Abstract
Background
Epidemiological studies have evaluated the association between 3801T>C polymorphism of CYP1A1 gene and the risk for idiopathic male infertility, but the results are inconclusive. We aimed to derive a more precise estimation of the relationship by conducting a meta-analysis of case-control studies.
Methods
This study conformed to Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines. PubMed, Embase and CNKI databases were searched through November 2013 to identify relevant studies. Pooled odds ratios with 95% confidence intervals were used to assess the strength of the association between CYP1A1 3801T>C polymorphism and idiopathic male infertility risk. Q-test was performed to evaluate between-study heterogeneity and publication bias was appraised using funnel plots. Sensitivity analyses were conducted to evaluate the robustness of meta-analysis findings.
Results
Six studies involving 1,060 cases and 1,225 controls were included in this meta-analysis. Overall, significant associations between 3801T>C polymorphism and idiopathic male infertility risk were observed in allelic comparison (OR = 1.36, 95% CI: 1.01–1.83), homozygous model (OR = 2.18, 95% CI: 1.15–4.12), and recessive model (OR = 1.86, 95% CI: 1.09–3.20), with robust findings according to sensitivity analyses. However, subgroup analyses did not further identify the susceptibility to idiopathic male infertility in all comparisons. Funnel plot inspections did not reveal evidence of publication bias.
Conclusions
The current meta-analysis provides evidence of a significant association between CYP1A1 3801T>C polymorphism and idiopathic male infertility risk. Considering the limitation inherited from the eligible studies, further confirmation in large-scale and well-designed studies is needed.
Introduction
Idiopathic male infertility (IMI) is a global health dilemma affecting approximately 10–15% of the male adults and up to one in six couples, which accounts for 40–50% male infertility problems [1]–[3]. The causes of IMI are the result of complex interaction of multi-factorial and environmental, behavioral and genetic factors [4]. Despite progresses in diagnosis of IMI, mainly in the field of genetics, etiology and pathogenesis are still unknown. Epidemiological data indicated that damage of xenobiotic to the genetic material might contribute to spermatogenetic failure for about 30% of infertility in males [5]. A number of genetic studies have been done to investigate the contribution of genes encoding to IMI, and some of these studies have revealed the direct relationship between the genotypes and the disease susceptibility [6], [7]. Much attention in the genetic studies has been paid to the cytochrome P450 1A1 (CYP1A1) that plays a curial role in phase I metabolism of polycyclic aromatic hydrocarbons to their ultimate biologically active intermediates that have potential reproductive toxicity in men [5], [8]. The CYP1A1 gene has been shown to be polymorphic, and several polymorphisms have been identified [9]. Among these polymorphisms, the most commonly studied is the 3801T>C polymorphism (also referred to as 2A, m1, or rs4646903), which is characterized by the T to C mutation at nucleotide 3801 in the 3′ flanking region of the CYP1A1 gene. The 3801T>C polymorphism can alter the level of gene expression or messenger RNA stability, resulting in a highly inducible activity of the enzyme [10], [11]. To date, several studies have investigated the relationship between the CYP1A1 3801T>C polymorphism and IMI risk. However, current results have been inconsistent. Moreover, no meta-analysis data on the correlation of the polymorphism with susceptibility to IMI is currently available. Therefore, to derive a more precise overall effect estimate, the present study aimed to evaluated the association between the CYP1A1 3801T>C polymorphism and susceptibility to IMI by performing a systematic review and meta-analysis of the literature.
Materials and Methods
Literature search
This systematic review and meta-analysis followed Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. The included studies were retrieved from the electronic databases including PubMed, Embase and China National Knowledge Infrastructure (CNKI) databases. We used the combination of the search terms: (“CYP1A1” or “cytochrome P450 1A1” or “rs4646903”), (“idiopathic male infertility” or “male infertility” or “oligozoospermia” or “azoospermia” or “teratospermia”) and (“polymorphism” or “allele” or “variant” or “mutation” or “gene” or “genotype”). No restrictions were imposed on the search in terms of language. The literature search was updated on 20 November 2013.
Study selection
The studies were selected according to the following inclusion criteria: (1) used a case-control design, (2) gave information on the distribution of CYP1A1 3801T>C genotypes in both cases and corresponding controls, (3) contained an evaluation of the 3801T>C polymorphism and IMI risk, and (4) consisted with Hardy-Weinberg equilibrium (HWE) in control groups. We excluded those studies that did not provide adequate information on selection criteria and in which allele frequencies in controls exhibited significant deviation from the HWE (P<0.05). Conference abstracts, case reports, editorials, review articles, and letters were also excluded.
Data extraction
Information was extracted from all eligible publications independently by two of the authors according to the above-listed inclusion criteria. An agreement was reached through a discussion between the two reviewers for cases with conflicting information. The following characteristics were collected from each study: the first author's name, publication year, country, population ethnicity, and genotype frequency for cases and controls. Populations from the included studies were stratified into Caucasian and Asian. Notably, Indians are mainly of Indo-European and Dravidian ancestries, which are quite different from East Asians [12]. From the point of view of history and geography, Iran lies on the route of major ancient movements of the Caucasian people towards the Mediterranean basin, Iranian should be descendants of European [13]. Moreover, Indian and Iranian were usually categorized into Caucasian in previous publications [14]. Thus, the two populations in the meta-analysis were also stratified into Caucasian.
