Abstract
We describe a detailed procedure to create photolabile, poly(ethylene glycol)-based (PEG) hydrogels and manipulate material properties in situ. The cytocompatible chemistry and degradation process enable dynamic, tunable changes for applications in 2D or 3D cell culture. The materials are created by synthesizing an o-nitrobenzylether-based photodegradable monomer that can be coupled to primary amines. Here, we provide coupling procedures to PEG-bis-amine to form a photodegradable crosslinker or to the fibronectin-derived peptide RGDS to form a photoreleasable tether. Hydrogels are synthesized with the photodegradable crosslinker in the presence or absence of cells, allowing direct encapsulation or seeding on surfaces. Cell-material interactions can be probed in 2D or 3D by spatiotemporally controlling the gel microenvironment, which allows unique experiments to be performed to monitor cell response to changes in their niche. Degradation is readily achieved with cytocompatible wavelengths of low intensity flood irradiation (365 to 420 nm) in minutes or with highintensity laser irradiation (405 nm) in seconds. In this protocol, synthesis and purification of the photodegradable monomers take approximately 2 weeks, but can be substantially shortened by purchasing the o-nitrobenzylether precursor. Preparation of the sterile solutions for hydrogel fabrication takes hours, while the reaction to form the final hydrogel is complete in minutes. Hydrogel degradation occurs on-demand, in seconds to minutes, with user-directed light exposure. This comprehensive protocol is useful for controlling peptide presentation and substrate modulus during cell culture on or within an elastic matrix. These PEG-based materials are useful for probing the dynamic influence of cell-cell and cell-material interactions on cell function in 2D or 3D. While other protocols are available for controlling peptide presentation or modulus, few allow manipulation of material properties in situ and in the presence of cells down to the micrometer scale.
Keywords: cell culture, tissue engineering, responsive materials, hydrogels, biomaterials, photolabile, poly(ethylene glycol), RGDS, mesenchymal stem cells
Introduction
Physiological processes are guided by interactions between cells and their extracellular matrix, and this understanding has led to a growing interest in the development of material systems for improved 3D cell culture. In particular, hydrogel systems based on both protein components (e.g., collagen and Matrigel) and highly-tunable synthetic chemistries (e.g., PEG) have evolved to address some of these needs1-3. However, as advances in real-time tracking of dynamic cellular functions have emerged4-6, complementary approaches to alter the surrounding extracellular environment in a user-defined and highly-controlled fashion are extremely limited. Such materials systems would have the potential to significantly improve our understanding of how cells receive information from their microenvironment and the role that these dynamic processes may play in biological questions ranging from directing stem cell differentiation4, 5 to understanding cancer metastasis6, 7. To realize this need for dynamically tunable culture systems, we recently developed an approach for in situ hydrogel property manipulation with light, allowing intimate control of a cell's microenvironment in both time and space8. These photoactive hydrogels afford unique user-defined manipulation of the biochemical and biomechanical nature of the extracellular microenvironment. The application of these photochemically tunable cell culture materials is of growing interest to a diverse audience of scientists and engineers9, 10. The goal of this manuscript is to provide a detailed protocol that makes synthesis of these photolabile gels and related degradation techniques simple and accessible.
Here, we provide a step-by-step protocol for synthesis of the photolabile group and photolabile monomer, which can be easily coupled to any primary amine-containing molecule. Subsequently, we provide the sequential protocol for synthesis of a photodegradable, PEG-based crosslinking monomer and photoreleasable peptide tether, allowing the synthesis and manipulation of hydrogel crosslinking density and modulus or peptide presentation, respectively, during 2D or 3D cell culture. Lastly, we provide detailed gel synthesis and degradation protocols, focusing on the manipulation of gel structure with photolithography or focused 405 nm light and subsequent verification of structural changes with a confocal microscope. While protocols for synthesizing the photolabile group for solid phase peptide synthesis or functional group (un)caging are available in the literature11-13, these protocols do not cover the details of synthesizing and degrading photolabile monomers and gels in the presence of cells. This manuscript provides a universal protocol for synthesizing photolabile gels from the ground up, and our goal is to facilitate the translation of these systems for a broad range of cell culture applications.
Development of the protocol
This protocol for synthesis and degradation of photolabile hydrogels under cytocompatible conditions was developed for precisely controlling the presentation of biophysical or biochemical cues within a cell's microenvironment8. The photolabile group, ethyl 4-(4-(1-hydroxyethyl)-2-methoxy-5-nitrophenoxy)butanoic acid, was selected as the degradable unit because of its previous use in live cell cultures14. Further, this moiety degrades under cytocompatible irradiation conditions, including longwave UV light (≥ 365 nm), visible (up to 420 nm), and two-photon irradiation12, 15. In addition, the photolabile group has been used previously in the uncaging of fluorophores for live cell imaging, indicating the cytocompatible nature of the photochemistry14. Similar nitrobenzyl photolabile molecules have been used in a number of different applications11, 16. These applications are growing and include the (un)caging of proteins17, reactive groups within hydrogels18, 19, or adhesive ligands on culture plates20, 21 to promote cell signaling, process extension, or control cell attachment, respectively; controlled degradation of hydrophobic, step-growth polymer networks22, 23; release of PEG from surfaces to modulate cell attachment24; and in situ tuning of poly(acrylamide) gel modulus during 2D cell culture25.
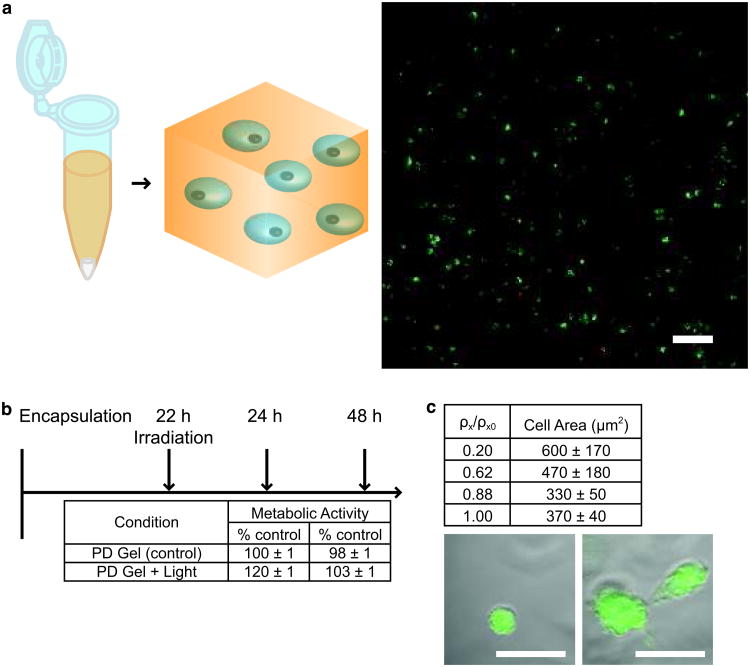
Recent work from our group8 demonstrates how this photolabile group can be incorporated within water-soluble macromolecular monomers to create a versatile platform that allows manipulation of the gel's physical or biochemical properties in 2D26 and 3D8, 27. To generate these photolabile hydrogels, a photodegradable acrylate monomer was synthesized from the o-nitrobenzylether-based photolabile group. This small molecule monomer was coupled to (i) PEG-bis-amine to create a photodegradable crosslinking monomer for tuning gel crosslinking density and modulus or (ii) to a cell adhesion peptide sequence, RGDS, to create photoreleasable pendant functional groups for modulating cell integrin binding and adhesion at any position during 2D or 3D cell culture8. The cytocompatibility of the material and the degradation process has been examined with two cell types, human mesenchymal stem cells (hMSCs) and porcine valve fibroblasts (valvular interstitial cells, VICs), where in both cases high viability was observed with or without irradiation and degradation. With hMSCs, encapsulation in (i) photodegradable gels or (ii) gels with a photoreleasable RGDS led to high survival, and subsequent irradiation and degradation did not affect viability as measured by membrane integrity and DNA assays8. Similar results are reported for VICs cultured on photodegradable hydrogel substrates26. Thus, using either the photodegradable crosslinker or the biofunctional monomer, an adaptable culture system can be fabricated that offers simultaneous manipulation and monitoring of cell-material interactions in the presence of cell in either two or three dimensions.
Applications of the method
To date, these synthetic approaches have been used to create photolabile hydrogels based on PEG with or without pendant peptide functionalities. However, the chemistry is quite diverse and could be easily coupled with other macromolecules to make densely or loosely crosslinked networks, neutral or charged gels, or even more hydrophobic or hydrophilic material systems, to achieve a broad range of properties. Beyond peptides, functional gels containing other small molecules, proteins, or biological signals are readily envisioned. Further, by varying the polymerization mechanism, materials can be designed with controlled structures, surface functionalization, or gradient properties. Because care was taken in the design of the monomer chemistry to insure cytocompatibility, we focus our discussion on how this protocol can be used for the creation of photodegradable or photoreleasing hydrogels for two-dimensional and three-dimensional cell culture and discuss how it might be exploited to answer a diverse array of biological questions. Such questions include investigating the influence of crosslinking density and modulus on cell morphology, migration, and differentiation; exploring how spatial and temporal control of integrin binding regulates cell survival, adhesion, and differentiation; or quantifying the cell response to dynamic changes in the pericellular mechanics. Furthermore, this versatile hydrogel platform enables investigations of cellular phenomena in 2D culture for subsequent translation to 3D for probing differences in cell function between 2D and 3D culture.
As cell culture systems, the photodegradable hydrogels presented in this protocol can be used for the real-time manipulation and monitoring of single and/or multiple cells. We have applied these photolabile hydrogels to (i) probe the influence of microenvironment modulus and dynamic changes in stiffness on cell differentiation26, (ii) investigate how matrix density influences cell morphology and process extension27, and (iii) examine how the temporal presentation of ligands in a stem cell niche influences cell differentiation8. There is a growing appreciation of the role of a cell's microenvironmental context in dictating how cells respond to biological cues (e.g., soluble growth factors). For example, substrate modulus has been shown to influence cell differentiation28, 29 and migration30, but in general, these materials have had static properties and less is known about how a cell would respond to dynamic changes25, 31, 32 in its microenvironment. Photodegradable hydrogels afford spatiotemporal control of modulus during 2D and 3D cell culture (Fig. 1a) and present the opportunity to conduct unique experiments. For example during 2D culture, valve fibroblasts were shown to activate into a wound healing phenotype on high modulus substrates, and subsequently, deactivate if the substrate modulus was made soft via photodegradation26. This finding provides unique insight into the broader issue of fibrosis and demonstrates how these novel material systems allow a better understanding of factors (e.g., matrix stiffness) that may be used to reverse myofibroblast activation and potentially disease progression. This 2D approach could be expanded to release cells from a culture substrate uniformly or in any region by erosion of the underlying hydrogel surface or photorelease of an adhesive ligand (e.g., RGDS tether). This capability may be broadly applicable in tissue engineering, as the photolabile gel strategy provides a spatially-unconfined, dynamic soft material complement to heat-releasable, poly(NIPAM)-based culture substrates for cell sheet tissue engineering33-36 or electrically-releasable, patterned RGDS hard culture substrates for examination of cell-cell and cell-material interactions37, 38. Here, the position of cell release is not predetermined and can be dictated by the user on-demand with irradiation, and the initial matrix modulus can be tuned with the initial crosslinker concentration and subsequently manipulated in situ with irradiation. This technique could also facilitate biological assays on subpopulations of cells in 2D or 3D culture where the user could release a subset of cells from the culture system for RT-PCR or FACS while leaving the rest of the population intact for further culture.
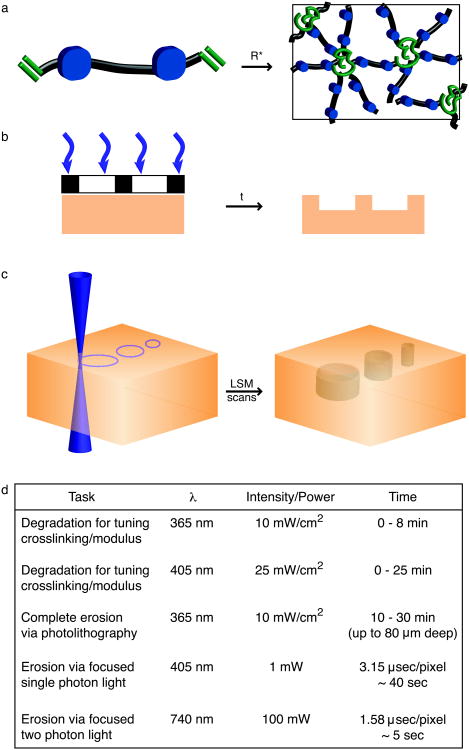
Figure 1. Photolabile hydrogels for cell culture.
Photolabile hydrogels are a dynamic class of cell culture materials that can be exploited to alter precisely and predictably the cellular microenvironment in both space and time. (a) Dynamic changes in the adhesive context influence cell morphology and cytoskeletal organization. Here, light-induced changes in polymer density or ligand concentration of the hydrogel (shown in yellow) are used to control cell spreading (shown in gray) and cytoskeletal organization (represented by red f-actin filaments and purple junctions). (b) Cells (gray) can be seeded within wells (white circles) of photoactive hydrogels (shown in yellow) and subsequent changes to the geometry of the cellular microenvironment can be introduced with light. Here, a channel is introduced between two wells, encouraging cells to migrate out of their niche. These patterning strategies can be extended easily to tune the cellular microenvironment around encapsulated cells or to connect encapsulated cells in 3D cell culture.
The ability to spatially and temporally regulate the hydrogel's crosslinking density and modulus allows one to explore many basic questions, including how cell morphology changes in response to the local polymer density or how cells sense local changes or gradients in modulus and migrate. Such information could prove beneficial to those seeking to expand or direct the differentiation of stem cells, understand mechanisms of migration or metastasis, or even regenerate tissues. For example, hMSC morphology and spreading have been controlled in 3D culture by controlled gel photodegradation and spatially-specific crosslinking density manipulation27. This strategy could be expanded to examine the effect of crosslinking density and modulus on more complex cell functions such as differentiation, process extension and signaling, and migration in three dimensions. Additionally, advanced patterning techniques can be used to degrade micron-scale voids or channels around, between, or under cells for directing cell process extension, migration, or detachment (Fig. 1b), and the resulting cell response can be imaged in real time. This same approach can be applied to cells cultured on the hydrogel surface (2D), where dynamic modulus changes can be used to study and direct stem cell differentiation or examine phenotype plasticity. Gels with a gradient in modulus can be used to examine cell migration in response to the modulus magnitude or gradient steepness, but more notably, the initial gradient can be eliminated or altered at any point during the experiment.
