Abstract
The accumulation of β-amyloid (Aβ) peptide in the brain is one of the pathological hallmarks of Alzheimer's disease and is thought to be of primary aetiological significance. In an unbiased genetic screen, we identified puromycin-sensitive aminopeptidase (PSA) as a potent suppressor of Aβ toxicity in a Drosophila model system. We established that coexpression of Drosophila PSA (dPSA) in the flies' brains improved their lifespan, protected against locomotor deficits, and reduced brain Aβ levels by clearing the Aβ plaque-like deposits. However, confocal microscopy and subcellular fractionation of amyloid-expressing 7PA2 cells demonstrated that PSA localizes to the cytoplasm. Therefore, PSA and Aβ are unlikely to be in the same cellular compartment; moreover, when we artificially placed them in the same compartment in flies, we could not detect a direct epistatic interaction. The consequent hypothesis that PSA's suppression of Aβ toxicity is indirect was supported by the finding that Aβ is not a proteolytic substrate for PSA in vitro. Furthermore, we showed that the enzymatic activity of PSA is not required for rescuing Aβ toxicity in neuronal SH-SY5Y cells. We investigated whether the stimulation of autophagy by PSA was responsible for these protective effects. However PSA's promotion of autophagosome fusion with lysosomes required proteolytic activity and so its effect on autophagy is not identical to its protection against Aβ toxicity.
Keywords: Puromycin-sensitive aminopeptidase, Amyloid, Alzheimer, Autophagy, Proteolysis
Highlights
-
•
PSA is a potent suppressor of Aβ toxicity and reduces brain Aβ levels in flies.
-
•
PSA is cytoplasmic but there is no direct epistatic interaction with Aβ in vivo.
-
•
Aβ is not a proteolytic substrate for PSA in vitro.
-
•
The enzymatic activity of PSA is not required for rescuing Aβ toxicity in cells.
-
•
PSA stimulates autophagy but this is unlikely to be the mechanism that suppresses Aβ toxicity.
1. Introduction
Many diseases in older people are characterized by the misfolding and aggregation of a wide range of proteins [1–3]. Amongst the most prevalent of these disorders is Alzheimer's disease (AD) in which the β-amyloid (Aβ) peptide self-associates in a concentration-dependent aggregation process [4] eventually forming amyloid plaques in the brain [5,6]. Elevated Aβ levels are observed in the earliest stages of AD [7] and most, if not all, mutations that cause familial Alzheimer's disease result in an increase in the production of aggregation-prone isoforms of the peptide [8,9].
The resting concentration of Aβ results from a dynamic equilibrium between its generation from the amyloid precursor protein (APP) and its clearance. While the production of Aβ has been studied extensively, less is known about how the peptide is cleared from the brain. A number of proteases have been characterized that directly cleave Aβ; the bulk of the proteolytic capacity appears to be derived from three enzymes, namely insulin-degrading enzyme (IDE) [10], endothelin-converting enzyme (ECE) [11], and neprilysin (NEP) [12]. While the bulk of the Aβ peptide is extracellular, these enzymes have been reported to be localized to the cytosol and vesicular lumena (IDE) [10], bound to membranes (ECE) [13] or extracellular (NEP) [14]. Overexpression or inhibition of these proteases in model systems has yielded the expected consequences for Aβ levels and toxicity, for example, the overexpression of neprilysin was found to suppress the rough eye and reduced longevity phenotypes in Aβ-expressing Drosophila [15]. Other clearance pathways are important for misfolded cytoplasmic proteins, including autophagy [16] and the ubiquitin–proteasome system [17]; both of these processes also involve a protease termed puromycin-sensitive aminopeptidase (PSA) [18,19]. Specifically, PSA has been shown to cooperate with the proteasome to degrade polyQ-containing peptides in Huntington's disease [18], appears to protect against tau toxicity in mouse and fly models of frontotemporal dementia (FTD) [20,21], and modulates SOD1 levels in amyotrophic lateral sclerosis (ALS) [22]. In each of these studies, the cleavage of the respective polypeptides by PSA has been assumed to be essential [18,20,22–24]. However, recent data suggests that tau is not directly degraded by PSA [25].
Our interest in PSA in the context of AD was prompted by the results of an unbiased genome-scale genetic modifier screen in which we searched for gene products that were able to rescue the reduced longevity observed in flies expressing Aβ. We identified PSA as one of the most powerful suppressors of Aβ toxicity in our model system and in this report, we investigate the protective mechanism.
2. Materials & methods
All reagents were purchased from Sigma-Aldrich unless otherwise stated.
2.1. Fly stocks
Drosophila melanogaster expressing Arctic Aβ42 have been previously described [27]. NSP (no signal peptide) Arctic Aβ42 overexpressing flies were created by site-directed mutagenesis of Arctic Aβ42 in the pUAST-AttB vector using the following primers: fwd 5′-AGGGAATTGGGAATTCATGGATGCGGAATTTCGCC-3′ and rev 5′-GGCGAAATTCCGCATCCATGAATTCCCAATTCCCT-3′. UAS-Arctic Aβ42 and UAS-NSP Arctic Aβ42 transgenic flies were created by site-directed insertion using the φC31 system at location 51D on the second chromosome [26]. Subsequently, the flies were backcrossed for six generations into w1118 flies to obtain isogenic lines. A 51D line without insert but crossed to the gal4 driver line was used as a negative control. UAS-dPSA RNAi constructs, dPSA RNAi 1 (NIG stock ID: 1009 R-2) and dPSA RNAi 2 (NIG stock ID: 1009 R-3), were obtained from the National Institute of Genetics Fly Stock Centre, Japan. The UAS-dicer 2 construct was obtained from the Vienna Drosophila Research Center. UAS-dPSA lines were created using pUAST transgenesis (PSA3-4, PSA3-7 and PSA8-10 were unique inserts, each on the third chromosome).
2.2. Drosophila longevity assays
Flies were mated for 24 h after eclosion and females were entered into longevity assays as described previously [27]. Unless otherwise stated the flies were cultured at 25 °C throughout the assay. Differences in the survival phenotype were analyzed using Kaplan–Meier survival plots and statistical significance was determined using the logrank test.
2.3. Drosophila PSA RNAi expression
Flies carrying transgenes for the various Aβ42 variants were crossed with the elavc155-gal4 driver line. The F1 offspring were crossed with the dPSA RNAi lines and subsequently, the F2 offspring of the forgoing cross with a UAS-dicer 2 line. One hundred mated female flies, expressing Aβ42 and Dicer 2, +/− PSA RNAi, were collected on the day of hatching. Longevity assays were performed at 29 °C.
2.4. Locomotor assays
Negative geotaxis assays were performed as previously described [28,29]. Fifteen flies were assessed in three trials separated by 1 min.
2.5. Quantitation of Aβ peptides by ELISA
Drosophila were cultured at 25 °C for five days after eclosion and then five heads were homogenized in 50 μl of 5 M guanidinium HCl, 5 mM EDTA, 50 mM HEPES, pH 7.3. Following centrifugation for 5 min at 13 krpm, 20 μl of the clear supernatant was removed and mixed with 180 μl of 25 mM HEPES, pH 7.3, 1 mM EDTA, and 0.1% (w/v) BSA with protease inhibitors (Complete, Roche). Triplicate 25 μl aliquots were mixed with an equal volume of PBS containing 2% (w/v) BSA, 0.2% (v/v) Tween-20, and protease inhibitors in the wells of a MESO microtiter plate (Meso Scale Discovery). The reaction was started by adding 25 μl of 4 μg/ml solutions of biotinylated 6E10 or 4G8 monoclonal antibodies (Covance). After mixing, a further 25 μl aliquot of 1 μg/ml Ruthenium-labeled G2-10, or G2-11 (Millipore), monoclonal antibody solution was added to each well. Following an overnight incubation at room temperature (RT), the plates were washed twice with PBS, 150 μl of MSD S Read Buffer (Meso Scale Discovery) was added, and measured in a MSD Sector PR instrument (Meso Scale Discovery).
2.6. Fly brain lysates
Lysates of fly heads were prepared as previously described [30].
2.7. Brain dissections, immunohistochemistry, and rough eye phenotyping
Brain dissections and immunohistochemistry were performed as described previously [31].
2.8. Quantitative PCR
Messenger RNA was extracted from 100 fly heads with the RNeasy Mini kit (Qiagen) and quantified using the NanoDrop (Thermo Scientific). Genomic DNA contamination was removed by treating 1 μg of RNA sample with DNase. Subsequently, a reverse transcription reaction was performed using an AMV Reverse Transcriptase kit (Promega). For quantitative PCR (qPCR), cDNA was diluted 1:100 in water and 10 μl was pipetted into a 96-well plate with a SYBR-GREEN PCR master mix (Bio-Rad). The quantitative PCR was performed using primers for Aβ and dPSA (Aβ: fwd 5′-CAGCGGCTACGAAGTGCATC-3′ and rev 5′-CGCCCACCATCAAGCCAATA-3′; dPSA: fwd 5′-AGAACTGCCTATGGGTGGTG-3′ and rev 5′-TGTCATCGATCAATCCCAGA-3′). Differences in gene expression levels between samples were normalized to expression levels of the housekeeping gene RP49 (RP49: fwd 5′-ATGACCATCCGCCCAGCATCAG-3′ and rev 5′-ATCTCGCCGAGTAAACG-3′). The reaction was carried out in an iQ5 iCycler (Bio-Rad) thermocycler. The SYBR green fluorescence was used to compare mRNA levels. For each sample, the fluorescence was averaged between the replicates and normalized to the respective RP49 fluorescence level (ΔΔCT) as well as to potential noise present in the non-template controls. At least five replicates of each sample were carried out and the standard deviation calculated.
2.9. Human PSA constructs
Expression constructs for full-length human PSA (GFP-PSA) and the enzymatically inactive mutant of PSA (GFP-ZBD) with N-terminal GFP tags in the pEGFP-C3 vector were prepared. A non-fluorescent (NF) version for each GFP construct (pEGFP-C3, GFP-PSA, and GFP-ZBD) was obtained by mutating two residues (T65S and Y66A) of the GFP fluorophore by site-directed mutagenesis. In addition, constructs (GFP-PSA and GFP-ZBD in the pTRE2hyg vector) for making the inducible HeLa Tet-On stable cell lines were generated. The GST-PSA/pET-41 Ek/LIC construct was generously provided by Dr. Stanislav Karsten (UCLA, USA) [20,23]. The inactive GST-ZBD construct was obtained by site-directed mutagenesis. The detailed cloning strategy may be found in the supplementary data.
2.10. Cell culture and transfection
7PA2 cells, kindly provided by Prof. Dennis Selkoe (Harvard Medical School, Cambridge, MA, USA), and the parental CHO (Chinese Hamster Ovary) cells were grown as described previously [32]. SH-SY5Y cells were grown in DMEM:nutrient mixture F-12 (DMEM/F-12) containing 10% (v/v) FBS. HeLa Tet-On cells (Clontech) were cultured in DMEM containing 10% (v/v) Tet System Approved FBS (Australia-sourced, Clontech) and G418 (200 μg/ml, Invitrogen). The set of constructs (GFP-PSA, and GFP-ZBD in the pTRE2hyg vector) was transfected using Fugene HD (Promega) and the single clones were selected by hygromycin B (Invitrogen) resistance (500 μg/ml) according to Clontech's instructions. The HeLa Tet-On stable cell lines (GFP-PSA and GFP-ZBD) were maintained in HeLa Tet-On cell medium containing 200 μg/ml hygromycin B. All cells were incubated at 37 °C and 5% (v/v) CO2.
For transient transfections, 7PA2 cells were plated in medium lacking antibiotics on poly-l-lysine coated surfaces and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
2.11. Autophagy
Expression of GFP-PSA and GFP-ZBD in HeLa Tet-On stable cell lines was induced by treatment with 1 μg/ml doxycycline for 48 h. To knock down PSA, parental HeLa Tet-On cells were transfected with 200 pmol ON-TARGETplus SMARTpool siRNA against human PSA/NPEPPS (Dharmacon) using Lipofectamine 2000 according to the manufacturer's protocol, and lysed 48 h post-transfection. For the mock transfection, the siRNA was replaced with Opti-MEM. Four hours before harvesting, cells were either left untreated or treated with 400 nM bafilomycin A1 [33]. Cell lysates were separated by SDS-PAGE and the bands on the immunoblots were quantified by densitometry using ImageJ.
