Abstract
Trinucleotide repeat disorders are a heterogeneous group of diseases caused by the expansion, beyond a pathogenic threshold, of unstable DNA tracts in different genes. Sequence interruptions in the repeats have been described in the majority of these disorders and may influence disease phenotype and heritability. Spinal bulbar muscular atrophy (SBMA) is a motor neuron disease caused by a CAG trinucleotide expansion in the androgen receptor (AR) gene. Diagnostic testing and previous research have relied on fragment analysis polymerase chain reaction to determine the AR CAG repeat size, and have therefore not been able to assess the presence of interruptions. We here report a sequencing study of the AR CAG repeat in a cohort of SBMA patients and control subjects in the United Kingdom. We found no repeat interruptions to be present, and we describe differences between sequencing and traditional sizing methods.
Keywords: Kennedy's disease, Spinal bulbar muscular atrophy, CAG, Trinucleotide, Interruptions
1. Introduction
Spinal bulbar muscular atrophy (SBMA), also known as Kennedy's disease, is a slowly progressive, adult-onset, motor neuron disease, inherited in an X-linked recessive manner and caused by a CAG repeat expansion in the first exon of the androgen receptor gene (AR) (La Spada et al., 1991). Although normal individuals carry 9 to 36 CAG repeats, SBMA patients carry 38 to 62 repeats (La Spada et al., 1991). SBMA is part of a wider group of neurological diseases in which the causative genetic defect is a coding CAG expansion and for which the pathogenic threshold is approximately 35 to 40 repeats (Table 1) (Atsuta et al., 2006; Orr and Zoghbi, 2007). Interestingly, although most frequently the CAG expansions are pure runs of CAGs, sequence interruptions have been documented in the majority of these disease genes (Table 1).
Table 1.
Summary of CAG characteristics in diseases caused by CAG coding expansions
| Disease | Gene | No. of CAG repeats |
Presence of interruptions in CAG repeat sequence | Reference | |||
|---|---|---|---|---|---|---|---|
| Normal allele | Intermediate allele | Reduced penetrance allele | Fully penetrant allele | ||||
| DRPLA | ATN1 | 6–35 | 36–47 | 48–93 | No | Koide et al., 1994 | |
| SCA1 | ATXN1 | 6–38 | 36–38 | 39–91 | Yes | Orr et al., 1993 | |
| SCA2 | ATXN2 | 14–31 | 32–270 | Yes | Pulst et al., 1996 | ||
| SCA3 | ATXN3 | 12–44 | 45–51 | 52–86 | Yes | Kawaguchi et al., 1994 | |
| SCA17 | TBP | 25–40 | 41–48 | 49–66 | Yes | Koide et al., 1999 | |
| HD | HTT | ≤26 | 27–35 | 36–39 | ≥40 | Yes | Huntington's Disease Collaborative Research Group, 1993 |
| SBMA | AR | 9–36 | 37 | 38–62 | No | La Spada et al., 1991; present work | |
Key: SBMA, spinal bulbar muscular atrophy.
Interruptions have been shown to play a role in determining the clinical phenotype and the disease heritability, and therefore carry potentially important information for genetic testing and counseling (Chong et al., 1995; Matsuyama et al., 1999; Yu et al., 2011). Pulic genomic databases report rare sequence variations (rs62636527, rs62636528, rs62636529) occurring with low frequency (0.013–0.043) in the AR CAG repeat, but SBMA diagnostic testing and previous studies have mostly used fragment sizing analysis to assess the presence and size of CAG repeats (Atsuta et al., 2006; La Spada et al., 1991), and have not been able to systematically address whether there are interruptions in SBMA expansions, thus warranting direct target sequence analysis.
2. Methods
Details of the patient cohorts, fragment sizing, and sequencing are given in the Supplementary text. The study was approved by the local ethical committee.
