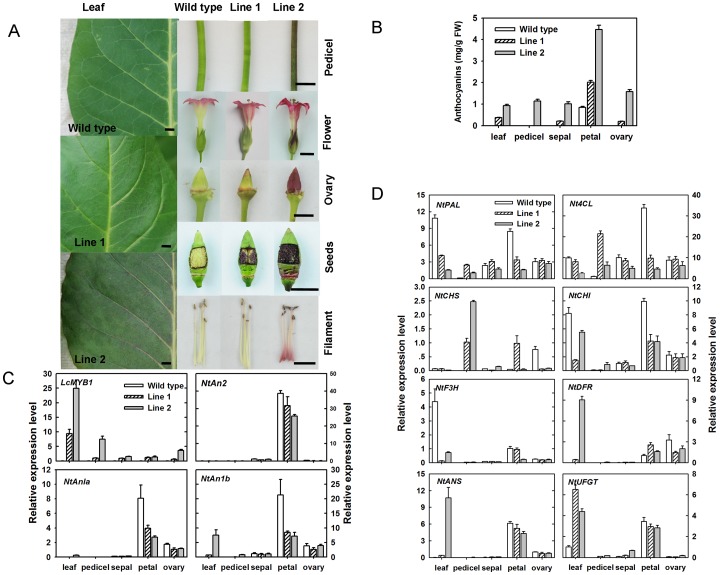
The LcMYB1 cDNA was fused with the CaMV 35S promoter and cloned into the binary pBI121 vector. To assess LcMYB1 function, Nicotiana tabacum cultivar K326 leaf strips were transformed with 35S:LcMYB1 and an Agrobacterium-mediated kanamycin selectable marker. Pigmented kanamycin-positive regenerated plantlets were noticed (Fig. S2). Pigmented leaf, pedicel, ovary, seed, filament, and highly pigmented petals were observed in the grown transformant lines, while the wide type plants demonstrated no obviously pigmented tissues except the petals (Fig. 6 A). The color of transformant line 2 was more intense than line 1. LcMYB1 overexpression in tobacco resulted in anthocyanin accumulation in many tissues. Anthocyanins were detected in the leaves, pedicels, sepals, and ovaries of the transformant lines, especially line 2 (Fig. 6 B). Anthocyanin contents in the petals of the transformant line 1 and line 2 were 2.01±0.09 and 4.46±0.20 mg g−1 FW, respectively, which were much higher than that in the wild type (0.84±0.05 mg g−1 FW).
Figure 6. Images of the transgenic tobacco lines and the anthocyanin content, and the expression of anthocyanin biosynthetic pathway structural and regulatory genes in transgenic tobacco lines.
A, Images of tobacco lines containing the empty vector (wild type) or the LcMYB1 allele with the CaMV 35S promoter (Line 1–2) (Scale bars for images = 1cm. B, Anthocyanin content in different organs of transgenic tobacco lines. C, Regulatory gene expression in the anthocyanin biosynthetic pathway in transgenic tobacco. D, Structural gene expression in the anthocyanin biosynthetic pathway in transgenic tobacco lines. The Lcactin gene was used to normalize gene expression of the genes under identical conditions. The vertical bars represent the standard error of triplicate experiments.
Furthermore, the expression levels of four regulatory factors were investigated, including LcMYB1, three tobacco anthocyanin biosynthesis regulators, and eight structural biosynthetic genes (Fig. 6 C–D). Distinct LcMYB1 expression was observed in all of the tested tissues of the transformant lines, but no expression was detected in the tissues of wild type (Fig. 6 C). This result confirms the successful transformation of LcMYB1. Higher LcMYB1 expression levels were observed in the leaves in comparison with the other tissues, and higher expression levels were observed in line 2. NtAn2, a R2R3-MYB transcription factor, interacts with bHLH regulator NtAn1a and NtAn1b to regulate the anthocyanin biosynthesis in tobacco flowers [41], [42]. In the present study, these endogenous regulatory factors (NtAn2, NtAn1a and NtAn1b) were specifically expressed in tobacco petals with extremely low or undetectable expression levels in other tissues (Fig. 6 C). The expression of NtAn2 in petals decreased in response to increased LcMYB1 expression. NtAn1a and NtAn1b were also downregulated in the petals of LcMYB1-overexpressing lines. In the leaf and pedicel, however, NtAn1b was upregulated especially in line 2.
In leaves, the expression levels of early and mid anthocyanin biosynthetic structural genes including NtPAL, Nt4CL, NtCHS, NtCHI and NtF3H, were all downregulated, whereas the expression levels of late anthocyanin biosynthetic genes, including NtDFR, NtANS, and NtUFGT, were all upregulated in transformant lines in comparison with wild type (Fig. 6 D). The expression levels of all structural genes in pedicel of wild type were extremely low, while upregulation of NtPAL, Nt4CL, and NtCHS was observed in the pedicels of the transformant lines. No obvious expression patterns, except for NtUFGT, were noticed in the sepals and ovaries of LcMYB1-overexpressing lines and wild type. In agreement with the anthocyanin content, the expression of NtUFGT was higher in the sepals and ovaries of the transformant lines, especially line 2, in comparison with wild type. Although the anthocyanin content was much higher in the petals of the transformant lines, the downregulation of most structural genes was noticeable. NtDFR was the only upregulated structural gene observed in both transformant lines.
