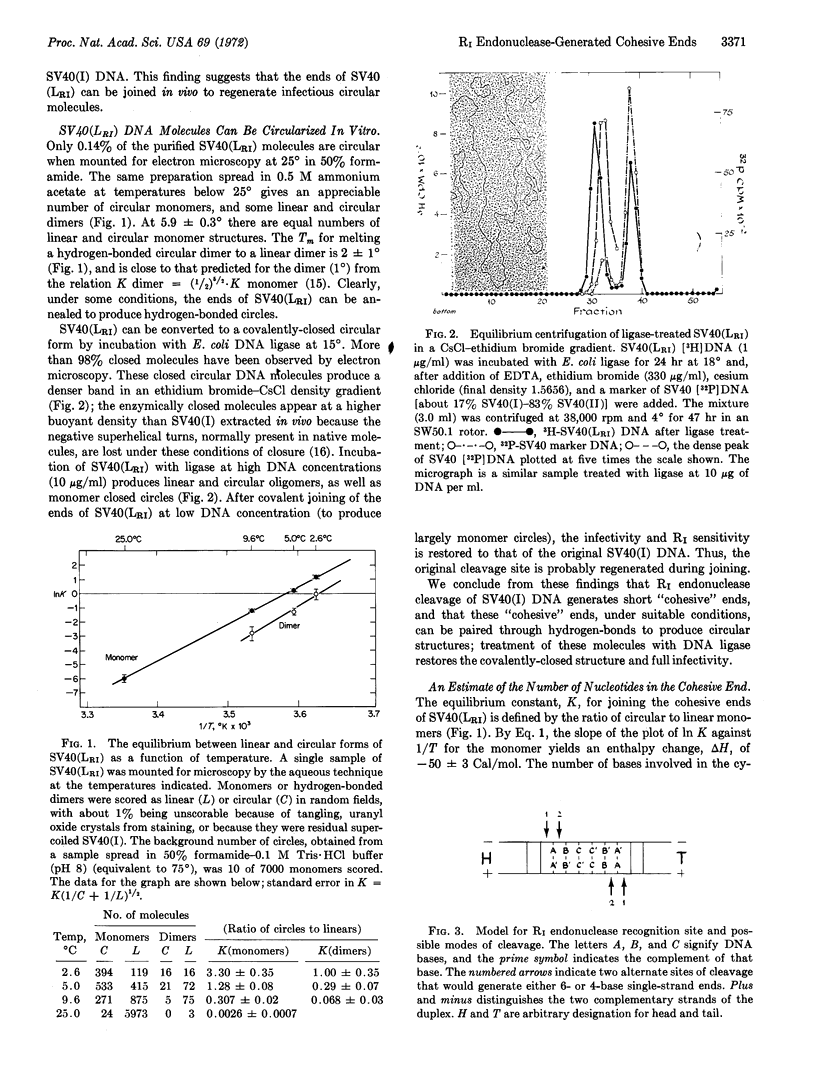
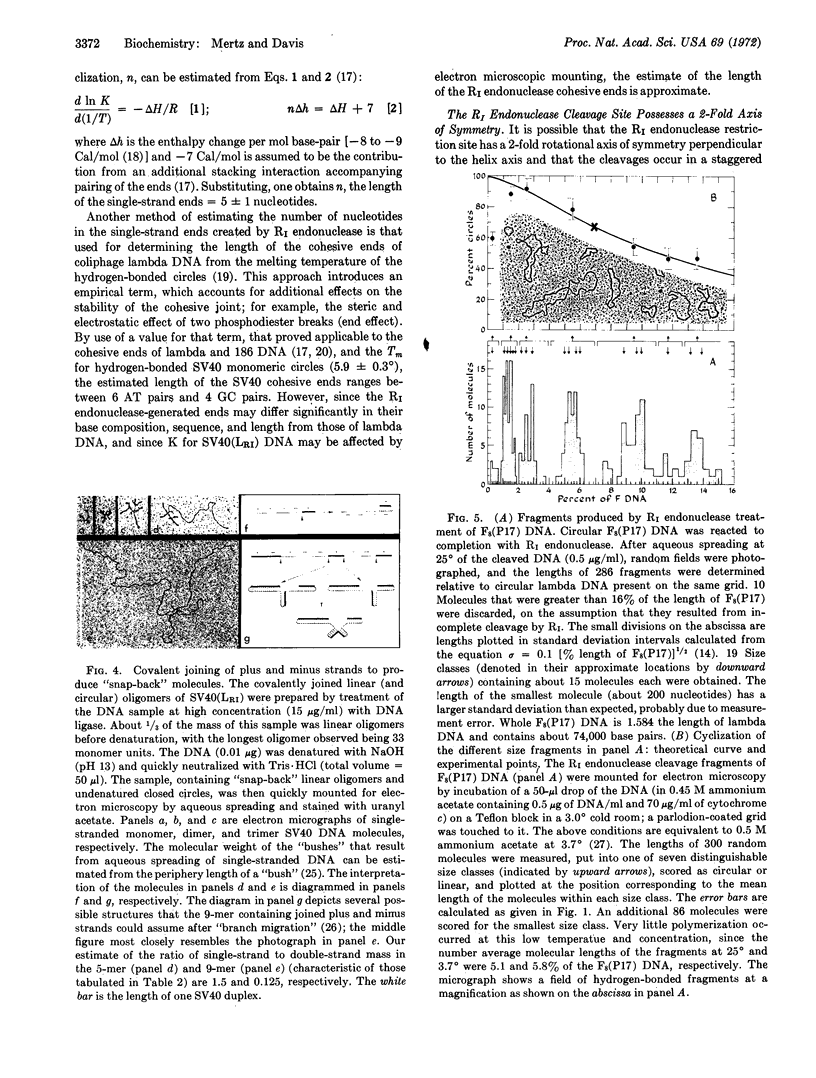
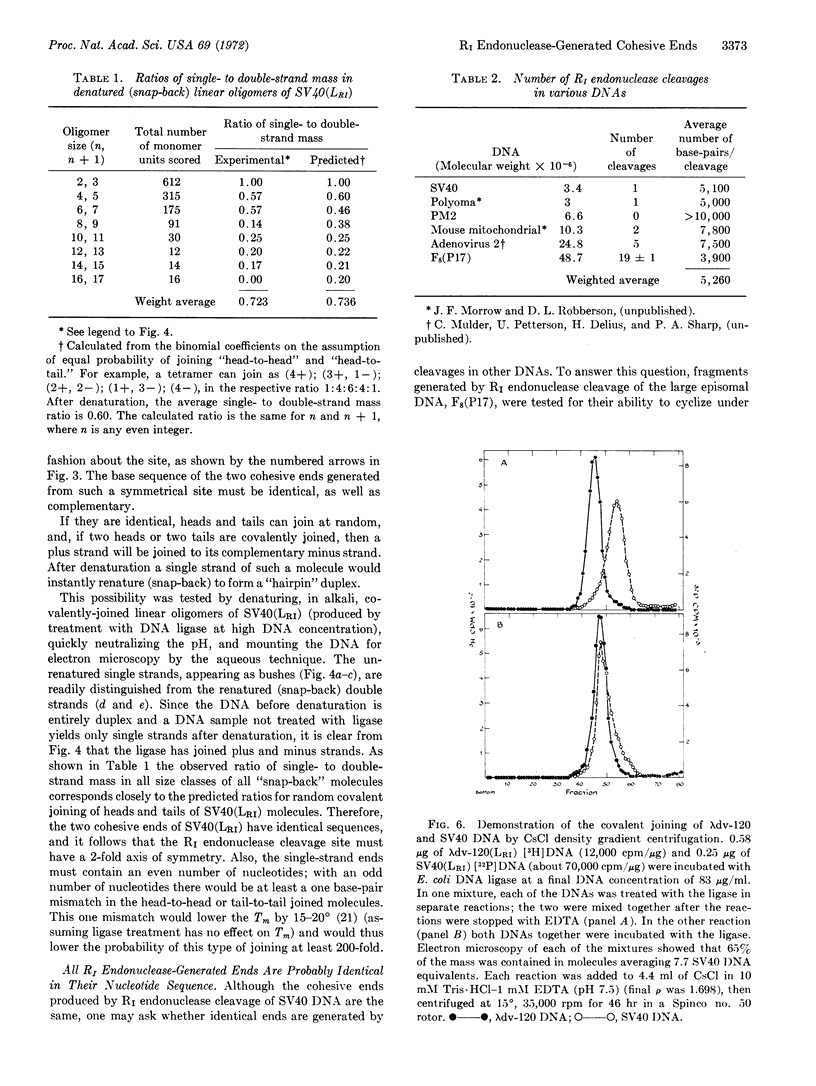
Abstract
R1 restriction endonuclease cleaves duplex DNA at a specific sequence, probably 6 nucleotide pairs in length, by making two single-strand staggered cleavages, generating 5′-phosphoryl and 3′-hydroxyl termini. The single-strand ends produced at each break have identical and complementary sequences of 4 or 6 nucleotides in length. Therefore, the cleavage site possesses a 2-fold rotational axis of symmetry perpendicular to the helix axis. The ends of full-length linear SV40 DNA, generated by R1 endonuclease cleavage, can be joined by Escherichia coli ligase to regenerate duplex, fully infectious, covalently-closed circular molecules. It was further found that all R1 endonuclease-generated ends are identical and complementary. Therefore, any two DNA molecules with R1 sites can be “recombined” at their restriction sites by the sequential action of R1 endonuclease and DNA ligase to generate hybrid DNA molecules.