Quality score evaluation
Quality assessment of the case-control studies in this meta-analysis was performed using the Newcastle Ottawa scale (NOS) recommended by the Cochrane Non-randomized Studies Methods Working Group [15], [16]. The NOS contains eight items that are categorized three categories: selection (four items, one star each), comparability (one item, up to two stars), and exposure (three items, one star each). A “star” presents a “high-quality” choice of individual study. Two independent reviewers discussed their evaluation, and any disagreements were resolved through discussion and consultation. Given the variability in quality of case-control studies found in our initial literature search, we considered studies as high quality if they met a score of six or more of the NOS criteria [17].
Statistical analysis
STATA version 11.0 (STATA Corporation, College Station, Texas) was used for all statistical analyses. All statistical tests were two sided. The combined odds ratio (OR), along with its corresponding 95% confidence interval (CI), was used to calculate and assess the strength of the association between polymorphisms of CYP1A1 and IMI risk. The allele model, co-dominant model (heterozygous carriers vs. “wild type” and homozygous carriers vs. wild type”), dominant model (heterozygous and homozygous carriers grouped together vs. “wild type”), and recessive model (homozygous carriers vs. “wild type” and heterozygous carriers grouped together) were estimated [18].
Heterogeneity assumption was examined using the Q-test [19]. A random-effects model (DerSimonian-Laird method) or fixed-effects model (Mantel-Haenszel method) was used to calculate the pooled effect estimates in the presence (P< = 0.10) or absence (P>0.10) of heterogeneity, respectively [20]. To explore the reasons of heterogeneity, subgroup analyses were performed by grouping studies that showed similar characteristics, such as ethnicity, sample size, and quality assessment score. Sensitivity analysis was also performed by omitting each individual study to reflect the influence of the individual dataset on the pooled OR using the “metaninf” STATA command. The appropriate Chi-square goodness-of-fit test [21] was performed using the “genhwcci” STATA command to assess the deviation from HWE only in control groups. Statistical significance for the interpretation of the chi-squared test was defined as P<0.05.
Publication bias was evaluated through the Begg's and the Egger's Asymmetry tests [22] and through visual inspection of funnel plots, in which the standard error was plotted against the log (OR) to form a simple scatterplot. Statistical significance for the interpretation of the Egger's test was defined as P<0.10.
Results
Eligible studies
Figure 1 illustrates the trial flow diagram for study inclusion. A total of 58 citations were identified during the initial search. Then 12 duplicate records were excluded. Of the 46 potential eligible records, 34 were excluded because of obvious irrelevance by reading their titles and abstracts (reasons for exclusion described in Table S1). After detailed evaluation, the following were excluded: one review [5], one not in a case-control design [23], one not of reporting 3801T>C polymorphism [24] and three articles that did not provide sufficient data for calculation of OR and 95% CI [8], [25], [26]. Finally, six articles [27]–[32] met the inclusion criteria involving 1,060 cases and 1,225 controls. In the articles eligible for the meta-analysis, three [28], [31], [32] were conducted in Caucasian populations, and three [27], [29], [30] involved Asian populations. For the determination of the genetic polymorphism, polymerase chain reaction-restriction fragment length polymorphism was used in all the studies. Semen specimens of cases were diagnosed primarily according to WHO guidelines. Control subjects were from hospital-based populations in all of the included studies. Five studies [27]–[31] stated that the controls were age matched, and one [32] was matched by smoking status. Noticeably, Lu et al. [27] didn't report age of cases and controls, but we read the author's academic dissertation published in Chinese and found that the population was age-matched. Salehi et al. [31] was age-matched too, although without detailed information about age. Examining the genotype frequencies in controls, no significant deviation from HWE was detected in all studies (all P>0.05). The studies were published between 2008 and 2013. A wide variation of the 3801C allele frequencies across different populations was observed, varying from 0.12 in Russians to 0.38 in Chinese. Four studies were of high quality with score greater than 6 (details of evaluation score for literature shown in Table S2). The characteristics of the case–control studies included for the polymorphism are summarized in Table 1 and S3.
Figure 1. Flow diagram of study selection and specific reasons for exclusion in the meta-analysis.
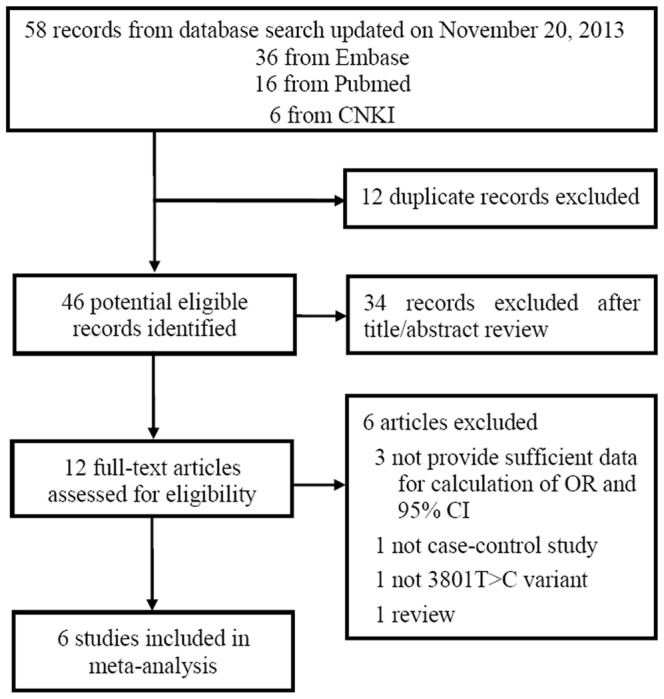
Table 1. CAYP1A1 3801T >C genotype distribution and allele frequencies in cases and controls.
