Abstract
A better understanding of the molecular mechanisms that regulate adipose tissue-derived stromal cell (ADSC) differentiation could provide new insight into some adipose-tissue-related disease. The differentiation of ADSCs into adipocytes is a complex physiological process that includes clonal expansion, growth arrest, and terminal differentiation. Here the role of microRNA-143 (miR-143) during ADSC adipogenic differentiation was systematically investigated. We found that miR-143 expression was transiently decreased after adipogenic induction while increased from day 3 and peaked on day 7 after induction. We show for the first time that the role of miR-143 is not consistent in the differentiation process. The regulatory role depends on the differentiation stage that miR-143 acts on. When miR-143 is overexpressed during the clonal expansion stage, the adipogenic differentiation of ADSCs is inhibited, whereas the overexpression of miR-143 during the growth arrest stage or terminal differentiation stage promotes differentiation. Further we firstly demonstrate that miR-143 plays the modulational role by directly repressing MAP2K5, a key member of the MAPKK family in the MAPK signaling pathway. These findings suggest that miR-143 plays an important role in adipose tissue formation, with special implications for some metabolic disease in which the amount and/or function of adipose tissue is altered.
The current obesity epidemic, which is partly induced by our increasingly sedentary lifestyles and high-fat diets, is expected to result in type 2 diabetes mellitus, considerable downstream morbidity, and mortality, and increasing costs to healthcare systems around the world1,2. The incidence of type 2 diabetes is increasing at a worrisome rate and is expected to double within the next few decades3. In recent years, despite advances in our understanding of obesity, the molecular mechanisms underlying individual differences in the predisposition to obesity and type 2 diabetes remain obscure.
Obesity is characterized clinically by an increased fat mass and enhanced energy storage in adipose tissue3. Increases in fat can be caused by adipocyte hypertrophy or adipocyte hyperplasia4. The adipocytes in adipose tissue are derived from adipose-tissue-derived stromal cells (ADSCs) via specialized cell-lineage differentiation processes. ADSCs are mesenchymal stem cells derived from adipose tissues, and play a key role in the generation and metabolism of adipose tissues. The processes by which ADSCs differentiate into adipocytes can be thought of as a cascade, with three primary stages. During the first stage, ADSCs are committed to become adipocyte precursor cells, and these cells then differentiate into preadipocytes. During the second stage, the preadipocytes reach confluence and undergo growth arrest, after which clonal expansion is induced. During the final stage, the cells undergo terminal differentiation, which is driven by the expression of adipocyte-specific genes, to produce the adipocyte phenotype5,6,7. Therefore, one possible way to reduce the fat mass is to therapeutically regulate the differentiation process at a certain stage or to block the transition between those stages.
MicroRNAs (miRNAs) are a class of short noncoding RNAs, about 19–22 nucleotides in length, that can bind to complementary target sites in mRNA molecules, causing translation repression or the cleavage of the target mRNAs8,9. Computational predictions suggest that 45,000 putative miRNA binding sites exist in human protein-coding genes10. However, many of these putative miRNA binding sites are yet to be validated experimentally. Over the past decade, miRNAs have been shown to play important regulatory roles in a variety of physiological processes11,12, and some miRNAs are reported to accelerate or inhibit the processes of adipocyte differentiation during adipogenesis. Thereinto, miR-143 is one of the miRNAs that has already been studied. However, although several previous studies have found that miR-143 can promote adipocyte differentiation, the details of the regulatory mechanisms are obscure, and the target gene of miR-143 has not been persuasively identified13,14.
In this study, we systematically investigated the role of miR-143 during the adipogenic differentiation of ADSCs, which naturally differentiate into adipocytes within adipose tissues. We analyzed the miRNA expression profile during ADSC differentiation and found that the expression of miR-143 decreased during the first 2 days and then increased markedly. Next, we varied the levels of miR-143 in ADSCs using a lentivirus and measured their lipid accumulation and the expression of specific adipogenesis-related genes. Cell numbers and the cell cycle were also monitored to measure cell proliferation after induction. The overexpression of miR-143 in the first 2 days after induction, i.e., during the clonal expansion stage, inhibited cell proliferation and adipogenic differentiation, whereas the overexpression of miR-143 in the days that followed, i.e., during the growth arrest stage and terminal differentiation stage, promoted adipogenesis. However, cell proliferation and the cell cycle were unaffected. Further experiments suggested that miR-143 regulates adipogenesis by directly repressing mitogen-activated protein kinase kinase 5 (MAP2K5), a key member of the mitogen-activated protein kinase kinase (MAPKK) family in the MAPK signaling pathway.
Results
Expression of miR-143 during ADSC adipogenesis and in ADSCs infected with an miR-143-expressing lentivirus
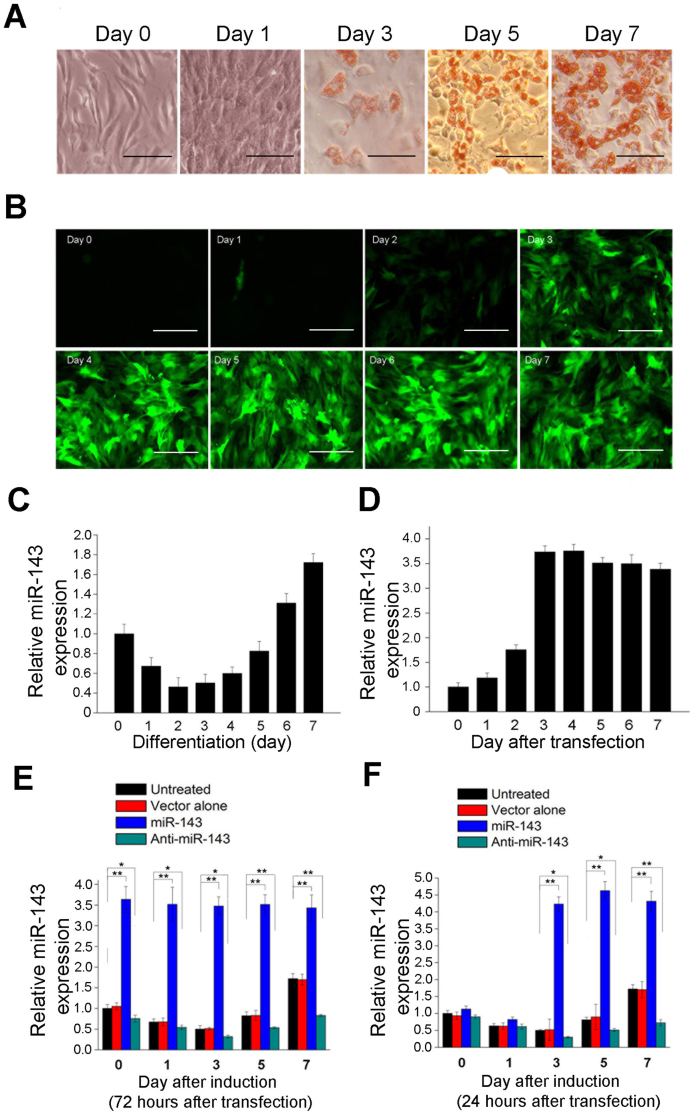
We selected miR-143 for analysis because its expression level showed a significant change between pre-induction and post-induction in a previous microRNA microarray study, and using quantitative real-time PCR (RT–qPCR), determined its expression during the whole adipogenic differentiation process of ADSCs. We found that the expression of miR-143 decreased during the first 2 days after adipogenic induction, but increased from day 3 and peaked on day 7 after induction (Figure 1A and 1C). To investigate whether miR-143 plays a role in the molecular regulation of ADSC adipogenesis, ADSCs were transfected with a lentivirus vector expressing either miR-143 or an miR-143 antagomir. The efficiency of lentivirus transduction was determined with fluorescence microscopy (Figure 1B). A preliminary test showed that the expression of miR-143 during the first 48 h was maintained at a low level, but began to increase 48 h after transduction, peaking at 72 or 96 h, and then remained at a high level until day 7 (Figure 1D). The transfected cells were then induced with a cocktail of 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone, and 5 μg/mL insulin (MDI) at 72 or 24 h after transduction. Accordingly, we found that the expression of miR-143 remained high from day 0 to day 7 after induction if the cells were induced 72 h after transduction, while the expression of miR-143 remained low in the first 2 days and increased from day 3 after induction if the cells were induced 24 h after transduction (Figure 1E and 1F).