Triggering and monitoring a change in a specific cell-material interaction is particularly important for understanding and controlling signals within the stem cell niche (e.g., the role of structure and modulus in directing cell fate via mechanotransduction). Cell adhesion within the microenvironment might also be probed by controlled peptide presentation using photoreleasable tethers for introduction, as well as removal, of biochemical cues (Fig. 1a). In particular, hMSC adhesion and chondrogenic differentiation were regulated within degradable PEG hydrogels when RGDS was temporally released at an appropriate time during 3D culture8. Such approaches can prove quite valuable for in situ regulation of cell adhesion in 2D or 3D culture. In general, we believe that these photolabile hydrogels will prove useful for testing hypotheses about the role of the cell environment in directing cell function. In addition to controlled presentation of peptides, this protocol can be used to create hydrogels that precisely release entrapped proteins via photodegradation39 or cleavage of a photoreleasable tether, as has been done previously with hydrolytically40 or enzymatically41 materials. The advantage here is the spatiotemporal control afforded by light-based degradation, which can be used to screen the effect of different release rates on biological function in vitro or tuning release rates in situ.
Comparison with other methods
The protocol used for synthesis of the photolabile molecule is based on protocols originally developed by Holmes12 and Akerblom et al.13 This photolabile molecule was then acrylated and used to form a photodegradable crosslinker and photoreleasable tether, which are readily polymerized to form light degradable materials. Complementary synthetic approaches to the formation of the photodegradable acrylate monomer and the photodegradable crosslinker were recently presented by Wong et al. with increased yield for the photodegradable acrylate42. These monomer syntheses are done in different solvent systems, improving the recovery of photodegradable acrylate, but currently, the resulting materials have not been demonstrated for use in cell culture.
For dynamic cell culture, several examples exist where cells are cultured on 2D surfaces and the underlying properties are manipulated. For example, electroresponsive hard substrates for controlled adhesive ligand presentation37, 38, 43 and thermoresponsive soft substrates for controlled swelling34, 44 have been used to temporally regulate cell adhesion and release cells in pre-defined geometries. These materials represent two important, useful classes of 2D culture substrates with externally-triggerable control of cell-material interactions. With the photolabile gels presented here, cells are cultured on an elastic substrate with moduli similar to that of a soft tissue type, such as collagenous bone and muscle, and the 2D substrate modulus or ligand presentation can be regulated at any position or time by in situ irradiation. Depending on the irradiation conditions, micron-scale resolution in the materials properties is achieved spatially, and the cleavage is complete in seconds to minutes affording nearly instant property manipulation. For property regulation in three-dimensions, few materials allow the tuning of properties in the presence of cells. One recent advance is temporally controlled cell spreading and morphology in three dimensions by the addition of crosslinks with photopatterning31, which mitigates cell spreading indefinitely; another is the removal of crosslinks with ion concentration32, 45, which facilitates cell spreading over a few days in culture. With the photolabile gels, cell spreading can be promoted at any position or time within 3D culture by controlled irradiation. These approaches provide alternative methods for externally-triggered manipulation of the cell microenvironment and offer varying levels of control.
For spatiotemporal property control, several photoresponsive polymer networks and gels have been developed. These systems include organic-solvent-based, photodegradable polymer networks for controlled degradation products22, 23, DNA-based polymer networks with photoreactive amino acids for increasing crosslinking and modulus with light46, photoreversible hydrogels for controlled protein release with low-wavelength UV light47, 48, and alginate-based hydrogels with photolabile group caged reactive groups for the spatiotemporal controlled addition of peptides18, 19. While interest and effort has grown in the development of photolabile materials, few photodegradable systems have been demonstrated for property manipulation during cell culture. Part of this relates to the challenges of developing material systems that are compatible with cell encapsulation and subsequent degradation mechanisms that can also be performed in the presence of cells. With respect to the latter, Frey and Wang recently demonstrated real-time modulus control by irradiation of polyacrylamide-based photodegradable hydrogels to study how real-time modulus changes influenced 3T3 cell morphology and migration in 2D culture25. Simultaneously, acrylate-based photodegradable monomers were developed and demonstrated for both cell encapsulation and controlled degradation of the physical or chemical structure of the cell microenvironment. These materials allow for real-time property control in 2D and 3D cell culture and are the materials presented in this protocol. Further, the PEG-based, photodegradable gels allow tuning of modulus over a larger range (up to 78% change from approximately 32 to 7 kPa) than the photolabile polyacrylamide gels (up to 30% change from approximately 7 to 5 kPa) so that cell differentiation can be probed in addition to cell morphology and migration. In addition to cell culture, Wong et al. recently demonstrated use of these photodegradable monomers and a new coumarin-based photodegradable monomer for positive and negative feature generation with single- and two-photon irradiation42. The coumarin-based, photodegradable gels allow degradation at lower irradiation intensities, which may be useful if a cellular system is observed to be particularly light sensitive; however, the coumarin moiety increases light absorbance within the gel, which may decrease the depth at which material properties can be tuned.
Limitations of the method
As with any light-based protocol for cell culture, there are limitations to the dose of light and the penetration depth of light. Depending on the wavelength of the light source, the total dose of light that a cell receives must be limited by controlling either irradiation intensity or time to avoid DNA damage and cell death. The light doses presented within this protocol allow cells to function, but as irradiation conditions are changed, cell viability and function must be assessed. Additionally, absorbance of light within the gel at irradiation wavelengths of interest must be calculated to determine the attenuation of light within the gel. One can exploit attenuation to control gel surface modulus, generate a z-gradient in modulus within a gel, or surface erode a gel by proper selection of irradiation parameters. However, with the gel composition and irradiation conditions presented here, light penetration is limited to approximately 100 μm at 365 nm and 450 μm at 405 nm, where a gel can be irradiated from both sides to affect cells within gels that are 200 μm and 900 μm thick, respectively. The photolabile monomer concentration can be adjusted to control light absorbance and attenuation based on the molar absorptivity of the photolabile group at the irradiation wavelength of interest.
To circumvent many of the issues with light attenuation, two-photon irradiation can be used to spatiotemporally tune gel properties with exquisite control. The main limitation of this approach is that it requires access to a two-photon laser scanning confocal microscope, which is an expensive piece of specialty equipment. Further, care must be taken to maintain sterility during irradiation, and fluorophores requiring two-photon excitation below 800 nm (i.e., DAPI) cannot be used without degrading the gel.
Finally, while the current photolabile material system provides many advantages for degrading hydrogel materials and removing selected cells signals, there are many cases where one might desire to add functionality or increase crosslinking density. The current system is limited to only removing gel material or biochemical signals. As such, advances in photoaddition chemistries, such as the sequential azide-alkyne and thiol-ene click reactions49, demonstrate strategies to add chemical functionality (e.g., peptide sequences) into PEG-based hydrogels. In a less controlled, but practical approach, interpenetrating networks of hydrogels can be created to increase polymer density and control material properties in 3D31, 50. These strategies could be applied to the photolabile hydrogels, where visible light photoinitiation at wavelengths greater than 405 nm could be used to add crosslinks while UV irradiation could be used to remove them.
Experimental Design
Monomer synthesis
In this protocol, several monomers are synthesized as precursors for making photolabile hydrogels. These monomers include (i) a base photodegradable acrylate monomer that can be coupled to primary amine containing molecules, (ii) a crosslinking macromolecular monomer for making photodegradable hydrogels, and (iii) an asymmetric monomer with a photolabile tether for releasing peptides or other molecules of interest from non-degradable gels. The synthesis and characterization of each of these will be covered.
Photodegradable acrylate. The base photolabile group is first synthesized, or can be purchased (photolabile precursor, EMD NovaBiochem, hydroxyethyl photolinker). The synthetic route presented here for the photolabile group8 is based on several other protocols12-14 and is described in detail within this manuscript for the synthesis of large, bench-scale batches. Since the synthetic procedure for this photolabile group requires several reaction steps, purchasing the photolabile group and proceeding with the acrylation procedure (starting at Step 20) may be preferred for some researchers and would decrease the protocol time by approximately 6 days. To create the photodegradable acrylate (PDA) monomer, the photolabile group is acrylated and purified to yield a small molecule monomer that cleaves in response to light and can be coupled to primary amine containing molecules, such as peptides, proteins, and PEG-bis-amine, for incorporating them within a hydrogel, or more generally within other polymer networks. The acrylate functionality could be replaced with a methacrylate or other reactive functionality for the creation of monomers with different polymerization rates or reaction mechanisms.
Photodegradable crosslinker. The photodegradable crosslinking monomer is synthesized using the base PDA and PEG-bis-amine (Mn ∼ 3400 g/mol, PEG3400). The carboxylic acid on the PDA is activated and coupled to the primary amine end groups of PEG-bis-amine using solid phase peptide synthesis (SPPS) chemistry. The resulting crosslinker is purified and can be used to create hydrogels for 2D or 3D cell culture whose modulus can be tuned or the gel eroded via light-controlled degradation. While PEG3400 is used here to create hydrogels for 2D and 3D cell culture, this crosslinker synthesis strategy could be used to create other photodegradable macromers (macromolecular monomers). For example, the PDA could be reacted with PEG-bis-amine of varying molecular weight or functionality (e.g., multi-arm PEG) or with other synthetic or natural-based polymers with pendant amine groups to create an array of crosslinking monomers.
Photoreleasable tether. The photoreleasable tether monomer is synthesized using the PDA and a peptide bound to a solid phase support (e.g., on resin). The adhesion peptide RGDS is synthesized on resin using standard SPPS techniques. The PDA is coupled to the terminal amine of the peptide on resin following SPPS procedures. Cleavage of the peptide from resin is short (< 2 h) to avoid cleavage of the ester bond within the PDA. The resulting peptide is purified and can be used for controlling peptide presentation and cell adhesion to a nondegradable hydrogel during 2D or 3D culture. While only synthesis of the fibronectin mimic RGDS is presented here, this tether strategy could be used for controlled release of other peptides, therapeutics, or proteins with light.
Hydrogel synthesis
Photoactive hydrogels can be formed from the photodegradable crosslinker or the photoreleasable tether via redox-initiated free radical chain polymerization (Fig. 2a). To fabricate photodegradable hydrogels, a macromer solution of the photodegradable crosslinker PEGdiPDA, PEG monoacrylate, and ammonium persulfate are prepared and aliquoted into individual tubes for each gel sample. To fabricate photoreleasable hydrogels, a base macromer solution of the non-degradable crosslinker PEG diacrylate (PEGDA), photoreleasable tether, and ammonium persulfate are prepared and aliquoted into individual tubes for each gel sample. Addition of tetraethylmethylenediamine (TEMED) initiates free radical chain polymerization of the macromer solution, and this polymerizing solution can be pipetted into defined molds to fabricate insoluble hydrogels of specified geometries. The initial biomechanical and biochemical properties of the photoactive hydrogels can be tuned by changing the initial composition of the macromer solution. For example, increasing the concentration of the crosslinker in the macromer solution will lead to an increase in the elasticity of the subsequently formed gel, and acrylated or methacrylated signaling peptides can be incorporated into the macromer solution to introduce biochemical signals. Further, the concentrations of ammonium persulfate and TEMED can be modified to control the time required for the macromer solution to reach complete gelation. Other initiation schemes, such as thermal initiation or photoinitiation, can also be used, and we present the redox-initiation as a simple and robust approach for hydrogel fabrication. In general, this protocol provides the basis to fabricate photodegradable or photoreleasable hydrogels with tunable mechanical or biochemical properties, which can be formed with sample dimensions and geometries of choice.
Figure 2. Photodegradable hydrogel synthesis and patterning.
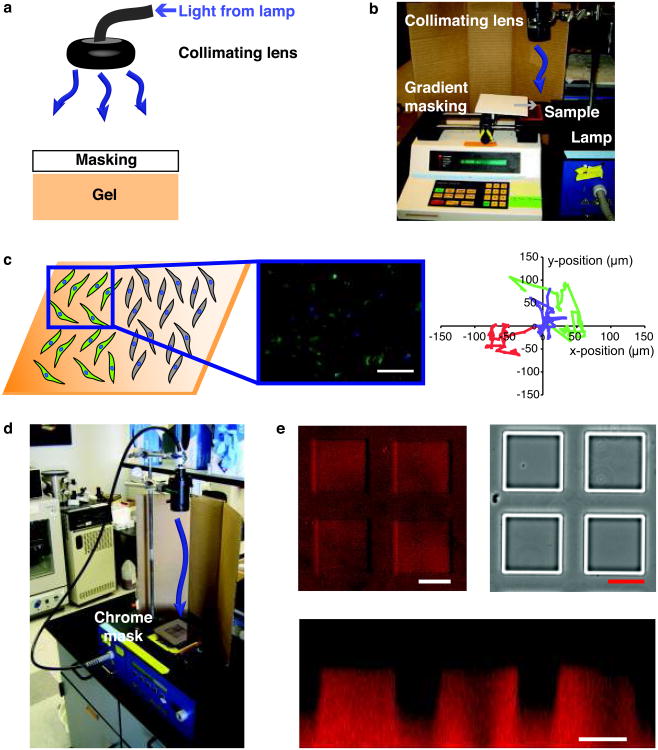
(a) Photodegradable hydrogels are formed by redox initiated, free radical (R*) chain polymerization of PEGdiPDA crosslinking molecules, rapidly forming an insoluble hydrogel. The resultant network structure is comprised of polyacrylate kinetic chains (green) connected by PEG-based crosslinks (black) with photocleavable o-nitrobenzylether moieties (blue) in the backbone. (b) Chrome masks (black and white layer) can be placed at the surface of photodegradable gels (orange) to selectively occlude incident light (purple). With sufficient irradiation time (t) features are formed at the surface of PEGdiPDA gels, where the depth of feature formation depends on the duration of exposure. This photolithographic degradation can be used to generate features in the surface of a gel with minutes of irradiation time. (c) The focal point of a laser can be used to pattern features within PEGdiPDA hydrogels. Here, a laser scanning microscope (LSM) with a 405 nm laser is used to draw regions of interest (ROI) in the material. The laser focal point is rastered through the ROI and into the z-dimension to completely erode 3D features within the hydrogel in seconds. Both of these patterning strategies can be translated for spatiotemporally controlled presentation of biochemical ligands with gels containing the photoreleasable tether. (d) An overview of irradiation parameters for degradation and complete erosion of photodegradable hydrogels. Ultimately, these patterning approaches allow the rapid, in situ creation of micron-scale features in the presence or absence of cells in 2D and 3D.
