Abstract
Overexpression of the adverse prognostic marker ERBB2 occurs in 30% of breast cancers and is associated with aggressive disease and poor outcomes. Our recent findings have shown that NR1D1 and the peroxisome proliferator-activated receptor-γ (PPARγ)-binding protein (PBP) act through a common pathway in upregulating several genes in the de novo fatty acid synthesis network, which is highly active in ERBB2-positive breast cancer cells. NR1D1 and PBP are functionally related to PPARγ, a well-established positive regulator of adipogenesis and lipid storage. Here, we report that inhibition of the PPARγ pathway reduces the aldehyde dehydrogenase (ALDH)-positive population in ERBB2-positive breast cancer cells. Results from in vitro tumorsphere formation assays demonstrate that the PPARγ antagonists GW9662 and T0070907 decrease tumorsphere formation in ERBB2-positive cells, but not other breast cells. We show that the mechanism by which GW9662 treatment causes a reduction in ALDH-positive population cells is partially due to ROS, as it can be rescued by treatment with N-acetyl-cysteine. Furthermore, global gene expression analyses show that GW9662 treatment suppresses the expression of several lipogenic genes, including ACLY, MIG12, FASN and NR1D1, and the stem-cell related genes KLF4 and ALDH in BT474 cells. Antagonist treatment also decreases the level of acetylation in histone 3 and histone 4 in BT474 cells, compared with MCF7 cells. In vivo, GW9662 pre-treatment inhibits the tumor-seeding ability of BT474 cells. Together, these results show that the PPARγ pathway is critical for the cancer stem cell properties of ERBB2-positive breast cancer cells.
Keywords: PPARγ, HER2/neu positive, stem cells
Introduction
Overexpression of the ERBB2 oncogene is one of the most clinically relevant genetic changes in breast cancer. Occurring in approximately 30% of breast cancers, it is strongly associated with increased disease recurrence and a worse prognosis.1 Trastuzumab, a monoclonal antibody that targets the extracellular domain of ERBB2, is used to treat cancers where ERBB2 is overexpressed. However, when used as single-agent therapy in ERBB2-positive breast cancer patients, response rates are only 11–26%.2
Cancer stem cells (CSCs) have been identified as subpopulations of cells within tumors that drive tumor growth and recurrence.3, 4, 5 CSCs have many features, including self-renewal and resistance to chemo- and radiation therapy, which lead to the failure of many current cancer treatments.6, 7, 8, 9 Studies have shown that the CD44+/CD24-low cell subpopulation, which is enriched with breast CSCs, are resistant to trastuzumab treatment.10, 11, 12 This may explain why the efficacy of trastuzumab therapy is limited, as this treatment does not kill CSCs, which survive to form a new tumor. For this reason, new drugs that selectively target CSCs, combined with trastuzumab therapy, may offer great promise for ERBB2-positive breast cancer treatment.
Recent work has shown that transcriptional regulators overexpressed in cells carrying the ERBB2 amplicon cooperatively change the metabolism of ERBB2-positive breast cancer cells inducing a unique, Warburg-like metabolism that is primed towards fat production.13 PBP and NR1D1, two genes tightly linked to ERRB2 on chromosome 17, have essential roles in ERBB2-positive breast cancer cells but not other breast cancer cells or normal breast epithelial cells.14 NR1D1 and PBP are tightly linked to ERBB2 and frequently reside on the 17q12-21 amplicons found in ERBB2-positive tumors.15, 16 Several studies have shown that irrespective of the amplicon size they are consistently co-overexpressed with ERBB217, 18 and are among the six genes that comprise the ‘ERBB2 gene expression signature' seen in breast cancers.19 PBP is a co-activator of PPARγ and plays a positive role in its transcription initiation activity. NR1D1 is a target of PPARγ and has also been shown to positively regulate PPARγ expression. At least one critical role of PPARγ in ERBB2-positive breast cancer cells is to prevent the palmitate-induced lipotoxicity20 that is a consequence of the high levels of lipids they synthesize. PPARγ is a member of the nuclear hormone transcription factor family that controls the expression of a large number of genes involved in adipogenesis, energy metabolism, proliferation and tumor progression.21, 22, 23, 24, 25 PPARγ is the major expressed subtype of its family in the mammary gland and in primary and metastatic breast cancer.26, 27, 28, 29 Although recent studies have noted interactions of PPARγ-targeting drugs with ERBB2 signaling14, 20, 30 and ERBB2-positve breast cancer survival,31 there is not, however, a definitive view of the interactions between these pathways and potential mechanisms. The effects of PPARγ activity in CSCs have been studied in a variety of cancers such as colorectal cancer, hepatocellular carcinoma, lung cancer, glioma and leukemia.32, 33, 34, 35, 36 Constitutively active PPARγ mutants in ERBB2-induced mammary tumor models enhanced tumor growth by increasing endothelial stem cells.37 However, the effects of inhibition of PPARγ on ERBB2-positive breast CSCs have not been investigated.
In this study, we report that PPARγ inhibition selectively removes CSC-like cells from ERBB2-positive breast cancer cell populations by increasing ROS and altering the expression of lipogenic and stem cell-related genes. We also show that the PPARγ antagonist, GW9662, effectively blocks tumor formation in an animal model. Our results support a potential therapeutic strategy for preventing human ERBB2-positive breast cancer progression.
Results
ERBB2-positive breast cancer cells possess high levels of fat and aldehyde dehydrogenase (ALDH)-positive cells
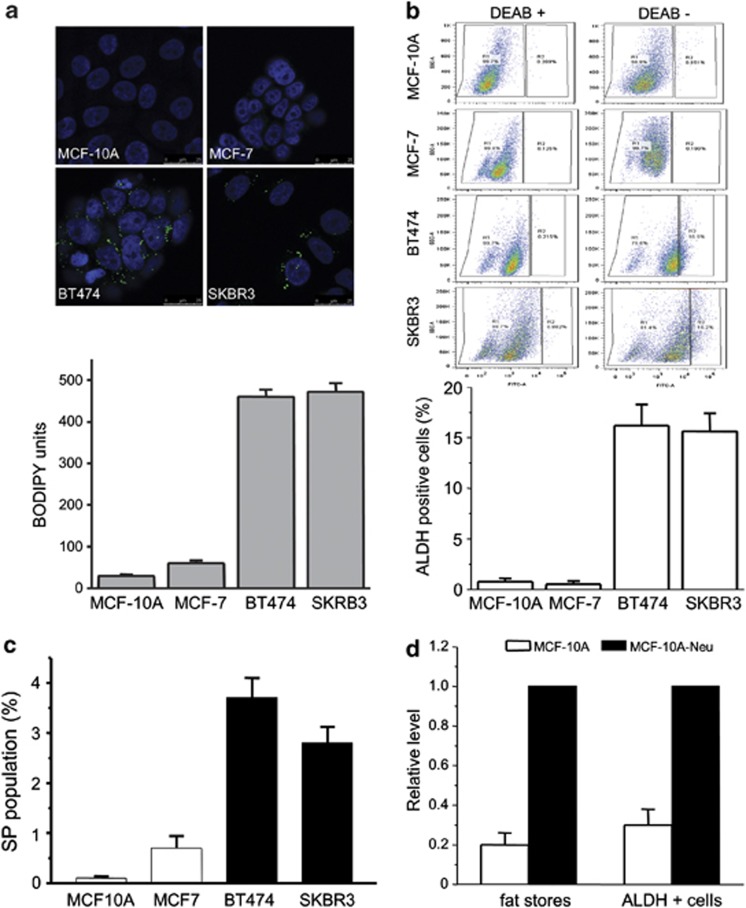
Metabolic regulators, NR1D1 and PBP, have been identified as novel survival factors for breast cancer cells with the ERBB2 signature. These two genes are involved in upregulating several genes in the de novo fatty acid synthesis network, which has been shown to be highly active in ERBB2-positive breast cancer cells.14 As shown in Figure 1a, stains of neutral fat show that ERBB2-positive breast cancer cells contain relatively high levels of neutral fats. These cells have an approximately 20-fold increased level of accumulated fat in lipid stores when compared with MCF-10A and a 10-fold increase when compared with MCF7 cells. ERBB2 is regarded as a breast cancer marker for aggressive tumor growth and metastasis, and as a gene that drives asymmetrical cell division. In addition, it has been shown that ERBB2 is an important regulator of subpopulations of breast cancer cells that display stem cell features and increases the frequency of tumor-initiating cells or CSCs.38 Staining ERBB2-positive breast cancer cells with Aldefluor39 shows that these cells have a higher percentage of cells in the CSC population. Cells from the ERBB2-positive breast cancer lines BT474 and SKBR3 exhibit an ∼18-fold increase of ALDH-positive cells when compared with MCF-10A or MCF7 cells (Figure 1b). In addition, quantification of the side populations (SPs) present in these lines shows that ERBB2-positive breast cancer cells contain more SP cells when compared with MCF-10A or MCF7 cells (Figure 1c and Supplementary Figure 1).