LcMYB1 promoter analysis
Anthocyanin content and MYB transcription can be affected by exogenous stimulations, such as light, temperature, and hormones. To investigate the elements that could be associated with these factors, we isolated and analyzed the promoter region of LcMYB1. A 1.5-kb region upstream from the start codon of the LcMYB1 promoter was predicted in PlantCARE database. The putative cis-elements identified in the LcMYB1 promoter are showed in Table 1. The promoter sequence displayed multiple light-regulating units. ACE, Box 4, G-Box and GA-motif are involved in light responsiveness. Two ABRE are involved in the abscisic acid responsiveness. The remaining cis-elements include two HSE elements involved in the heat stress response, CGTCA and TGACG motifs involved in MeJA-responsiveness, a gibberellin-responsive element (GARE) motif, and an auxin-responsive (TGA) element. In addition, two binding motifs for MYB proteins were also found in the upstream region.
Table 1. Putative cis-elements identified in the LcMYB1 promoter.
| Category | cis-acting element | Sequence | Distance from ATG |
| Light response | ACE | AAAACGTTTA | +716 |
| Box 4 | ATTAAT | −124, +585, +610 | |
| G-Box | CACGTT | +475, +714, +1440 | |
| G-Box | GACACGTAGT | −477 | |
| G-Box | CACGTC | +135 | |
| G-Box | TAAACGTG | −714 | |
| G-Box | CACGTGG | −476 | |
| GA-motif | ATAGATAA | +652, +1221 | |
| GAG-motif | AGAGAGT | −1475 | |
| Abscisic acid response | ABRE | GGACACGTGGC | −477 |
| ABRE | CACGTG | +475 | |
| MeJA response | CGTCA-motif | CGTCA | −112 |
| TGACG-motif | TGACG | +112 | |
| Gibberellin response | GARE-motif | TCTGTTG | +208 |
| TATC-box | TATCCCA | −1356 | |
| Auxin-response | TGA-element | AACGAC | −324 |
| MYBHv1-binding site | CCAAT-box | CAACGG | +427 |
| MYB-binding site involved in drought-inducibility | MBS | TAACTG | −1250 |
| Heat stress response | HSE | AAAAAATTTC | +493, +1071 |
| Defense and stress | TC-rich repeats | ATTTTCTTCA | −1246 |
+, distance from ATG in a positive DNA strand; −, distance from ATG in a negative DNA strand.
Discussion
LcMYB1 is homologous with other R2R3-MYBs involved in regulating anthocyanin biosynthesis
The pericarps of litchi demonstrate variations in anthocyanin accumulation depending on the cultivar, developmental stage, and environmental stimuli. It is possible that non-red pericarp lost the ability to produce anthocyanin due to a mutation in one or more biosynthetic or regulatory gene(s). As observed in white grapes [43], late biosynthetic genes, such as LcDFR and LcUFGT in particular, are coordinately expressed during the red coloration of litchi fruits [7]. It is well established that MYB transcriptional factors control the accumulation of anthocyanin in model plants [11], [44], [45]. In recent years, numerous MYBs responsible for anthocyanin biosynthesis in different kinds of fruits have been identified and characterized [6], [17], [20], [21], [24], [26], [29]. In the present study, the 940-bp full-length LcMYB1 gene was isolated from the red pericarp of ‘NMC’. Sequence analysis of the LcMYB1-encoded protein confirmed its homology with anthocyanin-regulating MYB transcription factors in other species (Fig. 1). The LcMYB1-encoded protein exhibited distinct R2 and R3 MYB repeat domains and one signature motif which was predicted to interacted with bHLH. LcMYB1 was clustered within the anthocyanin subgroup 6 MYBs and was highly orthologous with CsRuby, MrMYB1, MdMYB10, and MdMYB1 (Fig. 2). These results suggest that LcMYB1 maybe a member of the MYB anthocyanin activators.