Cell culture and encapsulation
Cells can be cultured on or encapsulated with either photolabile hydrogel formulation: (i) photodegradable hydrogel formed with the photodegradable crosslinking monomer or (ii) photoreleasing tether hydrogel formed with the photoreleasable tether monomer and a non-degradable PEG crosslinker. In this protocol, we show the case of encapsulation with the photodegradable hydrogel (i). For reference, we also have previously published the culture of cells on photodegradable gels26 and within photoreleasable gels8. The base photodegradable hydrogel formulation described in the previous section is prepared, and fibronectin is added to promote cell adhesion within the gel. While we demonstrate the entrapment of fibronectin, other large extracellular matrix proteins, such as laminin, can be added and entrapped within the hydrogel to promote specific cell interactions (e.g., selective integrin binding). With this monomer solution, cells are encapsulated based on methods described in other protocols in the literature51 but using a redox initiation system instead of a light-based initiation system: cells are isolated and counted, and an aliquot of cells for achieving the desired cell density within the gel (2×106 cells/mL here) is centrifuged, resuspended in the monomer solution, and polymerized within a mold. After polymerization (∼ 5 min), the cell-gel construct is placed in excess media, and the media is refreshed repeatedly over 24 h to remove any unreacted monomer and initiator. This procedure has been used to encapsulate several different cell lines and primary cells in our lab, including PC12s (pheochromocytoma cells, model cell line for neuronal differentiation)52, HT1080s (fibrosarcoma cells, model cell line for tumor metastasis)8, NIH 3T3s (fibroblast cell line) [unpublished], and human mesenchymal stem cells (bone marrow derived primary cells)8, 27. The protocol easily can be adapted for use with a broad array of cell types at a desired cell seeding density and gel formulation.
Hydrogel degradation and patterning
A major benefit of using photolabile hydrogels is that the incorporation of the o-nitrobenzylether moiety provides a phototunable handle through which the biomechanical or biochemical nature of the gel can be tuned with light. Since the user possesses both temporal and spatial control over light delivery, he/she can control spatiotemporally the mechanics or biochemistry of the hydrogel. In this protocol, we present the use of standard photolithography and confocal laser scanning microscopy (LSM) to pattern surface elasticity gradients, surface features, and bulk features within photodegradable hydrogels. The techniques described herein can be easily translated to pattern similar gradients or regions of defined biochemical signals with photoreleasable hydrogels. Furthermore, these degradation techniques are fully cytocompatible and can be performed in the presence of cells in the same manner described below with extra care taken when handling the samples to maintain sterility and cell viability.
Flood irradiation. Flood irradiation is irradiation of the entire gel without masking. With this technique and the photodegradable gel composition described in this protocol, the top 100 μm of the hydrogel is degraded, cleaving crosslinks and decreasing the modulus of the gel. Degradation is confined to the top ∼ 100 μm of the gel owing to light attenuation by the photodegradable group. This technique can be used to tune gel modulus during 2D cell culture or to create a z-gradient in crosslinking within a hydrogel during 3D cell culture (up to 8 min of 10 mW/cm2 at 365 nm).
Photolithography. Standard photolithography is a powerful technique employed to photopattern features in the x-y plane at the surface of photodegradable hydrogels with spatial resolution on the scale of 10 μm when using chrome masks (Fig. 2b). Features can be transferred into the zdimension, and the depth of feature formation depends on the irradiation conditions (intensity and wavelength) and the exposure time. When patterning photodegradable or photoreleasable hydrogels with standard photolithography, there are three critical aspects: (i) light source, (ii) patterning mask, and (iii) exposure time. In this protocol, we describe the use of standard photolithography with a 365 nm light source to generate gradients in elasticity and features at the surface of photodegradable hydrogels. To form the x-y gradient, an opaque mask is passed over the gel surface at a controlled rate during irradiation (up to 8 min at 10 mW/cm2), and to form patterned features, a chrome mask with opaque squares is used (> 10 min at 10 mW/cm2). The parameters described herein can be tuned depending on the experimental situation; however, we advocate using 365 nm light and chrome masks for the best results. Exposure times can be tuned readily to generate surface gradients of varying steepness or features of variable depths.
Single-photon confocal microscope. Laser scanning confocal microscopy (LSM) with 405 nm light can also be employed to pattern the surface of photodegradable hydrogels or to fabricate features within the bulk of photodegradable gels (Fig. 2c). LSM patterning offers micron-scale resolution in the x-y plane and z-dimension and readily tunable feature geometry. LSM patterning easily can be adapted to any confocal microscope with a 405 nm light source and region of interest (ROI) software. The user draws the desired geometry in the ROI software and subsequently irradiates the selected regions with the 405 nm light source. In this protocol, we employ a 30 mW, 405 nm laser at 50% transmission (power measured at the sample to be ∼ 1 mW) with a pixel dwell time of 3.15 μsec; however, these irradiation conditions can be tuned for other microscopes and/or needs. Furthermore, LSM pattern formation offers simultaneous imaging of changes in gel structure and cell response to changes in gel structure. Since the sample is already on the confocal stage, the irradiation parameters can be changed to excite fluorophores attached to the gel backbone (e.g., methacrylated rhodamine, 543 nm) to image changes in gel structure or cell labeling fluorophores (e.g., GFP, 488 nm) to image cellular responses.
Degradation parameter overview. Typical irradiation parameters and associated gel degradation times are given for each of these degradation strategies in Fig. 2d. While these are typical parameters used, nuances exist for degradation with 405 nm focused, single photon irradiation. For example, an increased “dose” of light is received with larger x-y features (e.g., a 50-μm versus a 5-μm diameter x-y circle), owing to overlapping regions of out of focus light, and results in increased non-specific z-direction degradation and patterned feature depth. These degradation parameter subtleties should be considered when planning advanced patterning experiments. If precise z-direction degradation is required within a gel (< 10 μm non-specific z-degradation), a focused, two photon irradiation source might be preferred8.
Materials
Reagents
CAUTION Most of the reagents listed below are hazardous (e.g., toxic, corrosive, flammable, irritant, lachrymator, sensitizer) and appropriate care as prescribed by the MSDS should be taken when handling them. All monomer synthesis steps should be performed within a chemical fume hood.
Monomer synthesis
Acetovanillone (Acros Organics, 102421000)
N,N-Dimethylformamide (DMF; Mallinckrodt, 492908)
Ethyl 4-bromobutyrate (Sigma-Aldrich, 167118-50G)
Potassium carbonate (anhydrous, Fisher, P208500)
Deionized water (DI water, 18 MΩ-cm, Barnstead NANOpure II)
Nitric acid (Mallinckrodt, 2704-46)
Ethanol (EtOH, absolute, Sigma-Aldrich, E7023 or similar)
Sodium borohydride (Sigma-Aldrich, 452882) CAUTION This reagent reacts violently with water and produces hydrogen gas. It should be handled with extreme caution in a fume hood.
Trifluoroacetic acid (TFA, Sigma-Aldrich, T62200) CAUTION This reagent is very corrosive and should be handled with extreme caution in a fume hood.
Deuterated chloroform (CDCl3, Cambridge Isotope Laboratory, DLM-7-100)
Deuterated dimethylsulfoxide ((CD3)2SO, Cambridge Isotope Laboratory, DLM-10-50)
Dichloromethane (DCM, anhydrous, Acros, AC348465000)
Triethylamine (TEA, Sigma-Aldrich, T0886)
Acryloyl chloride (AC, Sigma-Aldrich, A24109) CAUTION This reagent is very corrosive and a lachrymator and should be handled with extreme caution in a fume hood.
Alumina (adsorption, Fisher, A540500)
Sodium bicarbonate (Fisher, S233500)
Hydrochloric acid (HCl, Mallinckrodt, H613-45)
Sodium chloride (NaCl, Fisher, S271-3)
Acetone (Mallinckrodt, 244016)
N-methylpyrrolidone (NMP, Applied Biosystems, 400580)
2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU, Anaspec, 21001)
1-hydroxybenzotriazole (HOBt, Anaspec, 21003; may be no longer available but can be purchased through Fisher Scientific, NC9342953)
N,N-diisopropylethylamine (DIEA, Anaspec, 27221)
Poly(ethylene glycol)-bis-amine (average molecular weight (Mn) ∼ 3400 g/mol; Laysan Bio, NH2-PEG-NH2-3400-5g)
Ethyl ether (Fisher, E1384)
Amino acids for peptide synthesizer (Anaspec): glycine (G, 20128), arginine (R, 20312), aspartic acid (D, 20008), and serine (S, 20136)
DCM (standard, Mallinckrodt, 488108)
Triisopropylsilane (TIPS, Sigma-Aldrich, 233781)
TFA (spectrophotometric grade, Sigma-Aldrich, 302031; use with HPLC solvents)
Acetonitrile (HPLC grade, Sigma-Aldrich, 34851)
α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich, C8982-10×10)
Gel synthesis
Hydrogen peroxide (Fisher, H325100)
Sulfuric acid (Fisher, A298212)
3-acryloxypropyltrimethoxysilane (Gelest, SIA0200)
Poly(ethylene glycol) diacrylate (PEGDA, synthesized per [53]; can also be purchased from Laysan Bio, ACRL-PEG-ACRL)
Poly(ethylene glycol) monoacrylate (PEGA, Mn ∼ 400 g/mol, Monomer-Polymer & Dajac Labs, Inc, 9357)
Ammonium persulfate (AP, Acros, AC401165000)
Tetramethylene diamine (TEMED, Sigma-Aldrich, T9281-50mL)
PolyFluor™ 570 (Methacryloxyethyl Thiocarbonyl Rhodamine B) (MeRho, Polysciences, Inc., 23591)
Fibronectin (FN, Becton Dickinson, 354008)
Sterile Dulbecco's phosphate buffered saline (PBS, Invitrogen, 14190144)
Human mesenchymal stem cells (hMSCs, isolated from bone marrow)54
hMSC growth media54
LIVE/DEAD Cytotoxicity Assay, which contains calcein and ethidium homodimer (Invitrogen, L-3224)
Dish soap (for cleaning glassware)
Acetone
alamarBlue reagent (Invitrogen, DAL1100)
CellTracker green
Phenol-red-free Dulbecco's Modified Eagle Medium (Invitrogen, 11054-020)
Dulbecco's Phosphate Buffered Saline (PBS, Invitrogen, 14190)
Biopsy punch (Acuderm Acu-Punch 5 mm, Fisher, NC9151828)
Equipment
Round-bottomed flasks, one-necked
Addition funnel with gas purge
Temperature-controlled hotplate (e.g., IKA RCT Basic Safety Control Magnetic Stirrer, Fisher, 14-261-100)
Pipetteman with non-sterile and sterile plastic tips (P2, P20, P200, and P1000)
Glass funnel
Solid addition funnel
Metal cannula (18-gauge)
Separatory funnels
Glass frit filter (medium)
Rotary evaporator and water bath (e.g., Yamato RE47 and Yamato BM100)
Schlenk line and argon source
Desiccator
UV-visible (UV-vis) spectrophotometer (Lambda 40 UV/VIS Spectrophotometer, PerkinElmer)
NMR (Varian Inova 500 MHz NMR Spectrometer)
Centrifuge (e.g., Thermo Electron Corporation Centra CL3R)
Dialysis tubing (SpectraPor 7, molecular weight cutoff ∼ 1000 g/mol, Spectrum Labs, 132105)
Lyophilizer
Peptide synthesizer (433A Peptide Synthesizer, Applied Biosystems)
Glass reaction vial for peptide coupling (Fisher, CG-1861-13)
High performance liquid chromatograph (HPLC, e.g., Waters Delta Prep 4000)
Matrix-assisted laser desorption/ionization-mass spectrometer (MALDI-MS, PerSeptive Biosystems)
Staining chamber
Non-reactive plastic chamber
Rubber gasket/spacer (254-μm thick (0.01 inches), McMaster-Carr, 87315K62; 500 μm-thick, McMaster-Carr, 3788T21)
Glass microscope slides (25 mm × 75 mm, Corning, 2947-3×1; 50 mm × 75 mm, Corning, 2947-75×50)
Glass cover slips (22 mm × 22 mm, #2, Fisher, 12-546-1)
Binder clips
Sterile syringes and needles (3, 5, and 10 mL syringes; 18 gauge needles)
Sterile filters
UV-visible light source (mercury arc lamp, Novacure 1000 (or equivalent), EXFO) CRITICAL Use of the appropriate irradiation conditions, uniform intensity and wavelength, is important for achieving consistent gel property tuning under the conditions given in this protocol. For this reason, we recommend using the listed light source, or similar light source from EXFO, with appropriate filters, light guide, and collimating lens.
Band pass filters (365 nm, EXFO, 019-01006; 400-500 nm, EXFO, 019-01008; 320-500 nm, EXFO, 019-01011; 405 nm, MELLES GRIOT, 03FIM002) CRITICAL These filters easily fit within the EXFO lamp system or on the recommended collimating lens for irradiating samples with different wavelengths of light.
Liquid-filled light guide (5 mm × 1000 mm, EXFO, 805-00002) CRITICAL This light guide allows sample irradiation in different spatial orientations, for example from the top, side, or bottom of a sample in a glass slide chamber or tissue culture plate, and easy variation of the light intensity at the sample by varying the distance of the light guide from the sample surface.
Collimating lens (Large lens with adjustable aperture (shown here), EXFO, 810-00014; Small lens, EXFO, 810-00017) CRITICAL Light collimation is critical for uniform gel degradation over the irradiated sample area. This collimating lens simply fits on to the end of the light guide to achieve relatively even intensity over the spot area and uniform gel degradation in the x-y plane.