Figure 1.
Increased levels of fats found in ERBB2-positive breast cancer cells correlate with high levels of ALDH-positive cells. (a) Several breast cancer cell lines and normal MCF-10A cells were stained for fat stores with BODIPY 493/503 lipid stain. Hoechst 33342 was used for nuclei staining. Representative BODIPY-stained cell images (upper) and quantification of BODIPY signal (bottom). Error bars indicate the s.d. from three experiments. (b) MCF-10A, MCF-7, BT474 or SKBR3 cells were assayed for ALDH activity utilizing the ALDEFLUOR assay. Cells incubated with ALDEFLUOR substrate (BAAA) and the specific inhibitor of ALDH, diethylaminobenzaldehyde, were used to establish the baseline fluorescence of these cells (R1) and to define the ALDEFLUOR-positive region (R2). Incubation of cells with ALDEFLUOR substrate in the absence of diethylaminobenzaldehyde induces a shift in BAAA fluorescence defining the ALDEFLUOR-positive population. Quantification of ALDH-positive cells in each breast cell line is shown (bottom). Error bars indicate the s.d. from three experiments. (c) FACS for SP cells in MCF10A and breast cancer cell lines. (d) MCF-10A and MCF-10A-neu cells were stained with BODIPY and analyzed with ALDH-positive cell population; quantification of BODIPY staining and ALDH-positive cells is shown.
Although we have shown previously that increased expression of PBP and NR1D1 leads to increased fat production in these cells,14 part of the genetic basis of the CSC and fat content phenotypes may rest with ERBB2 itself. A number of studies have noted correlations between ERBB2 and several aspects of fat metabolism.25, 40, 41, 42 Although several groups have proposed potential regulatory interactions of fatty acid synthase (FASN) and ERBB2, a clear picture has not yet emerged.40, 43 In testing the contribution of ERBB2 itself to the CSC and fat content phenotypes, we find that MCF-10A cells carrying a retrovirus expressing constitutively active ERBB2 show a three- to fourfold increase in fats. In agreement with other studies,38 these cells exhibit a two- to threefold increase in the Aldefluor-positive population compared to vector control cells (Figure 1d and Supplementary Figure 2). Treatment with trastuzumab reduces the fat storage and ALDH-positive population in both ERBB2-positive BT474 and SKBR3 cell lines (Supplementary Figure 3). Although the fraction of cells in the CSC population and the levels of fat storage in MCF-10A-neu cells are significantly less than those seen in ERBB2-positive cell lines, the results suggest, in accord with other studies,25, 38, 40, 41, 42 that ERRB2 itself has some role in these processes.
PPARγ inhibition results in decreased CSCs in ERBB2-positive cancer cells
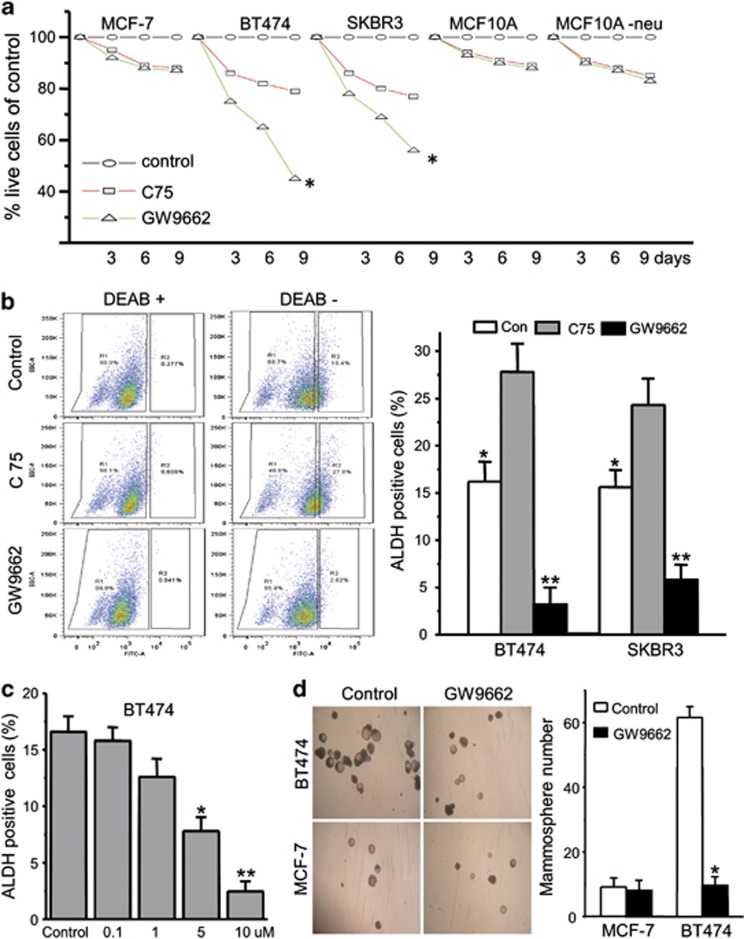
Although ERBB2-positive breast cancer cells have amplifications of the ERBB2 gene, they have also been shown to possess higher levels of PPARγ activity, which enable them to convert fatty acids to triglycerides, averting the apoptosis that results from lipotoxicity.20 PPARγ blockade using the PPARγ antagonist GW9662 results in increased apoptosis in the general population of ERBB2-positive BT474 and SKBR3 breast cancer cells, in a time- (Figure 2a) and dose-dependent manner20 (and data not shown). There is no significant effect on ERBB2-normal MCF7 (Figure 2a), normal MCF-10A or MCF-10A-neu cells at these drug concentrations. Genetic and pharmacological experiments point to the concerted effort of several PPARγ-regulated processes—the diversion of precursors, synthesis of fatty acids, and their processing and storage as triglycerides13, 20—that are important to these cells. Consistent with this notion, inhibition of the entire pathway has greater effects on ERBB2-positive cells. The FASN inhibitor C75 causes death of ERBB2-positive breast cancer cells when cells are treated with 40 μM C75 for the indicated times. At the concentrations tested, C75 is about twice as toxic to ERBB2-positive cells as the other cell lines, whereas GW9662 is about five times more toxic to the ERBB2-positive cells (Figure 2a).