2.12. Indirect immunofluorescence and confocal microscopy
Twenty-four hours post-transfection, cells were washed with PBS and fixed with 4% (v/v) paraformaldehyde (Electron Microscopy Sciences) in PBS for 20 min. After fixation, cells were washed three times with PBS, permeabilized with 0.2% (v/v) Triton X-100 for 5 min, and blocked in 1% (w/v) BSA in PBS for 30 min. Subsequently, cells were incubated with the following primary antibodies for 1 h at RT in a humidified chamber: a 1:50 dilution of goat polyclonal PSAP (N-20) (Santa Cruz Biotechnology) antibody against PSA and a 1:800 dilution of mouse monoclonal antibody against GFP (Abcam). After washing three times with PBS, cells were incubated with the appropriate secondary antibodies (Alexa Fluor 633 donkey-anti goat IgG and Alexa Fluor 546 donkey anti-mouse IgG, Invitrogen) for 45 min at RT in a humidified chamber. After washing the cover slips four to five times with PBS, they were mounted with ProLong Gold Antifade (Invitrogen). Single slices or z-stacks were acquired with a Zeiss LSM 510 confocal microscope (Carl Zeiss) using the 100 × objective and were processed with the Zeiss LSM Image Browser.
2.13. SDS-PAGE and Western blotting of cell lysates and conditioned media
∅10 cm plates of 7PA2 cells were conditioned with 10 ml DMEM, the conditioned medium (CM) collected after 18 h and cleared of cell debris. After adding protease inhibitors (Complete, EDTA-free, Roche), the CM was concentrated at least 20–30 times in Vivaspin 2 PES columns (3000 MWCO, Sartorius Stedim Biotech) for Aβ or Vivaspin 20 PES columns (30,000 MWCO, Sartorius Stedim Biotech) for PSA at 4 °C according to the manufacturer's instructions.
Twenty-four hours post-transfection, cell lysates were prepared in lysis buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 1% (v/v) NP-40 with protease inhibitors), boiled in SDS loading buffer, and separated by SDS-PAGE on 8%, 10%, 12% or 15% PAA gels depending on the size of the protein-of-interest. The protein bands were transferred on to PVDF or nitrocellulose membranes (both 0.45 μm) and blocked with 5% (w/v) fat-free milk powder in PBS-T (0.05% (v/v) Tween-20 in PBS) for 1 h at RT. The sensitivity of Aβ immunoblotting was increased by boiling the nitrocellulose membrane in PBS for 10 min [34–36], before blocking. The membranes were probed overnight at 4 °C with the following primary antibodies: goat polyclonal PSAP (N-20) against PSA (1:200, Santa Cruz Biotechnology), rabbit polyclonal to GFP (1:1000, Abcam), mouse monoclonal AC-15 against β-actin (1:5000, Sigma), mouse monoclonal against eIF2α (1:10,000, a gift from David Ron, Institute for Metabolic Science, Cambridge, UK) [37], mouse monoclonal 10C3 against KDEL (1:1000, Stressgen), mouse monoclonal 6E10 against APP (Aβ 1–16) (1:1000, Covance), mouse monoclonal 4G8 against APP (Aβ 17–24) (1:1000, Covance), rabbit monoclonal 139-5 against β amyloid 40 (1:2000, Covance), rabbit polyclonal to histone H3 (1:1000, Cell Signalling), or rabbit polyclonal against LC3 (1:2000, Novus Biologicals). The membranes were washed three times with PBS-T and then incubated with the corresponding HRP-conjugated secondary antibody for 1 h at RT. After washing the membrane three times with PBS-T and once with PBS, the protein bands were detected with the SuperSignal West Pico and/or Femto Chemiluminescent Substrate (Pierce Biotechnology) according to the manufacturer's protocol and exposed to X-ray film (Fujifilm).
2.14. Subcellular fractionation by density gradient ultracentrifugation
Forty-eight hours post-transfection, three Ø10 cm plates of 7PA2 cells (untransfected or transiently expressing pEGFP-N3, GFP-PSA, and PSA-GFP) were washed once with ice-cold PBS and then processed using a ball-bearing homogenizer with a 10 μm clearance (EMBL, Heidelberg, Germany) as previously described [38]. The homogenate was cleared of cell debris by centrifugation at 3000 ×g for 10 min at 4 °C (an “input” aliquot was taken) and adjusted to 35% (v/v) OptiPrep. Membranes were floated by centrifugation at 70,000 ×g for 2 h at 4 °C to the interface of a discontinuous 0%–30% (v/v) OptiPrep gradient buffered with 20 mM HEPES, pH 7.4 and 1 mM EDTA. An aliquot of the so-called depleted cytosol in 35% (v/v) OptiPrep was taken. The membranes were washed by diluting the membrane-enriched fraction 1:5 with homogenization buffer and repelleted by centrifugation at 100,000 ×g for 45 min at 4 °C. The pellets were pooled by solubilization in a total volume of 50 μl homogenization buffer.
2.15. Nuclear and cytoplasmic fractionation
One confluent ∅10 cm plate of 7PA2 cells was washed twice with ice-cold PBS, harvested in PBS with 1 mM EDTA, and collected by centrifugation at 1000 ×g for 5 min at 4 °C. The cell pellet was resuspended in 200 μl harvest buffer [10 mM HEPES pH 7.9, 50 mM NaCl, 500 mM sucrose, 0.1 mM EDTA, 0.5% (v/v) Triton X-100 supplemented with 1 mM DTT, 1 mM PMSF, protease inhibitors (Complete, EDTA-free, Roche)] and placed on ice for 5 min (an “input” aliquot was taken). After centrifugation at 100 ×g for 10 min at 4 °C, the supernatant was taken off and kept as the post-nuclear supernatant (PNS). The pellet corresponds to the nuclei and was washed four to five times with 500 μl low-salt wash buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA) to eliminate any cytoplasmic contamination. The nuclear pellet was recovered by centrifugation at 100 ×g for 5 min at 4 °C, resuspended in 100 μl high-salt extraction buffer [10 mM HEPES pH 7.9, 500 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1% (v/v) NP-40], and vortexed for 15 min at 4 °C. Centrifugation at 18,000 ×g for 10 min at 4 °C gives rise to the nuclear extract (NE) as the supernatant containing all soluble nuclear proteins and the pellet consisting of the remaining insoluble proteins, which was solubilized in 4 × SDS loading buffer. The input and nuclear insoluble fraction (NI) were sheared five times with a 25 G hypodermic needle.
2.16. Aminopeptidase activity assay
Forty-eight hours post-transfection, one confluent ∅10 cm plate of SH-SY5Y cells (untransfected or transiently expressing pEGFP-N3, GFP-PSA, or GFP-ZBD) was washed once with PBS and harvested in 500 μl PBS containing 0.5 mM EDTA. After centrifugation for 10 min at 3000 ×g, the cell pellet was washed once with 500 μl PBS, resuspended in 100 μl of 10 mM Tris, pH 7.4, and incubated on ice for 10 min. The cells were homogenized by ultrasonication and centrifuged for 30 min at 13,000 ×g at 4 °C. The supernatant containing the soluble fraction was collected and the protein concentration was measured at A280 nm using the NanoDrop (Thermo Scientific). The aminopeptidase activity assay was performed as described (http://www.sigmaaldrich.com/img/assets/18160/Aminopeptidase.pdf) with the following revisions: the reaction cocktail used a 2 mM l-leucine-p-nitroanilide solution; 150 μg of lysate was added to the reaction; the lysates were inhibited with 10 μM puromycin; the reaction was prepared in 1 ml and then 200 μl was immediately transferred to a 96-well plate in triplicate; the assay was carried out at RT. The A405 nm was monitored at RT every 5 s for 1 h in a ThermoMax microplate reader (Molecular Devices). Initial reaction rates (45 min) of substrate digestion were calculated and plotted in GraphPad Prism 4.
2.17. Aβ toxicity assay
Twenty-four hours post-transfection, SH-SY5Y cells were replated in triplicate into a 96-well plate. After the cells had reattached (~ 4 h), they were washed once with Opti-MEM and treated with 100 μl of 10 μM monomeric Aβ1–42 [39–41] in Opti-MEM for 48 h at 37 °C and 5% (v/v) CO2. All wells were washed once with 100 μl D-PBS. One hundred microliters of a 3 μM working solution of both dyes [calcein AM and ethidium homodimer-1 from the LIVE/DEAD Viability/Cytotoxicity kit (for mammalian cells), Invitrogen] in DMEM/F-12 medium (no phenol red) was added to each well, incubated at RT for 45 min, and the fluorescence signal was read using the EnVision XCite Multilabel Reader (Perkin Elmer). The numbers of live and dead cells were determined, converted to a calcein AM/ethidium homodimer-1 (Live/Dead cell) ratio, and displayed graphically in GraphPad Prism 4.
2.18. Purification of recombinant GST-tagged PSA and inactive GST-ZBD
The GST-PSA and GST-ZBD constructs were transformed into Rosetta2(DE3)pLysS bacterial cells and grown in LB medium at 37 °C until OD 0.8 at A600 nm, before induction with 1 mM IPTG and overnight expression at 18 °C in a fermenter. Cell pellets were resuspended in PBS containing 0.1 mg/ml lysozyme and 10 μg/ml DNaseI and incubated at 4 °C with stirring for 30 min. The cell suspension was disrupted by sonication and cleared by centrifugation at 25,000 ×g for 30 min at 4 °C. The clarified supernatant was incubated with Glutathione Sepharose 4B beads (GE Healthcare) for 2 h at 4 °C on a rotating platform. After washing the beads with PBS containing 0.5 M NaCl, the GST fusion proteins were eluted from the resin with 20 mM reduced l-glutathione, 100 mM Tris, pH 8, and 50 mM NaCl. GST-tagged PSA and the inactive GST-ZBD recombinant proteins were dialyzed into 20 mM Tris, pH 7.4 and 100 mM NaCl overnight using Slide-A-Lyzer Mini Dialysis Units (10,000 MWCO, Thermo Scientific). The protein concentration was determined with the NanoDrop using a calculated mass extinction coefficient of 14.45 l g− 1 cm− 1 for GST-PSA and 14.46 l g− 1 cm− 1 for GST-ZBD [42]. The aminopeptidase activity of GST-PSA and inactivity of GST-ZBD were determined using 2 μg enzyme in 10 mM Tris, pH 7.4 containing 1 mM DTT (required for PSA activity). The reaction was initiated with l-leucine-p-nitroanilide at a final concentration of 500 μM and monitored at A405 nm every 10 s for 5 min in a ThermoMax microplate reader (Molecular Devices) at RT.
2.19. In vitro digestion assay
Monomeric Aβ1–40 was dissolved at 1 μg/μl in 20 mM Tris, pH 7.4, 100 mM NaCl. The in vitro digestion assay (50 μl total volume) was set up as follows: 11.5 μM Aβ1–40 was pipetted into the reaction buffer (20 mM Tris, pH 7.4, 100 mM NaCl) containing 1 mM DTT and initiated by adding 0.8 μM of GST-PSA (the preparation's activity had been determined as described above). The reaction tubes were incubated at 30 °C for 0, 1/2, 2, 6, and 24 h and stopped at the designated time points by the addition of 2 × Tricine SDS Sample Buffer and snap-freezing on dry ice. The Aβ band from the 24 h timepoint was analyzed by N-terminal protein sequencing (Edman degradation). As negative controls, GST-PSA was incubated with Aβ1–40 in the presence of 100 μM puromycin (PSA inhibitor) for 24 h and the inactive mutant, GST-ZBD, was incubated with Aβ1–40 for 24 h.
3. Results
3.1. An overexpression screen of the Drosophila autosome identified puromycin-sensitive aminopeptidase as a suppressor of Aβ toxicity
Aβ toxicity was modeled in Drosophila melanogaster by fusing Aβ to an N-terminal secretion signal peptide and expressing it in the central nervous system of the fly [27]. This approach does not require the proteolytic processing of APP and permits the convenient study of pathological processes that are downstream of Aβ generation. In order to identify novel genetic modifiers of Aβ toxicity in the fly, a library of 3000 unique genomic insertions of the gene search (GS) mobile genetic element was generated and the effect of the consequent gal4-dependent modulation of neighboring gene transcription on the lifespan of flies expressing the Arctic variant (E22G) of Aβ42 [43] was assessed. Insertion 622 provided one of the most powerful suppressor effects resulting in an increase in median survival from 29 days in flies expressing Arctic Aβ42 (n = 72) to 36 days in flies expressing both Aβ42 and the GS622 element (n = 27 flies, p < 0.001 by logrank test). Inverse PCR demonstrated that the insertion of the GS element was on chromosome 3 in the 5′ untranslated regions of two genes. The first gene was the Drosophila ortholog of puromycin-sensitive aminopeptidase (dPSA, CG1009) and the second was cue, an ortholog of the low density lipoprotein receptor (CG12086).