Radiometer (International Light, Model IL1400A)
Syringe pump (e.g., Harvard Apparatus 44)
Chrome photomask (PhotoSciences, Inc.)
Optical microscope (e.g., Nikon Eclipse TE2000-S)
Confocal microscope with region of interest (ROI) scanning capability, high numerical aperture objective, and 405, 488, and 543 nm lasers (e.g., Zeiss LSM 710, N.A. > 0.75)
Buchner funnel
Qualitative filter paper
Glassware (round-bottomed flasks, Erlenmeyer flasks, beakers, dishes)
Magnetic stirrer and stir bars
Vacuum oven
Thermometer
Mortar and pestle
Glass frit filter
Rotary evaporator
Separatory funnel
Hemocytometer
Scintillation vial
Heat gun
Syringe pump
Foam board
Aluminum foil
ImageJ software
Equipment Setup
Glassware
All glassware used in described reactions are washed with soap and water, rinsed with water, cleaned in a base bath, rinsed with water, neutralized in an acid bath, rinsed with water and DI water, air dried, rinsed with acetone, blown dry with air, and dried in an 80°C oven for at least 1 h to overnight.
HPLC
Peptide purification is performed on a C18 preparatory column using a gradient of 5:95 acetonitrile:water to 95:5 with 0.1% (vol/vol) TFA over 70 min at 20 mL/min. Peptide elution is monitored with UV absorbance at 220 nm and 280 nm.
Procedure
Monomer synthesis (Timing ∼ 2 weeks)
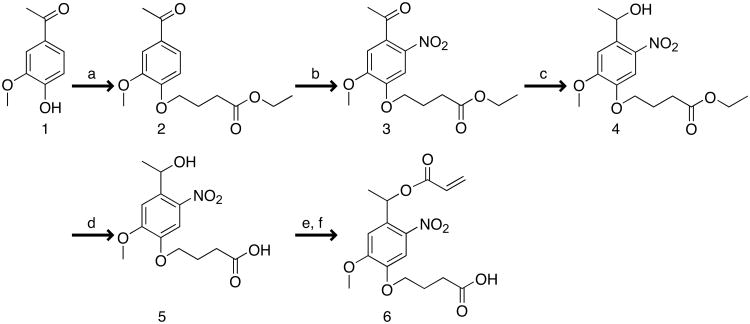
Photodegradable acrylate: synthesis of photolabile group (reaction a Fig. 3, Steps 1-3, Timing ∼ 24 h). The sequential reactions for synthesis of the photodegradable acrylate monomer are shown in Fig. 3 and are covered in Steps 1-34. Please note that the photolabile precursor also can be purchased (EMD NovaBiochem, hydroxyethyl photolinker) and Steps 1-19 omitted. Add acetovanillone (Compound 1, Fig. 3) (30 g, 180.5 mmol), DMF (150 mL), and ethyl 4-bromobutyrate (31 mL, 217 mmol) to a single-necked round-bottomed flask (500 mL) equipped with a gas adapter and magnetic stirrer. While stirring, purge the vessel with argon gas (10 min). Add potassium carbonate (37.4 g, 271 mmol), forming a suspension. Stir at room temperature (∼ 20°C) overnight under argon.
In a large beaker or Erlenmeyer flask (2000 mL) with magnetic stir bar, add DI water (1500 mL). While stirring, pour the reaction mixture into the DI water to dissolve remaining potassium carbonate and precipitate product. Stir at room temperature for 2 h. Stop stirring, and cool to 4°C for 4 h to overnight to fully precipitate the product.
Filter solution through a Buchner funnel with qualitative filter paper using a filter flask under vacuum. Pull air through the white powder product to dry (1-2 h). Transfer powder to a glass dish or weigh boat, and finish drying in a vacuum oven at 40°C overnight. Verify product (Compound 2, Fig. 3) purity with 1H NMR in CDCl3. Briefly, weigh out ∼ 6 mg of the sample, dissolve it in ∼ 0.6 mL of the deuterated solvent, pipette the solution into an NMR tube, and measure and analyze its contents55, 56. Typical reaction yield is 97%.
Photodegradable acrylate: synthesis of photolabile group (reaction b Fig. 3, Steps 4-12, Timing ∼48 h). Split the product from Step 3 in half (2× 25 g), and nitrate each portion in a separate reaction as described below.
-
For each portion, add nitric acid (70 mL) to a single-necked round-bottomed flask (1000 mL) with magnetic stir bar and thermometer. Cool the flask on ice until the nitric acid is less than 5°C.
CRITICAL STEP Heat transfer between the reaction vessel and the ice bath is important for maintaining the prescribed reaction temperature. Use of a round bottom flask that is at least 10 times the reaction volume.
-
Grind the powder product from Step 3 (24.5 g, 87 mmol) with a mortar and pestle to break apart any large clumps of powder. Add the powder in small portions (∼ 3 spatula scoops) to the nitric acid from Step 5. Allow each portion to dissolve in the nitric acid before adding the next portion (∼ 5 min between each addition for a total addition time of ∼ 1 h).
CRITICAL STEP Nitration of multiple positions of the aromatic ring can create an explosive material. Do not attempt to nitrate more than 50 g of material at a time. In addition, reaction yields have been best with 25 g batches as it is easier to maintain the reaction temperature at this scale.
-
Once all powder has been added, remove the ice bath, and heat the open reaction vessel with a water bath to 32°C. Check the reaction temperature every 5 min. When the reaction temperature reaches 33-35°C, place it back on ice until the mixture is cooled below 20°C. When the reaction temperature is below 20°C, return it to the 35°C water bath. Repeat this process until the total time of the reaction in the water bath is 1 h. The reaction should be yellow to orange/red.
CRITICAL STEP The reaction temperature is critical for nitration of the ortho position of the aromatic ring. Do not leave the reaction for more than 5 min without checking its temperature. If the reaction temperature is too low (never reaches 30°C), then the reaction will be incomplete. If the reaction temperature exceeds 35°C-40°C, then multiple positions will be nitrated on the ring, which makes purification difficult and is dangerous due to the explosive nature of trinitro aromatic compounds.
-
Add the reaction mixture drop-wise to chilled DI water (1050 mL at 4°C) on ice in a large beaker (2000 mL) with magnetic stir bar, stirring vigorously. To prevent further reaction during this time, keep the reaction mixture on ice during the precipitation.
CRITICAL STEP Dissolution of the nitric acid into water occurs slowly. Due to the density of the nitric acid, the reaction mixture can sink to the bottom of the beaker with precipitation subsequently occurring around the stir bar. Slowly add the reaction mixture to the DI water. If significant clumping begins, stop adding the reaction mixture for 30 min while the nitric acid phase dissipates; keep the reaction mixture on ice during this time.
Stir the precipitate mixture for 1 h at room temperature. The precipitate should be yellow to orange. Place the precipitate mixture at 4°C overnight.
Filter the precipitate mixture through a medium glass frit filter on a filter flask while pulling vacuum. Rinse recovered product on filter with cold DI water (4°C). Pull air through the filter for several hours to dry the product.
Re-crystallize the product from ethanol. Bring absolute EtOH (∼ 700 mL) to a boil in an Erlenmeyer flask. Add powder product to a beaker (1000 mL) with magnetic stir bar, and add boiling EtOH while stirring until powder dissolves (usually less than 500 mL of EtOH). If any brown clumps remain, carefully gravity filter (qualitative filter paper on glass funnel) the solution while hot to remove these insoluble impurities. Turn off the heat, stop the magnetic stirring, and let the solution and hot plate together cool to room temperature. Some crystal formation may be observed here. TROUBLESHOOTING?
-
Place re-crystallization solution at 4°C overnight for complete crystallization. Filter the suspension through a medium glass frit filter and pull air through the filter to dry the yellow fine powder product (∼ 1 h). Verify product (Compound 3 Fig. 3) purity with 1H NMR in CDCl3. Typical reaction yield is 60%.
CRITICAL STEP If little crystallization is observed, then too much EtOH may have been added; evaporate the EtOH using a rotary evaporator, and repeat the re-crystallization. In addition, if NMR shows an impure product (e.g., nitration of different ring positions), repeat re-crystallization (Steps 11-12). TROUBLESHOOTING?
Photodegradable acrylate: synthesis of photolabile group (reaction c Fig. 3, Steps 13-16, Timing ∼ 36 h). Add absolute EtOH (515 mL) and the nitrated product (30.9 g, 95 mmol) to a single-neck round-bottomed flask (1000 mL) with magnetic stir bar, gas adapter for argon purge, and water bath. Purge the reaction for 10 min with argon. Heat the reaction to 38°C using the water bath to improve dissolution of the powder in EtOH.
-
Add sodium borohydride (2.25 g, 59 mmol) in portions to the reaction solution (adding a portion every 5 min over 1 h). Bubbles of gas should be observed forming in the reaction solution. Stir at 38°C overnight. Reaction turns a deep orange/red.
CRITICAL STEP Hydrogen gas is evolved during this reaction. The argon purge should be vented through a Schlenk line. Do not have open flames in the fume hood and avoid significantly larger batches.
Add DI water to a large beaker or Erlenmeyer flask (5000 mL) with magnetic stir bar. Slowly pour the reaction solution into the beaker. Light yellow precipitate should form. After 30 min, place precipitate suspension at 4°C overnight.
Filter suspension to recover product. Dry in vacuum oven at 40°C overnight. Verify product (Compound 4 Fig. 3) purity with 1H NMR in (CD3)2SO. Typical reaction yield is 60%.
Photodegradable acrylate: synthesis of photolabile group (reaction d Fig. 3, Steps 17-19, Timing ∼ 36 h). Grind product from Step 16 to a fine powder (18 g, 55 mmol). Add finely ground powder and DI water (450 mL) to an Erlenmeyer flask with magnetic stir bar on a hot plate. Add TFA (45 mL), and heat suspension to ∼ 90°C, heating the reaction mixture but not boiling. CAUTION Use care and proper personal protective equipment when handling TFA. It is corrosive and causes severe burns.
Add additional TFA after 8 h (22.5 mL), and react overnight. Add additional TFA (22.5 mL), and react for 4 h, until brightening of the reaction mixture is not observed upon TFA addition. A light yellow powder in suspension should remain.
Cool to room temperature. Filter with a medium glass frit filter, and rinse with a small amount of chilled DI water (4°C). Dry in vacuum oven at 40°C overnight. Verify product (Compound 5 Fig. 3) purity with 1H NMR in (CD3)2SO. Typical reaction yield is 63%.
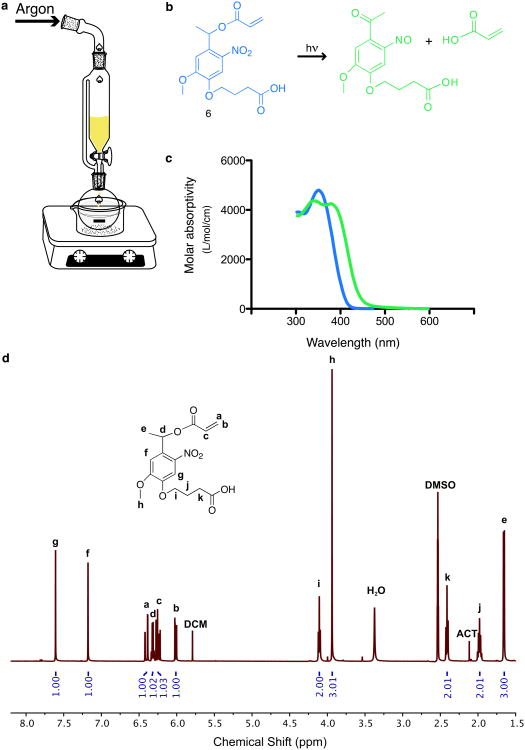
Photodegradable acrylate: acrylation of the photolabile group (reactions e and f Fig. 3, Steps 20-34, Timing ∼ 48 h). Add the photolabile group (4 g, 13 mmol, from Step 19 or purchased) to a single-neck round-bottomed flask (250 mL) with magnetic stir bar, addition funnel with gas purge, and septum (Fig. 4a).
Using two 18-gauge needles (one for gas inlet from a Schlenk line and one as gas outlet to the fume hood), purge the reaction vessel with argon (∼ 10 min).
Remove the argon inlet from the reaction vessel septum. Use argon administered from a Schlenk line to transfer anhydrous DCM (∼ 155 mL) air-free to the addition funnel through an 18-gauge metal cannula inserted into the reaction vessel septum.
Return the argon purge to the addition funnel, and use the addition funnel to add DCM (∼ 120 mL) to the round bottom flask.
Gently lift the addition funnel off of the round bottom flask and inject TEA (7.45 mL, 53 mmol). Quickly replace the addition funnel on the round bottom flask. The photolabile group should completely dissolve. Continue argon purge.
Inject AC through the addition funnel septum (3.26 mL, 43 mmol) into the remaining DCM.
-
Place the round bottom flask in an ice bath (∼ 15 min). Add the AC/DCM solution drop-wise to the round bottom flask (∼ 1 drop every 5 s). The acryloyl chloride will react with both the alcohol and carboxylic acid on the photolabile group. Keeping the reaction on ice reduces undesired side reactions.
CRITICAL STEP As the volume of solution in the addition funnel changes, the stopcock position may need to be adjusted to maintain the proper drop rate. If the reaction solution begins to turn brown, undesired side reactions may be occurring because the reaction is locally hot; to combat this, slow down the drop rate of AC solution.
React at room temperature overnight. Triethylamine salts, white crystals, may form in the reaction. Reaction mixture should be orange/brown.
Concentrate the reaction mixture by evaporating the DCM on a rotary evaporator (do not heat above 35°C), precipitating triethylamine salts.
Filter reaction mixture over alumina in a Buchner funnel with qualitative filter paper to remove triethylamine and triethylamine salts. Lightly rinse the solid residue (filtrand) with additional DCM.
Add filtered reaction solution (100 mL) to a separatory funnel and wash (1:1) sequentially with sodium bicarbonate solution (5 w/v% aqueous) to remove any unreacted photolabile group, dilute HCl (0.1 M aqueous) to remove any remaining triethylamine, and brine (10 % w/v NaCl aqueous) to remove water. After each wash, allow layers to fully separate (∼ 30 min); the DCM phase with product is the dense, lower layer. Brine can be added to water phase to improve separation. Add the intermediate product solution to a single-neck round-bottomed flask (500 or 1000 mL).