Figure 2.
GW9662 treatment causes cell death and decreases the CSC population of ERBB2-positive cancer cells. (a) Cell counts of MCF-7, BT474, SKBR3, MCF10A and MCF10A-neu cells treated with vehicle, 10 μM of the PPARγ antagonist GW9662 and 40 μM of FASN inhibitor C75 for 3, 6 and 9 days. Results are presented as percentage of control (vehicle). Error bars indicate the s.d. from three individual experiments. *P<0.001. (b) The ALDH-positive cells' FACS profiles are shown for BT474 cells treated as in a. Quantification of ALDH-positive cells in BT474 and SKBR3 cells treated as in a. Bars denote the s.e. for n=3. Comparison of control with C75 treatment: *P<0.05; comparison of control with GW9662 treatment: **P<0.001. (c) % of ALDH-positive cells of BT474 cells treated with different concentrations of GW9662 for 9 days. Bars denote the s.e. for n=3. *P<0.05, **P<0.001. (d) Quantification of tumorsphere-forming potential of BT474 and MCF7 cells treated with GW9662 for 9 days. Phase-contrast images of tumorspheres are shown (left). Bars denote the s.e. for n=3. *P<0.001.
We next examined the potential linkage of the increased CSC and increased fat storage phenotypes. The effect of PPARγ inhibition on CSC populations was tested by monitoring changes in ALDH expression or tumorsphere formation. Interestingly, we find that GW9662 drastically decreases the ALDH-positive population in both BT474 and SKBR3 cells (Figure 2b). In contrast, C75 causes a less than twofold increase in the ALDH-positive population of either BT474 or SKBR3 cells (Figure 2b, right panel). There is a stepwise decrease in the proportion of ALDH-positive BT474 cells from 16.6 to 2.4% when cells are treated with different concentrations of GW9662 (Figure 2c). These results suggest that GW9662 decreases the ALDH-positive cell population in a dose-dependent manner. To further confirm the ability of GW9662 treatment to decrease the CSC population in ERBB2-positive breast cancer cells, we performed tumorsphere culture experiments using both BT474 and MCF7 cells. We find that BT474, SKBR3 and UACC812 cells pre-treated with GW9662 form fewer tumorspheres compared with the cells treated with vehicle (Figure 2d and Supplementary Figure 4). In contrast, GW9662 has no effect on MCF7 tumorsphere formation (Figure 2d). Taken together, these results indicate that GW9662 selectively causes cell death in CSC-like ERBB2-positive breast cancer cell populations.
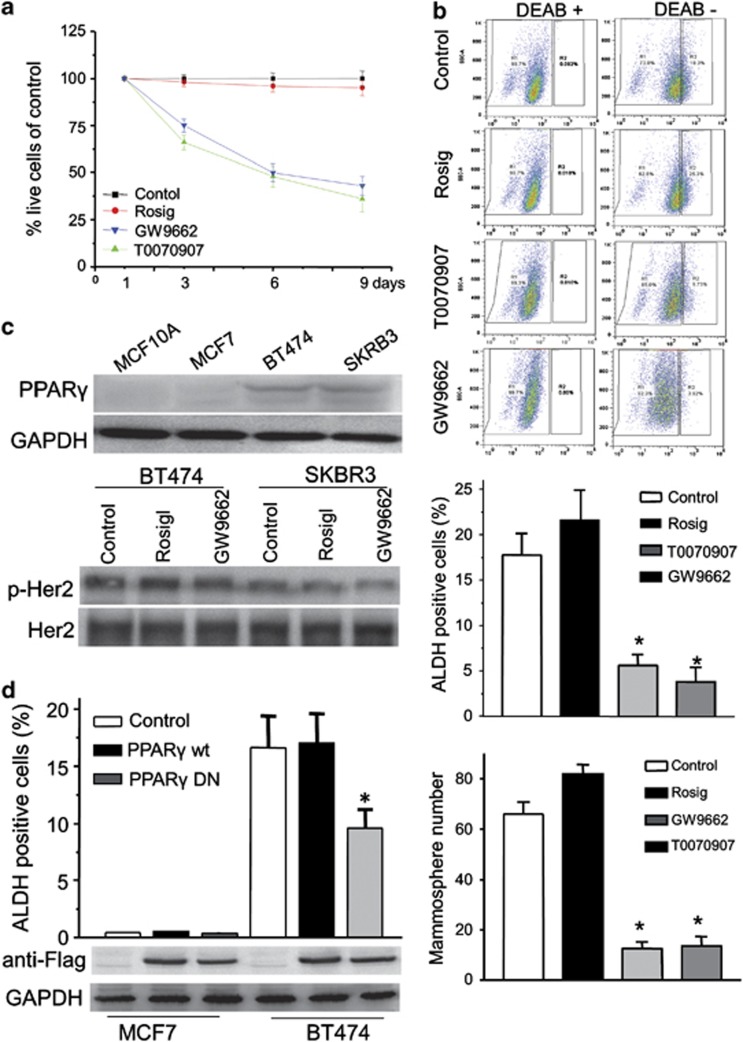
Previous studies have demonstrated that the importance of the fatty acid synthesis signaling pathway to the survival of ERBB2-positive cancer cells is not due to the amount of fat that is stored within the cell or its use as an energy source, but rather due to the synthetic process itself.14 PPARγ has been shown to regulate a number of metabolic processes related to this phenotype, including fatty acid β-oxidation, glucose utilization, cholesterol transport and energy balance.22 To extend our findings to other PPARγ antagonists or agonists, we examined the effect of another antagonist, T0070907, and an agonist rosiglitazone on the ALDH-positive population of ERBB2-positive breast cancer cells. Although the PPARγ agonist rosiglitazone has little effect on cell death, the PPARγ antagonist, T0070907, causes significant cell death and has essentially the same effect on cell survival as GW9662 (Figure 3a). We next determined the effects of rosiglitazone and T0070907 on the ALDH-positive population in BT474 cells. Rosiglitazone causes a slight increase in the ALDH-positive population compared with cells treated with vehicle. In contrast, T0070907, like GW9662, causes a significant decrease of the ALDH-positive population (Figure 3b). To further confirm this result, we performed tumorsphere assays. BT474 cells pre-treated with rosiglitazone form a slightly greater number of tumorspheres compared with cells pre-treated with vehicle. In contrast, T0070907 and GW9662 both reduce the number of tumorspheres (Figure 3b). As GW9662 is a PPARγ antagonist, it is likely that GW9662 directly regulates the activity of PPARγ in breast cancer cells. We find that PPARγ expression levels are higher when compared with MCF10A or MCF7 cells (Figure 3c). To exclude the possibility that GW9662 affects ERBB2 activity, we monitored ERBB2 phosphorylation by immunoblotting with a phosphotyrosine-specific antibody (p-Her2 Y1221/1222) after treatment of BT474 or SKBR3 cells with GW9662 or rosiglitazone. We find that neither GW9662 nor rosiglitazone affects ERBB2 activity (Figure 3c). To rule out potential off-target effects of GW9662, we overexpressed wild-type or mutant PPARγ-dominant negative (DN) gene constructs in MCF7 and BT474 cells, respectively. Consistent with the pharmacological results, we find that PPARγ-DN reduces the ALDH-positive population in BT474 cells (Figure 3d). These results demonstrate that PPARγ activity is important not only for cell survival,20, 44 but, specifically, also for CSC maintenance in ERBB2-positive breast cancer cells. Moreover, the contribution of PPARγ activity to fat synthesis and CSC maintenance appears to be significantly greater than the effects of ERRB2 in populations of ERBB2-positive breast cancer cells.