To gain an insight into which of these genes was more likely to be involved in modulating Aβ toxicity in the fly, an Affymetrix cDNA microarray screen was undertaken to search for genes that were up- or down-regulated in flies expressing Arctic Aβ42 as compared to flies expressing the non-toxic Aβ40 peptide [44]. In this study, the mRNA for dPSA exhibited a 2.7-fold upregulation in Arctic Aβ42 flies (false detection rate 1.5%), whereas the mRNA for CG12086 was not differentially regulated. To further define whether dPSA or CG12086 was responsible for the rescue of Aβ toxicity, flies were generated that overexpressed each gene individually. Three representative transgenic lines of flies for each gene were chosen and the effects of each transgene on longevity in flies coexpressing Arctic Aβ42 were measured. None of the overexpression constructs for CG12086 were able to replicate the suppression of Arctic Aβ42 toxicity; in contrast, all three independent transgenic fly lines coexpressing dPSA with Arctic Aβ42 demonstrated an increase in median survival (Fig. 1A, n = 100 flies for all lines, p < 0.001 by logrank test for all three UAS-dPSA lines with Arctic Aβ42 vs. Arctic Aβ42 alone). By contrast, dPSA expression in control flies caused a mild reduction in longevity (Supplementary Fig. 1). dPSA suppressed the retinal toxicity associated with Arctic Aβ42 expression resulting in normal eye development (Fig. 1B). The flies were also functionally protected as shown by improved locomotor function throughout their adult life as compared to flies expressing Arctic Aβ42 alone (Fig. 1C). Confocal microscopy of fly brains stained with the 6E10 antibody for Aβ42 revealed that Arctic Aβ42 flies exhibit profuse plaque-like deposits, whereas coexpression with dPSA resulted in a marked reduction in the number of Aβ deposits (Fig. 1D). An ELISA assay confirmed that total Arctic Aβ42 levels were reduced in brains of Arctic Aβ42 flies coexpressing dPSA compared to Arctic Aβ42 alone (Fig. 1E). This reduction in peptide accumulation is unlikely to be an artefactual genetic interaction, as there was no correlation between Aβ mRNA levels (as measured by qPCR) from fly brains and median survival (data not shown). Essentially identical results were observed when lifespan and locomotor assays were repeated with flies coexpressing dPSA with wild-type Aβ42 (data not shown).
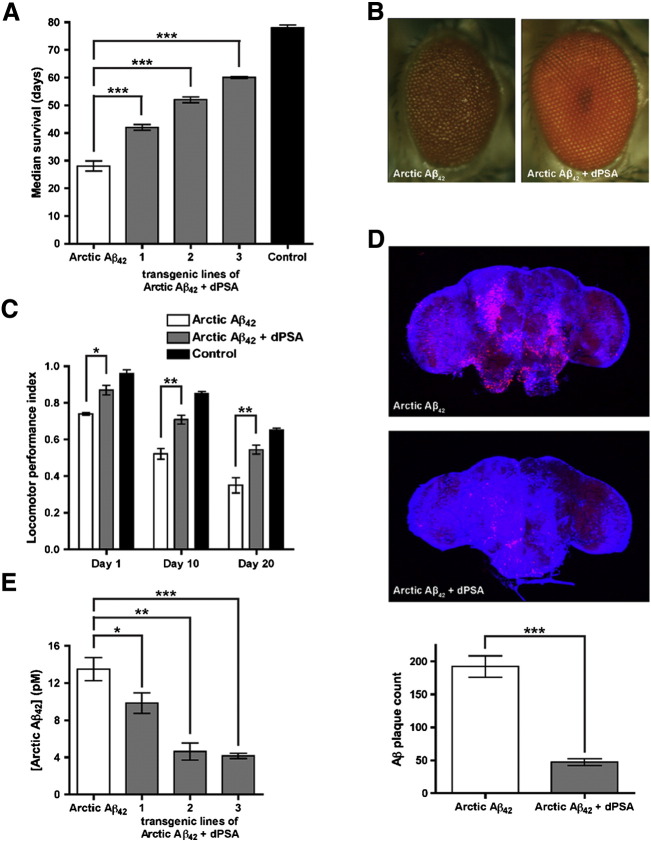
Fig. 1.
Coexpression of dPSA with Arctic Aβ42 in Drosophila melanogaster.
(A) Transgenic overexpression of dPSA in Arctic Aβ42 flies (gray, three independent transgenic lines) prolonged their lifespan compared to flies expressing Arctic Aβ42 alone (white) and almost completely rescued the longevity phenotype compared to control flies (black). n = 100 flies, mean ± S.E., logrank test. (B) Elavc155-gal4-driven expression of Arctic Aβ42 from two transgenes resulted in a rough eye phenotype that was suppressed by coexpression with dPSA (Arctic Aβ42 + dPSA). (C) Locomotor assays were performed on three independent groups of ten to fifteen flies for each of the genotypes on days 1, 10, and 20 of adult life. For each age group, the locomotor data for the three independent transgenic lines of dPSA were pooled. Throughout their lives flies coexpressing dPSA with Arctic Aβ42 (gray, three replicates for each of the three transgenic lines) were markedly more mobile than flies expressing only Arctic Aβ42 (white, three replicates). In comparison to control flies (black, three replicates), overexpression of dPSA alleviated the majority of the locomotor impairment caused by Arctic Aβ42 at all time points. n = 3, mean ± S.E., one-tailed unpaired t-test. (D) Elavc155-gal4-driven expression of Arctic Aβ42 from two transgenes resulted in profuse plaque-like deposits of Aβ that were cleared when dPSA was co-expressed (Arctic Aβ42 + dPSA). Aβ was detected with the 6E10 antibody (red) and DNA was stained with TOTO-3 (blue). Images were collected with a confocal microscope. Quantitation of the confocal slices revealed that dPSA clears 75% of the plaques as compared to Arctic Aβ42 flies alone. n = 4 ± S.D., two-tailed unpaired t-test. (E) Coexpression of dPSA with Arctic Aβ42 (gray) reduced the concentration of Aβ42 in the heads of all three transgenic fly lines compared to flies expressing Arctic Aβ42 alone (white) as measured by MSD ELISA. Representative of two repeats: n = 3 (three replicates), mean ± S.D., one-tailed unpaired t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
Despite these data, it is not clear whether Aβ is a proteolytic substrate for PSA due to uncertainty in the subcellular localization of the protease [24,45–47].
3.2. Mammalian PSA is localized in the cytoplasm and is not membrane-bound in 7PA2 cells
To precisely establish its intracellular distribution, human PSA was tagged with GFP at the N-terminus (GFP-PSA) and transiently transfected into CHO cells overexpressing human APP751 with the V717F familial AD linked mutation. These cells termed 7PA2 predominantly secrete Aβ42 [32]. GFP-PSA was localized to the cytoplasm of 7PA2 cells, colocalizing with antibodies against GFP and PSA by indirect immunofluorescence (Fig. 2A). As expected, the GFP control was distributed uniformly within both the cytoplasm and nuclei of cells transiently transfected with the control vector (pEGFP-C3) (Fig. 2A). Western blotting of the corresponding lysates with PSA and GFP antibodies confirmed that the full-length GFP-PSA fusion protein was expressed at the correct size (130 kDa) as was GFP from the control vector (pEGFP-C3) at ~ 30 kDa (Fig. 2B). In addition, the PSA antibody was able to detect endogenous PSA at 100 kDa in the 7PA2 cell lysates (Fig. 2B).
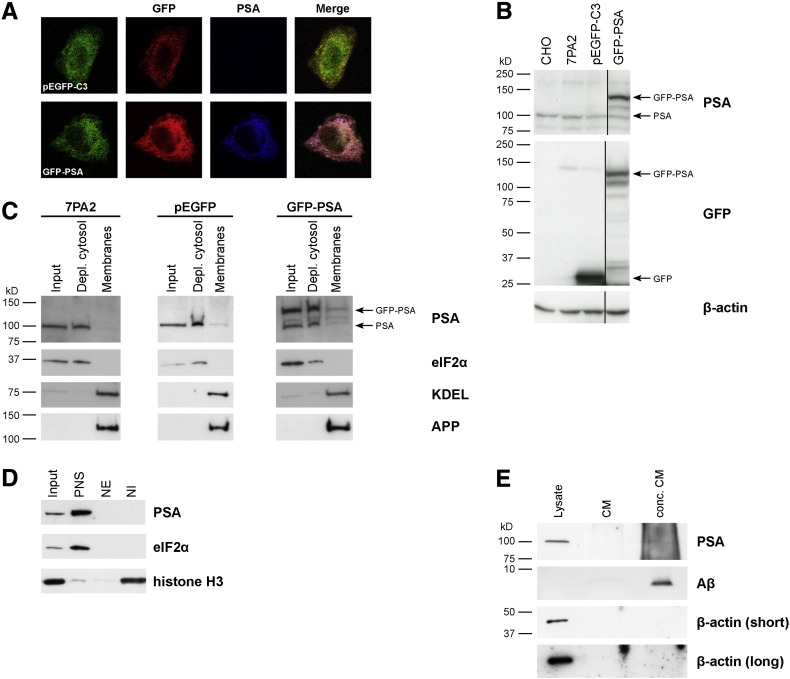
Fig. 2.
Intracellular localization of endogenous and GFP-tagged PSA in 7PA2 cells.
(A) Indirect immunofluorescence of 7PA2 cells transiently transfected with the control vector (pEGFP-C3) and a construct expressing PSA tagged with GFP (green) at the N-terminus (GFP-PSA). The GFP-PSA fusion protein colocalized with antibodies against GFP (red) and PSA (blue) in the cytoplasm. Images were collected with a confocal microscope and the merged panel is shown on the far right. (B) Lysates of untransfected cells (CHO and 7PA2), 7PA2 cells transiently transfected with pEGFP-C3 and GFP-PSA were prepared and separated by SDS-PAGE. The Western blot was probed with antibodies against PSA and GFP. The GFP-PSA fusion protein was expressed at the correct size of 130 kDa and endogenous PSA could be detected at 100 kDa. β-actin is shown as a loading control. (C and D) Subcellular fractionation of 7PA2 cells by density gradient ultracentrifugation. Membranes were prepared from untransfected 7PA2 cells and cells transiently transfected with pEGFP-N3 and GFP-PSA. Nuclei were isolated from 7PA2 cells. Samples were separated by SDS-PAGE and the immunoblot was probed with a PSA antibody. Endogenous PSA and overexpressed GFP-PSA were cytosolic and could not be detected in the membrane or nuclear fractions. The following loading controls were used: eIF2α (cytoplasmic), KDEL (membranes), APP (detected by the 6E10 antibody), and histone H3 (nuclear). PNS: post-nuclear supernatant, NE: nuclear extract, NI: nuclear insoluble. (E) 7PA2 cell lysates were prepared and the corresponding conditioned medium (CM) was collected and concentrated. The samples were separated by SDS-PAGE and the immunoblot was probed with PSA and 6E10 (detects the first sixteen amino acids of Aβ) antibodies. PSA could not be detected in the extracellular medium. β-actin is shown as a loading control.
Subcellular fractionation of 7PA2 cells by density gradient ultracentrifugation revealed that endogenous PSA and overexpressed GFP-PSA were mainly present in the depleted cytosol fraction and not associated with membranes (Fig. 2C). Transiently transfecting the control vector (pEGFP) expressing GFP into 7PA2 cells had no effect on the localization of endogenous PSA (Fig. 2C). When nuclei were isolated from 7PA2 cells, endogenous PSA was mainly located in the post-nuclear supernatant (confirming its cytoplasmic location) and was not present in the nuclear extract or nuclear insoluble fraction (Fig. 2D).
Since Aβ is secreted from cells, it remained a possibility that PSA was interacting with Aβ in the extracellular space. However, PSA could not be detected in concentrated conditioned medium of 7PA2 cells (Fig. 2E).