Evaporate the DCM using a rotary evaporator. Make an acetone:water mixture (50:50, 400 mL total), and add to round bottom flask with magnetic stir bar to dissolve product. Stir this mixture overnight at room temperature to cleave any anhydride linkages between the acrylate and the carboxylic acid end group that were formed during the acrylation reaction. The reaction mixture should become translucent.
Evaporate the acetone from the reaction solution using a rotary evaporator (do not heat above 45°C). The orange liquid product will begin to separate from the remaining aqueous solution.
Add the aqueous solution to a separatory funnel and extract the product with DCM (4× 100 mL), using brine to improve separation of the DCM and water phases.
Concentrate the product solution by rotary evaporation (∼ 100 mL). Wash this solution (1:1) sequentially with dilute HCl (0.1 M aqueous) and brine. Dry the product solution over magnesium sulfate (∼ 30 min or less), filter, and evaporate the solvent. Verify photodegradable acrylate monomer (Compound 6 Fig. 3, Fig. 4b) product absorbance with a UV-vis spectrophotometer (Fig. 4c) and purity with 1H NMR in (CD3)2SO (Fig. 4d). Typical reaction yield is 53%.
Photodegradable crosslinker: reaction (Steps 35-38, Timing ∼ 48 h). Add PDA (2.08 g, 5.9 mmol, from Step 34) to a scintillation vial with NMP (10 mL). Vortex and lightly heat with a heat gun to dissolve. Add HBTU (2.45 g, 6.5 mmol) and HOBt (0.87 g, 6.5 mmol) to the vial and rinse down with NMP (5 mL). Repeat vortexing and heating to begin dissolution.
Add PEG-bis-amine (2.5 g, 0.73 mmol) to a scintillation vial with NMP (15 mL). Vortex and lightly heat with a heat gun to dissolve.
Add PDA, HBTU, and HOBt in NMP from Step 35 to a single-neck round-bottomed flask with magnetic stir bar and gas adapter. Rinse scintillation vial with small amount of NMP (∼ 5 mL) to insure all is added. Add DIEA (2.05 mL, 11.8 mmol) and stir reaction solution, noting color change from orange to red/brown indicating activation of the PDA carboxylic acid (5 min). Add PEG-bis-amine in NMP from Step 36, rinsing scintillation vial with NMP (∼ 5 mL), and purge the reaction with argon (photodegradable reaction Fig. 5a).
Stir the reaction overnight. Precipitate in ice cold ethyl ether using eight 50 mL centrifuge tubes, each containing 40 mL of ether and evenly split reaction solution added dropwise. Vortex solution, balance weights, and centrifuge (5 min at 4°C with 1000 g). Decant off ether and repeat ether wash two times. Place centrifuge tubes under vacuum overnight to remove any remaining ether. Some NMP will remain with the precipitate, making it a soft solid.
Photodegradable crosslinker: purification (Step 39, Timing ∼ 3 days). Dissolve the product in DI water (100 mL), vortexing and stirring to break apart any clumps. A cloudy orange suspension will be formed. Centrifuge this mixture (4× 30 min at 4°C with 1400 g) to sediment out the precipitated HBTU/HOBt by-products. Decant off the translucent yellow/orange product solution. Dialyze the solution against DI water (5 L) for 3 days at room temperature, refreshing the DI water 3× per day. Additional precipitate may sediment over this time to the bottom of the dialysis tubing. TROUBLESHOOTING?
Photodegradable crosslinker: drying (Step 40, Timing ∼ 3 days). Recover the translucent, dialyzed product solution, freeze, and lyophilize until a dry powder is observed (∼ 2-3 days). Verify product purity (Compound 7 Fig. 5a) with 1H NMR in (CD3)2SO (modification of PEG with PDA ∼ 85-90%) (Fig. 5b). A few trace impurities may be observed in the NMR spectrum, such as residual water that is difficult to remove from PEG-based products or ether. Typical reaction yield is 70%.
-
Photoreleasable tether: SPPS reaction (Step 41, Timing ∼ 24 h). Using a peptide synthesizer or following standard SPPS procedures57, we recommend synthesizing the peptide sequence Glycine-Arginine-Glycine-Aspartic Acid-Serine-Glycine (GRGDSG) on resin. Do not cap the final amine terminus of the peptide.
PAUSE POINT Peptide on resin can be stored at -20°C for a few days prior to PDA coupling.
Photoreleasable tether: PDA coupling (Steps 42-46, Timing ∼ 24 h). In a small peptide reaction vessel or a scintillation vial, swell resin in DCM (10 mL) for 30 min.
Rinse with DCM (3× 5 mL). Rinse with NMP (3× 5 mL). Remove NMP.
Add the PDA (0.353 g, 1 mmol, from Step 34), HBTU (0.417 g, 1.1 mmol), HOBt (0.149 g, 1.1 mmol), and NMP (∼ 2 mL) to a scintillation vial. Intermittently vortex and gently heat to dissolve. Add DIEA (0.348 mL, 2 mmol). Vortex and allow to activate for 5 min; a color change should be observed. Add the reaction mixture to the reaction vial containing the resin. Rinse the scintillation vial several times with NMP to insure that all of the solution is added such that the total amount of NMP does not exceed 10 mL (photoreleasable reaction Fig. 5a).
Stir the reaction overnight at room temperature. Remove the reaction solution, retaining the resin. Wash the resin with NMP (3× 5 mL) and DCM (3× 5 mL).
Using the ninhydrin assay57, verify that all amines on the resin have been reacted. If amines are still present, repeat Steps 43-46.
-
Photoreleasable tether: cleavage from resin (Steps 47-48, Timing ∼ 24 h). Add 95% (vol/vol) TFA, 2.5% (vol/vol) TIPS, and 2.5% (vol/vol) DI water to the vial containing the resin and react for 1 h to cleave the peptide.
CRITICAL STEP This cleavage time is shorter than typically prescribed for peptide cleavage (> 2 h) and is used to maintain the ester bond of the PDA. This cleavage time may need to be adjusted for different peptide sequences, as amino acid deprotection times vary.
Remove the cleavage solution from the resin and precipitate in ice cold ethyl ether in two 50 mL centrifuge tubes. Pellet the precipitate by centrifugation (5 min at 4°C with 1000 g), and decant to remove the ether. Wash the peptide twice with cold ethyl ether and desiccate overnight under vacuum.
Photoreleasable tether: purification (Steps 49-50, Timing ∼ 24 h). Purify the peptide by HPLC using a gradient of 5:95 acetonitrile:water to 95:5 with 0.1% (vol/vol) TFA over 70 min at 20 mL/min on a C18 preparatory column. Monitor peptide elution with UV absorbance at 220 nm, where the peptide backbone strongly absorbs, and 280 nm, where the photolabile group strongly absorbs. Collect the product eluting at ∼ 35 min, where the collected effluent is a translucent yellow and strongly absorbs at 280 nm indicating the presence of the PDA. The exact elution time may vary by several minutes depending on the column type and exact amount of TFA added to the HPLC solvents.
Mix an aliquot of the purified product solution (∼ 1 μL) with the matrix molecule α-cyano-4-hydroxycinnamic acid solution (∼ 1 μL), and verify peptide purity with MALDI-MS (Fig. 5c [MP+H]). The MALDI laser can photolytically cleave the PDA, so a small amount of photodegraded peptide may also be observed in the MALDI-MS trace (Fig. 5c [MD+H]).
Photoreleasable tether: drying (Step 51, Timing ∼ 3 days). Freeze the purified peptide (Compound 8 (Fig. 5a)) solution in 50 mL centrifuge tubes and lyophilize until a dry powder is observed (2-3 days). Store the powder at -20°C until use. Typical reaction yield is ∼ 50%.
Figure 3. Photodegradable acrylate monomer synthetic scheme.
Synthetic route for preparing photodegradable acrylate. Reagents and conditions: (reaction a) DMF, ethyl-4-bromobutyrate, Ar, K2CO3; (reaction b) HNO3, ∼ 5 °C → ∼ 32 °C; (reaction c) EtOH, Ar, NaBH4, 38 °C; (reaction d) DI H2O, TFA, ∼ 90 °C; (reaction e) Ar, DCM, TEA, AC, 0 °C → room temperature; (reaction f) acetone, DI H2O. Each chemical compound is numbered (Compounds 1-6) for its identification in the experimental procedure.
Figure 4. Photodegradable acrylate.
(a) The reaction set-up for the acrylation of the photolabile group. The photolabile group is added to a round-bottomed flask with DCM and a stir bar and chilled to 0 °C in an ice bath. TEA is added to the round-bottomed flask while AC is mixed with DCM in an addition funnel under Argon purge. The AC/DCM (yellow) is added dropwise to the round-bottomed flask. (b) In response to light (hν), the photodegradable acrylate (blue, Compound 6) undergoes irreversible cleavage, producing the cleaved photolabile group and acrylic acid (green). (c) The molar absorptivities of photodegradable acrylate (blue) and the cleaved photolabile group (green) from 300 to 600 nm. (d)1H NMR shifts for the photodegradable acrylate monomer for synthetic verification. Peaks labeled a-k represent the parts of the molecule as shown in the inset molecular structure, and solvent impurities are noted by abbreviations DCM for dichloromethane, H2O for water, DMSO for dimethylsulfoxide, and ACT for acetone. To quantify acrylation of the photolabile group, protons associated with the acrylate group (a, b, and c) can be compared to those associated with the photolabile group, including the aromatic ring (f and g), the ether-linked carbon (h), and the carbon adjacent to the ester (d).
Figure 5. Macromolecular monomer synthetic schemes.
(a) Synthetic route for preparing the photodegradable crosslinker (Compound 7) and photoreleasable tether (Compound 8). (photodegradable) NMP, HBTU, HOBt, DIEA, and PEG-bis-amine. (photoreleasable) NMP, HBTU, HOBt, DIEA, and RGDS peptide. Each chemical compound is numbered (Compounds 6-8) for its identification in the experimental procedure. (b)1H NMR shifts for the photodegradable crosslinker for synthetic verification. A few minor impurities are observed, including residual water and traces of solvents such as ether. The modification of PEG-bis-amine with the photodegradable acrylate can be quantified by comparison of PEG backbone peak integration (peak m) with peaks specific to the PDA and its coupling, including the protons associated with the acrylate group and its nearest carbon neighbor (peaks a, b, c, and d), the aromatic ring (peaks f and g), the amide linkage (peak l), and the PEG repeat unit next to the PDA end groups (peak mend). (c) MALDI-MS spectrum for the photoreleasable tether for synthetic verification. MP is the mass of the photoreleasable tether, and MD is the mass of the photolytically-degraded photoreleasable tether, which is created during the measurement technique by irradiation and cleavage with the MALDI-MS ionizing laser.
Hydrogel synthesis (Timing ∼ 8 h to 48 h)
52. Acrylation of glass cover slips (Steps 52-55, 5-24 h). Clean glass cover slips (22 mm × 22 mm) in a solution of 75 mL of hydrogen peroxide and 75 mL of sulfuric acid for ∼ 1 h. CAUTION This solution of hydrogen peroxide and sulfuric acid is extremely reactive and should be prepared in an empty chemical fume hood free of solvents and flammables.
53. Rinse the cover slips well with DI H2O to remove the cleansing solution. Place the cover slips on a Kimwipe and rinse the first side with acetone. Let the acetone evaporate for ∼ 10 min; flip the cover slips and rinse the other side with acetone.
54. While the second dose of acetone evaporates, place two 20 mL scintillation vials in a Teflon chamber with a screw-on lid. Fill two ½ dram scintillation vials with 60 μL of acryloxypropyltrimethoxysilane each and place them within the 20 mL scintillation vials in the Teflon chamber. Once the acetone has evaporated, place the cover slips in the Teflon chamber standing vertically on a microscope slide holder.
55. Purge the Teflon chamber with argon for ∼ 5 min. Seal the chamber, and place it in an oven at 60 °C for 3 – 12 h.
56. Synthesis of photodegradable and photoreleasable gels for cell culture (Steps 56-58, Timing ∼ 3-24 h). Prepare sterile stock solutions of PEGdiPDA (20 w/w% in DI H2O), PEGDA (20 w/w% in DI H2O), PEGA (40 w/w% in DI H2O), photoreleasable tether (100 mM in DI H2O), ammonium persulfate (AP, 2 M in DI H2O assuming no volume contribution from the solid AP), and TEMED (2 M in DI H2O). If you will be fabricating photodegradable gels for cell encapsulation, prepare a stock of fibronectin (1 mg/mL in PBS), or if you will be imaging the gels, prepare a stock of methacrylated rhodamine (MeRho, 1.7 mM sterile filtered).
-
57. Prepare hydrogels using option A for photodegradable hydrogels and option B for photoreleasable hydrogels.
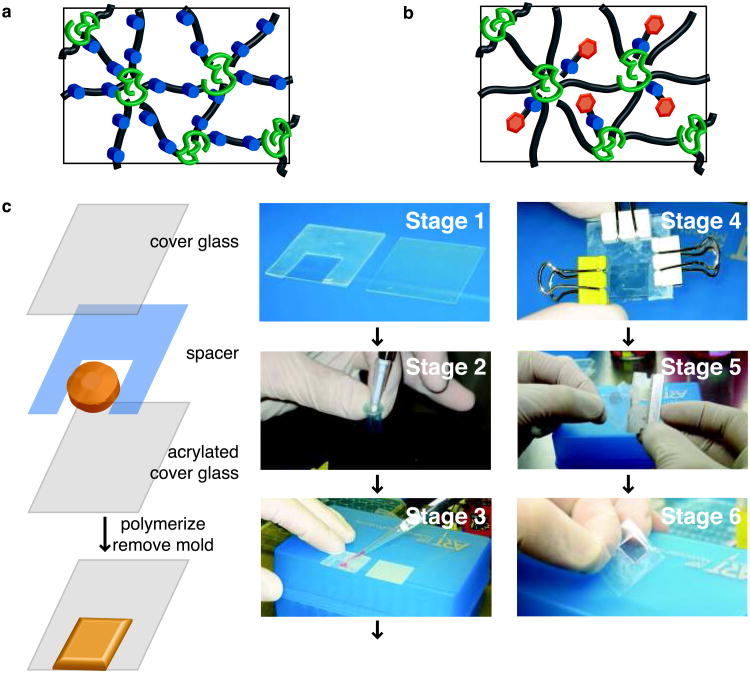
Option A: Photodegradable gels.- To prepare 40 μL photodegradable hydrogels, sufficient to fill a 1 cm × 1 cm × 254 μm geometry, combine 16.4 μL of the PEGdiPDA stock solution, 6.8 μL of the PEGA solution, and 10.8 μL PBS for each gel (Fig. 6a).