3. Results
To investigate the presence of interruptions in the AR CAG repeat, we sequenced the AR CAG repeat in a population of 40 SBMA patients and 93 control subjects in the United Kingdom. Overall, SBMA samples showed a median of 45 repeats (range, 40–53 repeats; Supplementary Fig. 1), which is comparable to the results reported in previous U.S. and Japanese series (Atsuta et al., 2006; La Spada et al., 1991). The quality of our direct sequencing of the CAG expansion allowed us to address the presence of interruptions by visual inspection in both forward and reverse sequences of the amplicon (Supplementary Fig. 2). We did not find interruptions in any of our SBMA case patients or control subjects (Table 1).
Interestingly, when comparing sizing obtained by sequencing to the fragment analysis used diagnostically, we observed a constant difference of 3 to 4 repeats, with estimation from fragment analysis being smaller. This was not dependent on the allele size, and was constant among pathological alleles and controls.
4. Discussion
Diagnostic testing and previous large-scale studies have relied on polymerase chain reaction fragment analysis for sizing the CAG repeat in AR, and have therefore not been able to address the presence of sequence interruptions in a large number of samples (Atsuta et al., 2006; La Spada et al., 1991). This is an important issue, because interruptions have been shown to be present in the majority of CAG expansion diseases and to play a role in clinical features of the disease, such as age of onset and presentation. In the case of ataxin 2 (ATXN2), the combination of presence of interruptions and repeat size determines very distinct clinical presentations that encompass SCA2, L-DOPA–responsive parkinsonism, and amyotrophic lateral sclerosis (Matsuyama et al., 1999; Yu et al., 2011). Interruptions have also been shown to influence the capability of the repeat to expand in the germ line, therefore carrying potential implications for genetic counseling (Chung et al., 1993).
Importantly, our results show that interruptions are not frequent in the AR CAG repeat and are therefore not likely to play a role in disease, making SBMA an exception among coding CAG expansion diseases.
Our results also show that fragment analysis, used routinely for the diagnostic testing of the SBMA and other CAG expansion diseases, underestimates the CAG expansion. This discrepancy is present also when analysing CAG expansions from other genes (Menon et al., 2013). In diagnostic laboratories, this bias is corrected for by calibrating the fragment analysis with amplicons of known sizes. Our results indicate that direct sequencing of the AR CAG repeat polymerase chain reaction amplicon in males is a reliable method for sizing evaluation, and therefore we suggest that this may be useful when analyzing size lengths of the CAG repeat near the disease threshold of 38 repeats. This possibility is unique to SBMA testing in males and differs from all other triplet expansion diseases in which the presence of sequence variability and the effect of having 2 alleles, makes direct polymerase chain reaction sequencing less informative.
Disclosure statement
The authors declare no actual of potential conflicts of interest.
Acknowledgements
This work was supported by Medical Research Council/Motor Neurone Disease Association Lady Edith Wolfson Fellowship (P.F.), and by grants from the Medical Research Council, the Motor Neurone Disease Association, and the Thierry Latran Foundation (E.F.).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.neurobiolaging.2013.07.015.
Appendix A. Supplementary data
Supplementary Fig. 1.
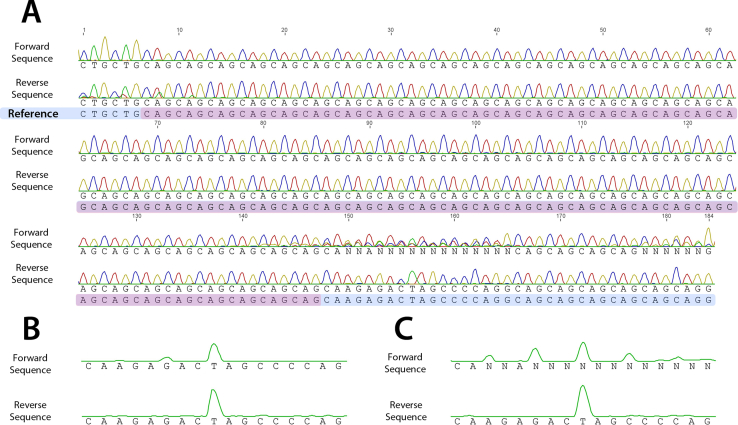
(A) Chromatogram illustrating forward and reverse sequencing of the AR CAG repeat. Control (B) and spinal bulbar muscular atrophy (SBMA) (C) chromatogram traces of only T nucleotides show an isolated T, 9 nucleotides downstream of the last CAG repeat, which is useful to size the repeat. Forward traces show upstream peaks due to polymerase chain reaction slippage (B and C) and downstream peaks due to somatic mosaicism (C).