3.3. Drosophila PSA does not interact with Arctic Aβ42 in the cytoplasm of fly neurons
One possible explanation for how cytoplasmic PSA can rescue phenotypes mediated by supposedly extracellular Aβ is that a highly toxic subpopulation of Aβ migrates into the cytoplasm where it exerts its deleterious effects. To test whether PSA can protect against a cytoplasmic Aβ challenge, we generated transgenic flies expressing Arctic Aβ42 without a secretion signal (NSP: no signal peptide; NSP Arctic Aβ42). Expression of secreted Arctic Aβ42 in the brain of flies resulted in the accumulation of peptides that appear as monomers following SDS-PAGE (Fig. 3A, Arctic Aβ42 lane), while NSP Arctic Aβ42 appeared as an oligomeric species (NSP Arctic Aβ42 lane). Despite similar expression levels as determined by qPCR (Fig. 3B), the lifespan of NSP Arctic Aβ42 flies did not differ from control flies indicating that cytoplasmic Aβ is not toxic (Fig. 3C). If dPSA is indeed responsible for protection against a toxic cytoplasmic fraction of Aβ, then dPSA knockdown should interact more strongly with NSP Aβ as compared to secreted Aβ. To detect whether a reduction in dPSA levels might enhance the toxicity of NSP Arctic Aβ42, we used two independent dPSA-RNAi lines (1 and 2) coexpressed with Dicer-2 (Fig. 3D). In control flies (Fig. 3E, Control), the knockdown of dPSA using both RNAi lines (Fig. 3E, Control + dPSA RNAi 1 & Control + dPSA RNAi 2) caused a reduction in longevity due to background genetic effects unrelated to Aβ toxicity. In flies expressing NSP Arctic Aβ42, the pattern of longevity reduction was remarkably similar to control flies, which is consistent with cytoplasmic Aβ not interacting with dPSA. In contrast, the pattern of longevity changes was quite distinct following dPSA knockdown in flies expressing secreted Arctic Aβ42, thereby supporting an interaction between secreted Aβ and dPSA (Fig. 3E). Although the nature of the interaction remains unclear, the particular conformation of Aβ in the neuronal environment may be important or else the flies may die of Arctic Aβ42 toxicity before the effect of dPSA RNAi can be observed. Similarly, an interaction between cytoplasmic Aβ and PSA could not be observed at the tissue level. In flies expressing NSP Arctic Aβ42, the appearance of plaque-like deposits could not be induced (Fig. 3F), even when endogenous PSA was knocked down using RNAi (NSP Arctic Aβ42 + Dicer-2 + dPSA RNAi). Our positive controls were flies expressing secreted Arctic Aβ42, which spontaneously formed plaque-like deposits (Fig. 3F).
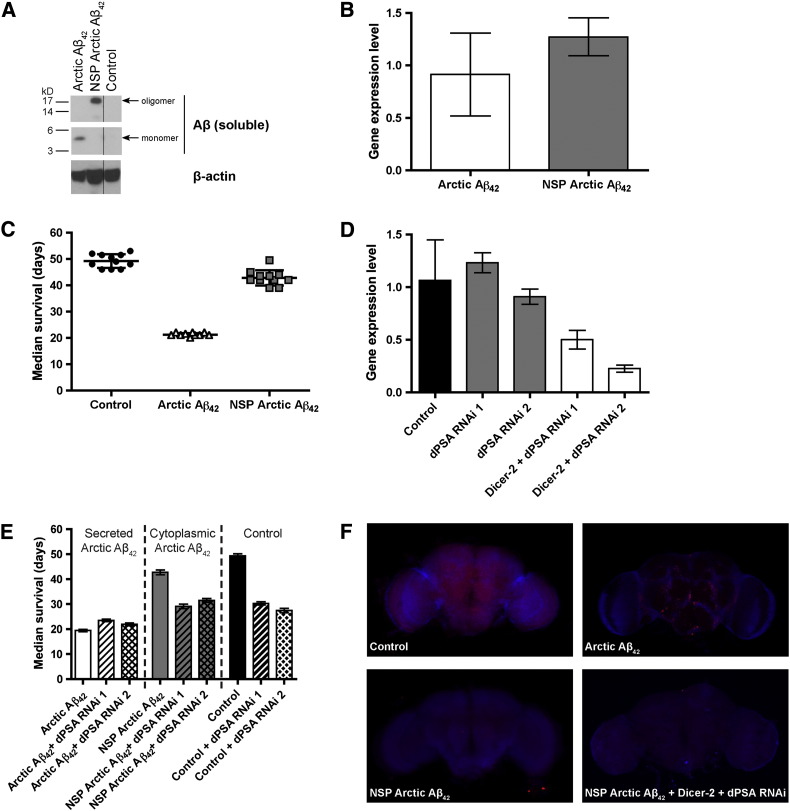
Fig. 3.
dPSA does not participate in the clearance of cytoplasmic Arctic Aβ42in vivo.
(A) Western blot analysis of the PBS-soluble fraction of control and transgenic fly brain extracts probed with the 6E10 antibody against Aβ. The material from Arctic Aβ42 flies is monomeric, whereas NSP (no signal peptide) Arctic Aβ42 flies form soluble oligomeric species. β-actin is shown as a loading control. (B) Quantitative real-time PCR (qPCR) demonstrating that the mRNA levels of Arctic Aβ42 (white) and NSP Arctic Aβ42 (gray) are similar. Gene expression values were normalized to the mRNA level of the housekeeping gene RP49. n = 5 (five replicates), mean ± S.D. (C) Transgenic overexpression of NSP Arctic Aβ42 (gray squares) in the cytoplasm of flies is non-toxic as their lifespan is similar to control flies (black circles). In comparison, expression of Arctic Aβ42 (white triangles) in the secretory pathway significantly reduces their longevity. n = 100 flies (separated into groups of ten), mean ± S.D. (D) qPCR of dPSA levels in two RNAi lines (dPSA RNAi 1 and dPSA RNAi 2) with (white) and without Dicer-2 (gray). The knockdown efficiency of dPSA in both RNAi lines is improved in the presence of Dicer-2. Gene expression values were normalized to the mRNA level of the housekeeping gene RP49. n = 6 (six replicates), mean ± S.D. (E) Control flies not expressing Aβ (Control) and flies expressing cytoplasmic NSP Arctic Aβ42 responded similarly to dPSA RNAi in the presence of Dicer-2. By contrast, PSA knockdown in flies expressing secreted Arctic Aβ42 gave a distinct pattern of longevity effects. n = 100 flies, mean ± S.D. (F) Dissections of fly brains illustrating plaque accumulation in Arctic Aβ42 flies, which appears to be absent in NSP Arctic Aβ42 with or without dPSA (+ Dicer-2 + dPSA RNAi). Aβ was detected with the 6E10 antibody (red) and DNA was stained with TOTO-3 (blue). Images were collected with a confocal microscope.
3.4. The proteolytic activity of PSA is not required for protection against Aβ toxicity
The aminopeptidase activity of neuronal SH-SY5Y cell lysates was determined by monitoring the hydrolysis of the chromogenic substrate, l-leucine-p-nitroanilide. To estimate the contribution that endogenous and overexpressed PSA made to the enzymatic activity, each lysate was assayed in the presence (+ P) and absence of puromycin (Fig. 4A). SH-SY5Y cells transfected with the pEGFP-C3 control vector or left untransfected had similar puromycin-sensitive activity, whereas the expression of GFP-PSA markedly increased this activity (Fig. 4A). In contrast, the single point mutation E309A [48] in the zinc-binding domain (ZBD) of PSA rendered the enzyme (GFP-ZBD, characterized in Supplementary Fig. 2A and B) catalytically inactive (Fig. 4A). Equal amounts of total protein were loaded in each reaction as demonstrated by immunoblotting (Supplementary Fig. 2C).
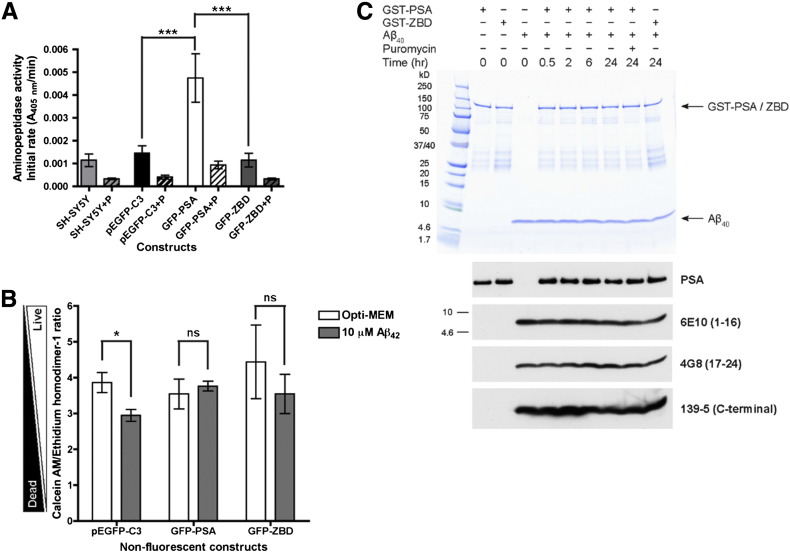
Fig. 4.
The rescue of Aβ42 toxicity in SH-SY5Y cells is independent of the proteolytic activity of PSA.
(A) Aminopeptidase activity assay of GFP-tagged PSA expressed in SH-SY5Y cells. SH-SY5Y cells (light gray) were transiently transfected with pEGFP-C3 (black), GFP-PSA (white), and the enzymatically inactive zinc-binding domain (ZBD) mutant (GFP-ZBD, dark gray). The proteolytic activity of PSA in each lysate was inhibited by 10 μM puromycin (P). Initial reaction rates of substrate digestion were plotted. n = 3, mean ± S.E., ***p < 0.001, repeated measures ANOVA followed by a Bonferroni's multiple comparison post-hoc test. (B) Rescue of Aβ42 toxicity in SH-SY5Y cells by overexpression of GFP-tagged PSA and the inactive mutant. SH-SY5Y cells were transiently transfected with non-fluorescent GFP constructs (pEGFP-C3, GFP-PSA, and GFP-ZBD) and treated with 10 μM monomeric Aβ42 in Opti-MEM medium for 48 h. The numbers of live and dead cells were measured using the LIVE/DEAD Viability/Cytotoxicity Kit; n = 3 (except for GFP-ZBD where n = 6), mean ± S.E.; ns, not significant; *p < 0.05, two-tailed paired t-test. (C) GST-tagged PSA does not digest Aβ40in vitro. Recombinant GST-PSA or GST-ZBD were incubated either alone or in the presence of monomeric Aβ40 with or without 100 μM puromycin (P) at pH 7.4 and 30 °C for up to 24 h. The samples were separated by SDS-PAGE and stained with Coomassie or the immunoblots were probed with an antibody against PSA and a selection of Aβ antibodies: 6E10 (detects 1–16 of Aβ), 4G8 (detects 17–24 of Aβ), and Aβ40-specific 139-5 (C-terminal region of Aβ40).
Subsequently, the effect of overexpressing active GFP-PSA and inactive GFP-ZBD in SH-SY5Y cells on the toxicity of exogenously applied Aβ42 was assessed. To make our constructs spectrometrically compatible with the LIVE/DEAD Cytotoxicity/Viability Kit, non-fluorescent (NF) versions of all constructs were generated by introducing two point mutations (T65S Y66A) in the GFP fluorophore (characterized in Supplementary Fig. 3A, B and C). SH-SY5Y cells expressing enzymatically active GFP-PSA NF were resistant to Aβ42 toxicity. In contrast, cells transfected with the control pEGFP-C3 NF vector remained sensitive to the toxic effects of Aβ42 (Fig. 4B). Surprisingly, and despite many repeats, the rescue was also observed in cells expressing the inactive mutant GFP-ZBD NF (n = 6, Fig. 4B and Supplementary Fig. 3D).
These data are compatible with a protective role for PSA against Aβ toxicity, yet the proteolytic activity of PSA appears to be dispensable. Indeed, we found that monomeric Aβ1–40 is not cleaved by recombinant human GST-tagged PSA in vitro over a 24 h time period (Fig. 4C). Specifically, we could detect proteolysis neither on a Coomassie stained SDS-PAGE gel nor by probing immunoblots with monoclonal anti-Aβ antibodies chosen to monitor trimming activity. The Aβ peptide remained intact under these conditions despite incubation with recombinant GST-PSA that retained > 83% of its initial aminopeptidase activity throughout the 24 h reaction. Moreover, N-terminal protein sequencing of the Aβ band after the 24 h-incubation with GST-PSA identified a single sequence (DAEFRHDS) corresponding to the first eight amino acids of the Aβ peptide with no detectable secondary species.
3.5. PSA regulates flux through the autophagy pathway
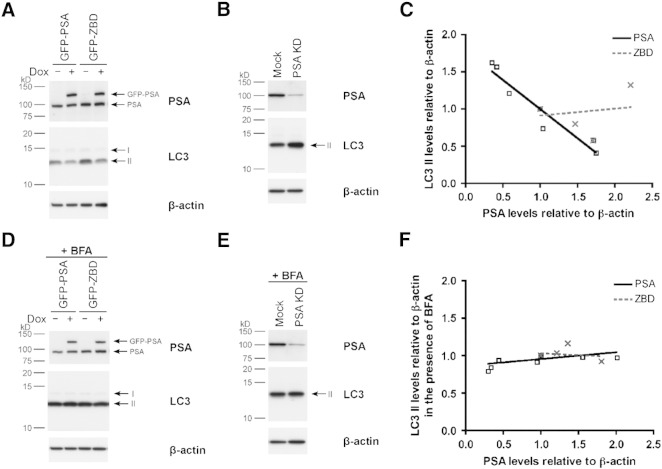
Previous work has shown that PSA is particularly important for the degradation of peptides containing polyglutamine stretches, which are otherwise resistant to proteolysis by the proteasome [18]. Menzies and colleagues have found in similar cell model systems that PSA can further accelerate the clearance of such peptides by stimulating autophagosome production [19]. To investigate the role of PSA in regulating autophagy, we generated HeLa Tet-On stable cell lines (see Supplementary Fig. 4) and measured LC3 II, a key marker of autophagosomes.