Figure 1. Expression of miR-143 during ADSC adipogenesis and in ADSCs infected with lentiviral vector expressing miR-143.
(A) Lipid accumulation during ADSC differentiation into adipocytes was monitored by Oil Red O staining. Bar, 100 μm. (B) GFP expression in ADSCs transfected with the lentiviral vectors. Bar, 100 μm. (C) Expression of miR-143 was monitored by RT–qPCR at the time points indicated during ADSC differentiation. (D) miR-143 expression was also monitored by RT–qPCR at the time points indicated after lentiviral vectors transduction. (E) and (F) The relative expression of miR-143 was analyzed by RT–qPCR at the time points indicated after adipogenic induction, which was initiated at 72 or 24 h after lentiviral transduction. All measurements shown are the means ± SD of three independent RT–qPCR experiments. *P < 0.05, **P < 0.01 vs. untreated cells or cells transfected with the empty vector.
miR-143 inhibits ADSC differentiation when it is overexpressed during the first 2 days after induction
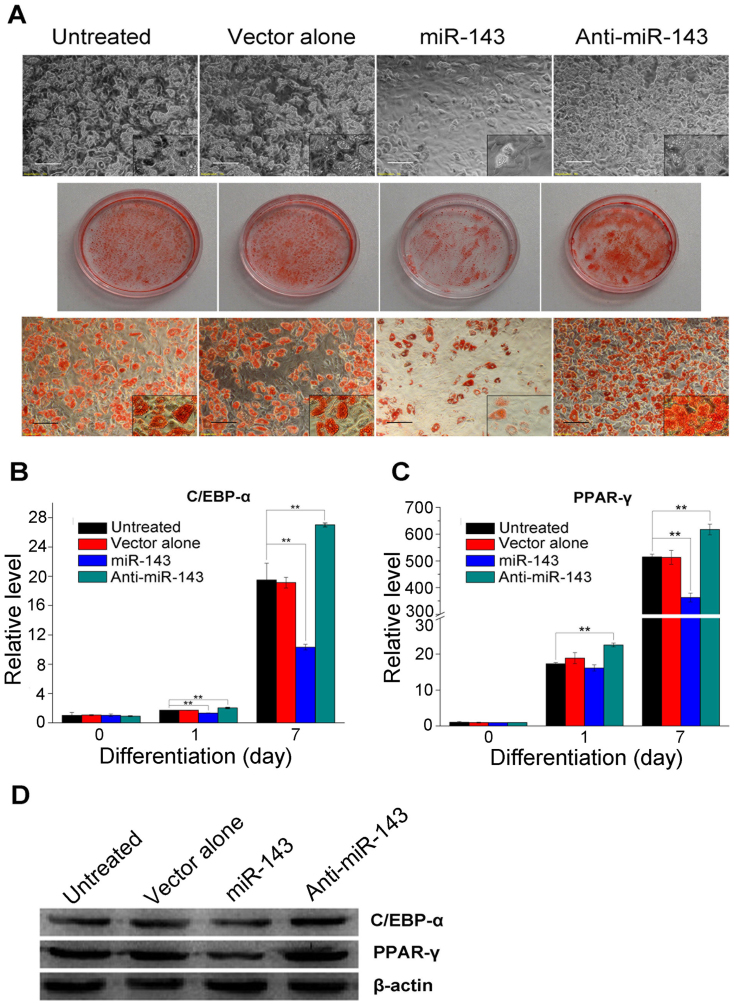
The ADSCs were adipogenic induced 72 h after lentivirus transduction, at which time the expression of miR-143 or the miR-143 antagomir peaked. Lipid accumulation was determined on day 7 after induction and the expression of two principal transcription factors involved in adipogenesis, CCAAT/enhancer binding protein α (C/EBP-α) and peroxisome proliferator-activated receptor γ (PPAR-γ), was detected with RT–qPCR on days 0, 1, and 7. These results indicate that the cells in the miR-143 group produced significantly fewer lipid droplets, whereas the cells in the anti-miR-143 group, in which the expression of miR-143 was downregulated, accumulated significantly more lipid droplets (Figure 2A). The RT–qPCR results showed that the levels of C/EBP-α and PPAR-γ mRNAs were significantly lower in the miR-143 group and significantly higher in the anti-miR-143 group (Figure 2B and 2C). Similar results were obtained with a western blot analysis on day 7 after induction (Figure 2D).
Figure 2. miR-143 inhibits ADSC differentiation when overexpressed during the first 2 days after adipogenic induction.
(A) Abundant lipid droplets formed on day 7 after adipogenic induction and were detected by Oil Red O staining. Bar, 100 μm. (B) and (C) The relative C/EBP-α and PPAR-γ mRNA levels were determined during differentiation by RT–qPCR at the time points indicated. (D) The protein levels of C/EBP-α and PPAR-γ were determined on day 7 after induction by western blot analysis. All measurements shown are the means ± SD of three independent experiments. **P < 0.01 vs untreated cells or cells transfected with the empty vector.