Figure 3.
PPARγ inhibition results in decreased numbers of CSCs in ERBB2-positive cancer cells. (a) Cell counts of BT474 cells treated with vehicle, 10 μM of PPARγ agonist rosiglitazone and 10 μM of PPARγ antagonist GW9662 or T0070907 for 3, 6 and 9 days. Results are presented as percentage of control. (b) FACS profiles identifying ALDH-positive cell populations are shown for BT474 cells treated as in a. Quantification of ALDH-positive cells (middle) and tumorsphere formation (bottom) in BT474 cells treated as in (a). Bars denote the s.e. for n=3, *P<0.001.(c) PPARγ expression levels determined by immunoblotting in MCF10A and breast cancer cell lines. GAPDH was used as loading control. BT474 and SKBR3 cells were treated with rosiglitazone, GW9662 or control for 9 days. Cell lysates were probed with anti-Her2 or anti-phospho-Her2 antibodies (bottom). (d) Expression of a DN mutant form of PPARγ reduces the ALDH-positive cells in BT474. MCF7 and BT474 cells were transfected with PPARγ-wild type, PPARγ-DN or control vector. ALDH-positive cells are shown (top). Bars denote the s.e. for n=3, *P<0.001. Immunoblotting was performed to test PPARγ-wild type or PPARγ-DN expression level using anti-Flag antibody. GAPDH was used as loading control.
GW9662 toxicity in ERBB2-positive CSCs is due in part to production of ROS
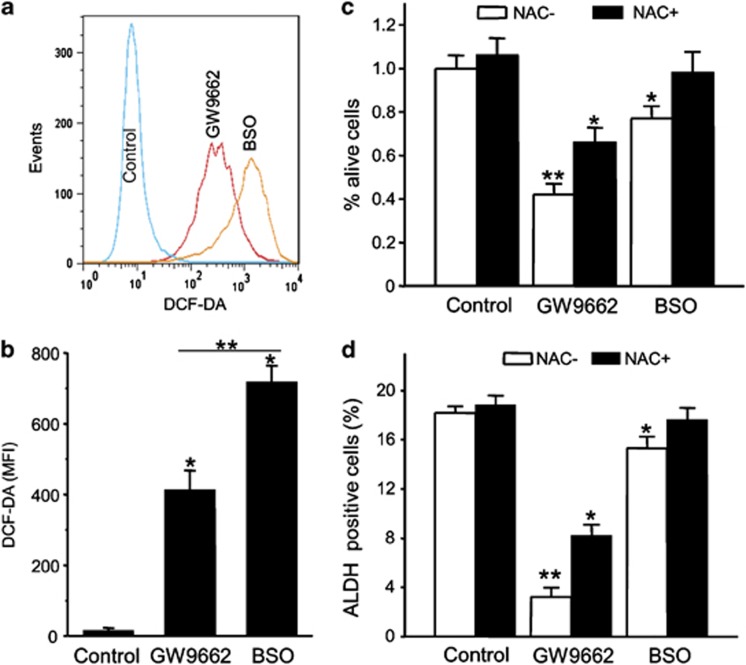
We next examined the mechanism of the reduction seen in the ALDH-positive population caused by inhibition of PPARγ activity. As PPARγ inhibition in ERBB2-positive breast cancer cells has been shown to result in ROS production and subsequent cell death,20 we determined whether ROS were involved in the selective loss of CSC-like cells. ROS production after cells were treated with GW9662 was assayed using DCF-DA staining. L-S,R-buthionine sulfoximine (BSO), which inhibits glutamate–cysteine ligase, was used as positive control for increasing ROS production. We observe increased ROS production following the GW9662 treatment in BT474 cells compared to cells treated with vehicle. The production of ROS induced by GW9662 was 60% of that caused by BSO (Figures 4a and b). As ROS have been linked to cell death and CSC survival, the potential involvement of ROS as the cause of cell death and decreased ALDH-positive population in ERBB2-positive breast cancer cells during PPARγ inhibition was tested. BT474 cells were treated with GW9662 or BSO together with the antioxidant N-acetyl-cysteine (NAC) or vehicle. NAC has been shown previously to rescue ERBB2-positive breast cancer cells from GW9662-induced apoptosis.20 As shown in Figure 4, NAC also reverses the loss of cells in the ALDH-positive population of ERBB2-positive breast cancer cells. However, as NAC almost completely reverses the effects of BSO on both cell death (Figure 4c) and decreases in the ALDH-positive population (Figure 4d), it seems likely that additional players must exist in the mechanism of toxicity of GW9662. Nevertheless, these results suggest that PPARγ inhibition by GW9662-induced cell death and the decrease in ALDH-positive population in ERBB2-positive breast cancer cells is partially due to ROS.
Figure 4.
GW9662 toxicity in ERBB2-positive CSCs is because of production of ROS. (a) Flow cytometry analysis of ROS production. BT474 cells treated with 10 μM GW9662 or 1 mM L-S,R- BSO for 9 days, and intracellular ROS concentration were assessed by DCF-DA staining. (b) DCF-DA fluorescence in BT474 cells presented as mean fluorescence intensity. Error bars indicate s.d. (n=3), *P<0.001, **P<0.05. (c) BT474 cells were treated with vehicle, 10 μM GW9662 or 1 mM BSO together with 1 mM of NAC for 9 days. Live cells were counted and presented as percentage of the vehicle control (*P<0.05, **P<0.001). (d) % of ALDH-positive cells of BT474 cells treated with vehicle, 10 μM GW9662 or 1 mM BSO together with 1 μM NAC for 9 days. Error bars indicate the s.d. from three individual experiments. *P<0.05, **P<0.001.