Controlled overexpression of GFP-PSA resulted in a reduction in the number of autophagosomes as shown by the decrease in basal LC3 II levels (Fig. 5A). In contrast, siRNA-mediated knockdown of PSA in parental HeLa Tet-On cells led to an accumulation of autophagosomes, which is evident from the increased LC3 II levels, under steady-state conditions compared to mock-transfected cells (Fig. 5B). When we pooled the data from the PSA-overexpression and PSA-RNAi experiments and quantified total PSA (GFP-PSA and endogenous PSA) versus LC3 II levels, we found a robust inverse correlation between the two (Fig. 5C, black solid line, R2 = 0.92). The absence of a similar correlation when we expressed inactive GFP-ZBD (Fig. 5C, gray dashed line) indicated that PSA activity is required to modulate the autophagy pathway and so reduce LC3 II levels.
Fig. 5.
PSA levels regulate autophagic flux.
(A and D) HeLa Tet-On stable cell lines were treated with (+) or without (–) 1 μg/ml doxycycline (Dox) for 48 h to induce expression of GFP-PSA and GFP-ZBD. (B and E) Endogenous PSA was knocked down (PSA KD) in parental HeLa Tet-On cells (a mock transfection is shown as a control) by siRNA and lysed 48 h post-transfection. The samples were separated by SDS-PAGE and the immunoblot was probed with antibodies against PSA and LC3. β-actin is shown as a loading control. (D and E) The cells were treated with 400 nM bafilomycin A1 (BFA) for 4 h before lysis. (C and F) Band intensities for total PSA comprised of endogenous PSA and GFP-PSA (black) or GFP-ZBD (gray), LC3 II, and β-actin were quantified by densitometry using ImageJ and normalized to the respective control: − Dox (A and D) or Mock (B and E). The results were displayed graphically by plotting PSA levels against the corresponding LC3 II levels both relative to β-actin. R2 = 0.92 for the black PSA correlation in (C).
Since LC3 II levels report the number of autophagosomes which in turn depends on the dynamic equilibrium between autophagosome synthesis and degradation, these data are insufficient to determine whether PSA is acting to slow autophagosome formation or alternatively to speed their onward progression and fusion with lysosomes. To address this uncertainty, we dissected the pathway using bafilomycin A1 (BFA) which blocks autophagosome–lysosome fusion [49]. Upon BFA treatment, we observed an invariant increase in LC3 II levels; remarkably the elevated levels of LC3 II did not change regardless of whether GFP-PSA or GFP-ZBD was overexpressed, or endogenous PSA was knocked down by siRNA (Fig. 5D, E, F & Supplementary Fig. 5). The absence of a correlation between PSA and LC3 II levels in the presence of BFA indicates that PSA does not inhibit the formation of autophagosomes, rather it (in the absence of BFA) relieves a block in autophagosome fusion with lysosomes.
Taken together, these data indicate that enzymatically active PSA can relieve a block in the fusion of autophagosomes with lysosomes. This requirement for PSA to be active to permit autophagy stimulation contrasts with PSA-mediated Aβ-protection where the active and inactive isoforms of PSA are equally effective.
4. Discussion
Our screen for genetic modifiers of Aβ toxicity in the fruit fly brain identified a transposon-like P-element insertion that markedly prolonged longevity. This enhancing genetic element was inserted between the gene for Drosophila PSA (dPSA, CG1009) and the fly ortholog of the low density lipoprotein receptor (CG12086). As a first approach to understanding which of the two genes was responsible for the rescue, we consulted the results of our previous transcriptomic analysis of flies expressing Aβ versus controls. In this way, we found that PSA – and not CG12086 – was upregulated in flies expressing Aβ as compared to control flies. Interestingly, similar transcriptional upregulation of PSA has also been reported in PC12 cells expressing huntingtin containing a polyQ expansion, but not for normal huntingtin [50]. Moreover, PSA is also induced in a mouse model expressing an FTD-linked variant of tau [20]. In our study, further benefits of PSA were investigated by generating transgenic flies that coexpress either dPSA or CG12086 with Aβ. We found that all three independent transgenic dPSA lines rescued Aβ toxicity as evidenced by their ability to restore median survival towards normal (Fig. 1A), suppress retinal toxicity (Fig. 1B), and slow the rate of locomotor deterioration (Fig. 1C). By contrast, none of the CG12086-overexpressing lines had beneficial effects. Drosophila PSA also led to clearance of Aβ plaque-like deposits (Fig. 1D) and reduced total Aβ levels in the brain (Fig. 1E) — an effect that could not be explained by any confounding reduction in Aβ mRNA levels as determined by qPCR.
While it is attractive to assume that clearance of Aβ from these fly brains is a result of the proteolytic activity of PSA, certain prerequisites must be fulfilled for this to be possible. The most important criterion is that PSA and Aβ are present in the same compartment, whether that be intra- or extra-cellular. It is generally believed that Aβ is generated in the lumina of vesicles or on the extracellular side of the plasma membrane. In our model system, the Aβ peptide is secreted under the control of a signal peptide, which is cleaved off in the process [51]. The subcellular localization of PSA has not been conclusively demonstrated, although there are reports claiming it is secreted [52] these have been widely discounted in favor of an intracellular distribution (cytoplasmic [45], Golgi apparatus [53], and (peri)nuclear [54]) with a possibility of being membrane-bound [47,55,56]. A crucial goal of this study was to precisely locate PSA in our cell cultures and to determine whether a significant amount might cross membrane to come into contact with extracellular Aβ (or vice versa). Both confocal microscopy of cells expressing GFP-PSA (Fig. 2A) and density gradient ultracentrifugation clearly show that PSA is cytoplasmic and neither secreted (Fig. 2E), nor associated with membranes (Fig. 2C), nor in the nucleus (Fig. 2D). The ability of PSA to modulate Aβ toxicity, despite not being in the same extracellular compartment, suggested another mechanistic possibility: that a toxic fraction of the Aβ peptide may be acting in the cytoplasm. While this requires the Aβ peptide to cross the plasma membrane, the same type of behavior has been described for a number of aggregation-prone proteins [57,58]. To test whether cytoplasmic Aβ could be toxic, we created flies that expressed Aβ without a secretion signal peptide. Surprisingly, these NSP flies exhibited no signs of toxicity (Fig. 3C). mRNA expression levels of the cytoplasmic Aβ peptide were essentially identical to its secreted counterpart (Fig. 3B), yet we observed only minimal accumulation of the peptide in fly brains (Fig. 3E). Hypothesizing that the lack of accumulation was the result of rapid Aβ clearance from neurons, we set out to determine whether RNAi-mediated knockdown of endogenous PSA could enhance the toxicity of non-secreted Aβ. However, we found that control flies and flies expressing cytoplasmic Aβ responded similarly to PSA RNAi indicating that there is no epistatic interaction between PSA knockdown and cytoplasmic Aβ when considering a measure of overall toxicity such as fly longevity (Fig. 3E). By contrast, there was a distinct pattern of longevity changes in response to PSA RNAi in the flies expressing secreted Aβ (Fig. 3E) indicating again an epistatic interaction between secreted Aβ and PSA knockdown.
Taken together, these data indicate that PSA and Aβ are unlikely to be in the same subcellular compartment (Fig. 2) and, when artificially placed in the same compartment, we cannot see a direct epistatic interaction (Fig. 3). Thus, it seems likely that PSA is not directly digesting Aβ, but instead may involve a more indirect mechanism of action. This hypothesis was supported by the in vitro finding that Aβ is not a proteolytic substrate for PSA (Fig. 4C). Furthermore, we demonstrated that the protective role of PSA against Aβ toxicity is independent of its enzymatic activity in SH-SY5Y cells (Fig. 4B). The finding that PSA has a cellular role that is distinct from its proteolytic activity is concordant with studies into its orthologues in the plant kingdom. In particular, the orthologous aminopeptidase M1 (APM1) in Arabidopsis thaliana possesses distinct proteolytic and protein–protein interaction domains, both of which are essential for normal development. Complementation analysis indicates that these two activities do not need to be present in the same polypeptide; indeed the plant enzyme can be complemented by expression, in trans, of orthologous domains of the human enzyme insulin-responsive aminopeptidase [59]. Concordant with a proposed non-proteolytic function for PSA, there is evidence from tauopathy model systems indicating that proteolytic degradation is not the mechanism by which PSA reduces protein deposition and rescues toxicity. In this respect, several groups have shown that PSA reduces tau levels in SH-SY5Y cells [24] and rescues a fly model of tau toxicity [20], yet PSA cannot cleave tau in vitro [25].
After discounting direct proteolysis, a strong candidate for PSA's protective mechanism of action was the ability to stimulate autophagy. Previous work has shown that, in cells expressing proteins with polyQ expansions (as well as other aggregation-prone proteins), PSA is able to cooperate with the proteasome to clear polyQ peptides [18], stimulates autophagosome formation, and reduces inclusion body formation [19]. For these reasons, we examined whether controlled expression of our PSA constructs in stable HeLa Tet-On cell lines could accelerate the formation of autophagosomes as evidenced by an increase in LC3 II levels. Under steady-state conditions, overexpression of GFP-PSA led to a decrease in this autophagy marker, signaling a reduction in the number of autophagosomes; the converse was true when PSA was knocked down. When the data were pooled we could observe a clear inverse linear correlation between the levels of enzymically active PSA and the number of autophagosomes (Fig. 5C). However, when the onward passage of autophagosomes was blocked by bafilomycin A1, LC3 II levels became insensitive to PSA levels under all circumstances (Fig. 5F). These observations indicate that PSA has proteolysis-dependent activity that promotes autophagosome–lysosome fusion; we have also seen that this activity can nonetheless be overwhelmed by BFA blockade.
In summary, we show that the rescue from Aβ toxicity is not the result of PSA proteolysis for two main reasons. Firstly, the two players are unlikely to meet in the same subcellular compartment and secondly, PSA cannot cleave Aβ in vitro. As is the case for tau, the protection of PSA from Aβ toxicity must be indirect. The regulation is unlikely to be transcriptional as the Aβ expression in our fly model is driven by a strong neuronal driver and mRNA levels are not affected by PSA. In cell culture, we could replicate the protective effects of PSA expression against Aβ toxicity but found to our surprise that the inactive ZBD variant of PSA was also beneficial. We hypothesized that PSA may protect Aβ-expressing flies and cells by upregulating autophagy as has been seen in models of Huntington's disease [19]. Pooling data from overexpression and knockdown experiments in SH-SY5Y cells revealed that PSA can modulate autophagic flux, however this effect requires the enzyme to be active. We conclude that PSA is a strong suppressor of Aβ toxicity, mediating its rescue by clearance of the peptide, but achieving this without direct proteolysis. The requirement for PSA to be active in order for it to stimulate autophagy makes this an unlikely explanation for the protection that PSA affords versus Aβ toxicity. The demonstration of important roles for the protein–protein interaction domains in plant orthologues of PSA indicates that a more indirect mechanism may be important. That similar aminopeptidases may be activated by dimerization provides a possible means by which our catalytically inactive PSA could stimulate other potentially protective proteases [59]. However, further work is required to understand the indirect mechanism(s) by which PSA has beneficial effects in multiple models of Alzheimer's disease and other neurodegenerative diseases.
Conflict of interest
Part of this project was funded by a research grant from Merck Sharp & Dohme. There was no commercial bias introduced into the generation or interpretation of the results.