Overexpression of miR-143 in the first 2 days after induction impairs the clonal expansion of ADSCs
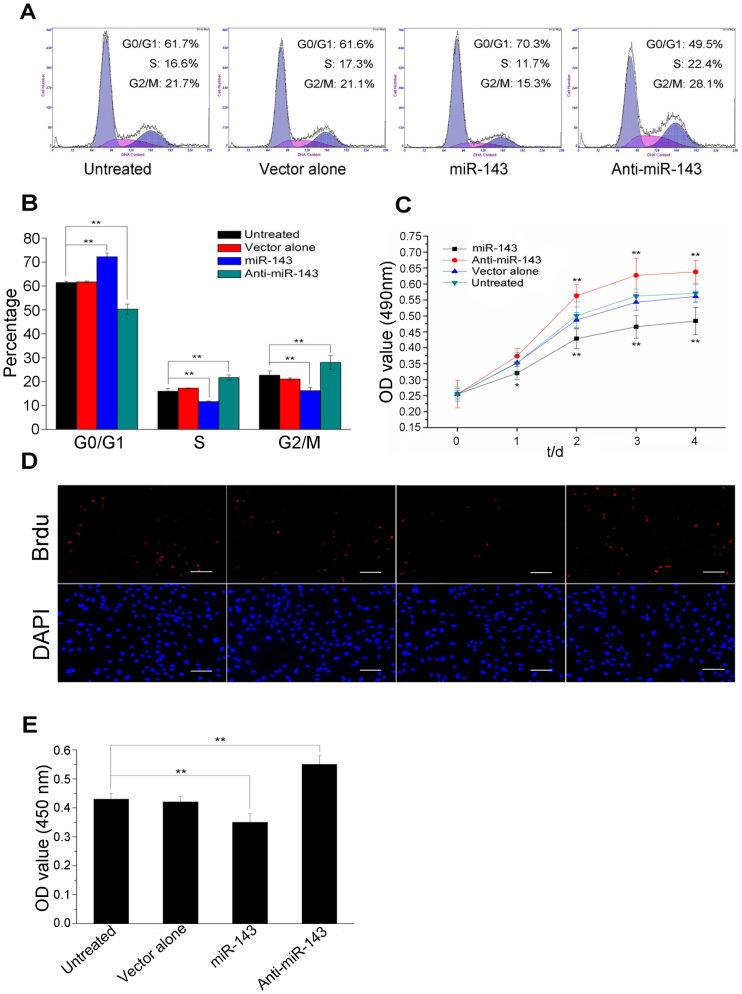
The treatment of ADSCs with an adipogenic stimulus initiates a complex sequence of events, including clonal expansion, growth arrest, and terminal differentiation. Because the overexpression of miR-143 during the first 2 days after stimulation inhibited ADSC differentiation, we postulated that miR-143 might play an important role during the clonal expansion stage. To test this hypothesis, ADSCs were transfected with a lentiviral vector expressing either miR-143 or an miR-143 antagomir and allowed to reach confluence, at which point the adipogenic program was initiated using the MDI cocktail. A cell-cycle analysis showed that the overexpression of miR-143 caused an increase in the fraction of cells in the G0/G1 phase and a reduction in the fraction in S phase (Figure 3A and 3B), indicating that miR-143 induced a cell-cycle delay. A 3-(4,5-dimethylthizol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay showed that the cell numbers decreased in the miR-143 group and increased in the anti-miR-143 group compared with the number of untreated cells or in the cells transfected with the empty vector (Figure 3C). The cell proliferation test based on bromodeoxyuridine (BrdU) incorporation showed corresponding results (Figure 3D and 3E). Taken together, these data support a role for miR-143 in blocking clonal expansion during adipogenesis.
Figure 3. Overexpression of miR-143 during the first 2 days after adipogenic induction impaires the clonal expansion of ADSCs.
(A) and (B) Cell-cycle analysis of ADSCs using flow cytometry 24 h after induction. (C) ADSC proliferation was monitored with an MTT assay for 4 days after induction. (D) Fluorescence attributable to BrdU incorporation by ADSCs. Bar, 50 μm. (E) Quantification of BrdU incorporation in cells 24 h after induction, determined with a cell-proliferation ELISA kit. All measurements shown are the means ± SD of three independent experiments. *P < 0.05, **P < 0.01 vs. untreated cells or cells transfected with the empty vector.
miR-143 promotes ADSC differentiation when miR-143 is overexpressed after clonal expansion stage
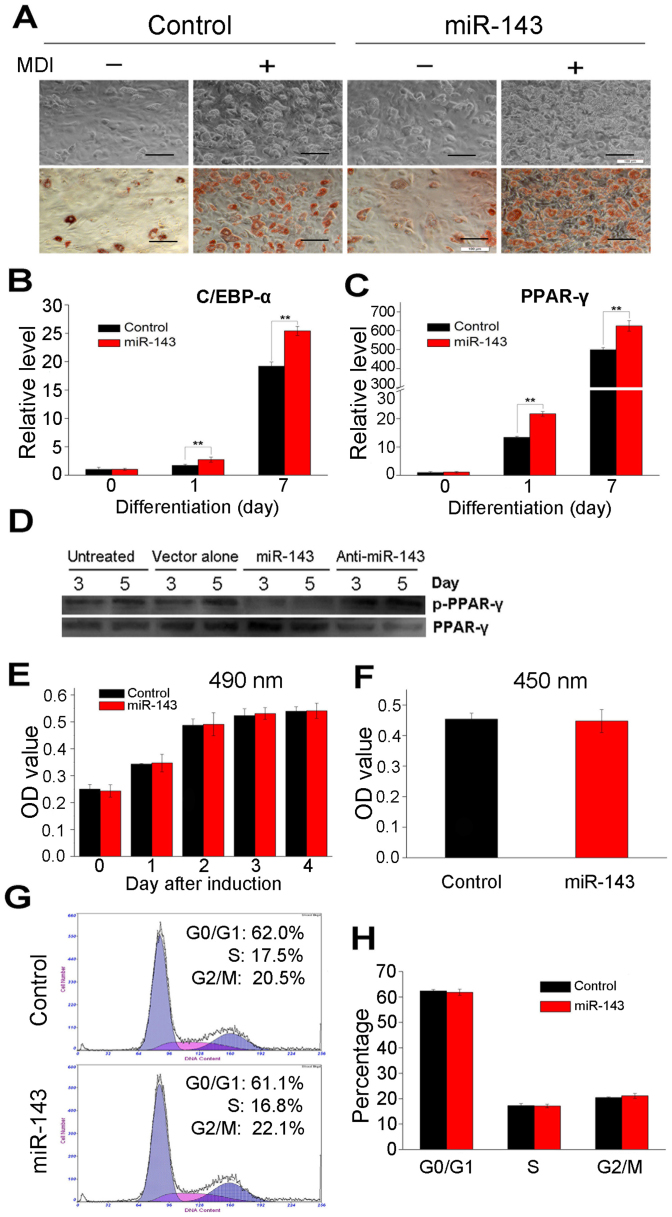
To investigate whether miR-143 plays a role after clonal expansion stage (2 days after induction), adipogenic induction was initiated 24 h after transduction. At this time, the expression of miR-143 did not increase, whereas 72 h later, it reached a peak. Interestingly, the introduction of ectopic miR-143 at this time promoted ADSC differentiation to adipocytes, measured with Oil Red O staining (Figure 4A), and the mRNA levels of C/EBP-α and PPAR-γ were significantly higher in the cells transfected with miR-143 than in the control cells (Figure 4B and 4C) on day 7 of differentiation. A western blot analysis also showed that the concentrations of active PPAR-γ were higher in the cells transfected with miR-143 than in the cells transfected with the empty vector or miR-143 antagomir, whereas the concentrations of inactive phosphorylated PPAR-γ (p-PPAR-γ) decreased (Figure 4D). To test whether miR-143 is also involved in the clonal expansion stage of adipocyte differentiation, cell proliferation and the cell cycle were monitored again. Cell numbers, BrdU incorporation, and the cell cycle were not influenced by the ectopic overexpression of miR-143 (Figure 4E, 4F, 4G, and 4H).