| Study | Year | Country | Ethnicity | Control source | Genotyping method | Cases/controls | Genotype data (cases) | Genotype data (controls) | MAF | HWE (P)a | Quality score | ||||||||
| T | C | TT | TC | CC | T | C | TT | TC | CC | ||||||||||
| Lu et al [27] | 2008 | China | Asian | HB | PCR-RFLP | 192/226 | 234 | 150 | 69 | 96 | 27 | 294 | 158 | 95 | 104 | 27 | 0.35 | 0.86 | 6 |
| Vani et al [28] | 2009 | India | Caucasian | HB | PCR-RFLP | 206/230 | 296 | 116 | 108 | 80 | 18 | 372 | 88 | 146 | 80 | 4 | 0.19 | 0.06 | 8 |
| Chen et al [29] | 2010 | China | Asian | HB | PCR-RFLP | 105/140 | 117 | 93 | 35 | 47 | 23 | 221 | 59 | 88 | 45 | 7 | 0.21 | 0.69 | 5 |
| Peng et al [30] | 2012 | China | Asian | HB | PCR-RFLP | 204/202 | 237 | 171 | 72 | 93 | 39 | 250 | 154 | 78 | 94 | 30 | 0.38 | 0.85 | 5 |
| Salehi et al [31] | 2012 | Iran | Caucasian | HB | PCR-RFLP | 150/200 | 188 | 112 | 58 | 72 | 20 | 261 | 139 | 85 | 91 | 24 | 0.35 | 0.96 | 6 |
| Yarosh et al [32] | 2013 | Russia | Caucasian | HB | PCR-RFLP | 203/227 | 365 | 41 | 165 | 35 | 3 | 400 | 54 | 176 | 48 | 3 | 0.12 | 0.89 | 8 |
HB hospital based, PCR-RFLP polymerase chain reaction-restriction fragment length polymorphism, MAF minor allele frequency, HWE Hardy–Weinberg equilibrium,
P value for HWE in control group.
Quantitative synthesis
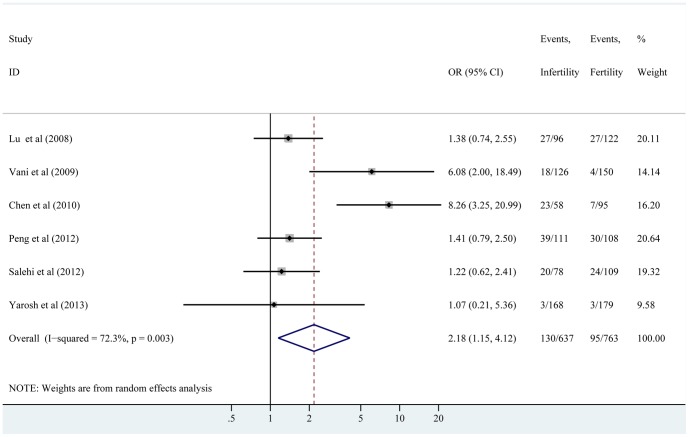
The pooled OR with its 95% CI, are presented in detail in Table 2. Overall, significant associations were demonstrated in allele model (C vs. T, OR = 1.36, 95% CI: 1.01–1.83), homozygous model (CC vs. TT, OR = 2.18, 95% CI: 1.15–4.12), and recessive model (CC vs. TT+TC, OR = 1.86, 95% CI: 1.09–3.20). However, null significant association was observed in heterozygous model (TC vs. TT, OR = 1.25, 95% CI: 0.95–1.65) and dominant model (CC+TC vs. TT, OR = 1.38, 95% CI: 0.99–1.92). A further analysis was performed on data stratified by ethnicity to determine possible factors that might have influenced the results. Table 2 shows that any association was not found in IMI patients with CYP1A1 3801T>C polymorphism among all comparisons. Moreover, similar results in the further subgroup analysis by quality score and sample size were observed on any genetic model because the associations did not reach a significant level. The forest plots for the overall association between the 3801T>C polymorphism of CYP1A1 and IMI risk are shown in Figure 2 and S1.
Table 2. Results of meta-analysis for 3801T>C polymorphism of CYP1A1 and idiopathic male infertility risk.