Acknowledgements
We thank Drs. Aileen Moloney, Mark S. Shearman and Matthew Townsend for their support and extremely valuable discussions. We gratefully acknowledge Prof. Dominic Walsh and Prof. Dennis Selkoe for their kind gift of the 7PA2 cells. This work was supported by the Wellcome Trust (DCC & DAL grant code: 082604/2/07/Z) and a Wellcome Trust PhD studentship (AJK), the Medical Research Council (DAL & DCC grant code: G0700990), Papworth NHS Trust (DAL), and an industrial collaboration grant (DCC) and a PhD studentship (RMP) as part of an educational agreement between Merck Sharp & Dohme Ltd. and the University of Cambridge. DCC is an Alzheimer's Research UK Senior Research Fellow (grant code: ART-SRF2010-2). SJM is an MRC Senior Clinical Fellow (grant code: G1002610).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbadis.2013.07.019.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Selkoe D.J. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 2.Jucker M., Walker L.C. Pathogenic protein seeding in Alzheimer disease and other neurodegenerative disorders. Ann. Neurol. 2011;70:532–540. doi: 10.1002/ana.22615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor R.C., Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knowles T.P., Waudby C.A., Devlin G.L., Cohen S.I., Aguzzi A., Vendruscolo M., Terentjev E.M., Welland M.E., Dobson C.M. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe D.J. Toward a comprehensive theory for Alzheimer's disease. Hypothesis: Alzheimer's disease is caused by the cerebral accumulation and cytotoxicity of amyloid beta-protein. Ann. N. Y. Acad. Sci. 2000;924:17–25. doi: 10.1111/j.1749-6632.2000.tb05554.x. [DOI] [PubMed] [Google Scholar]
- 6.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 7.Naslund J., Haroutunian V., Mohs R., Davis K.L., Davies P., Greengard P., Buxbaum J.D. Correlation between elevated levels of amyloid beta-peptide in the brain and cognitive decline. Jama. 2000;283:1571–1577. doi: 10.1001/jama.283.12.1571. [DOI] [PubMed] [Google Scholar]
- 8.Citron M., Vigo-Pelfrey C., Teplow D.B., Miller C., Schenk D., Johnston J., Winblad B., Venizelos N., Lannfelt L., Selkoe D.J. Excessive production of amyloid beta-protein by peripheral cells of symptomatic and presymptomatic patients carrying the Swedish familial Alzheimer disease mutation. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11993–11997. doi: 10.1073/pnas.91.25.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy J. Amyloid, the presenilins and Alzheimer's disease. Trends Neurosci. 1997;20:154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- 10.Farris W., Mansourian S., Chang Y., Lindsley L., Eckman E.A., Frosch M.P., Eckman C.B., Tanzi R.E., Selkoe D.J., Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckman E.A., Watson M., Marlow L., Sambamurti K., Eckman C.B. Alzheimer's disease beta-amyloid peptide is increased in mice deficient in endothelin-converting enzyme. J. Biol. Chem. 2003;278:2081–2084. doi: 10.1074/jbc.C200642200. [DOI] [PubMed] [Google Scholar]
- 12.Iwata N., Tsubuki S., Takaki Y., Shirotani K., Lu B., Gerard N.P., Gerard C., Hama E., Lee H.J., Saido T.C. Metabolic regulation of brain Abeta by neprilysin. Science. 2001;292:1550–1552. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- 13.Eckman E.A., Reed D.K., Eckman C.B. Degradation of the Alzheimer's amyloid beta peptide by endothelin-converting enzyme. J. Biol. Chem. 2001;276:24540–24548. doi: 10.1074/jbc.M007579200. [DOI] [PubMed] [Google Scholar]
- 14.Qiu W.Q., Walsh D.M., Ye Z., Vekrellis K., Zhang J., Podlisny M.B., Rosner M.R., Safavi A., Hersh L.B., Selkoe D.J. Insulin-degrading enzyme regulates extracellular levels of amyloid beta-protein by degradation. J. Biol. Chem. 1998;273:32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- 15.Finelli A., Kelkar A., Song H.J., Yang H., Konsolaki M. A model for studying Alzheimer's Abeta42-induced toxicity in Drosophila melanogaster. Mol. Cell. Neurosci. 2004;26:365–375. doi: 10.1016/j.mcn.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Imarisio S., Carmichael J., Korolchuk V., Chen C.W., Saiki S., Rose C., Krishna G., Davies J.E., Ttofi E., Underwood B.R., Rubinsztein D.C. Huntington's disease: from pathology and genetics to potential therapies. Biochem. J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 17.Sherman M.Y., Goldberg A.L. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 18.Bhutani N., Venkatraman P., Goldberg A.L. Puromycin-sensitive aminopeptidase is the major peptidase responsible for digesting polyglutamine sequences released by proteasomes during protein degradation. EMBO J. 2007;26:1385–1396. doi: 10.1038/sj.emboj.7601592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menzies F.M., Hourez R., Imarisio S., Raspe M., Sadiq O., Chandraratna D., O'Kane C., Rock K.L., Reits E., Goldberg A.L., Rubinsztein D.C. Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum. Mol. Genet. 2010;19:4573–4586. doi: 10.1093/hmg/ddq385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karsten S.L., Sang T.K., Gehman L.T., Chatterjee S., Liu J., Lawless G.M., Sengupta S., Berry R.W., Pomakian J., Oh H.S., Schulz C., Hui K.S., Wiedau-Pazos M., Vinters H.V., Binder L.I., Geschwind D.H., Jackson G.R. A genomic screen for modifiers of tauopathy identifies puromycin-sensitive aminopeptidase as an inhibitor of tau-induced neurodegeneration. Neuron. 2006;51:549–560. doi: 10.1016/j.neuron.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 21.Kudo L.C., Parfenova L., Ren G., Vi N., Hui M., Ma Z., Lau K., Gray M., Bardag-Gorce F., Wiedau-Pazos M., Hui K.S., Karsten S.L. Puromycin-sensitive aminopeptidase (PSA/NPEPPS) impedes development of neuropathology in hPSA/TAU(P301L) double-transgenic mice. Hum Mol Genet. 2011;20:1820–1833. doi: 10.1093/hmg/ddr065. [DOI] [PubMed] [Google Scholar]
- 22.Ren G., Ma Z., Hui M., Kudo L.C., Hui K.S., Karsten S.L. Cu, Zn-superoxide dismutase 1 (SOD1) is a novel target of Puromycin-sensitive aminopeptidase (PSA/NPEPPS): PSA/NPEPPS is a possible modifier of amyotrophic lateral sclerosis. Mol. Neurodegener. 2011;6:29. doi: 10.1186/1750-1326-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sengupta S., Horowitz P.M., Karsten S.L., Jackson G.R., Geschwind D.H., Fu Y., Berry R.W., Binder L.I. Degradation of tau protein by puromycin-sensitive aminopeptidase in vitro. Biochemistry. 2006;45:15111–15119. doi: 10.1021/bi061830d. [DOI] [PubMed] [Google Scholar]
- 24.Yanagi K., Tanaka T., Kato K., Sadik G., Morihara T., Kudo T., Takeda M. Involvement of puromycin-sensitive aminopeptidase in proteolysis of tau protein in cultured cells, and attenuated proteolysis of frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) mutant tau. Psychogeriatrics. 2009;9:157–166. doi: 10.1111/j.1479-8301.2010.00307.x. [DOI] [PubMed] [Google Scholar]
- 25.Chow K.M., Guan H., Hersh L.B. Aminopeptidases do not directly degrade tau protein. Mol. Neurodegener. 2010;5:48. doi: 10.1186/1750-1326-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crowther D.C., Kinghorn K.J., Miranda E., Page R., Curry J.A., Duthie F.A., Gubb D.C., Lomas D.A. Intraneuronal Ab, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer's disease. Neuroscience. 2005;132:123–135. doi: 10.1016/j.neuroscience.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Rival T., Soustelle L., Strambi C., Besson M.T., Iche M., Birman S. Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr. Biol. 2004;14:599–605. doi: 10.1016/j.cub.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 29.Crowther D.C., Page R., Chandraratna D., Lomas D.A. A Drosophila model of Alzheimer's disease. Methods Enzymol. 2006;412:234–255. doi: 10.1016/S0076-6879(06)12015-7. [DOI] [PubMed] [Google Scholar]
- 30.Speretta E., Jahn T.R., Tartaglia G.G., Favrin G., Barros T.P., Imarisio S., Lomas D.A., Luheshi L.M., Crowther D.C., Dobson C.M. Expression in Drosophila of tandem amyloid beta peptides provides insights into links between aggregation and neurotoxicity. J. Biol. Chem. 2012;287:20748–20754. doi: 10.1074/jbc.M112.350124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa R., Speretta E., Crowther D.C., Cardoso I. Testing the therapeutic potential of doxycycline in a Drosophila melanogaster model of Alzheimer disease. J. Biol. Chem. 2011;286:41647–41655. doi: 10.1074/jbc.M111.274548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podlisny M.B., Ostaszewski B.L., Squazzo S.L., Koo E.H., Rydell R.E., Teplow D.B., Selkoe D.J. Aggregation of secreted amyloid beta-protein into sodium dodecyl sulfate-stable oligomers in cell culture. J. Biol. Chem. 1995;270:9564–9570. doi: 10.1074/jbc.270.16.9564. [DOI] [PubMed] [Google Scholar]
- 33.Sarkar S., Ravikumar B., Rubinsztein D.C. Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods Enzymol. 2009;453:83–110. doi: 10.1016/S0076-6879(08)04005-6. [DOI] [PubMed] [Google Scholar]
- 34.Swerdlow P.S., Finley D., Varshavsky A. Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal. Biochem. 1986;156:147–153. doi: 10.1016/0003-2697(86)90166-1. [DOI] [PubMed] [Google Scholar]
- 35.Ida N., Hartmann T., Pantel J., Schroder J., Zerfass R., Forstl H., Sandbrink R., Masters C.L., Beyreuther K. Analysis of heterogeneous A4 peptides in human cerebrospinal fluid and blood by a newly developed sensitive Western blot assay. J. Biol. Chem. 1996;271:22908–22914. doi: 10.1074/jbc.271.37.22908. [DOI] [PubMed] [Google Scholar]
- 36.Shankar G.M., Welzel A.T., McDonald J.M., Selkoe D.J., Walsh D.M. Isolation of low-n amyloid beta-protein oligomers from cultured cells, CSF, and brain. Meth. Mol Biol. 2011;670:33–44. doi: 10.1007/978-1-60761-744-0_3. [DOI] [PubMed] [Google Scholar]
- 37.Scorsone K.A., Panniers R., Rowlands A.G., Henshaw E.C. Phosphorylation of eukaryotic initiation factor 2 during physiological stresses which affect protein synthesis. J. Biol. Chem. 1987;262:14538–14543. [PubMed] [Google Scholar]
- 38.Marciniak S.J., Yun C.Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H.P., Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teplow D.B. Preparation of amyloid beta-protein for structural and functional studies. Methods Enzymol. 2006;413:20–33. doi: 10.1016/S0076-6879(06)13002-5. [DOI] [PubMed] [Google Scholar]