Figure 4. miR-143 promotes ADSC differentiation when overexpressed 2 days after induction.
(A) After reaching confluence, ADSCs infected with miR-143-expressing lentiviral vector and untreated cells were incubated with (+) or without (−) MDI for 7 days and then stained with Oil Red O. Bar, 100 μm. (B) and (C) The relative C/EBP-α and PPAR-γ mRNA levels were determined with RT–qPCR during differentiation at the time points indicated. (D) Active PPAR-γ (PPAR-γ) and inactive phosphorylated PPAR-γ (p-PPAR-γ) concentrations were determined in each group with western blot analyses on days 3 and 5 after differentiation. (E) Proliferation of the ADSCs in (A) was tested for 4 days after induction with an MTT assay. (F) BrdU incorporation by the cells in (A) was quantified 24 h after induction. (G) and (H) Cell cycle of ADSCs was analyzed by flow cytometry 24 h after induction. Data shown are the means ± SD of three independent experiments. **P < 0.01 vs. untreated cells.
miR-143 may regulate ADSC differentiation by targeting the MAP2K5–ERK5 signaling pathway
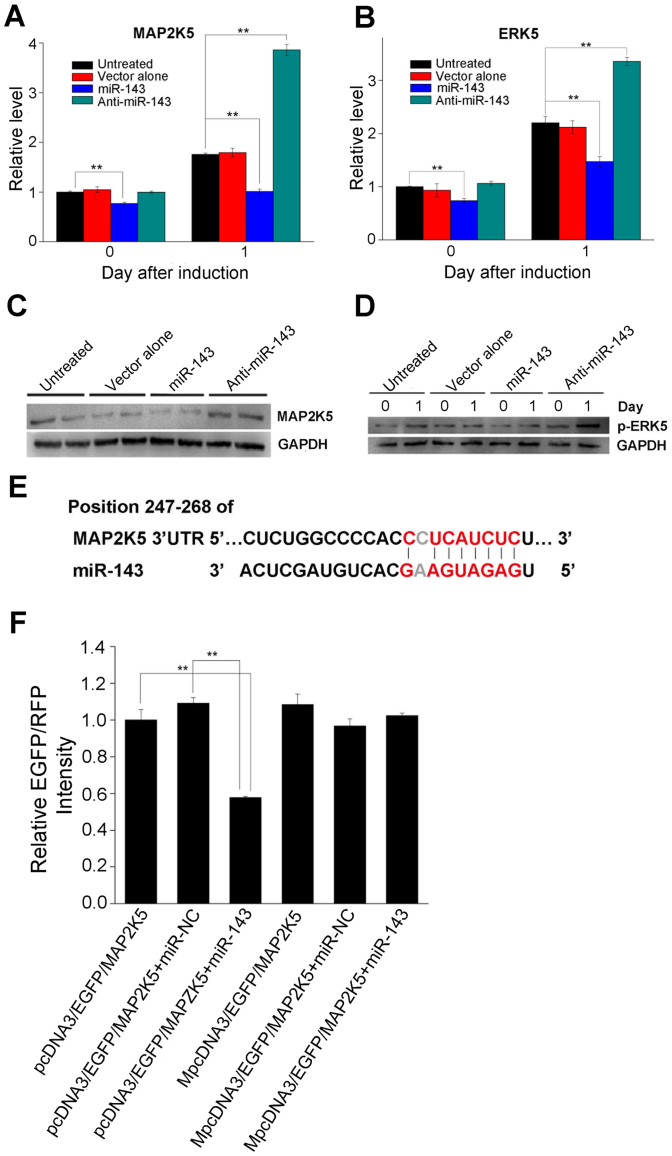
To identify the target genes through which miR-143 exerts its effects on adipocyte differentiation, bioinformatics was used to predict and rank the putative miR-143 target genes related to adipogenesis. MAP2K5, which is an activated kinase in the MAPK signaling pathway, was predicted to be a target gene of miR-143 with both the miRanda and MicroCosm target prediction programs. The mRNA levels and protein levels of MAP2K5 were determined 1 day after adipogenesis induced with the MDI cocktail. The MAP2K5 mRNA levels were significantly reduced in the miR-143 group compared with those in the untreated group or the empty-vector-treated group, whereas they were increased in the anti-miR-143 group (Figure 5A). This is consistent with the results of a western blot analysis of the protein concentrations (Figure 5C). In supplementary studies, we also examined the mRNA and protein levels of extracellular signal-regulated kinase 5 (ERK5), which is the direct downstream of MAP2K5 and known to promote cell growth and proliferation in previous studies15. Both RT–qPCR and western blot analyses indicated that ERK5 expression was downregulated in cells overexpressing miR-143 (Figure 5B and 5D). To confirm that MAP2K5 is a direct target gene of miR-143, we used an enhanced green fluorescent protein (EGFP) reporter assay to demonstrate that MAP2K5 expression is directly inhibited by miR-143. An alignment of miR-143 and the MAP2K5 3′-untranslated region (UTR) was constructed with bioinformatics and is illustrated in Figure 5E. When the 3′-UTR sequence of MAP2K5 was then cloned downstream from the EGFP-coding sequence, EGFP expression was strongly reduced when miR-143 was overexpressed. To confirm the direct interaction between miR-143 and the MAP2K5 3′-UTR, we introduced three mutations into the seed sequence of the MAP2K5 3′-UTR and found that EGFP expression from the mutated vector (MpcDNA3–EGFP–E2F3) was not affected by miR-143 overexpression (Figure 5F). Taken together, these results strongly suggest that miR-143 regulates the adipogenic differentiation of ADSCs, at least in part, by targeting the 3′-UTR of MAP2K5.