| Study group | No. of study | sample size (cases/controls) | Allele model (C vs. T) | Homozygous model (CC vs. TT) | Heterozygous model (TC vs. TT) | Dominant model (CC+TC vs. TT) | Recessive model (CC vs. TC+TT) | |||||
| OR (95% CI) | P h | OR (95% CI) | P h | OR (95% CI) | P h | OR (95% CI) | P h | OR (95% CI) | P h | |||
| All | 6 | 1060/1225 | 1.36(1.01,1.83) | <0.001 | 2.18(1.15,4.12) | 0.003 | 1.25(0.95,1.65) | 0.051 | 1.38(0.99,1.92) | 0.003 | 1.86(1.09,3.20) | 0.013 |
| Asian | 3 | 501/568 | 1.58(0.93,2.67) | <0.001 | 2.35(0.90,6.16) | 0.003 | 1.48(0.91,2.40) | 0.040 | 1.67(0.92,3.05) | 0.004 | 1.90(0.89,4.05) | 0.015 |
| Caucasian | 3 | 559/657 | 1.18(0.81,1.72) | 0.031 | 2.02(0.66,6.20) | 0.042 | 1.13(0.89,1.43) F | 0.353 | 1.16(0.79,1.71) | 0.086 | 1.89(0.64,5.58) | 0.046 |
| High quality | 4 | 751/883 | 1.19(0.93,1.53) | 0.073 | 1.75(0.89,3.43) | 0.028 | 1.15(0.92,1.42) F | 0.339 | 1.22(0.99,1.50) F | 0.170 | 1.58(0.83,3.00) | 0.084 |
| Low quality | 2 | 309/342 | 1.85(0.74,4.61) | <0.001 | 3.28(0.58,18.56) | 0.002 | 1.64(0.68,3.96) | 0.013 | 1.95(0.68,5.59) | 0.002 | 2.56(0.67,9.78) | 0.009 |
| Sample size >400 | 4 | 805/885 | 1.21(0.95,1.53) | 0.081 | 1.79(0.97,3.32) | 0.100 | 1.13(0.91,1.39) F | 0.330 | 1.20(0.92,1.56) F | 0.167 | 1.55(0.98,2.19) F | 0.107 |
| Sample size <400 | 2 | 255/340 | 1.81(0.69,4.73) | <0.001 | 3.09(0.47,20.14) | <0.001 | 1.71(0.77,3.82) | 0.027 | 1.86(0.59,55.89) | 0.001 | 2.37(0.52,10.89) | 0.005 |
Ph P value for heterogeneity based on Q test. All pooled ORs were derived from random-effects model except for cells marked with (fixed F).
Figure 2. Meta-analysis of the association between the 3801T>C polymorphism of CYP1A1 and risk for idiopathic male infertility under homozygous model.
The contribution of each study to the meta-analysis (its weight) is represented by the area of a box, the center of which represents the size of the OR estimated from that study. The 95% CI for the OR (extending lines) from each study is also shown. The overall OR is shown in the middle of a diamond, the left and right extremes of which represent the corresponding CI.
Evaluation of heterogeneity
We analyzed the heterogeneity of the studies selected according to the P-value for heterogeneity. Table 2 demonstrates that a significant heterogeneity was found in all genetic models (P heterogeneity<0.10). Therefore, a random-effects model was used for all the comparisons to calculate the overall OR estimates. After assessing the source of heterogeneity for all genetic models compared by subgroup analyses based on ethnicity, quality score, or sample size, the heterogeneity was partly decreased or removed.
Sensitivity analysis
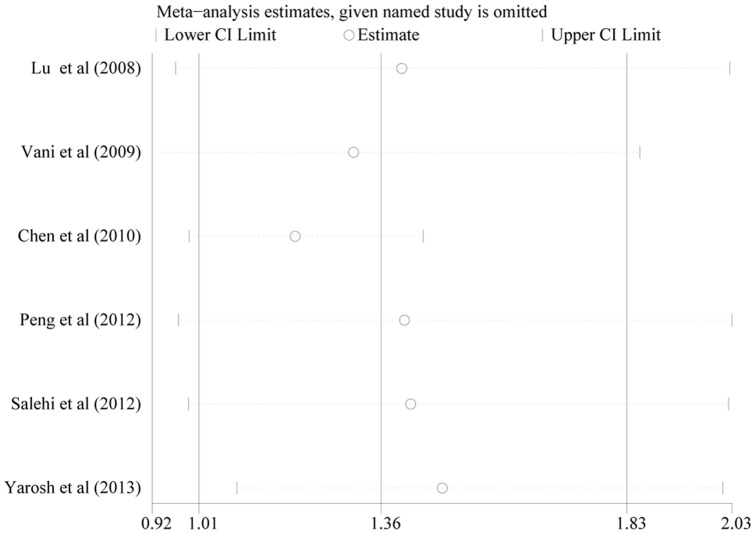
Separate meta-analyses were conducted to investigate the influence of each individual study on the overall meta-analytic estimate. Figure 3 demonstrates that no point estimate of the omitted individual study lay outside the 95% CI of the combined analysis on the allele model. Similarly, no significant influence was observed when an analysis was conducted on the other models (Figures were not shown). These analyses suggest that no individual study affected the results in the meta-analysis.
Figure 3. Effect of individual studies on the pooled OR under allele comparison for the 3801T>C polymorphism of CYP1A1 in idiopathic male infertility.
The vertical axis at 1.36 indicates the overall OR, and the two vertical axes at 1.01 and 1.83 indicate the 95% CI. Every hollow round indicates the pooled OR when the left study was omitted in a meta-analysis with a random model. The two ends of every broken line represent the respective 95% CI.
Publication bias
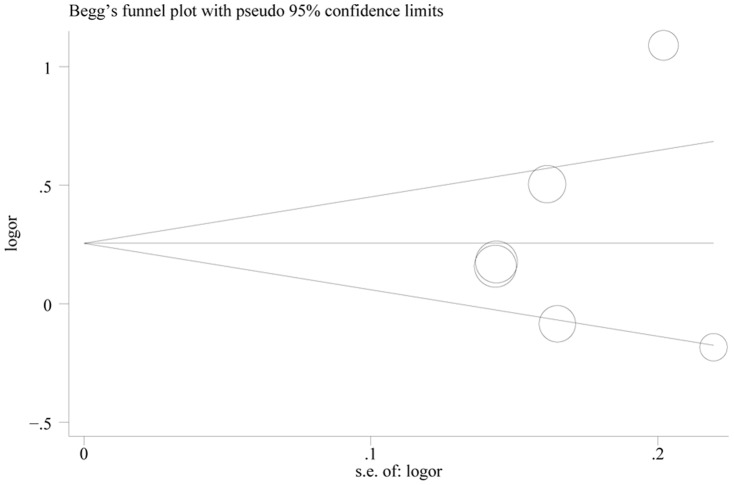
No publication bias on the overall OR analysis was detected at any comparison (Table 3, P>0.10). The shapes of the funnel plots appeared to be roughly symmetrical in all genetic models (Figure 4 and S2). In addition, neither the Begg's test nor the Egger's test provided any obvious evidence of publication bias in subgroup analysis (data was not shown).