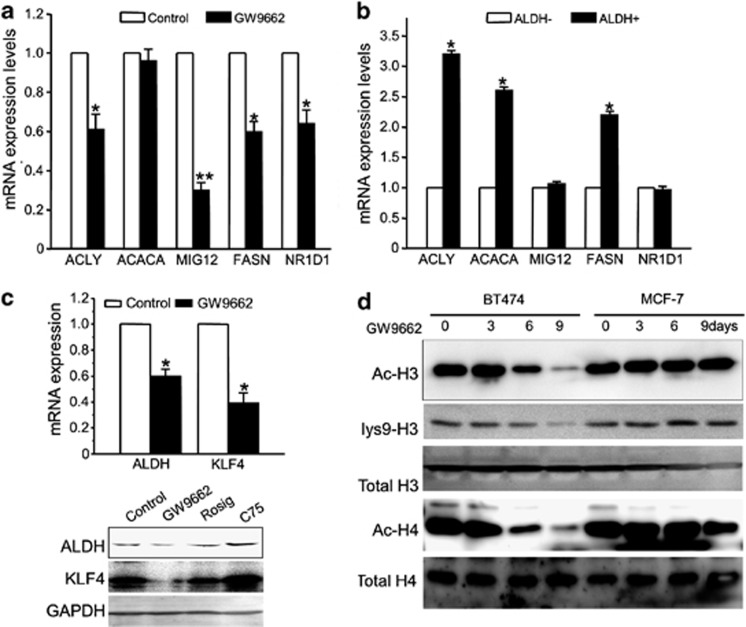
GW9662 treatment affects lipogenic and stem cell related gene expression in BT474 cells
PPARγ is a transcription factor that controls the expression of genes that regulate multiple cellular events. Microarray analysis was performed to identify PPARγ targets that might be involved in the GW9662-dependent decrease in the ALDH-positive populations of EBRR2-positive breast cancer cells. BT474 cells were treated with a sublethal concentration (10 μM) of GW9662 or vehicle for 9 days. This analysis showed that the expression levels of lipogenic genes (ACLY, MIG12, FASN and NR1D1) are low in BT474 cells treated with GW9662 compared with vehicle. Although the expression level of acetyl-CoA carboxylase-α, the first committed enzyme in the fatty acid synthesis pathway, does not change, its kinase activity modulator MIG12 was suppressed significantly by GW9662 (Figure 5a). These results suggest that PPARγ targets, including several related to fat synthesis and storage, have a role in maintaining CSC in ERBB2-positive breast cancer cells. To test the lipogenic gene expression profile in stem-like cells in ERBB2-positive breast cancer cells, we isolated ALDH-positive and ALDH-negative populations of BT474 cells. The expression of genes in isolated cells was confirmed by qRT–PCR. We find that ACLY, ACACA and FASN are overexpressed in BT474 ALDH-positive populations compared to ALDH-negative populations (Figure 5b). In addition, we find that the expression levels of KLF4 and ALDH are downregulated in BT474 cells treated with GW9662 compared with controls (Figure 5c upper panel). To further test the effects of GW9662, rosiglitazone and C75 on KLF4 and ALDH protein expression levels, we performed immunoblots. Rosiglitazone has little effect on levels of either protein, whereas C75 increases the expression of KLF4 and ALDH. However, GW9662 decreases their expression (Figure 5c lower panel). These results may suggest that GW9662 decreases the ALDH-positive population by reducing the expression of KLF4 and ALDH. Acetyl-CoA derived from glucose and ATP-citrate lyase (ACLY) activity is a critical reactant in chromatin modification and the resulting changes in gene expression that control cell lineage or fate.45 Acetyl-CoA is also a key metabolite in lipogenesis, and therefore represents a key link between lipogensis and chromatin modification. To test whether GW9662 has effects on chromatin modification, we analyzed the histone acetylation status of BT474 and MCF-7cells treated with GW9662 or vehicle (Figure 5d). We find that GW9662 decreases the amount of histone H3 and H4 acetylation in BT474 cells, which have high expression levels and activities of ACLY. In contrast, GW9662 has less of an effect on histone acetylation in MCF-7 cells. Taken together, these results suggest that inhibition of PPARγ activity decreases the ALDH-positive population of ERBB2-positive breast cancer cells by downregulating the stem-cell maintenance gene expression program through chromatin modification.
Figure 5.
GW9662 treatment affects lipogenic and stem cell-related gene regulation in BT474 cells. (a) The expression of various lipogenic genes in BT474 cells with or without GW9662 treatment was examined by qRT–PCR. GAPDH gene was used as an internal control. Error bars indicate three individual experiments; *P<0.05, **P<0.001. (b) ALDH-positive or negative cells were isolated from BT474 cells; RNA was extracted and qRT–PCR analysis performed to examine the expression levels of various lipogenic genes (*P<0.05). (c) The expression of ALDH and KLF4 gene in BT474 cells. BT474 cells were treated with vehicle and GW9662 for 9 days, and qRT–PCR was performed to examine the expression level of ALDH and KLF4 mRNAs. GAPDH expression was used as an internal control (*P<0.05). BT474 cells were treated with vehicle, GW9662, rosiglitazone and C75. After 9 days, cell lysates were used for immunoblotting to examine the expression of ALDH and KLF4, GAPDH was used as loading control (bottom). (d) Effects of GW9662 on histone acetylation. BT474 or MCF-7 cells were treated with GW9662 for different times as indicated. Immunoblot analysis of total protein extracts from cells was conducted using anti-Ac-H3, lys9-H3, H3, Ac-H4 and H4 antibodies.
Effects of GW9662 treatment on tumor seeding and growth in vivo
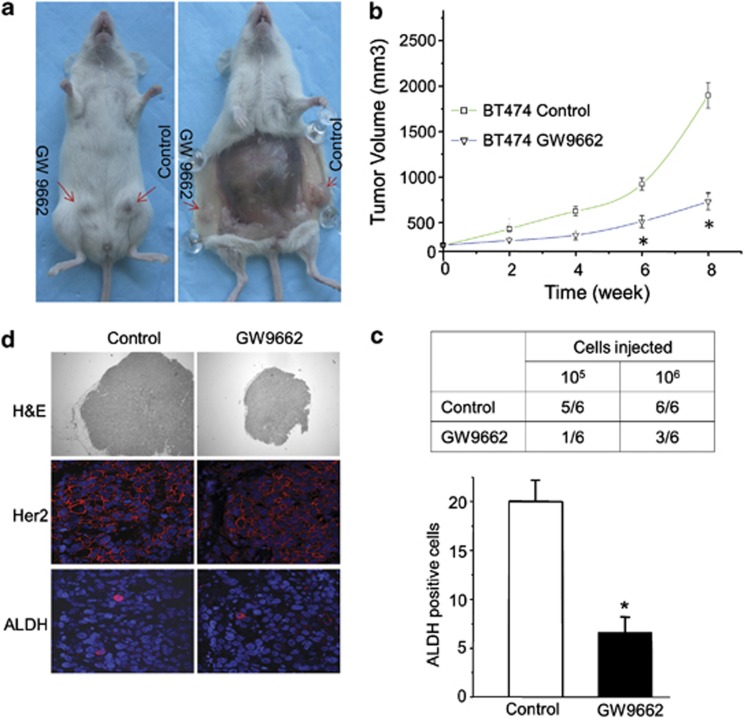
These molecular experiments carried out in vitro pointed to the possibility that GW9662 treatment might be useful in decreasing the CSC population in ERBB2-positive breast cancer cells and inhibiting tumor progression. To test this possibility, we assessed the functional presence of tumor stem-like cells by determining the tumor-seeding ability in vivo. For these experiments, BT474 cells were treated with the sublethal concentration of 10 μM GW9662 in vitro for 9 days, and then injected into mice. GW9662 pre-treatment results in a significant decrease in both BT474 cells' tumor-seeding ability and in ultimate tumor size compared to vehicle pre-treatment (Figures 6a–c). These results indicate that GW9662 pre-treatment decreases the number of functional tumor stem-like cells. In contrast, GW9662 had no effect on the tumor formation and growth of MCF-7 cells (data not shown). Tumors from animals orthotopically injected with GW9662 pre-treated or vehicle-treated cells have essentially the same levels of expression of ERRB2 as determined with immunofluorescence microscopy (Figure 6d). Importantly, tumors from animals receiving vehicle pre-treated cells have more ALDH-positive staining cells than tumors from animals receiving GW9662 pre-treated cells, although these cells are rare in either case. These results support our above findings that BT474 cells pre-treated with GW9662 form both fewer and smaller tumors in vivo. These results confirm the utility of GW9662 in selectively removing CSC-like cells from ERBB2-positive breast cancer cell populations, and thereby reducing tumor formation in vivo. These results also provide a strong rationale for the use of GW9662 for chemoprevention of ERBB2-positive breast cancer progression.