Figure 5. miR-143 regulates ADSC adipogenesis by inhibiting the MAP2K5–ERK5 signaling pathway.
(A) MAP2K5 mRNA levels were downregulated in cells infected with an miR-143-expressing lentiviral vector (miR-143 group) on days 0 and 1 following MDI induction. (B) As the direct downstream of MAP2K5, ERK5 mRNA levels were similarly downregulated in the miR-143 group. (C) MAP2K5 protein levels were evaluated by western blot analysis 1 day after incubation with MDI. (D) Phosphorylated ERK5 (p-ERK5) levels were determined by western blot analysis at the indicated time points after ADSCs were treated with MDI. (E) A sequence within the MAP2K5 mRNA that is complementary to miR-143 was identified using publicly available algorithms. (F) Cells were cotransfected with an EGFP reporter plasmid (pcDNA3–EGFP–MAP2K5) or the mutant vector (MpcDNA3–EGFP–MAP2K5) and the pDsRed–C1 plasmid, either alone or in combination with an miR-143 mimic. EGFP and RFP levels were measured 48 h after transduction with an F-4500 fluorescence spectrophotometer. The fluorescence value in the control group was set to 1. Histograms show the normalized mean ± SD fluorescence intensity of three independent experiments. All other data are shown as means ± SD. **P < 0.01 vs. untreated cells or cells transfected with the empty vector.
Discussion
Adipose tissue is not only a storage depot for triglycerides but also a functional organ with a role in regulating energy homeostasis. It is well known that abnormal fat accumulation is associated with adverse health outcomes, including obesity, type II diabetes, and cardiovascular and cerebrovascular diseases16. Therefore, a better understanding of the regulatory mechanisms of adipogenesis is crucial for the development of novel therapeutic strategies for these diseases. This is a complex physiological process, involving a large number of molecular events, and little is known about the precise mechanisms of adipogenesis. ADSCs, which act as somatic stem cells in adipose tissues, share many of the characteristics of their counterparts in the bone marrow, including extensive proliferative potential and the ability to differentiate into adipogenic, osteogenic, chondrogenic, and myogenic lineages17,18,19. Within the adipose tissues, adipocytes are derived from ADSCs via a specialized cell-lineage differentiation process, and ADSCs play a key role in the generation and metabolism of adipocyte tissues5,20.
In this study, the overexpression of miR-143 did not consistently promote or inhibit adipogenesis. When miR-143 was overexpressed during the clonal expansion stage, the adipogenic differentiation of ADSCs was inhibited. However, the expression of miR-143 increased after the clonal expansion stage, during the growth arrest stage and terminal differentiation, when differentiation was promoted. It is clear that during clonal expansion, growth-arrested cells re-enter the cell cycle and undergo several rounds of division, when DNA replication and changes in the chromatin structure increase the accessibility of cis-acting elements to trans-acting factors, triggering the expression of adipogenic-differentiation-related genes21,22. Therefore, the reason of miR-143 inhibiting adipogenic differentiation during clonal expansion stage is that the miR-143-mediated inhibition of adipogenesis is caused by a block at the clonal expansion stage. But why the overexpression of miR-143 promotes differentiation at the point at which cells switch from clonal expansion to growth arrest and terminal differentiation stage aroused our curiosity.
It is widely acknowledged that the MAPK signaling pathway plays a pivotal role in many essential cellular processes, including cell proliferation and differentiation. The role of MAPKs in the process of adipocyte differentiation is extremely complex and depends on many parameters. The adipogenic differentiation process involves several successive steps, and each step may be modulated by MAPKs. In the clonal expansion stage, DNA synthesis and cell mitotic division require the activation of the MAPK signaling pathway, but in the terminal differentiation stage, the activation of MAPKs inhibits cell differentiation via the phosphorylation of PPAR-γ23,24. Therefore, we examined whether miR-143 regulates adipogenesis by acting on the MAPK signaling pathway.
It has been suggested, although rarely, that miRNAs regulate adipogenesis by acting on the MAPK signaling pathway13,25,26. Strikingly, one study showed that the inhibition of miR-143 blocked adipogenesis and increased the protein levels of ERK5, suggesting that miR-143 targets the ERK5 gene13. However, those data showed that miRNAs regulate adipogenesis by predominantly regulating MAPKs during the growth-arrested stage after clonal expansion, whereas the clonal expansion stage is unaffected. Therefore, further studies were required to clarify the direct role of miR-143 in the clonal expansion stage. Furthermore, the real target gene of miR-143 in adipogenesis had not been persuasively identified.
ERK5 is a member of the MAPK family, and has a Thr–Glu–Tyr (TEY) sequence in its dual phosphorylation site, like ERK1/2, which plays a significant role in cell growth and differentiation. This suggests that the regulation and function of ERK5 are similar to those of ERK1/2. This was confirmed by Kato and colleagues, who demonstrated that ERK5 is even more essential than ERK1/2 for cell proliferation and cell-cycle progression in response to tyrosine kinase signaling15.The upstream kinase that directly phosphorylates ERK5 was identified as MAP2K527,28.
To determine whether miR-143 exerts its effects on adipocyte differentiation by targeting the MAPK signaling pathway, bioinformatics were used to predict and rank putative target genes related to both adipogenesis and the MAPK pathway in this study. MAP2K5 was predicted to be a target gene of miR-143 by both the miRanda and MicroCosm target prediction programs, and this was confirmed for the first time in an experimental assay in this study. Taken together, several results indicate that MAP2K5 mRNA is the direct target of miR-143. First, the 3′-UTR of MAP2K5 contains a seed region that is complementary to the miR-143 sequence. Second, the overexpression of miR-143 suppressed the expression of the MAP2K5 protein, whereas the inhibition of miR-143 enhanced MAP2K5 expression. Third, the overexpression of miR-143 in 293T cells resulted in the reduced expression of a fluorescent reporter containing the MAP2K5 3′-UTR. Fourth, specific mutations in the miR-143 target site within the MAP2K5 3′-UTR of the reporter abolished its regulation by miR-143. Therefore, in this study, we have identified the direct target gene of miR-143 and clarified the involvement of ERK5 observed in previous studies.