Table 3. Results of Egger's test and Begg's test.
| Comparison | Egger's test | Begg's test | ||||
| t | P | 95% CI | Z | P | ||
| Allele model | 0.42 | 0.697 | −16.480 | 22.339 | 0.00 | 0.999 |
| Homozygous model | 0.72 | 0.514 | −6.184 | 10.475 | 0.38 | 0.707 |
| Heterozygous model | 0.87 | 0.432 | −10.697 | 20.504 | 0.38 | 0.707 |
| Dominant model | 0.82 | 0.461 | −14.594 | 26.723 | 0.38 | 0.707 |
| Recessive model | 1.26 | 0.277 | −2.986 | 7.929 | 0.38 | 0.707 |
Figure 4. Funnel plot for the 3801T>C polymorphism of CYP1A1 under allele comparison in idiopathic male infertility.
The vertical axis represents log (OR), and the horizontal axis refers to the standard error of log (OR). The horizontal line indicates the pooled OR, and the sloping lines indicate the expected 95% CI for a given standard error. The area of each circle represents the contribution of the study to the pooled OR.
Discussion
Since the original identification of the CAYP1A1 polymorphisms, a number of studies have investigated the genetic effect of the polymorphisms on susceptibility to human complex diseases, such as various cancers [33], [34], polycystic ovary syndrome [35], chronic kidney disease [36], coronary artery disease [37], and systemic lupus erythematosus [38]. Accordingly, the association between polymorphisms of CPY1A1 gene and susceptibility to male infertility was also reported in different populations. For example, Vani et al. [28] and Chen et al. [29] found that individuals with CYP1A1 mutations had significantly increased risk for the development of IMI in Indian and Chinese populations, whereas Salehi et al [31] and Yarosh et al. [32] revealed that CYP1A1 variants had no effect on the genetic susceptibility to the disease in Iranian and Russian populations. This inconsistency could be due to many factors, including various recruitment procedures of the study populations and differences in the genetic and environmental backgrounds. Considering the limitations from individual study, the meta-analysis of the published studies was conducted.
To the best of our knowledge, this is the first meta-analysis which comprehensively assessed the association between CYP1A1 3801T>C polymorphism and IMI risk. The principal result showed that the 3801T>C variant was associated with IMI risk in allele comparison, homozygous and recessive genetic models. In addition, the sensitivity analyses and publication bias results confirmed the robustness of these conclusions. Thus, the positive findings indicated that the CYP1A1 3801T>C polymorphism might be a potential risk factor for IMI. However, the reason for the negative findings in further subgroup analyses may be caused by the limited studies and population numbers of Caucasians and Asians included in the meta-analysis, this may have been insufficient statistical power to detect slight associations in stratified analyses.
Up to now, the underlying biological mechanism by which CYP1A1 exerts its effects on the pathogenesis of IMI is still not clearly elucidated. Recently, the important influence of estrogen in the development of male infertility has been acknowledged [32], [39]. It is well recognized that estrogens are metabolized by CYP1A1 (as an estrogen-metabolizing gene) and converted into catecholestrogens 2-hydroxyestradiol and 4-hydroxyestradiol [40]. CYP1A1 is also involved in inactivation of xenobiotic metabolism, and activation of environmental toxins. There is a complex interaction circuit between CYP1A1, estrogen receptor alpha and aryl hydrocarbone receptor with anti-estrogenic properties [41], [42]. CYP1A1 is induced by diverse exogenous and endogenous chemicals through the aryl hydrocarbone receptor [43]. Moreover, CYP1A1 expression interacts with the aryl hydrocarbone receptor and estrogen receptor alpha expression [44]. Notably, the CYP1A1 3801T>C polymorphism can alter activity and expression of the enzyme [10], [11], further regulate the expression level of aryl hydrocarbone receptor and estrogen receptor alpha, resulting in male reproduction disorders. In addition, association of CYP1A1 and estrogen polymorphisms with impaired spermatogenesis implies that both genetic and environmental factors contribute to testicular dysfunction, which can lead to sperm damage, deformity, and eventually male infertility [25]. This may be the underlying mechanism by which the CYP1A1 3801C allele resulted in male infertility.
Our study had several important strengths. Only studies in HWE among controls were included, which guaranteed quality control. Moreover, all relevant studies published in both English and Chinese were recruited for the meta-analysis, which would reduce language biases. In addition, the methodological issues for meta-analysis, such as subgroup analysis, publication bias, and stability of results were all well investigated.