Key role of LcMYB1 in anthocyanin accumulation
Most MYBs demonstrate tissue-specific expression characteristics. In apple, MdMYB10 can be detected only in the leaf, cortex, and skin where containing large amount of anthocyanins [21], [40]. Similarly LcMYB1 transcripts can be detected only in anthocyanin-rich tissues in litchi, i.e., red pericarp and young leaf (Fig. 3 A). In addition, the transcription level of LcMYB1 is undetectable or extremely low in non-red pericarp, but is upregulated during fruit development and pigmentation (Fig. 3 B). The LcMYB1 expression patterns in different tissues and fruit development stages are consistent with MrMYB1 in bayberry and VvMYBA in wine grape [6], [17]. The expression levels of LcMYB1 were weak in non-red cultivars (‘KXQPT’, ‘XQML’, ‘YML’ and ‘YX2’), but notable in red cultivars (‘FZX’, ‘SYH’, ‘MGL’, ‘BL’, ‘GW’, ‘NMC’ and ‘GN’) (Fig. 4 A). In sweet potato, IbMYB1 is also expressed in purple-fleshed cultivars, but not in those of orange-, yellow-, or white-fleshed cultivars [46]. LcMYB1 expression was positively correlated with anthocyanin accumulation in the pericarp of the 12 litchi cultivars (Fig. 4 B).
In non-climacteric strawberry, ABA is a signal molecule that promotes fruit ripening including the accumulation of anthocyanin [47]. However, the mechanisms in which ABA enhance anthocyanin synthesis in fruit have not yet been established. In grape berry, exogenous ABA rapidly induces the accumulation of anthocyanin and the expression of the corresponding structural genes [48]. Litchi is a typical non-climacteric fruit, and the endogenous ABA concentrations in the pericarp increases along with the synthesis of anthocyanins [49]. ABA significantly enhances anthocyanin accumulation in pericarp (while cytokinins inhibits accumulation), and decrease in endogenous ABA caused by cytokinins has also been observed [49]. Applying ABA 1 month before commercial harvest enhanced (whereas CPPU inhibited) LcUFGT expression and anthocyanin synthesis in the pericarp of litchi [7]. In the present study, ABA promoted, while CPPU reduced, the expression of LcMYB1 (Fig. 4 C). It is possible that ABA affects the upstream gene LcMYB1, which regulates LcUFGT expression and subsequently the accumulation of anthocyanin in the pericarp of litchi. CPPU inhibits anthocyanin accumulation probably by decreasing endogenous ABA. Putative ABA responsive elements (ABRE) are identified in the promoter region of LcMYB1 (Table 1). The mechanism that ABA enhances MYB expression and anthocyanin accumulation requires further study.
Bagging experiments have revealed that sunlight is essential for anthocyanin synthesis in apple [26], pear [19], peach [30], bayberry [6] and litchi [7]. Sunlight affects both anthocyanin accumulation and the expression of both anthocyanin biosynthetic genes and/or regulatory genes in these fruits. In the present study, LcMYB1 expression is reduced in the pericarp of bagged fruit, but markedly upregulates when re-exposed to sunlight (Fig. 4 D). It is likely that sunlight regulates anthocyanin synthesis via LcMYB1 expression. Constitutive Photomorphogenic 1 (COP1) is a RING-finger type ubiquitin E3 ligase involved in proteolysis during light signaling [50]. Recent studies have shown that COP1 can influence the stability of MYB transcription factors in the dark, thereby reducing anthocyanin accumulation in Arabidopsis and apple [51], [52]. Numerous putative light responsive elements have also been identified in the promoter region of LcMYB1 (Table 1).
We use both transient transformation in tobacco leaves and stable tobacco transformants to assess the function of LcMYB1 in the anthocyanin synthesis pathway. Microscopic images and HPLC data show the significant accumulation of anthocyanin compounds in transient expression tobacco leaves, confirming the functional presence of anthocyanin biosynthetic enzymes in tobacco leaves and the presence of the cellular machinery needed to transport anthocyanins to the vacuole (Fig. 5). The transient expression of LcMYB1 in tobacco results in two anthocyanin peaks, representing cyanidin-3-glucoside and cyanidin-3-rutinoside, which is consistent with the peaks identified in the pericarps of red litchi cultivars [7].
When LcMYB1 was stably transformed in tobacco, anthocyanin accumulation is noticed not only in the reproductive organ but also in the leaf and pedicel (Fig. 6 A). Considerable amounts of anthocyanins were detected in the leaves, pedicels, sepals, petals and ovaries of transgenic lines, especially line 2 (Fig. 6 B). These results suggested that LcMYB1 is responsible for controlling anthocyanin biosynthesis in litchi.
Coordinately regulation of litchi anthocyanin biosynthesis
Retrotransposon-induced insertion into the grape anthocyanin regulatory gene VvmybA1 resulted in a lack of anthocyanin biosynthesis and the development of white grape cultivars [18]. Sicilian blood orange arose by insertion of a Copia-like retrotransposon adjacent to a gene encoding Ruby, a MYB transcriptional activator of anthocyanin production [53].