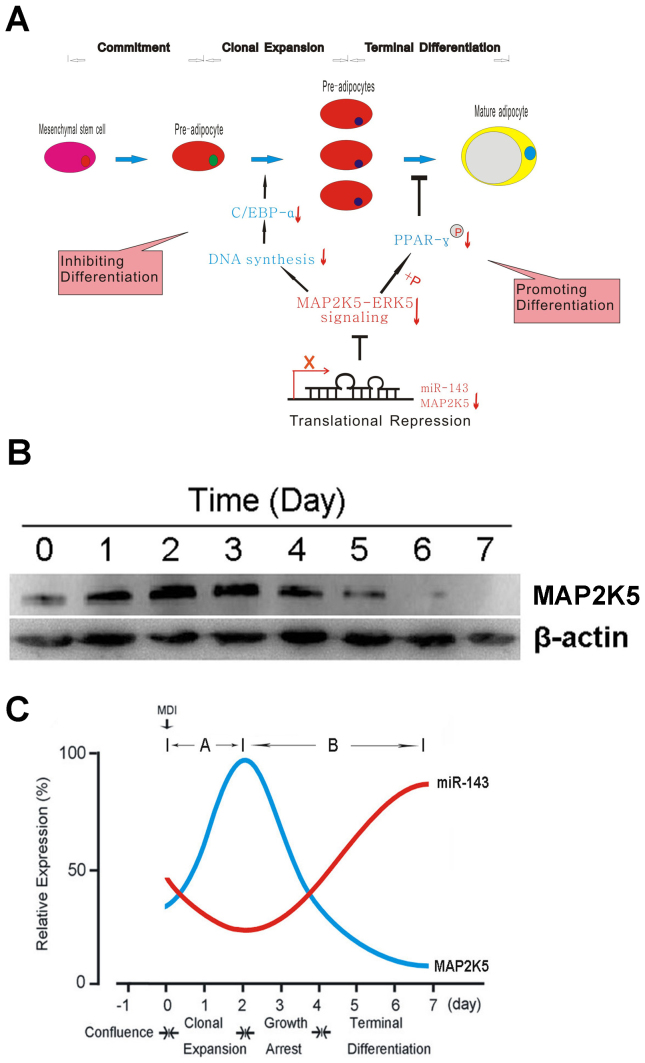
The arguments discussed above suggest an explanation of the results of this study. During the clonal expansion stage, the overexpression of miR-143 suppresses the expression of MAP2K5. As the phosphorylation substrate for MAP2K5, activated ERK5 decreases, thus blocking clonal expansion. In rapid succession, C/EBP-α that triggered by DNA synthesis and changes in the chromatin structure during clonal expansion, is suppressed, and eventually leads to the inhibition of adipogenic differentiation. During the growth arrest and terminal differentiation stages, the overexpression of miR-143 blocks the MAP2K5–ERK5 signaling pathway, so the ERK5-mediated phosphorylation of PPAR-γ is reduced. Consequently, more activated nonphosphorylated PPAR-γ protein remains, so adipogenic differentiation is ultimately promoted (Figure 6A).
Figure 6. Schematic model of miR-143 and MAP2K5 regulation during ADSC adipogenesis.
(A) ERK5, as the substrate of MAP2K5, triggers clonal expansion and activates C/EBP-α, a pleiotropic transactivator of numerous adipocyte-specific genes that contribute to terminal differentiation. Thus, the overexpression of miR-143 in this phase blocks the clonal expansion stage and then terminal differentiation by inhibiting MAP2K5. However, blocking MAP2K5-ERK5 signaling pathway by inhibiting MAP2K5 during terminal differentiation stage reduces phosphorylation of PPAR-γ and thus promotes adipogenic differentiation. (B) Protein levels of MAP2K5 during ADSC differentiation were evaluated by western blot analysis at the indicated time points after the ADSCs were treated with MDI. (C) Induction of adipocyte differentiation with MDI initially resulted in cell growth and clonal expansion, which require MAP2K5 expression and miR-143 downregulation. However, when the cells switch from clonal expansion to the growth-arrest and terminal differentiation stages, MAP2K5 expression must be inhibited, so the expression of miR-143 increases.
In a complementary study, we also examined the protein levels of MAP2K5 during ADSC differentiation. MAP2K5 was markedly induced 1 day after MDI treatment and peaked on days 2 or 3, slowly returning to the baseline level on day 5 (Figure 6B). Thus, when the data discussed above are considered together, we can see that MAP2K5 and miR-143 expression (Figure 1C) are inversely correlated. Induction of the adipogenic differentiation of ADSCs initially results in cell growth and clonal expansion, which require increased MAP2K5 expression, so the expression of miR-143 is reduced. However, when the cells switch from clonal expansion to the growth arrest and terminal differentiation stages, MAP2K5 expression must be inhibited, so the expression of miR-143 increases (Figure 6C).
In summary, we have provided evidence that miR-143 regulates adipocyte differentiation, and propose that miR-143 plays a partial role by directly targeting MAP2K5 mRNA, thereby promoting the transition from clonal expansion to terminal differentiation. Our findings suggest that miR-143 plays an important role in adipose tissue formation, with special implications for some metabolic disease in which the amount and/or function of adipose tissue is altered.
Methods
Cell isolation, culture, and differentiation
Normal female Sprague-Dawley rats (2 weeks old) were cared for and used in accordance with the International Guiding Principles for Animal Research. ADSCs were isolated from the inguinal fat tissues and validated by their multilineage differentiation potential, flow cytometry (CD29+, CD31−, and CD45−), plastic adherence, and colony-forming ability, according to the methods described in our previous studies29,30. The cells were then cultured in α-MEM with 10% fetal bovine serum in a humidified 5% CO2 atmosphere. When the cells reached 80% confluence, a cocktail of 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone, and 5 μg/mL insulin (MDI) was added to the culture medium to induce adipogenic differentiation. The induction medium was replaced every 72 h until the ADSCs had differentiated into mature adipocytes.
Oil Red O staining
When the ADSCs differentiated into mature adipocytes on day 7 following adipogenic induction, the cells were stained with Oil Red O. Briefly, the cells were washed twice with PBS and fixed in 4% paraformaldehyde at room temperature for 30 min. They were then washed three times with 70% isopropanol. Finally, the cells were stained with an Oil Red O solution (60% Oil Red O stock solution [0.5% Oil Red O in isopropanol] and 40% H2O) for 15 min and then washed three times with PBS. The stained lipid droplets in the adipocytes were observed under an inverted microscope.