- 40.Bolognesi B., Kumita J.R., Barros T.P., Esbjorner E.K., Luheshi L.M., Crowther D.C., Wilson M.R., Dobson C.M., Favrin G., Yerbury J.J. ANS binding reveals common features of cytotoxic amyloid species. ACS Chem. Biol. 2010;5:735–740. doi: 10.1021/cb1001203. [DOI] [PubMed] [Google Scholar]
- 41.Brorsson A.C., Bolognesi B., Tartaglia G.G., Shammas S.L., Favrin G., Watson I., Lomas D.A., Chiti F., Vendruscolo M., Dobson C.M., Crowther D.C., Luheshi L.M. Intrinsic determinants of neurotoxic aggregate formation by the amyloid beta peptide. Biophys. J. 2010;98:1677–1684. doi: 10.1016/j.bpj.2009.12.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasteiger E., Hoogland C., Gattiker A., Davaud S., Wilkins M.R., Appel R.D., Bairoch A. Identification and analysis tools on the ExPASy Server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press Inc.; New Jersey: 2005. pp. 571–607. [Google Scholar]
- 43.Nilsberth C., Westlind-Danielsson A., Eckman C.B., Condron M.M., Axelman K., Forsell C., Stenh C., Luthman J., Teplow D.B., Younkin S.G., Naslund J., Lannfelt L. The ‘Arctic’ APP mutation (E693G) causes Alzheimer's disease by enhanced Abeta protofibril formation. Nat. Neurosci. 2001;4:887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 44.Rival T., Page R.M., Chandraratna D.S., Sendall T.J., Ryder E., Liu B., Lewis H., Rosahl T., Hider R., Camargo L.M., Shearman M.S., Crowther D.C., Lomas D.A. Fenton chemistry and oxidative stress mediate the toxicity of the beta-amyloid peptide in a Drosophila model of Alzheimer's disease. Eur. J. Neurosci. 2009;29:1335–1347. doi: 10.1111/j.1460-9568.2009.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tobler A.R., Constam D.B., Schmitt-Graff A., Malipiero U., Schlapbach R., Fontana A. Cloning of the human puromycin-sensitive aminopeptidase and evidence for expression in neurons. J. Neurochem. 1997;68:889–897. doi: 10.1046/j.1471-4159.1997.68030889.x. [DOI] [PubMed] [Google Scholar]
- 46.Huber G., Thompson A., Gruninger F., Mechler H., Hochstrasser R., Hauri H.P., Malherbe P. cDNA cloning and molecular characterization of human brain metalloprotease MP100: a beta-secretase candidate? J. Neurochem. 1999;72:1215–1223. doi: 10.1046/j.1471-4159.1999.0721215.x. [DOI] [PubMed] [Google Scholar]
- 47.Matsas R., Stephenson S.L., Hryszko J., Kenny A.J., Turner A.J. The metabolism of neuropeptides. Phase separation of synaptic membrane preparations with Triton X-114 reveals the presence of aminopeptidase N. Biochem. J. 1985;231:445–449. doi: 10.1042/bj2310445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson M.W., Govindaswami M., Hersh L.B. Mutation of active site residues of the puromycin-sensitive aminopeptidase: conversion of the enzyme into a catalytically inactive binding protein. Arch. Biochem. Biophys. 2003;413:236–242. doi: 10.1016/s0003-9861(03)00123-1. [DOI] [PubMed] [Google Scholar]
- 49.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., Ahn H.J., Ait-Mohamed O., Ait-Si-Ali S., Akematsu T., Akira S., Al-Younes H.M., Al-Zeer M.A., Albert M.L., Albin R.L., Alegre-Abarrategui J., Aleo M.F., Alirezaei M., Almasan A., Almonte-Becerril M., Amano A., Amaravadi R., Amarnath S., Amer A.O., Andrieu-Abadie N., Anantharam V., Ann D.K., Anoopkumar-Dukie S., Aoki H., Apostolova N., Arancia G., Aris J.P., Asanuma K., Asare N.Y., Ashida H., Askanas V., Askew D.S., Auberger P., Baba M., Backues S.K., Baehrecke E.H., Bahr B.A., Bai X.Y., Bailly Y., Baiocchi R., Baldini G., Balduini W., Ballabio A., Bamber B.A., Bampton E.T., Banhegyi G., Bartholomew C.R., Bassham D.C., Bast R.C., Jr., Batoko H., Bay B.H., Beau I., Bechet D.M., Begley T.J., Behl C., Behrends C., Bekri S., Bellaire B., Bendall L.J., Benetti L., Berliocchi L., Bernardi H., Bernassola F., Besteiro S., Bhatia-Kissova I., Bi X., Biard-Piechaczyk M., Blum J.S., Boise L.H., Bonaldo P., Boone D.L., Bornhauser B.C., Bortoluci K.R., Bossis I., Bost F., Bourquin J.P., Boya P., Boyer-Guittaut M., Bozhkov P.V., Brady N.R., Brancolini C., Brech A., Brenman J.E., Brennand A., Bresnick E.H., Brest P., Bridges D., Bristol M.L., Brookes P.S., Brown E.J., Brumell J.H., Brunetti-Pierri N., Brunk U.T., Bulman D.E., Bultman S.J., Bultynck G., Burbulla L.F., Bursch W., Butchar J.P., Buzgariu W., Bydlowski S.P., Cadwell K., Cahova M., Cai D., Cai J., Cai Q., Calabretta B., Calvo-Garrido J., Camougrand N., Campanella M., Campos-Salinas J., Candi E., Cao L., Caplan A.B., Carding S.R., Cardoso S.M., Carew J.S., Carlin C.R., Carmignac V., Carneiro L.A., Carra S., Caruso R.A., Casari G., Casas C., Castino R., Cebollero E., Cecconi F., Celli J., Chaachouay H., Chae H.J., Chai C.Y., Chan D.C., Chan E.Y., Chang R.C., Che C.M., Chen C.C., Chen G.C., Chen G.Q., Chen M., Chen Q., Chen S.S., Chen W., Chen X., Chen Y.G., Chen Y., Chen Y.J., Chen Z., Cheng A., Cheng C.H., Cheng Y., Cheong H., Cheong J.H., Cherry S., Chess-Williams R., Cheung Z.H., Chevet E., Chiang H.L., Chiarelli R., Chiba T., Chin L.S., Chiou S.H., Chisari F.V., Cho C.H., Cho D.H., Choi A.M., Choi D., Choi K.S., Choi M.E., Chouaib S., Choubey D., Choubey V., Chu C.T., Chuang T.H., Chueh S.H., Chun T., Chwae Y.J., Chye M.L., Ciarcia R., Ciriolo M.R., Clague M.J., Clark R.S., Clarke P.G., Clarke R., Codogno P., Coller H.A., Colombo M.I., Comincini S., Condello M., Condorelli F., Cookson M.R., Coombs G.H., Coppens I., Corbalan R., Cossart P., Costelli P., Costes S., Coto-Montes A., Couve E., Coxon F.P., Cregg J.M., Crespo J.L., Cronje M.J., Cuervo A.M., Cullen J.J., Czaja M.J., D'Amelio M., Darfeuille-Michaud A., Davids L.M., Davies F.E., De Felici M., de Groot J.F., de Haan C.A., De Martino L., De Milito A., De Tata V., Debnath J., Degterev A., Dehay B., Delbridge L.M., Demarchi F., Deng Y.Z., Dengjel J., Dent P., Denton D., Deretic V., Desai S.D., Devenish R.J., Di Gioacchino M., Di Paolo G., Di Pietro C., Diaz-Araya G., Diaz-Laviada I., Diaz-Meco M.T., Diaz-Nido J., Dikic I., Dinesh-Kumar S.P., Ding W.X., Distelhorst C.W., Diwan A., Djavaheri-Mergny M., Dokudovskaya S., Dong Z., Dorsey F.C., Dosenko V., Dowling J.J., Doxsey S., Dreux M., Drew M.E., Duan Q., Duchosal M.A., Duff K., Dugail I., Durbeej M., Duszenko M., Edelstein C.L., Edinger A.L., Egea G., Eichinger L., Eissa N.T., Ekmekcioglu S., El-Deiry W.S., Elazar Z., Elgendy M., Ellerby L.M., Eng K.E., Engelbrecht A.M., Engelender S., Erenpreisa J., Escalante R., Esclatine A., Eskelinen E.L., Espert L., Espina V., Fan H., Fan J., Fan Q.W., Fan Z., Fang S., Fang Y., Fanto M., Fanzani A., Farkas T., Farre J.C., Faure M., Fechheimer M., Feng C.G., Feng J., Feng Q., Feng Y., Fesus L., Feuer R., Figueiredo-Pereira M.E., Fimia G.M., Fingar D.C., Finkbeiner S., Finkel T., Finley K.D., Fiorito F., Fisher E.A., Fisher P.B., Flajolet M., Florez-McClure M.L., Florio S., Fon E.A., Fornai F., Fortunato F., Fotedar R., Fowler D.H., Fox H.S., Franco R., Frankel L.B., Fransen M., Fuentes J.M., Fueyo J., Fujii J., Fujisaki K., Fujita E., Fukuda M., Furukawa R.H., Gaestel M., Gailly P., Gajewska M., Galliot B., Galy V., Ganesh S., Ganetzky B., Ganley I.G., Gao F.B., Gao G.F., Gao J., Garcia L., Garcia-Manero G., Garcia-Marcos M., Garmyn M., Gartel A.L., Gatti E., Gautel M., Gawriluk T.R., Gegg M.E., Geng J., Germain M., Gestwicki J.E., Gewirtz D.A., Ghavami S., Ghosh P., Giammarioli A.M., Giatromanolaki A.N., Gibson S.B., Gilkerson R.W., Ginger M.L., Ginsberg H.N., Golab J., Goligorsky M.S., Golstein P., Gomez-Manzano C., Goncu E., Gongora C., Gonzalez C.D., Gonzalez R., Gonzalez-Estevez C., Gonzalez-Polo R.A., Gonzalez-Rey E., Gorbunov N.V., Gorski S., Goruppi S., Gottlieb R.A., Gozuacik D., Granato G.E., Grant G.D., Green K.N., Gregorc A., Gros F., Grose C., Grunt T.W., Gual P., Guan J.L., Guan K.L., Guichard S.M., Gukovskaya A.S., Gukovsky I., Gunst J., Gustafsson A.B., Halayko A.J., Hale A.N., Halonen S.K., Hamasaki M., Han F., Han T., Hancock M.K., Hansen M., Harada H., Harada M., Hardt S.E., Harper J.W., Harris A.L., Harris J., Harris S.D., Hashimoto M., Haspel J.A., Hayashi S., Hazelhurst L.A., He C., He Y.W., Hebert M.J., Heidenreich K.A., Helfrich M.H., Helgason G.V., Henske E.P., Herman B., Herman P.K., Hetz C., Hilfiker S., Hill J.A., Hocking L.J., Hofman P., Hofmann T.G., Hohfeld J., Holyoake T.L., Hong M.H., Hood D.A., Hotamisligil G.S., Houwerzijl E.J., Hoyer-Hansen M., Hu B., Hu C.A., Hu H.M., Hua Y., Huang C., Huang J., Huang S., Huang W.P., Huber T.B., Huh W.K., Hung T.H., Hupp T.R., Hur G.M., Hurley J.B., Hussain S.N., Hussey P.J., Hwang J.J., Hwang S., Ichihara A., Ilkhanizadeh S., Inoki K., Into T., Iovane V., Iovanna J.L., Ip N.Y., Isaka Y., Ishida H., Isidoro C., Isobe K., Iwasaki A., Izquierdo M., Izumi Y., Jaakkola P.M., Jaattela M., Jackson G.R., Jackson W.T., Janji B., Jendrach M., Jeon J.H., Jeung E.B., Jiang H., Jiang J.X., Jiang M., Jiang Q., Jiang X., Jimenez A., Jin M., Jin S., Joe C.O., Johansen T., Johnson D.E., Johnson G.V., Jones N.L., Joseph B., Joseph S.K., Joubert A.M., Juhasz G., Juillerat-Jeanneret L., Jung C.H., Jung Y.K., Kaarniranta K., Kaasik A., Kabuta T., Kadowaki M., Kagedal K., Kamada Y., Kaminskyy V.O., Kampinga H.H., Kanamori H., Kang C., Kang K.B., Kang K.I., Kang R., Kang Y.A., Kanki T., Kanneganti T.D., Kanno H., Kanthasamy A.G., Kanthasamy A., Karantza V., Kaushal G.P., Kaushik S., Kawazoe Y., Ke P.Y., Kehrl J.H., Kelekar A., Kerkhoff C., Kessel D.H., Khalil H., Kiel J.A., Kiger A.A., Kihara A., Kim D.R., Kim D.H., Kim E.K., Kim H.R., Kim J.S., Kim J.H., Kim J.C., Kim J.K., Kim P.K., Kim S.W., Kim Y.S., Kim Y., Kimchi A., Kimmelman A.C., King J.S., Kinsella T.J., Kirkin V., Kirshenbaum L.A., Kitamoto K., Kitazato K., Klein L., Klimecki W.T., Klucken J., Knecht E., Ko B.C., Koch J.C., Koga H., Koh J.Y., Koh Y.H., Koike M., Komatsu M., Kominami E., Kong H.J., Kong W.J., Korolchuk V.I., Kotake Y., Koukourakis M.I., Kouri Flores J.B., Kovacs A.L., Kraft C., Krainc D., Kramer H., Kretz-Remy C., Krichevsky A.M., Kroemer G., Kruger R., Krut O., Ktistakis N.T., Kuan C.Y., Kucharczyk R., Kumar A., Kumar R., Kumar S., Kundu M., Kung H.J., Kurz T., Kwon H.J., La Spada A.R., Lafont F., Lamark T., Landry J., Lane J.D., Lapaquette P., Laporte J.F., Laszlo L., Lavandero S., Lavoie J.N., Layfield R., Lazo P.A., Le W., Le Cam L., Ledbetter D.J., Lee A.J., Lee B.W., Lee G.M., Lee J., Lee J.H., Lee M., Lee M.S., Lee S.H., Leeuwenburgh C., Legembre P., Legouis R., Lehmann M., Lei H.Y., Lei Q.Y., Leib D.A., Leiro J., Lemasters J.J., Lemoine A., Lesniak M.S., Lev