MYBs control the biosynthesis of anthocyanins by activating the expression of structural genes [8]. Structural genes and MYBs are usually coordinately expressed during anthocyanin synthesis. In Chinese bayberry, the relative intensities of MrF3H, MrF3′H, MrDFR1, MrUFGT and MrMYB1 mRNA are strongly associated with anthocyanin content [6]. In apple, the expression of MdANS, MdUFGT, and MdMYB1 demonstrate a positive correlation with anthocyanin synthesis in cultivars that show different colors [26]. Our previous study on different fruit color genotypes of litchi demonstrates that late genes in the anthocyanin biosynthetic pathway, LcDFR and LcUFGT in particular, are coordinately expressed during red coloration [7]. In the present study, LcUFGT expression was highly correlated with LcMYB1 expression in different tissues and different fruit developmental stages in litchi (Fig. 3 A–B). The other genes were less influenced by tissue and fruit developmental stage. Relatively high expression levels of these genes were observed in non-red mature leaf and green pericarp. This result is consistent with studies on grapes that report UFGT as the only gene that makes the difference in coloration between white type and its red sport [42], [54].
Response of tobacco endogenous genes in anthocyanin biosynthesis to LcMYB1 overexpression
In apple and Chinese bayberry, without co-infiltrate with a bHLH transcription factor, the functions of MdMYB10, MdMYB110a and MrMYB1 were quite weak in transient expression tobacco leaf [6], [21]. In the present study, LcMYB1 alone effectively induced anthocyanin accumulation in tobacco leaf (Fig. 5). Interestingly, pigmented organs varied between tobacco lines that were transformed with MYBs from different species. When PAP1 or NtAn2 are overexpressed in tobacco, the whole plant produces anthocyanins [41], [55]. However, only petals and seeds accumulate anthocyanins in MdMYB1 and MrMYB1 transformed tobacco [19], [20]. This difference might result from the abilities of the exogenous MYBs and the interactions with endogenous bHLH. When CsRuby alone is overexpressed in tobacco, no anthocyanins are observed; when combined with different bHLH transcription factors, tobacco leaves accumulate different concentrations of anthocyanins [53].
The whole plant of LcMYB1 overexpression line 2 produces anthocyanins (Fig. 6 A–B). The responses of endogenous MYB and bHLH transcription factors and anthocyanin biosynthetic genes to the overexpression of LcMYB1 were investigated in tobacco in the present study (Fig. 6 C–D). Different tissues responded differently to LcMYB1 overexpression in terms of endogenous gene expression. Higher expression levels of endogenous leaf NtAn1b and late anthocyanin biosynthetic genes, including NtDFR, NtANS, and NtUFGT, were observed in both transgenic tobacco lines. In the pigmented pedicel of line 2, the upregulation of NtAn1b and NtUFGT was noticable. The expression of NtAnb1 seems to be essential for the accumulation of anthocyanins in tobacco leaf and pedicel. In apple, the efficient induction of anthocyanin biosynthesis by MdMYB10 is dependent on the coexpression of a bHLH protein [21]. In MrMYB1-overexpressing tobacco leaves, both anthocyanin accumulation and the expression of NtAn1b are absent, despite the upregulation of late anthocyanin biosynthetic genes [19]. In petunia, PhAn1, a bHLH transcription factor, reportedly directly activates the expression of the DFR gene and is specifically regulated by the MYB factor PhAn2 [56]. In Arabidopsis, R2R3-MYB transcription factor TT2 or PAP1 regulate the expression of the bHLH reportedly TT8 [57]. Therefore, in the vegetative organs of tobacco, it can be concluded that exogenous MYB induces the expression of bHLH transcription factor NtAn1b and late anthocyanin biosynthetic genes, resulting in the accumulation of anthocyanin.
The mechanisms in reproductive organs are quite different from those in vegetative organs. Downregulated or unchanged expression levels in the three endogenous MYB and bHLH transcription factors were observed in the sepals, petals and ovaries of the transgenic tobacco lines (Fig. 6 C). These results are consistent with those of MrMYB1-overexpressing Arabidopsis, but inconsistent with MrMYB1 transgenic tobacco [19]. In MrMYB1- overexpressing Arabidopsis, the expression of AtPAP1, a MYB transcription factor, was suppressed in the roots and leaves but not obviously affected the seeds. However, in tobacco petal, MrMYB1 overexpression stimulated the expression of endogenous NtAn2, NtAn1a, and NtAn1b. Furthermore, the responses of the structural genes to the overexpression of LcMYB1 were inconsistent with MrMYB1-overexpressing tobacco. Most structural anthocyanin genes were significantly upregulated in the reproductive tissues of the transgenic MrMYB1 lines in comparison with wild type plants [19]. In the present study, the expression of NtUFGT in the transgenic tobacco was upregulated in most of the organs except for petal (Fig. 6 D). Coincident to the much higher expression of LcMYB1 in leaves of transgenic lines, the upregulation of NtUFGT was much obvious in leaf than in pedicel, sepal and ovary. In the petals of transgenic lines, however, only upregulated NtDFR expression was observed. These results suggested that the effects of exogenous MYBs appear to differ considerably between species as well as tissues.