Lentiviral vector construction
The miR-143 (MIMAT0000849) sequence was obtained from the miRBase database. The pGCsil-H1–CMV–GFP vector (Genechem, Shanghai, China) was linearized with AgeI and EcoRI (New England Biolabs). To create the pGC-LV recombinant vector, a fragment containing miR-143 and a fragment containing an miR-143 antagomir were individually introduced into the pGCsil-H1–CMV–GFP vector. Competent cells were then transformed with the pGC-LV recombinant vector, which was confirmed by PCR and sequencing. 293T packaging cells were concurrently transfected with the pGC-LV recombinant vector and the lentiviral packaging vectors pHelper1.0 and pHelper2.0 using Lipofectamine 2000, to produce the virus. The viral titer was determined using the hole dilution method. The lentiviral vectors were stored at −80°C until use. The following primers were used for vector construction: mir-143-AgeI, CGGGTACCGGTGACCAGTATAGGAGGCAGAC (forward); mir-143-EcoRI, CGGAATTCAAAAACCTGAGTCAAGGAACATCAC (reverse); anti-miR-143-AgeI, CCGGTTGAGCTACAGTGCTTCATCTCATTTTTG (forward); anti-mir-143-EcoRI, AATTCAAAAATGAGATGAAGCACTGTAGCTCAA (reverse); pGCsil, CGCGTGGATAACCGTATTAC (reverse); HI, GGAAAGAATAGTAGACATAATAGC (forward).
Cell transduction assay
Second-passage ADSCs were plated in 12-well plates. When the ADSCs reached the optimal infection phase (30–50% confluence), the cells of the miR-143 group were infected with 4.2 μL of miR-143-lentivirus (titer, 1.2 × 109 TU/mL; multiplicity of infection [MOI] = 50), the cells of the anti-miR-143 group with 2 μL of anti-miR-143-lentivirus (titer, 2.5 × 109 TU/mL; MOI = 50), and the cells in the empty-vector group were infected with 2.5 μL of the empty lentivirus vector (titer, 2 × 109 TU/mL; MOI = 50) in individual wells. The untreated group was not infected with lentivirus. The cell culture medium was replaced 12 h after infection. Fluorescent protein expression in the ADSCs was observed using an inverted fluorescence microscope 24, 48, 72, and 96 h after infection to assess the infection efficiency. The highest level of fluorescent protein expression occurred 72 h after transduction. Each experiment in this study was performed after the transduction of the cells, and the adipogenic induction agent (MDI) was added when the confluence of the infected cells reached 80% (after 3–4 days in culture; the cells were not passaged).
Quantitative real-time PCR
mRNA was quantified using MMLV reverse transcriptase (Epicentre) and SYBR-Green-based quantitative PCR (qPCR) with an ABI PRISM 7500 system (Applied Biosystems). The qPCR reactions were performed with initial incubation at 95°C for 10 min followed by 40 cycles of 95°C for 10 s and 60°C for 60 s. The relative expression level of each miRNA was normalized to the level of GAPDH mRNA and calculated using the ΔΔCt method. The following primer pairs were used for RT–qPCR: C/EBP-α, 5′-CGGGAACGCAACAACATC-3′ (forward) and 5′-TCCAGCGACCCTAAACCAT-3′ (reverse); PPAR-γ, 5′-CAGAGTCTGCTGATCTGCGAG-3′ (forward) and 5′-TGCAGGGGGGTGATATGTT-3′ (reverse); MAP2K5, 5′-GGAAGCAGCCCAAGGAA-3′ (forward) and 5′-CCAATCAGGCAGCAGACAG-3′ (reverse); ERK5, 5′-TAGTGGGTTGCCTGGTTCTG-3′ (forward) and 5′-GTCTGCCTGGCTATCCTGTG-3′ (reverse); and GAPDH, 5′-GGAAAGCTGTGGCGTGAT-3′ (forward) and 5′-AAGGTGGAAGAATGGGAGTT-3′ (reverse).
The miRNA concentrations were determined using TaqMan RT–qPCR. The PCR reactions were performed in a final volume of 20 μL using an ABI PRISM 7900 system (Applied Biosystems). Each reaction contained PCR primers, gene-specific stem-loop primers, and probes. The PCRs were performed with an initial incubation at 95°C for 10 min followed by 40 cycles of 95°C for 10 s and 60°C for 60 s. The absolute miRNA expression levels were normalized to an internal control (small nuclear U6) and calculated using the ΔΔCt method. The primers and probes used were as follows: miR-143, 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTGAGCT-3′ (RT primer), 5′-GGCTGAGATGAAGCACTGT-3′ (gene-specific stem-loop primer), and 5′-CAGTGCGTGTCGTGGAG-3′ (probe); and U6, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse).
Western blot analysis
Confluent ADSCs were treated under the appropriate conditions and lysed. The protein content of the lysate was determined using a protein assay kit (Bio-Rad Laboratories). The proteins were separated with 10% SDS-PAGE and transferred to cellulose membranes. The membranes were incubated with the following primary antibodies: anti-MAP2K5 (1:500; Bioss), anti-phospho-ERK5 (1:200; Bioss), anti-CEBP-α (1:200; Sigma-Aldrich), anti-PPAR-γ and anti-phospho-PPAR-γ (1:500; Sigma-Aldrich), anti-β actin (1:1000; Santa Cruz Biotechnology), and anti-GAPDH (1:1000; Santa Cruz Biotechnology). Following overnight incubation with the primary antibodies, the membranes were immunoblotted with a horseradish-peroxidase-conjugated secondary antibody (1:5000; Santa Cruz Biotechnology). Immunoreactivity was visualized with enhanced chemiluminescence (ECL kit; Santa Cruz Biotechnology) and analyzed using the Quantity One system (Bio-Rad).
MTT assay and BrdU incorporation assay
An MTT assay was used to detect cell proliferation. On days 0, 1, 2, 3, and 4, ADSCs were transferred to 96-well plates and incubated for 2 h with MTT (5.0 mg/mL) at 37°C, followed by centrifugation. The supernatants were then removed, and 200 μL of dimethyl sulfoxide was added. The absorbance at 490 nm was measured. BrdU labeling was used to detect DNA synthesis. ADSCs from each group were cultured in 96-well plates and treated with BrdU (30 μg/mL) 24 h after induction. After the cells were labeled with BrdU for 2 h, the medium was removed and BrdU incorporation was measured using the BrdU Cell Proliferation ELISA Kit (Abcam). The reactions were stopped with 1 M H2SO4. Finally, the cells were examined under an inverted microscope, and the absorbance was measured by spectrophotometry at 450 nm.
Cell-cycle analysis
ADSCs from each group were harvested and washed with PBS. The cells were fixed with cold 70% ethanol for 2 h, treated with 1 mg/mL RNase A for 30 min at 37°C, and then stained with 20 mg/mL propidium iodide and subjected to cell-cycle analysis using flow cytometry (Beckman FC500). The results were analyzed with the WinMDI software.