CRITICAL STEP: If you plan to fluorescently image these gels (e.g., for examining gel patterning), substitute 7.5 μL of the PBS with MeRho solution (for a final concentration of 300 μM), and if you plan to seed cells on or encapsulate cells within these gels, substitute 5.3 μL of the PBS with fibronectin solution. If you plan to both image and encapsulate or seed, substitute the PBS with 5.3 μL of fibronectin solution and 5.8 μL of MeRho solution.- ii) Add 4 μL of the AP solution for each gel to the macromer solution prepared in Step 57Ai and aliquot the macromer solution into 38 μL volumes into individual 0.7 mL Eppendorf tubes for each gel to be made.
CRITICAL STEP Once the AP has been added to the macromer solution, it should be used within ∼ 30 min. Further, at this point the initial properties of the gel, e.g., crosslinking density and biofunctionality, can be tuned by altering the concentrations of the components in the macromer solution, entrapping other proteins than fibronectin, or incorporating other acrylated/methacrylated moieties. If the amount of photodegradable monomer is altered, the final concentrations of AP and TEMED will need to be adjusted since the o-nitrobenzylether moiety is an effective radical scavenger. As the concentration of photodegradable crosslinker is increased, the concentration of AP and TEMED should be increased in order to achieve gelation, and as the concentration of photodegradable crosslinker is decreased, the concentration of AP and TEMED should be decreased so that the solution does not gel too rapidly. As adjustments are made, the changes in volumes should be supplemented with PBS to maintain the total volume at 40 μL per gel. TROUBLESHOOTING?
Option B: Photoreleasable gels.- To prepare 40 μL photoreleasable gels, combine 16.4 μL of the PEGDA solution, 6.8 μL of the PEGA solution, 4 μL of the photoreleasable tether solution, and 5.4 μL of PBS for each gel (Fig. 6b).
CRITICAL STEP: If you plan to fluorescently image the presence of the photoreleasable tether within these gels (e.g., for examining spatially-specific tether removal), a fluorescently-tagged photoreleasable tether should be used as described in [8].- ii) Add 2 μL of the AP solution for each gel to the macromer solution prepared in Step 57Bi and aliquot the macromer solution into 39 μL volumes into individual 0.7 mL Eppendorf tubes for each gel to be made.
CRITICAL STEP Once the AP has been added to the macromer solution, it should be used within ∼ 30 min. Further, at this point the initial properties of the gel, e.g., crosslinking density and biofunctionality, can be tuned by altering the concentrations of the components in the macromer solution. If the amount of photodegradable monomer is altered, the final concentrations of AP and TEMED will need to be adjusted since the o-nitrobenzylether moiety is an effective radical scavenger. As the concentration of photoreleasable tether is increased the concentration of AP and TEMED should be increased in order to achieve gelation and as the concentration of photoreleasable tether is decreased the concentration of AP and TEMED should be decreased so that the solution does not gel too rapidly. As adjustments are made, the changes in volumes should be supplemented with PBS to maintain the total volume at 40 μL per gel. TROUBLESHOOTING?
-
58. Prepare hydrogels without encapsulated cells using Option A, which can be utilized for gel patterning without cells or for 2D cell culture, or with encapsulated cells, which can be utilized for 3D cell culture, using Option B.
Option A: Hydrogel formation without encapsulated cells.- Prepare an acrylated cover slip with a 254 μm silicon gasket (Fig. 6c Stage 1), a non-acrylated cover slip, three small binder clips, and a P200 pipetteman set to 40 μL with a tip.
- Whether making photodegradable or photoreleasable gels (Step 57 Option A or B), take one of the 0.7 mL Eppendorf tubes with an aliquot of macromer solution. While mixing the macromer solution vigorously, add 2 μL of TEMED for photodegradable crosslinker gels and 1 μL of TEMED for photoreleasable tether gels (Fig. 6c Stage 2). Take the solution to the prepared cover slip mold from Step 58Ai and pipette the 40 μL solution into the center of the void in the silicon gasket (Fig. 6c Stage 3). Cover the solution with the non-acrylated cover slip, close the mold, and clamp with the binder clips (Fig. 6c Stage 4).
CRITICAL STEP Once the TEMED is added, the macromer solution will begin to gel within ∼ 30 s, so the transfer to the prepared mold should be rapid.
iii) Allow the solution to gel in the mold for ∼ 5 min. Upon gelation, open the mold carefully with a razor blade (Fig. 6c Stage 5) and place the formed gel (Fig. 6c Stage 6) into ∼ 4 mL of PBS (or enough to fully cover the gel) to allow unreacted monomer and initiator to diffuse out. Refresh the PBS after ∼ 30 min, 4 h, and 12-24 h. To study how dynamic cell-material interactions influence cell function in 2D culture, cells of interest can be seeded on photodegradable gels with entrapped fibronectin at this step as described in [26] and Step 59Bvi or on photoreleasable tether gels [unpublished AMK and MWT]
PAUSE POINT The gels should remain attached to the cover slip for 10-14 days and can be used for degradation or cell seeding at any time. To maintain sterility, keep the gel submerged in sterile PBS in an incubator for the duration of use.
Option B: Hydrogel formation with encapsulated cells and cell viability assessment.- All operations described in this section should be performed using sterile technique. Prepare the base monomer solution as described in Step 57A and add fibronectin (225 nM) to promote cell adhesion within photodegradable hydrogels, or prepare the base monomer solution described in Step57B without the addition of fibronectin for photoreleasable tether hydrogels. Prepare a rectangular mold from two glass microscope slides and a 254-μm-thick medical-grade silicon rubber gasket.
- Aspirate media from hMSCs on 100-mm dishes. Rinse the plated cells with PBS (5 mL per plate) to remove any remaining media and incubate with 1× trypsin (4 mL per plate for ∼ 6 min at 37°C) to detach the cells from the plate. Quench the trypsin with fresh media (2 mL per plate); collect the detached cells; and count an aliquot of cells (2× 10 μl) with a hemacytometer while the collected cells are pelleted by centrifugation (5 min at room temperature with 200 g).
- Resuspend the cells in a small amount of medium (∼1 ml), and take an appropriate aliquot to achieve a cell density of 2 × 106 cells per ml in the monomer solution. Pellet this aliquot of cells by centrifugation (5 min at room temperature with 200g) in an Eppendorf tube (1.7 ml), remove the medium and resuspend in the monomer solution (Fig. 7a).
CRITICAL STEP Take care to gently triturate cells in the monomer solution to break apart any cell clusters and suspend cells individually within the gel for probing cell-material interactions.- iv) Similar to Step 58Aii, add TEMED to the monomer-cell solution while vortexing, and pipette the polymerizing solution quickly into the glass mold.
CRITICAL STEP Since redox initiation is used, the polymerization begins as soon as TEMED is added, and molding gels within this time constraint (< 1 min before the onset of gelation) may take practice.- v) After 5 min of polymerization, submerge the mold in media and open it with a sterile razor blade. Remove the top glass slide, and transfer to a well of a 6-well plate (∼ 4 mL media per well) using sterile metal spatulas. Replace the media in the wells after 30 min, 4 h, and 24 h to ensure that all unreacted monomer and initiators components are removed.
Option A: LIVE/DEAD cytotoxicity assay.- Briefly, prepare solution with 1 mL serum-free, phenol-red-free DMEM, 0.5 μL calcein solution, and 2 μL ethidium homodimer.
- Remove media from the gel, and cover the gel with the dye solution (∼ 0.5 mL per gel in a 48-well plate).iii. Incubate the gel in the dye solution for 25 min at 37°C.
- Remove the dye solution, replace with fresh serum-free, phenol-red-free DMEM, and incubate the gel for 5 min at 37°C.
- Remove the media from the gel, replace it with PBS, and image the gel immediately using a confocal microscope equipped with 488 nm and 543 nm lasers and 10× or 20× magnification objective, where live cells are fluorescently stained green throughout their cytoplasm and dead cells red in their nuclei. Typically, image a 200+ μm thick z-stack with slices every ∼ 5 μm so that the overall viability of cells within the gel is sampled and that a cell body ∼ 10 μm in diameter is captured within 2 z-slices. For easy analysis and comparison of the cell viability within the imaged volume, create a single z-projection of the entire image stack (Fig 7a).
Option B: alamarBlue metabolic activity assay.- Punch at least 3 5-mm diameter disks out of the gel slab formed in Step 58Bv using a biopsy punch, and transfer each disk to a well of a 96-well plate with 150 μL of media.
- Dilute the alamarBlue reagent 1:10 in media.
- Exchange the media on each disk with 150 μL of the 1:10 alamarBlue:media solution, and incubate overnight under standard cell culture conditions, a sterile 37°C incubator with 5% CO2.
- Transfer 100 μL of the digested alamarBlue solution from each well to the well of a black 96-well plate.
- Refresh the media on each disk, and return to the incubator.
- Measure the fluorescence in each well of the black 96-well plate using a 560 nm excitation filter and a 590 nm emission filter on a plate reader.
- Repeat steps iii-vi for each metabolic activity time point, typically on Day 0 or 1, every 3 days in culture, and 24 h after any perturbation in the culture conditions, such as sample irradiation, to assess initial cell metabolism/viability, on-going viability, and viability in response to an acute insult, respectively.
CRITICAL STEP Please note that metabolic activity decreases when cells are encapsulated within a three-dimensional gel as compared to their activity on a culture plate. This is likely due to the confining nature of the gel environment decreasing proliferation and related cell functions. A typical result for cell metabolic activity in photodegradable hydrogels with and without irradiation is shown in Fig. 7b, where irradiation appears to slightly increase metabolic activity at 2 h post irradiation but not significantly at 24 h post irradiation.
-
59. Hydrogel degradation and patterning (Timing ∼ 1 to 3 h). Spatiotemporal hydrogel degradation and patterning can be achieved as follows, using option A (flood irradiation for z-gradient formation), option B (photolithography for pattern formation), option C (photolitography for gradient formation), or option D (degradation with a confocal microscope):
Option A: Hydrogel degradation via flood irradiation for z-gradient formation.- Set-up a 365 nm light source, such as an EFOS/EXFO NovaCure or OmniCure, with a liquid filled light guide and collimating lens. Tune the intensity such that the intensity is 10 mW/cm2 at the sample height. Collimate the light spot with the collimating lens so that the intensity is even over the sample.
CRITICAL STEP It is important that the light intensity is even over the entire sample, and this light setup is a simple, flexible way to achieve this.- ii) Put a gel with encapsulated cells between two glass slides (50 mm × 75 mm) spaced by a 500 μm rubber gasket and fill the void with PBS or serum-free, phenol-red-free media. Place the chamber under the light source such that the sample is directly below the center of the light spot.
- iii) Turn the light source on and irradiate the sample for up to 8 min to create a z-direction gradient in crosslinking density and modulus up to 100 μm deep within the gel.
CRITICAL STEP The length of irradiation dictates the steepness of the crosslinking density gradient as described in [27], thus the user should tune the exposure time based on the desired crosslinking density range or gradient steepness.- iv) Submerge the gel in PBS or media to allow the degradation products to diffuse out of the gel. After ∼ 30 min, refresh the PBS or media.
- v) Cell spreading within the gel can be imaged in brightfield or in fluorescence, if the cells are labeled with CellTracker Green prior to encapsulation or if the cells are stained in situ with calcein per Step 58Bvi. Cell spreading in response to crosslinking density can be quantified using ImageJ and correlated with the z-position-dependent crosslinking density (ρx normalized to its undegraded, initial value ρxo, table, Fig. 7c), as described in [27].
Option B: Hydrogel degradation with photolithography for gradient formation.- Repeat Steps 59Ai-ii except using a gel without encapsulated cells (Fig. 8a).
CRITICAL STEP If cells are attached to the surface of the gel, special care should be taken when transferring the gel between liquids.- ii) Attach an opaque rectangle, such as a piece of foam board (Fig. 8b), to the drive shaft of a syringe pump, such that the foam board will move with the drive shaft. Align the edge of the foam board to be flush with one edge of the gel so that all of the gel is initially exposed to the light source.
- iii) Set the syringe pump diameter to 1.1283 mm such that an infuse rate of 1 μL/min will be equal to movement of 1 mm/min. Start the syringe pump at the same time as the light source shutter is opened. Irradiate the sample for up to 8 min; this will insure that 8 mm of the gel will be covered by the end of the irradiation and that a gradient in degradation will be created over the first 8 mm of the gel.
- iv) Repeat Step 59Aiv.
- v) Cells can be seeded on the gels before or after (Fig. 8c) gradient generation to probe the effect of modulus and/or modulus gradient steepness on cell function.
- vi) To seed cells on gels, place each hydrogel of interest from Step 58A in an individual well of a 6-well sterile culture plate (non-tissue culture treated or ultra-low adhesion) and incubate with medium. Trypsinize cells from a standard tissue culture plate as described in Step 58Bii, and count the number of cells within the cell suspension during centrifugation. Remove the media-trypsin mixture from the cell pellet by gentle aspiration, and re-suspend the pellet in 1 ml of media. Take an aliquot of this concentrated cell suspension, and dilute it in media so that ∼ 3 mL of cell suspension will be added to each well of the 6-well plate and achieve a desired cell seeding density, which varies by cell type and experiment from ∼ 1,000 to 50,000 cells/cm2. Aspirate off media from gel samples, and cover each with 3 mL of diluted cell suspension. Incubate cells under standard tissue culture conditions, allowing cells to attach overnight before additional sample processing.