Materials and Methods
Plant materials and treatments
Twelve different litchi cultivars were used in this study: four non-red skin cultivars [‘Kuixingqingpitian’ (‘KXQPY’), ‘Xinqiumili’ (‘XQML’), ‘Yamulong’ (‘YML’), and ‘Yongxing No. 2’ (‘YX2’)]; two unevenly red cultivars [‘Feizixiao’ (‘FZX’) and ‘Sanyuehong’ (‘SYH’)]; and six evenly red cultivars [‘Meiguili’ (‘MGL’), ‘Baila’ (‘BL’), ‘Baitangying’ (‘BTY’), ‘Guiwei’ (‘GW’), ‘Nuomici’ (‘NMC’) and ‘Guinuo’ (‘GN’)]. These trees were grown in the experimental orchard of South China Agricultural University (Guangzhou, China) received standard horticultural practices, and disease and insect control. Pericarp discs were collected at commercial maturity as reflecting by aril Brix-acid ratio.
Double-layer kraft paper bagging of the clusters on ‘FZX’ commenced at 1 month after full bloom, and the bags were removed at color break. Pericarp samples were taken on the day of bag removal and on the 7 day later. The growth regulators were applied 4 weeks before harvest. Triplicate lots with 3 ‘FZX’ trees were sprayed with abscisic acid (ABA: 25 mg L−1), forchlorofenuron (CPPU: 4 mg L−1) and tap water (control), respectively. Pericarp discs were sampled when growth regulator spray was applied (day 0) and 1, 3, 7, 14, and 28 days after treatments.
Samples were used to determine anthocyanin content and the transcription levels of the anthocyanin biosynthetic genes and litchi MYBs. Thirty exposed fruit were randomly sampled and the pericarps from 10 individual fruits were pooled into a single replication. Mature leaves, young leaves, young stems, and arils were collected from cultivar ‘NMC’ on July 2, 2011, and pericarps were also collected between May 29 and July 2 at 7-day intervals. All samples were immediately frozen in liquid nitrogen and stored at −80°C until use.
Tobacco (Nicotiana tabacum) was used in all genetic transformation. All plants were grown in green houses at 28°C using natural light.
Anthocyanin analysis
The total anthocyanin content was determined according to the method developed by Fuleki and Francis [58], which involves measuring the absorbance (520 nm) of extracts that have been diluted with pH 1.0 and 4.5 buffers. HPLC analysis of anthocyanins were extracted and determined as previously described [7].
RNA extraction and cDNA synthesis
Total RNA was extracted from the different tissues of litchi and tobacco using the RNAOUT kit (Tiandz, Beijing, China). DNase I (TaKaRa, Japan) was added to remove genomic DNA, and RNase-free columns (Tiandz,) were used to purify the total RNA. Then, cDNA was synthesized from total RNA (2 μg) using oligo (dT) primers according to the manufacturer's instructions of PrimeScript™ RT-PCR Kit (TaKaRa).
Cloning of transcription factor genes
Degenerate primers were designed based on the highly conservative amino acid regions of MYB [6]. The cDNAs were synthesized from the total RNA of the mature pericarp of cultivar ‘NMC’ and used as the PCR templates. PCR-amplified products of appropriate length were cloned into T/A cloning vector pMD®20-T (TaKaRa, Japan) and then transformed into E.coli DH5α Max Efficiency® Chemically Competent Cells (TaKaRa). Plasmid DNA was isolated from positive E. coli cells and digested with EcoRI and the final DNA sample was sent to Beijing Genomics Institute for sequencing.
Rapid amplification of cDNA ends (RACE) was performed to obtain the 3′and 5′ ends of the two genes from mature pericarp ‘NMC’ using SMART RACE (Clontech, USA). The primers are shown in Table 2.
Table 2. LcMYB1 3′ RACE, 5′ RACE, ORF and RT-PCR primers used in this study.
| Primer name | Sequence(5′→3′) | function |
| LcMYB1-3′race | TGGCATCAAGTTCCTGTTAGAG | 3′RACE |
| LcMYB1-5′race | CTAACAGGAACTTGATGCCATTTTTGTTCGCCAT | 5′RACE |
| LcMYB1-F | ATGTCGCATTTACTTGGTG | ORF |
| LcMYB1-R | TTACTTTGCATTGTCTTCTTC | |
| Q LcMYB1-F | GTTGGTCCCTTCAATCTTATC | RT-PCR |
| Q LcMYB1-R | GAAGACGAGGACTCCAACAC |
The promoter region was isolated according to the instructions of the genome walking kit (Takara, Japan). The primer sequences used in the three rounds of amplification are shown in Table 3.