In spite of several important strengths of the present meta-analysis, potential limitations should be considered. First, the results should be interpreted with caution because of obvious heterogeneity in most comparisons, although some stratified analyses by quality score and sample size did partly identify the source of heterogeneity for the 3801T>C polymorphism under homozygous model and dominant model. However, it is widely accepted that potential heterogeneity from genetic backgrounds may interfere with the conclusions of meta-analyses [45]. In our meta-analysis, there is a differential population impact depending on the 3801C allele frequency in the background population. In particular, it is evident that 3801C allele frequency is highest in Chinese [27] and Iranian [31], with much lower frequency being recorded in Russian [32] and Indian [28]. Clearly, this discrepancy may have interfered with our results. Second, lacking information of other confounding factors (e.g. age, family history, environmental factors and lifestyles) in the eligible studies limited our further stratified analysis to explore the possible sources of heterogeneity. Third, other ethnic decent studies were absent in our study, for example, Africans and African-Americans, which may have biased our results. Thus, we are not sure whether there is a significant association between the CYP1A1 3801T>C polymorphism and increased male infertility risk in the whole population. Fourth, for each selected study, our results were based primarily on unadjusted effect estimates and confidence intervals, while a more precise analysis should be conducted if all individual raw data were available. Fifth, diagnostic criteria of semen specimens in cases and definition of control subjects (including matching variables, such as age, smoking status, and alcohol intake) were differed in the meta-analyzed studies, which is of concern as inconsistent phenotype definitions may weaken the validity of meta-analysis. Therefore, more precisely defined case–control phenotypes in future studies may not only improve efficiency and validity in individual studies, but also for the overarching meta-analysis, and should be a consideration in study design. Sixth, the corresponding authors of these studies [8], [25], [26] without adequate reporting of genotype frequency were contacted for additional information, but unfortunately did not respond by the time of analysis and writing, and the papers were thus excluded, which also limits the ability to synthesize all available evidence. Noticeably, all the three previous studies were reported that the increased frequencies of CYP1A1 3801C allele contributed to risk of IMI. In a future study, we would conduct a new meta-analysis that includes the studies to further strengthen the robustness of the current meta-analysis findings. Finally, all included studies were case-control design, which precludes further comments on cause-effect relationship. The results of long-term prospective, designed for the investigation of gene-gene and gene-environment interactions, in different ethnic populations might produce more conclusive claims about the association between CYP1A1 and IMI risk.
Conclusion
In summary, the present meta-analysis provides evidence of a significant association between CYP1A1 3801T>C polymorphism and IMI risk. However, given the limitation of the eligible studies, larger well-designed case-control or cohort studies are necessary to draw comprehensive and true conclusions.
Supporting Information
Forest plots for the association between the 3801T>C polymorphism of CYP1A1 and risk of idiopathic male infertility. The contribution of each study to the meta-analysis (its weight) is represented by the area of a box, the center of which represents the size of the OR estimated from that study. The 95% CI for the OR (extending lines) from each study is also shown. The overall OR is shown in the middle of a diamond, the left and right extremes of which represent the corresponding CI. A: allele model, B: heterozygous model, C: dominant model, D: recessive model.
(TIF)
Funnel plots for the 3801T>C polymorphism of CYP1A1 in idiopathic male infertility. The vertical axis represents log (OR), and the horizontal axis refers to the standard error of log (OR). The horizontal line indicates the pooled OR, and the sloping lines indicate the expected 95% CI for a given standard error. The area of each circle represents the contribution of the study to the pooled OR. a: homozygous model, b: heterozygous model, c: dominant model, d: recessive model.
(TIF)
Details of reasons for exclusion of studies from meta-analysis.
(DOC)
Main characteristics of studies included in the meta-analysis.
(DOC)
Methodological quality of included case–control studies based on the Newcastle–Ottawa Scale.
(DOC)
Prisma checklist.
(DOC)
Acknowledgments
The authors thank Wenkai Chen at the library of Guangdong Medical College for literature searches.
Funding Statement
This study was supported by Guangdong Natural Science Foundation (Grant No. S2011010004146). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. de Kretser DM (1997) Male infertility. Lancet 349 (9054) 787–790 doi:S0140673696083419 [DOI] [PubMed] [Google Scholar]