- vii) To change the sample modulus during culture, irradiate 2 samples at the time within the 6-well culture plate for up to 5 min. To maintain sterility during irradiation, leave the plate lid on during irradiation and increase the light intensity by ∼ 1.5 to 3 mW/cm2 (i.e., instead of 10 mW/cm2 use 11.5 to 13 mW/cm2) to compensate for light absorption by the culture plate lid. Cover wells not receiving light at that moment with aluminum foil. If the modulus was changed prior to cell seeding, proceed with planned culture experiment without additional irradiation. For example, valve fibroblasts, VICs, can be seeded at 40,000 cells/cm2 on gradient moduli samples (right Fig. 8c), their activation in response to modulus assessed with immunostaining (middle Fig. 8c), and their migration in response to the modulus gradient examined with real-time tracking (left Fig. 8c) as described in [26].
Option C: Hydrogel degradation with photolithography for pattern formation.- Repeat Step 59Ai (Fig. 8a).
- Place a gel on a glass slides (25 mm × 75 mm) and pipette ∼ 150 μL of PBS or serum-free, phenol-red-free media onto the gel surface. Place three cover slips on either side of the gel to support the chrome mask. Place the sample directly below the center of the light spot.
CRITICAL STEP If cells are attached to the surface of the gel, special care should be taken when transferring the gel between liquids.- iii) Rest a chrome mask with the desired pattern on the stacks of cover slips adjacent to the sample such that the pattern is covering the surface of the gel.
- iv) Turn the light source on and irradiate the sample for at least 8 min to create features at the surface of the gel (Fig. 8d).
CRITICAL STEP The length of irradiation dictates the depth of feature formation as described in [27], thus the user should tune the exposure time based on the desired depth of feature formation. Feature depth can be quantified with profilometry or confocal microscopy (Fig. 8e).- v) Repeat Step 59Aiv.
Option D: Hydrogel degradation with a confocal microscope.- Place a gel between two glass slides (25 mm × 75 mm) separated by 500 μm rubber gasket and fill the void with PBS or serum-free, phenol-red-free media (Fig. 9a left).
- Place the chamber with the sample on the stage of a confocal microscope with a 405 nm light source (Fig. 9a right). Find the surface of the gel by imaging the first plane that fluoresces with MeRho or by looking for dust particles or surface aberrations.
CRITICAL STEP Due to the nature of the confocal process, there is out of plane irradiation causing a loss in the resolution of the pattern. This problem exacerbates with increasing feature size (Fig. 9b, fluorescent cross-section) and the user should take this into account when generating features with the ROI software.- iv) Irradiate these samples with a 30 mW, 405 nm laser at 50% transmission (∼ 1 mW measured at sample) with a pixel dwell time of 3.15 μs (total time to scan 512 × 512 pixel2 frames in 1-μm intervals over a 50-μm stack is ∼ 40 s). The sample surface (Fig. 9b) or interior (Fig. 9c) can be irradiated for pattern formation on or within a gel.
- v) When the pattern is formed, the sample can be imaged using transmitted light or with fluorescent confocal microscopy on the same stage.
- vi) Upon completion of patterning and imaging, the sample should be removed from the confocal microscope and submerged in PBS or media.
Figure 6. Hydrogel synthesis.
(a) A schematic of the network structure of photodegradable hydrogels formed via free-radical chain polymerization of the photodegradable crosslinker: polyacrylate kinetic chains (green coils), PEG-based crosslinks (black lines), and photolabile moieties (blue circles). (b) A schematic of the network structure of photoreleasing hydrogels formed via free-radical chain co-polymerization of the non-degradable crosslinker with the photoreleasable tether: polyacrylate kinetic chains (green coils), PEG-based crosslinks (black lines), photolabile moieties (blue circles), and pendant peptides (orange hexagons). (c) Photolabile hydrogels are formed between an acrylated cover glass and a non-acrylated cover glass in a silicone rubber mold (spacer) in 6 stages. The silicon mold is placed on the acrylated cover glass (Stage 1) and set-up next to the non-acrylated cover glass. The macromer solution is initiated by adding TEMED to the solution while mixing (Stage 2), and this solution is quickly pipetted onto the acrylated cover glass (Stage 3) in the void of the mold. The non-acrylated cover glass is used to cover the solution, and the sandwich is clamped shut with binder clips (Stage 4). After ∼ 5 min, the sandwich is opened with a razor blade (Stage 5), and the formed gel (Stage 6) is placed in PBS.
Figure 7. Cell encapsulation within photodegradable hydrogels.
(a) Cells are encapsulated within photolabile hydrogels by resuspending a cell pellet in the macromer solution just before the addition of TEMED. Cell viability is verified post-polymerization using a LIVE/DEAD Cytotoxicity Assay and confocal imaging. Here, hMSCs exhibit good viability within the photodegradable hydrogel after encapsulation (live cells stained green, dead cells stained red, Day 1, confocal z-projection). Scale bar, 100 μm. (b) Cell function after irradiation and gel degradation can be assessed by measuring metabolic activity with the alamarBlue assay. Metabolic activity of hMSCs encapsulated in a photodegradable gel increases slightly upon irradiation and degradation (8 min of 10 mW/cm2 at 365 nm) as compared to photodegradable gels without irradiation (control) (p < 0.001), but the two conditions are similar ∼ 24 h after irradiation (48 h in 3D culture). (c) Irradiation of the hydrogels for up to 8 min (10 mW/cm2 at 365 nm) creates a z-gradient in degradation and gel crosslinking density (ρx)27. Cells were imaged on Day 4 in culture after irradiation for 8 min on Day 1. This 3D gradient in crosslinking density can be used to study how gel density influences cell function in 3D culture. hMSCs labeled with CellTracker Green are observed spreading in lower crosslinking density regions of the gel (right, confocal slice overlaid with brightfield) as compared to regions with high crosslinking density (left), and increasing cell area in response to this z-direction degradation gradient is observed and quantified with image analysis (table, ρx normalized to its initial, non-degraded value, ρxo)27. Scale bars, 50 μm. Copyright Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission.
Figure 8. Photopatterning with photolithography.
(a) To pattern photolabile hydrogels with standard photolithography, a mask is placed between a collimated light source and the gel surface. For proper pattern transfer, it is important that the light be well collimated with a collimating lens and that the mask be appropriately placed above the hydrogel surface. (b) Photopatterning can be used to form gradients in elasticity or biochemical signals in photolabile hydrogels via gradient masking. Briefly, the sample is placed in a PBS or medium-filled chamber, and a syringe pump is used to slowly cover the sample so that the sample receives a gradient of light, which induces a gradient in photocleavage. (c) Gradients substrate modulus in 2D can be used to screen its effect on cell functions such as differentiation (left). With modulus gradients created via photodegradation, the minimum modulus for activation of valve fibroblasts into myofibroblasts, a wound healing cell phenotype, was established and assayed by immunostaining for α-smooth muscle actin (αSMA green; nuclei blue) (middle)26. Cell migration in response to modulus can also be assayed on these substrates (right, traces of the x-y position of individual cells taken in 15-min intervals over 3 days)26. While a static gradient is shown here, the steepness and direction of the gradient can be changed temporally in the presence of cells with irradiation. Findings from 2D can subsequently be translated to 3D culture with the same gel formulation for comparison of cell response in 2D and 3D. Scale bar, 100 μm. Copyright Elsevier. Reproduced with permission. (d) Photopatterning can also be employed to generate features at the surface of photolabile hydrogels using a chrome mask. To ensure pattern transfer, use a well-collimated light source and rest the chrome mask just above the gel surface on glass slides or cover glass. (e) Here, we used a mask with square features to pattern positive square pillars on the surface of photodegradable hydrogels. Features are shown in a confocal surface image (upper left), brightfield image (upper right), and confocal cross section image (bottom). Scale bars, 100 μm.
Figure 9. 3D photopatterning with a confocal microscope.
Focused, 405 nm laser light can be used on a confocal microscope to completely erode features within photodegradable hydrogels. (a) To maintain sterility, the prepared gel is sealed within a liquid filled chamber (left, glass microscope slide with gasket cut to hold the gel; gasket is filled with PBS and the chamber sealed with a glass cover slip). The chamber keeps the gel hydrated and sterile, fits onto the stage of a confocal microscope, and allows the appropriate working distance for patterning over 100 μm into the hydrogel (right, sample ready for patterning in chamber on a confocal microscope stage). ROI software is used to raster the focal point of the 405 nm laser through defined geometries within photolabile hydrogels. (b) Here, cylindrical wells (5, 25, and 50 μm diameter, left) are degraded into the surface of a photodegradable gel and imaged in brightfield (middle, scale bar 100 μm) and confocal fluorescence (right, y-cross section, scale bar 50 μm). Such surface features can be used for cell aggregation, studying cell connectivity and migration, and manipulation of cell adhesion. (c) Further, channels can be created within photodegradable hydrogels on size scales relevant to mammalian cells to study 3D migration and connectivity. Here, we show a bifurcating channel patterned 50 μm below the gel surface as imaged in brightfield (middle, scale bar 100 μm) and confocal fluorescence (right, z-cross section, scale bar 50 μm). Note that there is non-specific degradation in the z-dimension owing to overlapping regions of out-of-focus light.
Timing
Steps 1-3, Synthesis of photolabile group (reaction a), ∼ 24 h
Steps 4-12, Synthesis of photolabile group (reaction b), ∼ 48 h
Steps 13-16, Synthesis of photolabile group (reaction c), ∼ 36 h
Steps 17-19, Synthesis of photolabile group (reaction d), ∼ 36 h
Steps 20-24, Synthesis of photodegradable acrylate (reaction e and f), ∼ 48 h
Steps 35-38, Synthesis of photodegradable crosslinker (reaction), ∼ 48 h
Step 39, Synthesis of photodegradable crosslinker (dialysis), ∼ 3 d
Step 40, Synthesis of photodegradable crosslinker (lyophilization), ∼ 3 d
Step 41, Synthesis of photoreleasable tether (peptide synthesis), ∼ 24 h
Steps 42-46, Synthesis of photoreleasable tether (coupling reaction), ∼ 24 h
Steps 47 and 48, Synthesis of photoreleasable tether (cleavage), ∼ 24 h
Steps 49 and 50, Synthesis of photoreleasable tether (HPLC purification), ∼ 24 h
Step 51, Synthesis of photoreleasable tether (lyophilization), ∼ 3 d
Steps 52-55, Hydrogel synthesis (acrylation of cover slips), 5-24 h
Steps 56-58, Hydrogel synthesis for cell culture (hydrogel fabrication without or with encapsulated cells and cell viability assessment), 3-24 h
Steps 59 Option A, Hydrogel degradation (degradation for crosslinking density z-gradient), 90 min
Steps 59 Option B, Hydrogel degradation (surface gradient formation), 90 min
Steps 59 Option C, Hydrogel degradation (surface feature formation), 2 h
Steps 59 Option D, Hydrogel degradation (confocal photopatterning), 3 h
Troubleshooting
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 11 | Fine white powder emerges upon mixing of the product with boiling EtOH | Reaction temperature was too low, and incomplete nitration occurred. | Re-start the procedure at Step 4. |
| 12 | Multiple re-crystallizations do not lead to pure product | Reaction became too hot and lead to over half of the product being nitrated in the wrong position or multiple positions on the benzyl ring. | Product can be purified by other methods, such as column chromatography, or procedure can be re-started at Step 4. |
| 39 | By-product precipitate does not completely settle during centrifugation | Particle size is small and/or remaining NMP in combination with PEG is forming a stable suspension. | Leave at least half of the volume of the dialysis tubing empty and allow dialysis to proceed for longer until solution istranslucent. |
| 57 | Gelation occurs before the solution can be pipetted into the mold | Initiator (AP & TEMED) concentrations are too high in the final macromer solution. | Decrease the final concentration of AP & TEMED in the macromer solution while maintaining a 2:1 ratio, respectively. |
| 57 | Gelation fails to occur after 5 min. | Initiator (AP & TEMED) concentrations are too low in the final macromer solution. | Increase the final concentration of AP & TEMED in the macromer solution while maintaining a 2:1 ratio, respectively. |
Anticipated Results
This protocol provides a detailed procedure for the synthesis of photolabile monomers and hydrogels and their use in 2D and 3D cell culture for dynamically controlling cell-material interactions. Anticipated NMRs for the monomer syntheses are provided below (Fig. 3, reactions a-f). Examples of 3D and 2D cell culture results are shown in Fig. 7 and Fig. 8, where in situ manipulation of gel modulus and crosslinking density has been explored for examining their effect on cell differentiation and morphology, respectively. In addition, viability and metabolic activity of hMSCs after encapsulation and irradiation are shown in Fig. 7. Last, typical examples of gel degradation in 2D and in 3D via controlled irradiation are shown in Fig. 8 and Fig. 9.
Synthesis of photolabile group (reaction a)
White solid, 1H NMR (500 MHz, CDCl3) δ 7.6 (d, 1H), δ 7.58 (s, 1H), δ 6.92 (d, 1H), δ 4.2 (tm, 4H), δ 3.95 (s, 3H), δ 3.6 (s, 3H), δ 2.58 (t, 2H), δ 2.2 (m, 2H), and δ 1.3 (t, 3H).
Synthesis of photolabile group (reaction b)
Yellow solid, 1H NMR (500 MHz, CDCl3) δ 7.62 (s, 1H), δ 6.75 (s, 1H), δ 4.18 (tm, 4H), δ 3.95 (s, 3H), δ 2.55 (t, 2H), δ 2.5 (s, 3H), δ 2.2 (m, 2H), and δ 1.25 (t, 3H).
Synthesis of photolabile group (reaction c)
Yellow solid, 1H NMR (500 MHz, (CD3)2SO) δ 7.55 (s, 1H), δ 7.4 (s, 1H), δ 5.55 (s, 1H), δ 5.3 (m, 1H), δ 4.1 (tm, 4H), δ 3.95 (s, 3H), δ 2.5 (t, 2H), δ 2.0 (m, 2H), δ 1.4 (d, 3H), and δ 1.2 (t, 3H).
Photolabile group (reaction d)
Yellow solid, 1H NMR (500 MHz, (CD3)2SO) δ 12.2 (s, 1H), δ 7.6 (s, 1H), δ 7.4 (s, 1H), δ 5.5 (s, 1H), δ 5.3 (m, 1H), δ 4.1 (t, 2H), δ 3.9 (s, 3H), δ 2.4 (t, 2H), δ 2.0 (m, 2H), and δ 1.4 (d, 3H).