Table 3. Primers used to isolated the LcMYB1 promotor region.
| Primer name | Sequence(5′→3′) | Function |
| MYBSP1-1 | GGAAATAGAATCAAATGGATAACGA | First round |
| MYBSP1-2 | CTGCTCTAACAGGAACTTGATGCCA | |
| MYBSP1-3 | TAACAGGAACTTGATGCCATTTTTG | |
| MYBSP2-1 | ACTCCTATGTAACCCTCCGCAGATG | Second round |
| MYBSP2-2 | TCCTATGTAACCCTCCGCAGATGTG | |
| MYBSP2-3 | AAGTGTAGTTTCCACATTCTTTCGT | |
| MYBSP3-1 | TAAGGCAGTTTCACTGTGAGCAGCAC | Third round |
| MYBSP3-2 | TGGACTGTTGAGATGCTATAACCC | |
| MYBSP3-3 | GGTTTACAGGGCTTTGTGCGGAA |
Sequence analysis
Multiple sequence alignment was performed using ClustalX 1.83 (http://www.ebi.ac.uk) and MEGA5 [59]. Cis-acting elements were predicted using the PlantCARE program (http://www.bioinformatics.psb.ugent.be/webtools/plantcare/html/) [60].
Quantitative real-time PCR analysis
Total RNA from the pericarps of litchi and tobacco was extracted and first strand cDNA was synthesized as described above. The transcription levels of both the litchi and tobacco anthocyanin biosynthetic genes were analyzed using quantitative real-time PCR (qRT-PCR),) with THUNDERBIRD qPCR Mix (TOYOBO, Japan) and ABI 7500 Real-Time PCR Systems (Applied Biosystems, USA) according to the manufacturers' instructions. The primers are shown in Table S1. Each reaction (20 μL final volume,) contained 9.96 μL 2×SYBR® qPCR Mix (TOYOBO), 0.04 μL 50×ROX reference dye, 1.0 μL of each the forward and reverse primers (0.25 μM), 2.0 μL of the cDNA template (corresponding to 50.0 ng of total RNA), and 7.0 μL of RNase-free water. The reaction mixtures were heated to 95°C for 30 s, followed by 40 cycles at 95°C for 10 s, 56°C for 15 s, and 72°C for 35 s. A melting curve was generated for each sample at the end of each run to ensure the purity of the amplified products. The specific qRT-PCR primers were designed using a Primer 5.0 program (PREMIER Biosoft International, Canada) (Table 2). Using these gene-specific primers, each assay amplified a single product of the correct size and demonstrated an acceptable PCR efficiency (approximately 90%). qRT-PCR reactions were normalized to the Ct values for LcActin (HQ615689) and NtACT (GQ281246) in litchi and tobacco, respectively. The relative expression levels of the target genes were calculated using the formula 2−ΔΔCT [61]. All biological replicates were measured in triplicate.
Induction of anthocyanins by transient transformation in tobacco
The plasmids used in the transient expression assay were constructed by ligating full-length LcMYB1 to pEAQ-HT using Nru I and Xho I. The primers used to amplify the encoding region were: Trans-F: TTCTGCCCAAATTCGCGAATGTCGCATTTACTTGGTGC and Trans-R: AGTTAAAGGCCTCGAGTTACTTTGCATTGTCTTCTTC. The product was recombined with the linearized vector pEAQ-HT (In-Fusion™ Advantage PCR Cloning Kits; Clontech). The constructed vector (pEAQ-LcMYB1) was maintained in Agrobacterium tumefaciens strain GV3101. Agrobacterium cultures containing pEAQ-LcMYB1 infiltrated into the abaxial leaf surface of N. tabacum, as described in Sainsbury et al. [62]. Control infiltrations comprised of pEAQ-HT (empty vector) were pressure infiltrated at the same time. Digital photographs were taken 5 days after infiltration.
Constructing vector and stable tobacco transformants
To obtain full-length LcMYB1, forward (LcMYB1-F: CGGGATCCCGCATGTCGCATTTACTTGGTG) and reverse (LcMYB1-R: CGAGCTCGGTCCATTAAATTACTTTGCATTGTC) primers were designed to amplify the coding regions. Fragments were digested with BamHI and SacI and integrated into pBI121 in place of the gusA gene sequence. The resulting construct (pBI121- LcMYB1) was introduced into A. tumefaciens strain EHA105. The recombinant strains were used to transform Nicotiana tabacum K326 using the leaf disk method [63]. Transgenic plants were selected based on kanamycin resistance. Two CaMV 35S:LcMYB1 lines were selected.
Supporting Information
Expression of LcMYB2 in different tissues (A) and in the pericarp of developmental stages (B) of litchi cultivar ‘NMC’. Lcactin gene was used to normalize expression of the genes under identical conditions. The vertical bars represent standard error of three replicates.
(TIF)
Images of tobacco lines containing LcMYB1 allele with CaMV 35S promoter. A-E: pigment formation during transformed process; F and G: pigmented leaves during seedling domestication.
(TIF)
Primers for real-time PCR analysis.
(DOC)
Acknowledgments
We thank Ruo Ouyang and Cheng-Ming Liu for providing information about the plant material and Jie-Tang Zhao for technical assistance.