D., Levenson V.V., Levine B., Levy E., Li F., Li J.L., Li L., Li S., Li W., Li X.J., Li Y.B., Li Y.P., Liang C., Liang Q., Liao Y.F., Liberski P.P., Lieberman A., Lim H.J., Lim K.L., Lim K., Lin C.F., Lin F.C., Lin J., Lin J.D., Lin K., Lin W.W., Lin W.C., Lin Y.L., Linden R., Lingor P., Lippincott-Schwartz J., Lisanti M.P., Liton P.B., Liu B., Liu C.F., Liu K., Liu L., Liu Q.A., Liu W., Liu Y.C., Liu Y., Lockshin R.A., Lok C.N., Lonial S., Loos B., Lopez-Berestein G., Lopez-Otin C., Lossi L., Lotze M.T., Low P., Lu B., Lu Z., Luciano F., Lukacs N.W., Lund A.H., Lynch-Day M.A., Ma Y., Macian F., MacKeigan J.P., Macleod K.F., Madeo F., Maiuri L., Maiuri M.C., Malagoli D., Malicdan M.C., Malorni W., Man N., Mandelkow E.M., Manon S., Manov I., Mao K., Mao X., Mao Z., Marambaud P., Marazziti D., Marcel Y.L., Marchbank K., Marchetti P., Marciniak S.J., Marcondes M., Mardi M., Marfe G., Marino G., Markaki M., Marten M.R., Martin S.J., Martinand-Mari C., Martinet W., Martinez-Vicente M., Masini M., Matarrese P., Matsuo S., Matteoni R., Mayer A., Mazure N.M., McConkey D.J., McConnell M.J., McDermott C., McDonald C., McInerney G.M., McKenna S.L., McLaughlin B., McLean P.J., McMaster C.R., McQuibban G.A., Meijer A.J., Meisler M.H., Melendez A., Melia T.J., Melino G., Mena M.A., Menendez J.A., Menna-Barreto R.F., Menon M.B., Menzies F.M., Mercer C.A., Merighi A., Merry D.E., Meschini S., Meyer C.G., Meyer T.F., Miao C.Y., Miao J.Y., Michels P.A., Michiels C., Mijaljica D., Milojkovic A., Minucci S., Miracco C., Miranti C.K., Mitroulis I., Miyazawa K., Mizushima N., Mograbi B., Mohseni S., Molero X., Mollereau B., Mollinedo F., Momoi T., Monastyrska I., Monick M.M., Monteiro M.J., Moore M.N., Mora R., Moreau K., Moreira P.I., Moriyasu Y., Moscat J., Mostowy S., Mottram J.C., Motyl T., Moussa C.E., Muller S., Munger K., Munz C., Murphy L.O., Murphy M.E., Musaro A., Mysorekar I., Nagata E., Nagata K., Nahimana A., Nair U., Nakagawa T., Nakahira K., Nakano H., Nakatogawa H., Nanjundan M., Naqvi N.I., Narendra D.P., Narita M., Navarro M., Nawrocki S.T., Nazarko T.Y., Nemchenko A., Netea M.G., Neufeld T.P., Ney P.A., Nezis I.P., Nguyen H.P., Nie D., Nishino I., Nislow C., Nixon R.A., Noda T., Noegel A.A., Nogalska A., Noguchi S., Notterpek L., Novak I., Nozaki T., Nukina N., Nurnberger T., Nyfeler B., Obara K., Oberley T.D., Oddo S., Ogawa M., Ohashi T., Okamoto K., Oleinick N.L., Oliver F.J., Olsen L.J., Olsson S., Opota O., Osborne T.F., Ostrander G.K., Otsu K., Ou J.H., Ouimet M., Overholtzer M., Ozpolat B., Paganetti P., Pagnini U., Pallet N., Palmer G.E., Palumbo C., Pan T., Panaretakis T., Pandey U.B., Papackova Z., Papassideri I., Paris I., Park J., Park O.K., Parys J.B., Parzych K.R., Patschan S., Patterson C., Pattingre S., Pawelek J.M., Peng J., Perlmutter D.H., Perrotta I., Perry G., Pervaiz S., Peter M., Peters G.J., Petersen M., Petrovski G., Phang J.M., Piacentini M., Pierre P., Pierrefite-Carle V., Pierron G., Pinkas-Kramarski R., Piras A., Piri N., Platanias L.C., Poggeler S., Poirot M., Poletti A., Pous C., Pozuelo-Rubio M., Praetorius-Ibba M., Prasad A., Prescott M., Priault M., Produit-Zengaffinen N., Progulske-Fox A., Proikas-Cezanne T., Przedborski S., Przyklenk K., Puertollano R., Puyal J., Qian S.B., Qin L., Qin Z.H., Quaggin S.E., Raben N., Rabinowich H., Rabkin S.W., Rahman I., Rami A., Ramm G., Randall G., Randow F., Rao V.A., Rathmell J.C., Ravikumar B., Ray S.K., Reed B.H., Reed J.C., Reggiori F., Regnier-Vigouroux A., Reichert A.S., Reiners J.J., Jr., Reiter R.J., Ren J., Revuelta J.L., Rhodes C.J., Ritis K., Rizzo E., Robbins J., Roberge M., Roca H., Roccheri M.C., Rocchi S., Rodemann H.P., Rodriguez de Cordoba S., Rohrer B., Roninson I.B., Rosen K., Rost-Roszkowska M.M., Rouis M., Rouschop K.M., Rovetta F., Rubin B.P., Rubinsztein D.C., Ruckdeschel K., Rucker E.B., III, Rudich A., Rudolf E., Ruiz-Opazo N., Russo R., Rusten T.E., Ryan K.M., Ryter S.W., Sabatini D.M., Sadoshima J., Saha T., Saitoh T., Sakagami H., Sakai Y., Salekdeh G.H., Salomoni P., Salvaterra P.M., Salvesen G., Salvioli R., Sanchez A.M., Sanchez-Alcazar J.A., Sanchez-Prieto R., Sandri M., Sankar U., Sansanwal P., Santambrogio L., Saran S., Sarkar S., Sarwal M., Sasakawa C., Sasnauskiene A., Sass M., Sato K., Sato M., Schapira A.H., Scharl M., Schatzl H.M., Scheper W., Schiaffino S., Schneider C., Schneider M.E., Schneider-Stock R., Schoenlein P.V., Schorderet D.F., Schuller C., Schwartz G.K., Scorrano L., Sealy L., Seglen P.O., Segura-Aguilar J., Seiliez I., Seleverstov O., Sell C., Seo J.B., Separovic D., Setaluri V., Setoguchi T., Settembre C., Shacka J.J., Shanmugam M., Shapiro I.M., Shaulian E., Shaw R.J., Shelhamer J.H., Shen H.M., Shen W.C., Sheng Z.H., Shi Y., Shibuya K., Shidoji Y., Shieh J.J., Shih C.M., Shimada Y., Shimizu S., Shintani T., Shirihai O.S., Shore G.C., Sibirny A.A., Sidhu S.B., Sikorska B., Silva-Zacarin E.C., Simmons A., Simon A.K., Simon H.U., Simone C., Simonsen A., Sinclair D.A., Singh R., Sinha D., Sinicrope F.A., Sirko A., Siu P.M., Sivridis E., Skop V., Skulachev V.P., Slack R.S., Smaili S.S., Smith D.R., Soengas M.S., Soldati T., Song X., Sood A.K., Soong T.W., Sotgia F., Spector S.A., Spies C.D., Springer W., Srinivasula S.M., Stefanis L., Steffan J.S., Stendel R., Stenmark H., Stephanou A., Stern S.T., Sternberg C., Stork B., Stralfors P., Subauste C.S., Sui X., Sulzer D., Sun J., Sun S.Y., Sun Z.J., Sung J.J., Suzuki K., Suzuki T., Swanson M.S., Swanton C., Sweeney S.T., Sy L.K., Szabadkai G., Tabas I., Taegtmeyer H., Tafani M., Takacs-Vellai K., Takano Y., Takegawa K., Takemura G., Takeshita F., Talbot N.J., Tan K.S., Tanaka K., Tang D., Tanida I., Tannous B.A., Tavernarakis N., Taylor G.S., Taylor G.A., Taylor J.P., Terada L.S., Terman A., Tettamanti G., Thevissen K., Thompson C.B., Thorburn A., Thumm M., Tian F., Tian Y., Tocchini-Valentini G., Tolkovsky A.M., Tomino Y., Tonges L., Tooze S.A., Tournier C., Tower J., Towns R., Trajkovic V., Travassos L.H., Tsai T.F., Tschan M.P., Tsubata T., Tsung A., Turk B., Turner L.S., Tyagi S.C., Uchiyama Y., Ueno T., Umekawa M., Umemiya-Shirafuji R., Unni V.K., Vaccaro M.I., Valente E.M., Van den Berghe G., van der Klei I.J., van Doorn W., van Dyk L.F., van Egmond M., van Grunsven L.A., Vandenabeele P., Vandenberghe W.P., Vanhorebeek I., Vaquero E.C., Velasco G., Vellai T., Vicencio J.M., Vierstra R.D., Vila M., Vindis C., Viola G., Viscomi M.T., Voitsekhovskaja O.V., von Haefen C., Votruba M., Wada K., Wade-Martins R., Walker C.L., Walsh C.M., Walter J., Wan X.B., Wang A., Wang C., Wang D., Wang F., Wang G., Wang H., Wang H.G., Wang H.D., Wang J., Wang K., Wang M., Wang R.C., Wang X., Wang Y.J., Wang Y., Wang Z., Wang Z.C., Wansink D.G., Ward D.M., Watada H., Waters S.L., Webster P., Wei L., Weihl C.C., Weiss W.A., Welford S.M., Wen L.P., Whitehouse C.A., Whitton J.L., Whitworth A.J., Wileman T., Wiley J.W., Wilkinson S., Willbold D., Williams R.L., Williamson P.R., Wouters B.G., Wu C., Wu D.C., Wu W.K., Wyttenbach A., Xavier R.J., Xi Z., Xia P., Xiao G., Xie Z., Xu D.Z., Xu J., Xu L., Xu X., Yamamoto A., Yamashina S., Yamashita M., Yan X., Yanagida M., Yang D.S., Yang E., Yang J.M., Yang S.Y., Yang W., Yang W.Y., Yang Z., Yao M.C., Yao T.P., Yeganeh B., Yen W.L., Yin J.J., Yin X.M., Yoo O.J., Yoon G., Yoon S.Y., Yorimitsu T., Yoshikawa Y., Yoshimori T., Yoshimoto K., You H.J., Youle R.J., Younes A., Yu L., Yu S.W., Yu W.H., Yuan Z.M., Yue Z., Yun C.H., Yuzaki M., Zabirnyk O., Silva-Zacarin E., Zacks D., Zacksenhaus E., Zaffaroni N., Zakeri Z., Zeh H.J., III, Zeitlin S.O., Zhang H., Zhang H.L., Zhang J., Zhang J.P., Zhang L., Zhang M.Y., Zhang X.D., Zhao M., Zhao Y.F., Zhao Y., Zhao Z.J., Zheng X., Zhivotovsky B., Zhong Q., Zhou C.Z., Zhu C., Zhu W.G., Zhu X.F., Zhu X., Zhu Y., Zoladek T., Zong W.X., Zorzano A., Zschocke J., Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kita H., Carmichael J., Swartz J., Muro S., Wyttenbach A., Matsubara K., Rubinsztein D.C., Kato K. Modulation of polyglutamine-induced cell death by genes identified by expression profiling. Hum. Mol. Genet. 2002;11:2279–2287. doi: 10.1093/hmg/11.19.2279. [DOI] [PubMed] [Google Scholar]
- 51.Iijima K., Liu H.P., Chiang A.S., Hearn S.A., Konsolaki M., Zhong Y. Dissecting the pathological effects of human A{beta}40 and A{beta}42 in Drosophila: a potential model for Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6623–6628. doi: 10.1073/pnas.0400895101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hui K.S. Brain-specific aminopeptidase: from enkephalinase to protector against neurodegeneration. Neurochem. Res. 2007;32:2062–2071. doi: 10.1007/s11064-007-9356-3. [DOI] [PubMed] [Google Scholar]
- 53.Huber A.B., Brosamle C., Mechler H., Huber G. Metalloprotease MP100: a synaptic protease in rat brain. Brain Res. 1999;837:193–202. doi: 10.1016/s0006-8993(99)01693-5. [DOI] [PubMed] [Google Scholar]
- 54.Constam D.B., Tobler A.R., Rensing-Ehl A., Kemler I., Hersh L.B., Fontana A. Puromycin-sensitive aminopeptidase. Sequence analysis, expression, and functional characterization. J. Biol. Chem. 1995;270:26931–26939. doi: 10.1074/jbc.270.45.26931. [DOI] [PubMed] [Google Scholar]
- 55.McLellan S., Dyer S.H., Rodriguez G., Hersh L.B. Studies on the tissue distribution of the puromycin-sensitive enkephalin-degrading aminopeptidases. J. Neurochem. 1988;51:1552–1559. doi: 10.1111/j.1471-4159.1988.tb01124.x. [DOI] [PubMed] [Google Scholar]
- 56.Dyer S.H., Slaughter C.A., Orth K., Moomaw C.R., Hersh L.B. Comparison of the soluble and membrane-bound forms of the puromycin-sensitive enkephalin-degrading aminopeptidases from rat. J. Neurochem. 1990;54:547–554. doi: 10.1111/j.1471-4159.1990.tb01906.x. [DOI] [PubMed] [Google Scholar]
- 57.Munch C., O'Brien J., Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3548–3553. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren P.H., Lauckner J.E., Kachirskaia I., Heuser J.E., Melki R., Kopito R.R. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat. Cell Biol. 2009;11:219–225. doi: 10.1038/ncb1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hosein F.N., Bandyopadhyay A., Peer W.A., Murphy A.S. The catalytic and protein–protein interaction domains are required for APM1 function. Plant Physiol. 2010;152:2158–2172. doi: 10.1104/pp.109.148742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.