Photodegradable acrylate (reactions e and f)
Orange liquid to crystalline solid (crystallizes upon storage at 4°C), 1H NMR (500 MHz, (CD3)2SO) δ 12.2 (s, 1H), δ 7.6 (s, 1H), δ 7.2 (s, 1H), δ 6.4 (d, 1H), δ 6.35 (m, 1H), δ 6.25 (m, 1H), δ 6.05 (d, 1H), δ 4.1 (t, 2H), δ 3.9 (s, 3H), δ 2.4 (t, 2H), δ 2.0 (m, 2H), and δ 1.4 (d, 3H).
Photodegradable crosslinker
Light yellow powder, 1H NMR (500 MHz, (CD3)2SO) δ 8.0 (t, 1H), δ 7.6 (s, 1H), δ 7.2 (s, 1H), δ 6.4 (d, 1H), δ 6.35 (m, 1H), δ 6.25 (m, 1H), δ 6.05 (d 1H), δ 4.1 (t, 2H), δ 3.9 (s, 3H), δ 3.5 (m, ∼ 304H), δ 2.75 (t, 2H), δ 2.4 (t, 2H), δ 2.0 (m, 2H), and δ 1.4 (d, 3H).
Acknowledgments
The authors acknowledge the NIH (DEO12998 and DEO16523) and HHMI for funding this work. M.W. Tibbitt acknowledges the US Department of Education's Graduate Assistantships in Areas of National Need program and the National Institutes of Health (T32 GM-065103) for fellowship assistance. A.M. Kloxin acknowledges the NASA Graduate Student Researchers Program and the US Department of Education's Graduate Assistantships in Areas of National Need program for fellowship assistance. The authors thank Professor A.M. Kasko for initial guidance and contributions towards the photolabile molecule synthesis, A. A. Aimetti for discussions and training related to peptide synthesis, and J. A. Benton for assistance with VIC culture. Additionally, the authors thank the MediaLab in the Department of Biochemistry at the University of Wisconsin at Madison for use of an Adobe Illustrator template.
Footnotes
Author contributions: All authors contributed extensively to the work presented in this paper.
Competing Financial Interests: The authors declare that they have no competing financial interests.
Contributor Information
April M. Kloxin, Email: april.kloxin@colorado.edu.
Mark W. Tibbitt, Email: mark.tibbitt@colorado.edu.
Kristi S. Anseth, Email: kristi.anseth@colorado.edu.
References
- 1.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Liu SQ, et al. Synthetic hydrogels for controlled stem cell differentiation. Soft Matter. 2010;6:67–81. [Google Scholar]
- 3.Tibbitt MW, Anseth KS. Hydrogels as Extracellular Matrix Mimics for 3D Cell Culture. Biotechnol Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guilak F, et al. Control of Stem Cell Fate by Physical Interactions with the Extracellular Matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang NS, Varghese S, Elisseeff J. Controlled differentiation of stem cells. Adv Drug Deliv Rev. 2008;60:199–214. doi: 10.1016/j.addr.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf K, et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–U839. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 7.Tayalia P, Mendonca CR, Baldacchini T, Mooney DJ, Mazur E. 3D Cell-Migration Studies using Two-Photon Engineered Polymer Scaffolds. Adv Mater. 2008;20:4494–4498. [Google Scholar]
- 8.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jay SM, Saltzman WM. Shining light on a new class of hydrogels. Nat Biotechnol. 2009;27:543–544. doi: 10.1038/nbt0609-543. [DOI] [PubMed] [Google Scholar]
- 10.Lutolf MP. Spotlight on hydrogels. Nat Mater. 2009;8:451–453. doi: 10.1038/nmat2458. [DOI] [PubMed] [Google Scholar]
- 11.Yu HT, Li JB, Wu DD, Qiu ZJ, Zhang Y. Chemistry and biological applications of photo-labile organic molecules. Chem Soc Rev. 2010;39:464–473. doi: 10.1039/b901255a. [DOI] [PubMed] [Google Scholar]
- 12.Holmes CP. Model studies for new o-nitrobenzyl photolabile linkers: Substituent effects on the rates of photochemical cleavage. Journal Of Organic Chemistry. 1997;62:2370–2380. doi: 10.1021/jo961602x. [DOI] [PubMed] [Google Scholar]
- 13.Akerblom EB, Nygren AS, Agback KH. Six new photolabile linkers for solid-phase synthesis. 1. Methods of preparation. Mol Divers. 1998;3:137–148. doi: 10.1023/a:1009660431758. [DOI] [PubMed] [Google Scholar]
- 14.Zhao YR, et al. New caged coumarin fluorophores with extraordinary uncaging cross sections suitable for biological imaging applications. Journal Of The American Chemical Society. 2004;126:4653–4663. doi: 10.1021/ja036958m. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez M, et al. Single-Photon and Two-Photon Induced Photocleavage for Monolayers of an Alkyltriethoxysilane with a Photoprotected Carboxylic Ester. Adv Mater. 2008;20:4563–4567. [Google Scholar]
- 16.Pelliccioli AP, Wirz J. Photoremovable protecting groups: reaction mechanisms and applications. Photochem Photobiol Sci. 2002;1:441–458. doi: 10.1039/b200777k. [DOI] [PubMed] [Google Scholar]
- 17.Deiters A. Principles and Applications of the Photochemical Control of Cellular Processes. ChemBioChem. 2010;11:47–53. doi: 10.1002/cbic.200900529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Shoichet MS. A photolabile hydrogel for guided three-dimensional cell growth and migration. Nat Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 19.Wosnick JH, Shoichet MS. Three-dimensional chemical Patterning of transparent hydrogels. Chem Mat. 2008;20:55–60. [Google Scholar]
- 20.Ohmuro-Matsuyama Y, Tatsu Y. Photocontrolled cell adhesion on a surface functionalized with a caged arginine-glycine-aspartate peptide. Angew Chem -Int Edit. 2008;47:7527–7529. doi: 10.1002/anie.200802731. [DOI] [PubMed] [Google Scholar]
- 21.Nakanishi J, et al. Spatiotemporal control of cell adhesion on a self-assembled monolayer having a photocleavable protecting group. Anal Chim Acta. 2006;578:100–104. doi: 10.1016/j.aca.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 22.Johnson JA, Finn MG, Koberstein JT, Turro NJ. Synthesis of photocleavable linear macromonomers by ATRP and star macromonomers by a tandem ATRP-Click reaction: precursors to photodegradable model networks. Macromolecules. 2007;40:3589–3598. [Google Scholar]
- 23.Johnson JA, Baskin JM, Bertozzi CR, Koberstein JT, Turro NJ. Copper-free click chemistry for the in situ crosslinking of photodegradable star polymers. Chemical Communications. 2008:3064–3066. doi: 10.1039/b803043j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kikuchi Y, et al. Grafting Poly(ethylene glycol) to a Glass Surface via a Photocleavable Linker for Light-induced Cell Micropatterning and Cell Proliferation Control. Chem Lett. 2008;37:1062–1063. [Google Scholar]
- 25.Frey MT, Wang YL. A photo-modulatable material for probing cellular responses to substrate rigidity. Soft Matter. 2009;5:1918–1924. doi: 10.1039/b818104g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloxin AM, Benton JA, Anseth KS. In situ elasticity modulation with dynamic substrates to direct cell phenotype. Biomaterials. 2010;31:1–8. doi: 10.1016/j.biomaterials.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloxin AM, Tibbitt MW, Kasko AM, Fairbairn JF, Anseth KS. Tunable hydrogels for external manipulation of cellular microenvironments through controlled photodegradation. Adv Mater. 2010;22:61–66. doi: 10.1002/adma.200900917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhury F, et al. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat Mater. 2010;9:82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong JY, Velasco A, Rajagopalan P, Pham Q. Directed movement of vascular smooth muscle cells on gradient-compliant hydrogels. Langmuir. 2003;19:1908–1913. [Google Scholar]
- 31.Khetan S, Katz JS, Burdick JA. Sequential crosslinking to control cellular spreading in 3-dimensional hydrogels. Soft Matter. 2009;5:1601–1606. [Google Scholar]
- 32.Gillette BM, Jensen JA, Wang MX, Tchao J, Sia SK. Dynamic Hydrogels: Switching of 3D Microenvironments Using Two-Component Naturally Derived Extracellular Matrices. Adv Mater. 2010;22:686–+. doi: 10.1002/adma.200902265. [DOI] [PubMed] [Google Scholar]
- 33.Isenberg BC, et al. A thermoresponsive, microtextured substrate for cell sheet engineering with defined structural organization. Biomaterials. 2008;29:2565–2572. doi: 10.1016/j.biomaterials.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams C, et al. Aligned Cell Sheets Grown on Thermo-Responsive Substrates with Microcontact Printed Protein Patterns. Adv Mater. 2009;21:2161–+. [Google Scholar]
- 35.Yang J, et al. Reconstruction of functional tissues with cell sheet engineering. Biomaterials. 2007;28:5033–5043. doi: 10.1016/j.biomaterials.2007.07.052. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, et al. Tissue Engineering Using Laminar Cellular Assemblies. Adv Mater. 2009;21:3404–3409. doi: 10.1002/adma.200801990. [DOI] [PubMed] [Google Scholar]
- 37.Yeo WS, Yousaf MN, Mrksich M. Dynamic interfaces between cells and surfaces: Electroactive substrates that sequentially release and attach cells. Journal Of The American Chemical Society. 2003;125:14994–14995. doi: 10.1021/ja038265b. [DOI] [PubMed] [Google Scholar]
- 38.Yeo WS, Mrksich M. Electroactive self-assembled monolayers that permit orthogonal control over the adhesion of cells to patterned substrates. Langmuir. 2006;22:10816–10820. doi: 10.1021/la061212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murayama S, Kato M. Photocontrol of Biological Activities of Protein by Means of a Hydrogel. Analytical Chemistry. 2010;82:2186–2191. doi: 10.1021/ac1003757. [DOI] [PubMed] [Google Scholar]
- 40.Benoit DSW, Nuttelman CR, Collins SD, Anseth KS. Synthesis and characterization of a fluvastatin-releasing hydrogel delivery system to modulate hMSC differentiation and function for bone regeneration. Biomaterials. 2006;27:6102–6110. doi: 10.1016/j.biomaterials.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 41.Salinas CN, Anseth KS. The enhancement of chondrogenic differentiation of human mesenchymal stem cells by enzymatically regulated RGD functionalities. Biomaterials. 2008;29:2370. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong DY, Griffin DR, Reed J, Kasko AM. Photodegradable Hydrogels to Generate Positive and Negative Features over Multiple Length Scales. Macromolecules. 2010;43:2824–2831. [Google Scholar]
- 43.Sniadecki N, Desai RA, Ruiz SA, Chen CS. Nanotechnology for cell-substrate interactions. Ann Biomed Eng. 2006;34:59–74. doi: 10.1007/s10439-005-9006-3. [DOI] [PubMed] [Google Scholar]
- 44.Yamato M, et al. Temperature-responsive cell culture surfaces for regenerative medicine with cell sheet engineering. Prog Polym Sci. 2007;32:1123–1133. [Google Scholar]
- 45.Mohammed JS, Murphy WL. Bioinspired Design of Dynamic Materials. Adv Mater. 2009;21:2361–2374. [Google Scholar]
- 46.Nowatzki PJ, Franck C, Maskarinec SA, Ravichandran G, Tirrell DA. Mechanically tunable thin films of photosensitive artificial proteins: Preparation and characterization by nanoindentation. Macromolecules. 2008;41:1839–1845. [Google Scholar]
- 47.Andreopoulos FM, Persaud I. Delivery of basic fibroblast growth factor (bFGF) from photoresponsive hydrogel scaffolds. Biomaterials. 2006;27:2468–2476. doi: 10.1016/j.biomaterials.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 48.Trenor SR, Shultz AR, Love BJ, Long TE. Coumarins in polymers: From light harvesting to photo-cross-linkable tissue scaffolds. Chem Rev. 2004;104:3059–3077. doi: 10.1021/cr030037c. [DOI] [PubMed] [Google Scholar]
- 49.DeForest CA, Polizzotti BD, Anseth KS. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat Mater. 2009;8:659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hahn MS, Miller JS, West JL. Three-dimensional biochemical and biomechanical patterning of hydrogels for guiding cell behavior. Adv Mater. 2006;18:2679–+. [Google Scholar]
- 51.Nuttelman CR, Tripodi MC, Anseth KS. In vitro osteogenic differentiation of human mesenchymal stem cells photoencapsulated in PEG hydrogels. J Biomed Mater Res Part A. 2004;68A:773–782. doi: 10.1002/jbm.a.20112. [DOI] [PubMed] [Google Scholar]
- 52.Kloxin AM. Department of Chemical and Biological Engineering, Photolabile hydrogels for dynamic tuning of physical and chemical properties to probe cell-cell and cell-material interactions. University of Colorado at Boulder; Boulder, Colorado: 2010. [Google Scholar]
- 53.Salinas CN, Anseth KS. The influence of the RGD peptide motif and its contextual presentation in PEG gels on human mesenchymal stem cell viability. J Tissue Eng Regen Med. 2008;2:296–304. doi: 10.1002/term.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salinas CN, Cole BB, Kasko AM, Anseth KS. Chondrogenic differentiation potential of human mesenchymal stem cells photoencapsulated within poly(ethylene glycol)-arginine-glycine-aspartic acid-serine thiol-methacrylate mixed-mode networks. Tissue Eng. 2007;13:1025–1034. doi: 10.1089/ten.2006.0126. [DOI] [PubMed] [Google Scholar]
- 55.Pretsch E, Buhlmann P, Affolter C. Structure determination of organic compounds. 3. Springer; Berlin: 2000. [Google Scholar]
- 56.Pavia DL, Lampman GM, Kriz GS. Introduction to spectroscopy: a guide for students of organic chemistry. 3. Thomson Learning, Inc.; 2001. [Google Scholar]
- 57.Chan WC, White PD, editors. Fmoc Solid Phase Peptide Synthesis. Oxford University Press, Inc.; New York: 2000. [Google Scholar]