- 2. O'Flynn O'Brien KL, Varghese AC, Agarwal A (2010) The genetic causes of male factor infertility: a review. Fertil Steril 93 (1) 1–12 10.1016/j.fertnstert.2009.10.045 [DOI] [PubMed] [Google Scholar]
- 3. Iammarrone E, Balet R, Lower AM, Gillott C, Grudzinskas JG (2003) Male infertility. Best Pract Res Clin Obstet Gynaecol 17 (2) 211–229 doi:S1521693402001475 [DOI] [PubMed] [Google Scholar]
- 4. Singh K, Jaiswal D (2011) Human male infertility: a complex multifactorial phenotype. Reprod Sci 18 (5) 418–425 10.1177/1933719111398148 [DOI] [PubMed] [Google Scholar]
- 5. Schuppe HC, Wieneke P, Donat S, Fritsche E, Kohn FM, et al. (2000) Xenobiotic metabolism, genetic polymorphisms and male infertility. Andrologia 32 (4–5) 255–262. [DOI] [PubMed] [Google Scholar]
- 6. Finotti AC, Costa ESRC, Bordin BM, Silva CT, Moura KK (2009) Glutathione S-transferase M1 and T1 polymorphism in men with idiopathic infertility. Genet Mol Res 8 (3) 1093–1098. [DOI] [PubMed] [Google Scholar]
- 7. Tang K, Xue W, Xing Y, Xu S, Wu Q, et al. (2012) Genetic polymorphisms of glutathione S-transferase M1, T1, and P1, and the assessment of oxidative damage in infertile men with varicoceles from northwestern China. J Androl 33 (2) 257–263 10.2164/jandrol.110.012468 [DOI] [PubMed] [Google Scholar]
- 8. Fritsche E, Schuppe HC, Dohr O, Ruzicka T, Gleichmann E, et al. (1998) Increased frequencies of cytochrome P4501A1 polymorphisms in infertile men. Andrologia 30 (3) 125–128. [DOI] [PubMed] [Google Scholar]
- 9. Nebert DW, McKinnon RA, Puga A (1996) Human drug-metabolizing enzyme polymorphisms: effects on risk of toxicity and cancer. DNA Cell Biol 15 (4) 273–280. [DOI] [PubMed] [Google Scholar]
- 10. Shah PP, Saurabh K, Pant MC, Mathur N, Parmar D (2009) Evidence for increased cytochrome P450 1A1 expression in blood lymphocytes of lung cancer patients. Mutat Res 670 (1–2) 74–78. [DOI] [PubMed] [Google Scholar]
- 11. Agundez JA (2004) Cytochrome P450 Gene Polymorphism and Cancer. Curr Drug Metab 5 (3) 211–224. [DOI] [PubMed] [Google Scholar]
- 12. Xi B, Zeng T, Liu W (2011) Catechol-O-methyltransferase Val158Met polymorphism in breast cancer risk. Breast Cancer Res Treat 126 (3) 839–840. [DOI] [PubMed] [Google Scholar]
- 13. Alibakhshi R, Moradi K, Mohebbi Z, Ghadiri K (2013) Mutation analysis of PAH gene in patients with PKU in western Iran and its association with polymorphisms: identification of four novel mutations. Metab Brain Dis 10.1007/s11011-013-9432-0 [DOI] [PubMed] [Google Scholar]
- 14. Xi B, Zeng T, Liu L, Liang Y, Liu W, et al. (2011) Association between polymorphisms of the renin-angiotensin system genes and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 130 (2) 561–568. [DOI] [PubMed] [Google Scholar]
- 15. Maxwell L, Santesso N, Tugwell PS, Wells GA, Judd M, et al. (2006) Method guidelines for Cochrane Musculoskeletal Group systematic reviews. J Rheumatol 33 (11) 2304–2311. [PubMed] [Google Scholar]
- 16.Wells GA, Shea B, O'Connell D, Peterson J, Welch V et al. (2012) The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses, Ottawa Health Research Institute Web site.
- 17. Wang J, Li C, Tao H, Cheng Y, Han L, et al. (2013) Statin use and risk of lung cancer: a meta-analysis of observational studies and randomized controlled trials. PLoS One 8 (10) e77950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thakkinstian A, McElduff P, D'Este C, Duffy D, Attia J (2005) A method for meta-analysis of molecular association studies. Stat Med 24 (9) 1291–1306 10.1002/sim.2010 [DOI] [PubMed] [Google Scholar]
- 19. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327 (7414) 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22 (4) 719–748. [PubMed] [Google Scholar]
- 21. Rohlfs RV, Weir BS (2008) Distributions of Hardy-Weinberg Equilibrium Test Statistics. Genetics 180 (3) 1609–1616 10.1534/genetics.108.088005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Egger M, Davey SG, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315 (7109) 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Messaros BM, Rossano MG, Liu G, Diamond MP, Friderici K, et al. (2009) Negative effects of serum p,p'-DDE on sperm parameters and modification by genetic polymorphisms. Environ Res 109 (4) 457–464 10.1016/j.envres.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 24. Aydos SE, Taspinar M, Sunguroglu A, Aydos K (2009) Association of CYP1A1 and glutathione S-transferase polymorphisms with male factor infertility. Fertil Steril 92 (2) 541–547 10.1016/j.fertnstert.2008.07.017 [DOI] [PubMed] [Google Scholar]
- 25. Su MT, Chen CH, Kuo PH, Hsu CC, Lee IW, et al. (2010) Polymorphisms of estrogen-related genes jointly confer susceptibility to human spermatogenic defect. Fertil Steril 93 (1) 141–149 10.1016/j.fertnstert.2008.09.030 [DOI] [PubMed] [Google Scholar]
- 26. Lee IW, Kuo PH, Su MT, Kuan LC, Hsu CC, et al. (2011) Quantitative trait analysis suggests polymorphisms of estrogen-related genes regulate human sperm concentrations and motility. Hum Reprod 26 (6) 1585–1596. [DOI] [PubMed] [Google Scholar]
- 27. Lu N, Wu B, Xia Y, Wang W, Gu A, et al. (2008) Polymorphisms in CYP1A1 gene are associated with male infertility in a Chinese population. Int J Androl 31 (5) 527–533. [DOI] [PubMed] [Google Scholar]