Funding Statement
The project was supported by the China Litchi and Longan Industry Technology Research System (Project No. CARS-33), the National Natural Science Fund of China (Project No. 30971985), and the Key Laboratory of Innovation and Utilization for Germplasm Resources in Horticultural Crops in Southern China of Guangdong Higher Education Institutes, South China Agricultural University (No. KBL11008). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, et al. (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiology 140: 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology 126: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Springob K, Nakajima J, Yamazaki M, Saito K (2003) Recent advances in the biosynthesis and accumulation of anthocyanins. Natural Product Reports 20: 288–303. [DOI] [PubMed] [Google Scholar]
- 4. Boss PK, Davies C, Robinson SP (1996) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiology 111: 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Honda C, Kotoda N, Wada M, Kondo S, Kobayashi S, et al. (2002) Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiology and Biochemistry 40: 955–962. [Google Scholar]
- 6. Niu SS, Xu CJ, Zhang WS, Zhang B, Li X, et al. (2010) Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry Myrica rubra fruit by a R2R3 MYB transcription factor. Planta 231: 887–899. [DOI] [PubMed] [Google Scholar]
- 7. Wei YZ, Hu FC, Hu GB, Li XJ, Huang XM, et al. (2011) Differential expression of anthocyanin biosynthetic genes in relation to anthocyanin accumulation in the pericarp of Litchi chinensis Sonn. PLoS One 6: e19455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Petroni K, Tonelli C (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Science 181: 219–229. [DOI] [PubMed] [Google Scholar]
- 9. Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends in Plant Science 13: 99–102. [DOI] [PubMed] [Google Scholar]
- 10. Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, et al. (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. Journal of Experimental Botany 62: 2465–2483. [DOI] [PubMed] [Google Scholar]
- 11. Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. The Plant Cell 12: 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, et al. (2010) MYB transcription factors in Arabidopsis . Trends in Plant Science 15: 573–581. [DOI] [PubMed] [Google Scholar]
- 13. Takahashi R, Benitez ER, Oyoo ME, Khan NA, Komatsu S (2011) Nonsense mutation of an MYB transcription factor is associated with purple-blue flower color in soybean. Journal of Heredity 102: 458–463. [DOI] [PubMed] [Google Scholar]
- 14. Takahashi R, Yamagishi N, Yoshikawa N (2013) A MYB transcription factor controls flower color in Soybean. Journal of Heredity 1041: 149–153. [DOI] [PubMed] [Google Scholar]
- 15. Chiu LW, Li L (2012) Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 236: 1153–1164. [DOI] [PubMed] [Google Scholar]
- 16. Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K (2010) Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower Tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiology 51: 463–474. [DOI] [PubMed] [Google Scholar]
- 17. Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape Vitis labruscana regulate anthocyanin biosynthesis. Planta 215: 924–933. [DOI] [PubMed] [Google Scholar]
- 18. Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- 19. Huang Y, Song S, Allan AC, Liu X, Yin X, et al. (2013) Differential activation of anthocyanin biosynthesis in Arabidopsis and tobacco over-expressing an R2R3 MYB from Chinese bayberry. Plant Cell, Tissue and Organ Culture 113: 491–499. [Google Scholar]
- 20. Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, et al. (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiology 48: 958–970. [DOI] [PubMed] [Google Scholar]
- 21. Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, et al. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10 . The Plant Journal 49: 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. The Plant Journal 55: 954–967. [DOI] [PubMed] [Google Scholar]
- 23. Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG (2009) CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis . Molecular Plant 2: 790–802. [DOI] [PubMed] [Google Scholar]
- 24. Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, et al. (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal 28: 319–332. [DOI] [PubMed] [Google Scholar]
- 25. Paolocci F, Robbins MP, Passeri V, Hauck B, Morris P, et al. (2011) The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in Lotus corniculatus leaves. Journal of Experimental Botany 62: 1189–1200. [DOI] [PubMed] [Google Scholar]
- 26. Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, et al. (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiology 142: 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimizu Y, Maeda K, Kato M, Shimomura K (2010) Isolation of anthocyanin-related MYB gene, GbMYB2, from Gynura bicolor leaves. Plant Biotechnology 27: 481–487. [Google Scholar]
- 28. Chiu LW, Zhou X, Burke S, Wu X, Prior RL, et al. (2010) The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiology 154: 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feng S, Wang Y, Yang S, Xu Y, Chen X (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10 . Planta 232: 245–255. [DOI] [PubMed] [Google Scholar]
- 30. Jia H, Araki A, Okamoto G (2005) Influence of fruit bagging on aroma volatiles and skin coloration of ‘Hakuho’ peach Prunus persica Batsch. Postharvest Biology and Technology 35: 61–68. [Google Scholar]
- 31. Lin-Wang K, Micheletti D, Palmer J, Volz R, Lozano L, et al. (2011) High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environment 34: 1176–1190. [DOI] [PubMed] [Google Scholar]
- 32. Xie XB, Li S, Zhang RF, Zhao J, Chen YC, et al. (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environment 35: 1884–1897. [DOI] [PubMed] [Google Scholar]
- 33.Hiratsuka S, Onodera H, Kawai Y, Kubo T, Itoh H, et al.. (2001) ABA and sugar effects on anthocyanin formation in grape berry cultured in vitro. Scientia Horticulturae 90, 121–130.