Figure 6.
Effects of GW9662 treatment on tumor seeding and growth in vivo. (a) Images of tumors formed in animals after mammary fat pad injection of BT474 cancer cells pre-treated with GW9662 or DMSO. (b) Tumor growth curves obtained following fat pad injection of control or pre-treated BT474 cells. The data represent the mean±s.e. (n=12, *P<0.001). (c) Tumor-seeding ability of BT474 breast cancer cells treated with GW9662 or DMSO. (d) Histological analysis of tumors from BT474 cells treated with GW9662 or vehicle. Shown are H&E, Her2 and ALDH staining. Quantification of positive ALDH-staining cells per 50 fields. Error bars denote the s.e. (n=5, *P<0.05).
Discussion
In this report, we have shown that ERBB2-positive breast cancer cells have increased fat stores and ALDH-positive cell populations compared with other breast cancer cells or normal breast epithelial cells. Inhibition of PPARγ leads to a significant decrease of the ALDH-positive cell population, specifically in ERBB2-positive breast cancer cells, which have as a general feature increased expression of the adipogenesis-related transcription factors NR1D1 and PBP in addition to ERBB2.14 The results suggest that PPARγ has a larger role in maintaining CSC populations than ERRB2 in these cells. Our results also indicate that this phenomenon is explained by downregulation of stem cell-related gene expression and involves ROS-mediated cell death. Finally, we have provided evidence that the tumor-seeding ability of ERBB2-positive breast cancer cells is dramatically decreased by PPARγ antagonist treatment. This work has revealed a potential clinically significant application of GW9662 in targeting ERBB2-positive breast CSCs.
Increased activity of the lipogenesis pathway is a hallmark of cancer cells. This is especially true in ERBB2-positive human breast cancer cells, which have a high degree of fat storage.14, 46 We have previously shown that PPARγ is essential for the survival of ERBB2-positive breast cancer cells. Its targets function in the unique metabolism of these cells preventing the high levels of palmitate produced from becoming toxic.20 PPARγ is expressed in many tissues and regulates lipid metabolism, glucose homoeostasis and tumor progression. It has emerged as a potential target for cancer therapy although its utility is highly tumor specific.47 Several studies have shown that its activation promotes cell cycle arrest, apoptosis and differentiation in many human tumors and inhibits invasion and metastasis of human breast cancer cells.48, 49 Still other studies have demonstrated that PPARγ antagonists have more potent anticancer effects than agonists in some epithelial cancer cell lines.50 In the case of ERBB2-positive breast cancer cells, especially those with amplification of the 17q12-q21 region that contains ERBB2, NR1D1 and PBP (Supplementary Table 1), it appears that PPARγ has a critical role in the Warburg-like glycolytic metabolism operative in these cells. To maintain a high flux through glycolysis, NAD+ must be regenerated. Many highly proliferative stem and cancer cells meet this need by converting pyruvate to lactate at high rates. Our recent findings have shown that the overexpression of PBP and NR1D1 genes found in ERBB2-positive breast cancer cells causes the upregulation of de novo fatty acid synthesis, enabling these cells to regenerate NAD+ indirectly by oxidizing NADPH during fatty acid synthesis.13, 14
The PPARγ-signaling pathway has been shown to be involved in fate determination in a wide range of stem cells. Regulation of PPARγ activation inhibits growth and expansion of brain tumor stem cells.32 PPARγ also regulates KLF4 expression, a key gene for pluripotent stem cells.51, 52 Recent animal studies have shown that GW9662 enhances the chemopreventive effect of anti-estrogen therapy in mammary tumors, which suggests that GW9662 promotes breast CSCs' differentiation into tumors that exhibit higher ER expression and resulting responsiveness to the ER antagonist fulvestrant.53 Our studies have shown that GW9662 supresses KLF4 expression (Figure 5c). This may lead to stem cell program gene expression changes in ERBB2-positive breast CSCs and sensitize the cells to ROS. It will be intriguing to test how GW9662 cooperates with traditional therapy to treat ERBB2-positive breast cancer. Our ongoing experiments are investigating the effect of GW9662 with trastuzumab on ERBB2-positive breast CSCs.
The increased activity of the lipogenesis pathway in ERBB2-positive human breast CSCs has parallels in other stem cell types. Developmental studies in embryos have underscored the involvement of lipogenesis in the proliferation of stem cells. The cytoplasm of oocytes and embryos of some mammals are rich with lipids and fatty acids54 that serve as precursors for membrane biosynthesis and substrates for energy production. The concentration of triglycerides decreases significantly during embryo development because of increased utilization of endogenous stored lipids via β-oxidation.55 During tumor metastasis, CSCs seed distal organs after extravasation and produce new tumors. The rapid proliferation in an unstructured cellular environment is reminiscent of early steps in embryo development. In addition, studies have shown that many lipid molecules are present at different levels in the ESCs compared to neurons and cardiomyocytes. Various lipids and fatty acids can regulate the proliferation and differentiation of pluripotent stem cells and adult progenitors. Our results showed that GW9662 decrease the ALDH-positive cell population in ERBB2-positive breast cancer cells. These exciting results may provide evidence that glycolytic flux or metabolites can regulate the CSC phenotype.
Finally, our results indicate that increased flux through the fatty acid synthetic pathway may have direct mechanistic implications for ERBB2-positive breast CSCs. ACLY, which generates acetyl-CoA from mitochondria-derived citrate for lipogenesis, is a key enzyme for tumor cells.56 ACLY and its production of acetyl-CoA have been shown to regulate histone acetylation and chromatin remodeling.45 ACLY is a downstream target of PPARγ.14 We predicted that PPARγ antagonist GW9662 can inhibit ACLY expression and regulate histone acetylation status. Our results have shown that GW9662 indeed affects histone H3 and H4 acetylation levels in ERBB2-positive breast cancer cells but not in other cells. These results suggest that chromatin remodeling by histone acetylation status is one potential reason that changes in gene expression occur after GW9662 treatment. Transcription profile changes in ERBB2-positive breast cancer cells may trigger reprogramming of CSCs, which ultimately results in a loss of cells from the CSC population after GW9662 treatment. Clearly, more work will be required to establish this linkage.
In summary, we have reported that GW9662 treatment induced decrease of CSCs in ERBB2-positive human breast cancer cells by upregulation of intracellular ROS and inhibition of the lipogenesis pathway. We have also shown that GW9662 pre-treatment can inhibit tumor formation in an animal model. Our results provide a rationale for using PPARγ antagonist GW9662 as a new treatment for ERBB2-positive human breast cancer.
Materials and methods
Cell culture and chemicals.
Breast cancer cell lines and MCF-10A were obtained from American Type Culture Collection. All cell lines were cultured in DMEM (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone) and 100 U/μl of penicillin-streptomycin (Cellgro, Manassas, VT, USA), except for MCF-10A cells cultured as indicated in Debnath et al.57 To generate the MCF-10A cell line that expresses Her2, pLXSN-neu or vector control retroviruses were used to infect MCF-10A cells. The Flag-PPARγ (plasmid # 8895) and Flag-PPARγ E499Q (plasmid # 8896) are from Addgene.58 The PPARγ antagonists GW9662 and T0070907, agonist rosiglitazone, and the FASN inhibitor C75 were obtained from Sigma-Aldrich, (St Louis, MO, USA). Cell counts were performed after growing cells in 96-well plates, fixing with 4% formaldehyde and counterstaining nuclei with Hoechst 33342. Images of cells were acquired using an In Cell Analyzer 1000 (GE Healthcare, Piscataway, NJ, USA) high-content imaging system. At least 30 fields were imaged per well for cell number determination. Statistics were performed using the In Cell Investigator 3.4 image analysis software (GE Healthcare).