Keywords: SV40, restriction site, cyclization, electron microscopy, DNA joining
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W., Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- Boyer H. W. DNA restriction and modification mechanisms in bacteria. Annu Rev Microbiol. 1971;25:153–176. doi: 10.1146/annurev.mi.25.100171.001101. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Davidson N. Electron-microscopic visualization of deletion mutations. Proc Natl Acad Sci U S A. 1968 May;60(1):243–250. doi: 10.1073/pnas.60.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray H. B., Jr, Hearst J. E. Flexibility of native DNA from the sedimentation behavior as a function of molecular weight and temperature. J Mol Biol. 1968 Jul 14;35(1):111–129. doi: 10.1016/s0022-2836(68)80041-5. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Upholt W. B., Vinograd J. A buoyant method for the determination of the superhelix density of closed circular DNA. J Mol Biol. 1971 Nov 28;62(1):1–19. doi: 10.1016/0022-2836(71)90127-6. [DOI] [PubMed] [Google Scholar]
- Howard B. V., Estes M. K., Pagano J. S. The uptake of SV40 DNA by nonpermissive cells in the presence of DEAE-dextran. Biochim Biophys Acta. 1971 Jan 1;228(1):105–116. doi: 10.1016/0005-2787(71)90550-8. [DOI] [PubMed] [Google Scholar]
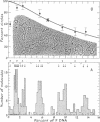
- Jackson D. A., Symons R. H., Berg P. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2904–2909. doi: 10.1073/pnas.69.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Smith H. O. A restriction enzyme from Hemophilus influenzae. II. J Mol Biol. 1970 Jul 28;51(2):393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- Lee C. S., Davis R. W., Davidson N. A physical study by electron microscopy of the terminally reptitious, circularly permuted DNA from the coliphage particles of Escherichia coli 15. J Mol Biol. 1970 Feb 28;48(1):1–22. doi: 10.1016/0022-2836(70)90215-9. [DOI] [PubMed] [Google Scholar]
- Meselson M., Yuan R., Heywood J. Restriction and modification of DNA. Annu Rev Biochem. 1972;41:447–466. doi: 10.1146/annurev.bi.41.070172.002311. [DOI] [PubMed] [Google Scholar]
- Morrow J. F., Berg P. Cleavage of Simian virus 40 DNA at a unique site by a bacterial restriction enzyme. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3365–3369. doi: 10.1073/pnas.69.11.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder C., Delius H. Specificity of the break produced by restricting endonuclease R 1 in Simian virus 40 DNA, as revealed by partial denaturation mapping. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3215–3219. doi: 10.1073/pnas.69.11.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. C. Enzymes in DNA metabolism. Annu Rev Biochem. 1969;38:795–840. doi: 10.1146/annurev.bi.38.070169.004051. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Sturtevant J. M. Thermodynamics of the helix-coil transition of the alternating copolymer of deoxyadenylic acid and deoxythymidylic acid. J Mol Biol. 1969 Jun 28;42(3):577–580. doi: 10.1016/0022-2836(69)90244-7. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Martin F. H., Doty P. Self-complementary oligoribonucleotides: effects of helix defects and guanylic acid-cytidylic acid base pairs. J Mol Biol. 1971 Apr 28;57(2):217–229. doi: 10.1016/0022-2836(71)90342-1. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Cyclization of coliphage 186 DNA. J Mol Biol. 1967 Sep 28;28(3):403–411. doi: 10.1016/s0022-2836(67)80089-5. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Davidson N. On the probability of ring closure of lambda DNA. J Mol Biol. 1966 Aug;19(2):469–482. doi: 10.1016/s0022-2836(66)80017-7. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Davidson N. Thermodynamic and kinetic studies on the interconversion between the linear and circular forms of phage lambda DNA. J Mol Biol. 1966 Jan;15(1):111–123. doi: 10.1016/s0022-2836(66)80213-9. [DOI] [PubMed] [Google Scholar]