- 28. Vani GT, Mukesh N, Siva Prasad B, Rama Devi P, Hema Prasad M, et al. (2009) Association of CYP1A1*2A polymorphism with male infertility in Indian population. Clin Chim Acta 410 (1–2) 43–47. [DOI] [PubMed] [Google Scholar]
- 29. Chen WC, Kang XX, Huang ZS, Wei YS, Pan Y (2010) CYP1A1 gene polymorphism in oligozoospermic infertile patients of Zhuang population in Guangxi area. Guangdong Med ica l Journal 31 (13) 1669–1671. [Google Scholar]
- 30. Peng L, Wang G, Jiao HY, Li YJ, Dang J (2012) Relationship between CYP1A1 rs4646903 gene polymorphism and male infertility in Ningxia. Journal of Ningxia Medical University 34 (3) 208–210. [Google Scholar]
- 31. Salehi Z, Gholizadeh L, Vaziri H, Madani AH (2012) Analysis of GSTM1, GSTT1, and CYP1A1 in idiopathic male infertility. Reprod Sci 19 (1) 81–85. [DOI] [PubMed] [Google Scholar]
- 32. Yarosh SL, Kokhtenko EV, Starodubova NI, Churnosov MI, Polonikov AV (2013) Smoking Status Modifies the Relation Between CYP1A1*2C Gene Polymorphism and Idiopathic Male Infertility: The Importance of Gene-Environment Interaction Analysis for Genetic Studies of the Disease. Reprod Sci 20 (11) 1302–1307. [DOI] [PubMed] [Google Scholar]
- 33. Juarez-Cedillo T, Vallejo M, Fragoso JM, Hernandez-Hernandez DM, Rodriguez-Perez JM, et al. (2007) The risk of developing cervical cancer in Mexican women is associated to CYP1A1 MspI polymorphism. Eur J Cancer 43 (10) 1590–1595. [DOI] [PubMed] [Google Scholar]
- 34. Pandey SN, Choudhuri G, Mittal B (2008) Association of CYP1A1 Msp1 polymorphism with tobacco-related risk of gallbladder cancer in a north Indian population. Eur J Cancer Prev 17 (2) 77–81. [DOI] [PubMed] [Google Scholar]
- 35. Akgul S, Derman O, Alikasifoglu M, Aktas D (2011) CYP1A1 polymorphism in adolescents with polycystic ovary syndrome. Int J Gynaecol Obstet 112 (1) 8–10. [DOI] [PubMed] [Google Scholar]
- 36. Siddarth M, Datta SK, Ahmed RS, Banerjee BD, Kalra OP, et al. (2013) Association of CYP1A1 gene polymorphism with chronic kidney disease: a case control study. Environ Toxicol Pharmacol 36 (1) 164–170. [DOI] [PubMed] [Google Scholar]
- 37. Taspinar M, Aydos S, Sakiragaoglu O, Duzen IV, Yalcinkaya A, et al. (2012) Impact of genetic variations of the CYP1A1, GSTT1, and GSTM1 genes on the risk of coronary artery disease. DNA Cell Biol 31 (2) 211–218 10.1089/dna.2011.1252 [DOI] [PubMed] [Google Scholar]
- 38. Zhang J, Deng J, Zhang C, Lu Y, Liu L, et al. (2010) Association of GSTT1, GSTM1 and CYP1A1 polymorphisms with susceptibility to systemic lupus erythematosus in the Chinese population. Clin Chim Acta 411 (11–12) 878–881. [DOI] [PubMed] [Google Scholar]
- 39. Lucas TF, Pimenta MT, Pisolato R, Lazari MF, Porto CS (2011) 17beta-estradiol signaling and regulation of Sertoli cell function. Spermatogenesis 1 (4) 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eugster HP, Probst M, Wurgler FE, Sengstag C (1993) Caffeine, estradiol, and progesterone interact with human CYP1A1 and CYP1A2. Evidence from cDNA-directed expression in Saccharomyces cerevisiae. Drug Metab Dispos 21 (1) 43–49. [PubMed] [Google Scholar]
- 41. Kisselev P, Schunck WH, Roots I, Schwarz D (2005) Association of CYP1A1 polymorphisms with differential metabolic activation of 17beta-estradiol and estrone. Cancer Res 65 (7) 2972–2978. [DOI] [PubMed] [Google Scholar]
- 42. Lai KP, Wong MH, Wong CK (2004) Modulation of AhR-mediated CYP1A1 mRNA and EROD activities by 17beta-estradiol and dexamethasone in TCDD-induced H411E cells. Toxicol Sci 78 (1) 41–49. [DOI] [PubMed] [Google Scholar]
- 43. Denison MS, Nagy SR (2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43: 309–334. [DOI] [PubMed] [Google Scholar]
- 44. MacPherson L, Lo R, Ahmed S, Pansoy A, Matthews J (2009) Activation function 2 mediates dioxin-induced recruitment of estrogen receptor alpha to CYP1A1 and CYP1B1. Biochem Biophys Res Commun 385 (2) 263–268. [DOI] [PubMed] [Google Scholar]
- 45. Neupane B, Loeb M, Anand SS, Beyene J (2012) Meta-analysis of genetic association studies under heterogeneity. Eur J Hum Genet 20 (11) 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plots for the association between the 3801T>C polymorphism of CYP1A1 and risk of idiopathic male infertility. The contribution of each study to the meta-analysis (its weight) is represented by the area of a box, the center of which represents the size of the OR estimated from that study. The 95% CI for the OR (extending lines) from each study is also shown. The overall OR is shown in the middle of a diamond, the left and right extremes of which represent the corresponding CI. A: allele model, B: heterozygous model, C: dominant model, D: recessive model.
(TIF)
Funnel plots for the 3801T>C polymorphism of CYP1A1 in idiopathic male infertility. The vertical axis represents log (OR), and the horizontal axis refers to the standard error of log (OR). The horizontal line indicates the pooled OR, and the sloping lines indicate the expected 95% CI for a given standard error. The area of each circle represents the contribution of the study to the pooled OR. a: homozygous model, b: heterozygous model, c: dominant model, d: recessive model.
(TIF)
Details of reasons for exclusion of studies from meta-analysis.
(DOC)
Main characteristics of studies included in the meta-analysis.
(DOC)
Methodological quality of included case–control studies based on the Newcastle–Ottawa Scale.
(DOC)
Prisma checklist.
(DOC)