Supplementary Fig. 2.
Frequency distribution of CAG repeat size in our control and Spinal bulbar muscular atrophy (SBMA) populations.
References
- Atsuta N., Watanabe H., Ito M., Banno H., Suzuki K., Katsuno M., Tanaka F., Tamakoshi A., Sobue G. Natural history of spinal and bulbar muscular atrophy (SBMA): a study of 223 Japanese patients. Brain. 2006;129:1446–1455. doi: 10.1093/brain/awl096. [DOI] [PubMed] [Google Scholar]
- Chong S.S., McCall A.E., Cota J., Subramony S.H., Orr H.T., Hughes M.R., Zoghbi H.Y. Gametic and somatic tissue–specific heterogeneity of the expanded SCA1 CAG repeat in spinocerebellar ataxia type 1. Nature Genetics. 1995;10:344–350. doi: 10.1038/ng0795-344. [DOI] [PubMed] [Google Scholar]
- Chung M.Y., Ranum L.P., Duvick L.A., Servadio A., Zoghbi H.Y., Orr H.T. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat. Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I., Kimura J., Narumiya S., Kakizuka A. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- Koide R., Ikeuchi T., Onodera O., Tanaka H., Igarashi S., Endo K., Takahashi H., Kondo R., Ishikawa A., Hayashi T., Saito M., Tomoda A., Miike T., Naito H., Ikuta F., Tsuji S. Unstable expansion of CAG repeat in hereditary dentatorubral–pallidoluysian atrophy (DRPLA) Nat Genet. 1994;6:9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- Koide R., Kobayashi S., Shimohata T., Ikeuchi T., Maruyama M., Saito M., Yamada M., Takahashi H., Tsuji S. A Neurological Disease Caused By an Expanded CAG Trinucleotide Repeat in The TATA-Binding Protein Gene: A New Polyglutamine Disease? Hum. Mol. Genet. 1999;8:2047–2053. doi: 10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- La Spada A.R., Wilson E.M., Lubahn D.B., Harding A.E., Fischbeck K.H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Matsuyama Z., Izumi Y., Kameyama M., Kawakami H., Nakamura S. The effect of CAT trinucleotide interruptions on the age at onset of spinocerebellar ataxia type 1 (SCA1) J. Med. Genet. 1999;36:546–548. [PMC free article] [PubMed] [Google Scholar]
- Menon R.P., Nethisinghe S., Faggiano S., Vannocci T., Rezaei H., Pemble S., Sweeney M.G., Wood N.W., Davis M.B., Pastore A., Giunti P. The role of interruptions in polyQ in the pathology of SCA1. PLoS Genet. 2013;9:e1003648. doi: 10.1371/journal.pgen.1003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Orr H.T., Chung M., Banfi S., Kwiatkowski T.J., Servadio A., Beaudet A.L., McCall A.E., Duvick L.A., Ranum L.P.W., Zoghbi H.Y. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- Pulst S.M., Nechiporuk A., Nechiporuk T., Gispert S., Chen X.N., Lopes-Cendes I., Pearlman S., Starkman S., Orozco-Diaz G., Lunkes A., DeJong P., Rouleau G.A., Auburger G., Korenberg J.R., Figueroa C., Sahba S. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- Yu Z., Zhu Y., Chen-Plotkin A.S., Clay-Falcone D., McCluskey L., Elman L., Kalb R.G., Trojanowski J.Q., Lee V.M.-Y., Van Deerlin V.M., Gitler A.D., Bonini N.M. PolyQ repeat expansions in ATXN2 associated with ALS are CAA interrupted repeats. PLoS ONE. 2011;6:e17951. doi: 10.1371/journal.pone.0017951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.