Measurement of ROS production
ROS was measured as previously described using H2DCF-DA (Invitrogen, Carlsbad, CA, USA). The fluorescence signal was detectable at an excitation wavelength of 488 nm and emission wavelength of 535 nm with FACS Canto flow cytometer (BD Biosciences, San Jose, CA, USA). For treatment with GW9662 and BSO, cells were incubated with GW9662 (Sigma-Aldrich) at 10 μM or BSO at 1 mM for 9 days.
Aldefluor assay
The ALDEFLUOR kit (StemCell Technologies, Vancouver, BC, Canada) was used to isolate the population with a high ALDH enzymatic activity. Cells were suspended in ALDEFLUOR assay buffer containing ALDH substrate (BAAA, 1 μmol/l per 1 × 106 cells) and incubated during 45 min at 37 °C. As a negative control for each sample of cells an aliquot was treated with 50 mmol/l diethylaminobenzaldehyde, a specific ALDH inhibitor. The gates were established using as negative controls the ALDEFLUOR-stained cells treated with diethylaminobenzaldehyde.
SP and cell sorting
The SP analysis based on Hoechst 33342 (Sigma-Aldrich) staining was performed as previously described.59, 60 Briefly, cells were collected and re-suspended in warm culture medium (106 cells/ml). Hoechst 33342 was added at a final concentration of 5 μg/ml; as control reactions, Verapamil (Sigma-Aldrich) was added at a final concentration of 50 μM in the presence of Hoechst 33342. Cells were incubated at 37 °C for 90 min with intermittent mixing. After incubation, cells were washed and re-suspended in ice-cold PBS containing 2% fetal bovine serum (Invitrogen). Cells were stained with propidium iodide (2 μg/ml, Sigma-Aldrich) to determine viability, then analyzed and sorted by flow cytometry.
Tumorsphere assay
Mammosphere culture was performed as previously described. Cells were treated with GW9662 or vehicle for the indicated days. Single cells were plated in ultra-low attachment plates (StemCell Technologies), at a density of 5000 cells/ml. For counting mammospheres, the content of all wells was collected, pooled and transferred on a collagen-coated dish, in differentiating medium (see below). Mammospheres adhered in these conditions in approximately 48 h, after which they were counted under low magnification.
Metabolic assays
For detection of neutral fat stores, cells were stained with 1 μg/ml4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacence (BODIPY 493/503; Molecular Probes, Grand Island, NY, USA). Cells were grown on 96-well plates, fixed with 4% formaldehyde, stained with 1 μg/ml BODIPY 493/503 and counterstained for nuclei with Hoechst 33342. Cells were imaged and analyzed using the In Cell analyzer 1000-In Cell Investigator 3.4 system.
Immunofluoresence
Mouse breast cancer tissue sections were baked for 1 h at 62 °C, subjected to serial alcohol rehydration and microwaved in 0.01 M sodium citrate for 20 min for antigen retrieval. The sections were serum blocked for 30 min, incubated overnight at 4 °C with first antibodies in phosphate-buffered saline and subsequently with Cy3-labeled secondary antibodies for 30 min; the nucleus was stained with Hoechst 33342. The stained sections were mounted with anti-fade solution for microscopy.
Animal experiments
NOD/SCID mice were purchased from Jackson Lab (The Jackson Laboratory, Bar Harbor, ME, USA). All mouse procedures were approved by the Animal Care and Use Committees of SUNY Albany and performed in accordance with institutional policies.
For xenograft tumor-seeding studies, the indicated numbers of BT474 cells pre-treated with vehicle or 10 μM GW9662 for 9 days were suspended in 50 μl Matrigel (BD Biosciences) diluted 1:2 with DMEM and injected into mammary fat pad. Tumor formation was assayed by palpation. The tumor volume in mm3 is calculated by the formula: volume=(width)2 × length/2.
Quantitative real time reverse transcription-PCR
These assays were carried out essentially as described previously.61 Primers used for qRT–PCR were as follows: for ATP citrate lyase (ACLY), 5′-AAGATCTCGTGGCCAATGGA-3′ (forward) and 5′-AGGTTTGCGGATCAAACCAA-3′ (reverse); for acetyl-CoA carboxylase-α (ACACA), 5′-CTTTGTGCCCACGGTTATCA-3′ (forward) and 5′-AGTGGTCCCTGTTTGTCTCCA-3′ (reverse); for midline-1-interacting G12-like protein (MIG12), 5′-AACGTCCGCGCGGCCACC-3′ (forward) and 5′-GTGGCCCCAATTGCCGAAG-3′ (reverse); for fatty acid synthase (FASN), 5′-GAACTCCTTGGCGGAAGAGAA-3′ (forward) and 5′-GCGAGAAGTCAACACGAGCTT-3′ (reverse); for nuclear receptor subfamily 1, group D, member 1 (NR1D1), 5′-CCGTGACCTTTCTCAGCATGA-3′ (forward) and 5′-CACTGTCTGGTCCTTCACGTT-3′ (reverse); for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-GCAAATCCATGGCACCGT-3′(forward) and 5′-TCGCCCCACTTGATTTTGG-3′ (reverse). Product levels were calculated after normalization with GAPDH control.
Immunoblotting
Immunoblotting was performed essentially as described previously.61 Equal amounts of proteins were used. Antibodies used were anti-ALDH (1:1000, BD Biosciences), anti-KLF4 (1:1000, Sigma), anti-Histone H3, H4 antibody, anti-acetyl-Histone H3 (lys9) (1:1000, Cell Signaling, Technology, Danvers, MA ,USA), anti-acetyl-Histone H3, H4 antibodies (1:1000, Millipore, Billerica, MA, USA), anti-Flag antibody (1:1000, Sigma), anti-rabbit IgG-HRP and mouse IgG-HRP (1:5000, Jackson ImmunoResearch, West Grove, PA, USA).
Acknowledgments
We thank Jan Baumann and other members of the UAlbany CRC for helpful suggestions. This work was supported by NCI 1R01CA136658 to DSC.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Menard S, Fortis S, Castiglioni F, Agresti R, Balsari A. HER2 as a prognostic factor in breast cancer. Oncology. 2001;61 (Suppl 2:67–72. doi: 10.1159/000055404. [DOI] [PubMed] [Google Scholar]
- Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatmentof HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley M, Ashworth A. Stem cells and breast cancer: a field in transit. Nat Rev Cancer. 2003;3:832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839–2845. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Clarke MF. Cancer stem cells and radiotherapy: new insights into tumor radioresistance. J Natl Cancer Inst. 2006;98:1755–1757. doi: 10.1093/jnci/djj505. [DOI] [PubMed] [Google Scholar]
- Bedard PL, Cardoso F, Piccart-Gebhart MJ. Stemming resistance to HER-2 targeted therapy. J Mammary Gland Biol Neoplasia. 2009;14:55–66. doi: 10.1007/s10911-009-9116-x. [DOI] [PubMed] [Google Scholar]
- Oliveras-Ferraros C, Vazquez-Martin A, Martin-Castillo B, Cufi S, Del Barco S, Lopez-Bonet E, et al. Dynamic emergence of the mesenchymal CD44(pos)CD24(neg/low) phenotype in HER2-gene amplified breast cancer cells with de novo resistanceto trastuzumab (Herceptin) Biochem Biophys Res Commun. 2010;397:27–33. doi: 10.1016/j.bbrc.2010.05.041. [DOI] [PubMed] [Google Scholar]
- Fumagalli D, Michiels S, Sotiriou C. CD44+ CD24-/low phenotype and resistance to trastuzumab in HER2-positive breast cancer cell lines. Pharmacogenomics. 2011;12:12–13. [PubMed] [Google Scholar]
- Baumann J, Karch C, Kourtidis A, Conklin DS. The electronics of HER2/neu positive breast cancer cells. Breast Cancer Recent Adv Biol Imaging Ther. 2012. pp. 17–36.