Fluorescent reporter assay
The MicroCosm and miRanda target prediction programs recommended by miRBase were used to predict the adipogenesis-related target genes of miR-143. MAP2K5 was predicted as a target gene with a high probability score by both algorithms. The 3′-UTR sequence of the MAP2K5 mRNA was acquired from the National Center for Biotechnology Information and the sequence of miR-143 from the miRBase. EGFP/RFP reporter vectors were constructed based on these data. 293T cells were then seeded in a 24-well plate at 30,000 cells per well the day before transduction. The cells were cotransfected with an miR-143 mimic or negative control oligonucleotide, and the following day with the reporter vector pcDNA3–EGFP–MAP2K5 3′-UTR or the mutant pcDNA3–EGFP–MAP2K5 3′-UTR. The reactions were spiked with the red fluorescent protein (RFP)-expressing vector pDsRed2–C1, which was used for normalization. The EGFP and RFP activities were measured 48 h after transduction with an F-4500 fluorescence spectrophotometer (Hitachi).
Statistical analysis
All quantitative data are expressed as means ± standard deviations. One-way analysis of variance (ANOVA) was used to compare two groups in a single experiment and multiple comparisons, with the SPSS software program for Windows. A value of P < 0.05 or P < 0.01 was considered to be statistically significant. The statistically significance of microarray result was analyzed by fold change and the Student t test. The threshold value we used to screen differentially expressed miRNAs is a fold change ≥ 1.5 (P < 0.01, FDR < 0.05).
Author Contributions
L.C.: conception and design, collection of data and analysis and interpretation, manuscript writing; L.F.Y. and Y.W.C.: design, data analysis and interpretation; J.H.C. and J.H.: collection and assembly of data; C.L.: conception and design, data analysis and interpretation, manuscript writing; W.D.T.: supervising the study design and vouching for the data; L.L.: conception and design, data analysis and interpretation, manuscript writing, financial support and final approval of manuscript.
Acknowledgments
We thank the members in State Key Laboratory of Oral Diseases in Sichuan University for their excellent assistance. We particularly thank to Dr. Shuting Wang and Yongchun Yu for their critical comments and stimulating discussions. This study was supported by National Natural Foundation of China (81070802, 81271096), Fundamental Research Funds for the Central Universities of China, and The National High Technology Research and Development Program of China (2011AA030107).
References
- Lean M., Han T. & Seidell J. Impairment of health and quality of life in people with large waist circumference. Lancet 351, 853–856 (1998). [DOI] [PubMed] [Google Scholar]
- Zimmet P., Alberti K. G. & Shaw J. Global and societal implications of the diabetes epidemic. Nature 414, 782–787 (2001). [DOI] [PubMed] [Google Scholar]
- Cummings D. E. & Schwartz M. W. Genetics and pathophysiology of human obesity. Annu Rev Med 54, 453–471 (2003). [DOI] [PubMed] [Google Scholar]
- Rosen E. D. & MacDougald O. A. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7, 885–896 (2006). [DOI] [PubMed] [Google Scholar]
- Gregoire F. M., Smas C. M. & Sul H. S. Understanding adipocyte differentiation. Physiol Rev 78, 783–809 (1998). [DOI] [PubMed] [Google Scholar]
- Billon N., Monteiro M. C. & Dani C. Developmental origin of adipocytes: new insights into a pending question. Biol Cell 100, 563–575 (2008). [DOI] [PubMed] [Google Scholar]
- Chen L. et al. MicroRNAs regulate adipocyte differentiation. Cell Biol In 37, 533–546 (2013). [DOI] [PubMed] [Google Scholar]
- Zarnore P. D. & Haley B. The big world of small RNAs. Science 309, 1519–1524 (2005). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Friedman R. C., Farh K. K., Burge C. B. & Bartel D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19, 92–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K., Pal B. M., Girschick H. J. & Bhadra U. MicroRNAs-micro in size but macro in function. FEBS J 275, 4929–4944 (2008). [DOI] [PubMed] [Google Scholar]
- Rottiers V. & Näär A. M. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13, 239–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C. et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279, 52361–52365 (2004). [DOI] [PubMed] [Google Scholar]
- Takanabe R. et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun 376, 728–732 (2008). [DOI] [PubMed] [Google Scholar]
- Kato Y. et al. Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature 395, 713–716 (1998). [DOI] [PubMed] [Google Scholar]
- Kopelman P. Obesity as a medical problem. Nature 404, 635–643 (2000). [DOI] [PubMed] [Google Scholar]
- Zuk P. A. et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell 13, 4279–4295 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A. M., Elabd C., Amri E. Z., Ailhaud G. & Dani C. The human adipose tissue is a source of multipotent stem cells. Biochimie 87, 125–128 (2005). [DOI] [PubMed] [Google Scholar]
- Gimble J. M. & Guilak F. Differentiation potential of adipose derived adult stem (ADAS) cells. Curr Top Dev Biol 58, 137–160 (2003). [DOI] [PubMed] [Google Scholar]
- Schäffler A. & Büchler C. Concise review: adipose tissue-derived stromal cells–basic and clinical implications for novel cell-based therapies. Stem Cells 25, 818–827 (2007). [DOI] [PubMed] [Google Scholar]
- Tang Q. Q., Otto T. C. & Lane M. D. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc Natl Acad Sci USA 100, 44–49 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feve B. Adipogenesis: cellular and molecular aspects. Best Pract Res Clin Endocrinol 19, 483–499 (2005). [DOI] [PubMed] [Google Scholar]
- Hu E., Kim J. B., Sarraf P. & Spiegelman B. M. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPAR gamma. Science 274, 100–103 (1996). [DOI] [PubMed] [Google Scholar]
- Akaike M. et al. The hinge-helix 1 region of peroxisome proliferator-activated receptor γ1 (PPARγ1) mediates interaction with extracellular signal-regulated kinase 5 and PPARγ1 transcriptional activation: involvement in flow-induced PPARγ activation in endothelial cells. Mol Cell Biol 24, 8691–8704 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H. Y. et al. MicroRNA-375 promotes 3T3-L1 adipocyte differentiation through modulation of extracellular signal-regulated kinase signaling. Clin Exp Pharmacol Physiol 38, 239–246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P. Z., Vernooy S. Y., Guo M. & Hay B. A. The drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 13, 790–795 (2003). [DOI] [PubMed] [Google Scholar]
- Lee J. D., Ulevitch R. J. & Han J. Primary structure of BMK1: a new mammalian MAP kinase. Biochem Biophys Res Commun 213, 715–724 (1995). [DOI] [PubMed] [Google Scholar]
- Zhou G., Bao Z. Q. & Dixon J. E. Components of a new human protein kinase signal transduction pathway. J Biol Chem 270, 12665–12669 (1995). [DOI] [PubMed] [Google Scholar]
- Lin Y. F. et al. Multilineage differentiation of adipose-derived stromal cells from GFP transgenic mice. Mol Cell Biochem 285, 69–78 (2006). [DOI] [PubMed] [Google Scholar]
- Lin Y. F. et al. Pluripotency potential of human adipose-derived stem cells marked with exogenous green fluorescent protein. Mol Cell Biochem 291, 1–10 (2006). [DOI] [PubMed] [Google Scholar]