- 34. Gagne S, Cluzet S, Merillon JM, Geny L (2011) ABA initiates anthocyanin production in grape cell cultures. Journal of Plant Growth Regulation 30: 1–10. [Google Scholar]
- 35. Peppi MC, Fidelibus MW, Dokoozlian N (2006) Abscisic acid application timing and concentration affect firmness, pigmentation, and color of ‘flame seedless’ grapes. HortScience 41: 1440–1445. [Google Scholar]
- 36. Qi T, Song S, Ren Q, Wu D, Huang H, et al. (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana . The Plant Cell 23: 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee HS, Wicher L (1991) Anthocyanin pigments in the skin of lychee fruit. Journal of Food Science 56: 466–468. [Google Scholar]
- 38. Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. The Plant Journal 40: 22–34. [DOI] [PubMed] [Google Scholar]
- 39. Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana . Current Opinion in Plant Biology 4: 447–456. [DOI] [PubMed] [Google Scholar]
- 40. Chagne D, Lin-Wang K, Espley RV, Volz RK, How NM, et al. (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiology 161: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, et al. (2010) Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 231: 1061–1076. [DOI] [PubMed] [Google Scholar]
- 42. Bai Y, Pattanaik S, Patra B, Werkman JR, Xie CH, et al. (2011) Flavonoid-related basic helix-loop-helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta 234: 363–375. [DOI] [PubMed] [Google Scholar]
- 43. Boss PK, Davies C, Robinson SP (1996) Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Molecular Biology 32: 565–569. [DOI] [PubMed] [Google Scholar]
- 44. Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. The EMBO Journal 6: 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, et al. (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. The Plant Cell 11: 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mano H, Ogasawara F, Sato K, Higo H, Minobe Y (2007) Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiology 143: 1252–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jia HF, Chai YM, Li CL, Lu D, Luo JJ, et al. (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiology 157: 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koyama K, Sadamatsu K, Goto-Yamamoto N (2010) Abscisic acid stimulated ripening and gene expression in berry skins of the Cabernet Sauvignon grape. Functional & Integrative Genomics 10: 367–381. [DOI] [PubMed] [Google Scholar]
- 49. Wang H, Huang H, Huang X (2007) Differential effects of abscisic acid and ethylene on the fruit maturation of Litchi chinensis Sonn. Plant Growth Regulation 52: 189–198. [Google Scholar]
- 50. Jiao YL, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nature Reviews Genetics 8: 217–230. [DOI] [PubMed] [Google Scholar]
- 51. Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, et al. (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiology 160: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maier A, Schrader A, Kokkelink L, Falke C, Welter B, et al. (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis . The Plant Journal 74: 638–51. [DOI] [PubMed] [Google Scholar]
- 53. Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, et al. (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. The Plant Cell 24: 1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kobayashi S, Ishimaru M, Ding CK, Yakushiji H, Goto N (2001) Comparison of UDP-glucose, flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Science 1603: 543–550. [DOI] [PubMed] [Google Scholar]
- 55. Malone LA, Barraclough EI, Lin-Wang K, Stevenson DE, Allan AC (2009) Effects of red-leaved transgenic tobacco expressing a MYB transcription factor on two herbivorous insects, Spodoptera litura and Helicoverpa armigera . Entomologia Experimentals Et Applicata 133: 117–127. [Google Scholar]
- 56. Spelt C, Quattrocchio F, Mol J, Koes R (2000) ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. The Plant Cell 14: 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, et al. (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana . The Plant Journal 39: 366–380. [DOI] [PubMed] [Google Scholar]
- 58. Fuleki T, Francis FJ (1968) Quantitative methods for anthocyanins. 2. Determination of total anthocyanin and degradation index for cranberry juice. Journal of Food Science 33: 78–83. [Google Scholar]
- 59. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5, molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, et al. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-Delta Delta CT Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 62. Sainsbury F, Thuenemann EC, Lomonossoff GP (2009) pEAQ, versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnology Journal 7: 682–693. [DOI] [PubMed] [Google Scholar]
- 63. Horsch R, Fry J, Hoffmann N, Eichholtz D, Rogers S, et al. (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of LcMYB2 in different tissues (A) and in the pericarp of developmental stages (B) of litchi cultivar ‘NMC’. Lcactin gene was used to normalize expression of the genes under identical conditions. The vertical bars represent standard error of three replicates.
(TIF)
Images of tobacco lines containing LcMYB1 allele with CaMV 35S promoter. A-E: pigment formation during transformed process; F and G: pigmented leaves during seedling domestication.
(TIF)
Primers for real-time PCR analysis.
(DOC)