- Kourtidis A, Jain R, Carkner RD, Eifert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Res. 2010;70:1783–1792. doi: 10.1158/0008-5472.CAN-09-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Kauraniemi P, Kallioniemi A. Activation of multiple cancer-associated genes at the ERBB2 amplicon in breast cancer. Endocr Relat Cancer. 2006;13:39–49. doi: 10.1677/erc.1.01147. [DOI] [PubMed] [Google Scholar]
- Dressman MA, Baras A, Malinowski R, Alvis LB, Kwon I, Walz TM, et al. Gene expression profiling detects gene amplification and differentiates tumor types in breast cancer. Cancer Res. 2003;63:2194–2199. [PubMed] [Google Scholar]
- Zhu Y, Qi C, Jain S, Le Beau MM, Espinosa R, 3rd, Atkins GB, et al. Amplificationand overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc Natl Acad Sci USA. 1999;96:10848–10853. doi: 10.1073/pnas.96.19.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F, Borie N, Ginestier C, Groulet A, Charafe-Jauffret E, Adelaide J, et al. Identification and validation of an ERBB2 gene expression signature in breast cancers. Oncogene. 2004;23:2564–2575. doi: 10.1038/sj.onc.1207361. [DOI] [PubMed] [Google Scholar]
- Kourtidis A, Srinivasaiah R, Carkner RD, Brosnan MJ, Conklin DS. Peroxisome proliferator-activated receptor-gamma protects ERBB2-positive breast cancer cells from palmitate toxicity. Breast Cancer Res. 2009;11:R16. doi: 10.1186/bcr2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- Lehrke M, Lazar MA. The many faces of PPARgamma. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nat Rev Cancer. 2004;4:61–70. doi: 10.1038/nrc1254. [DOI] [PubMed] [Google Scholar]
- Glazer RI, Yuan H, Xie Z, Yin Y. PPARgamma and PPARdelta as modulators of neoplasia and cell fate. PPAR Res. 2008;2008:247379. doi: 10.1155/2008/247379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli M, Cortes-Canteli M, Lai C, Santos A, Perez-Castillo A. The peroxisome proliferator-activated receptor gamma is an inhibitor of ErbBs activity in human breast cancer cells. J Cell Sci. 2001;114:4117–4126. doi: 10.1242/jcs.114.22.4117. [DOI] [PubMed] [Google Scholar]
- Kim KY, Kim SS, Cheon HG. Differential anti-proliferative actions of peroxisome proliferator-activated receptor-gamma agonists in MCF-7 breast cancer cells. Biochem Pharmacol. 2006;72:530–540. doi: 10.1016/j.bcp.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Mueller E, Sarraf P, Tontonoz P, Evans RM, Martin KJ, Zhang M, et al. Terminal differentiation of human breast cancer through PPAR gamma. Mol Cell. 1998;1:465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- Yin F, Wakino S, Liu Z, Kim S, Hsueh WA, Collins AR, et al. Troglitazone inhibits growth of MCF-7 breast carcinoma cells by targeting G1 cell cycle regulators. Biochem Biophys Res Comm. 2001;286:916–922. doi: 10.1006/bbrc.2001.5491. [DOI] [PubMed] [Google Scholar]
- Elstner E, Williamson EA, Zang C, Fritz J, Heber D, Fenner M, et al. Novel therapeutic approach: ligands for PPARgamma and retinoid receptors induce apoptosis in bcl-2-positive human breast cancer cells. Breast Cancer Res Treat. 2002;74:155–165. doi: 10.1023/a:1016114026769. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bagheri-Yarmand R, Balasenthil S, Hortobagyi G, Sahin AA, Barnes CJ, et al. HER2 regulation of peroxisome proliferator-activated receptor gamma (PPARgamma) expression and sensitivity of breast cancer cells to PPARgamma ligand therapy. Clin Cancer Res. 2003;9:3198–3203. [PubMed] [Google Scholar]
- He X, Esteva FJ, Ensor J, Hortobagyi GN, Lee MH, Yeung SC. Metformin and thiazolidinediones are associated with improved breast cancer-specific survival of diabetic women with HER2+ breast cancer. Ann Oncol. 2012;23:1771–1780. doi: 10.1093/annonc/mdr534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chearwae W, Bright JJ. PPARgamma agonists inhibit growth and expansion of CD133+ brain tumour stem cells. Br J Cancer. 2008;99:2044–2053. doi: 10.1038/sj.bjc.6604786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao T, Kornblau S, Safe S, Watt JC, Ruvolo V, Chen W, et al. Role of peroxisome proliferator-activated receptor-gamma and its coactivator DRIP205 in cellular responses to CDDO (RTA-401) in acute myelogenous leukemia. Cancer Res. 2010;70:4949–4960. doi: 10.1158/0008-5472.CAN-09-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Yan C, Czader M, Foreman O, Blum JS, Kapur R, et al. Inhibition of PPARgamma in myeloid-lineage cells induces systemic inflammation, immunosuppression, and tumorigenesis. Blood. 2012;119:115–126. doi: 10.1182/blood-2011-06-363093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reka AK, Kurapati H, Narala VR, Bommer G, Chen J, Standiford TJ, et al. Peroxisome proliferator-activated receptor-gamma activation inhibits tumor metastasis by antagonizing Smad3-mediated epithelial-mesenchymal transition. Mol Cancer Ther. 2010;9:3221–3232. doi: 10.1158/1535-7163.MCT-10-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuniyasu H. The roles of dietary PPARgamma ligands for metastasis in colorectal cancer. PPAR Res. 2008;2008:529720. doi: 10.1155/2008/529720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Zhou J, Casimiro MC, Liang B, Ojeifo JO, Wang M, et al. Activating peroxisome proliferator-activated receptor gamma mutant promotes tumor growth in vivo by enhancing angiogenesis. Cancer Res. 2009;69:9236–9244. doi: 10.1158/0008-5472.CAN-09-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez JA, Vellon L, Mehmi I, Oza BP, Ropero S, Colomer R, et al. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in cancer cells. Proc Natl Acad Sci USA. 2004;101:10715–10720. doi: 10.1073/pnas.0403390101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani V, Botti C, Di Tondo U, Visca P, Pizzuti L, Santeusanio G, et al. Tissue microarray analysis of FAS, Bcl-2, Bcl-x, ER, PgR, Hsp60, p53 and Her2-neu in breast carcinoma. Anticancer Res. 2006;26:2983–2987. [PubMed] [Google Scholar]
- Silva SD, Agostini M, Nishimoto IN, Coletta RD, Alves FA, Lopes MA, et al. Expression of fatty acid synthase, ErbB2 and Ki-67 in head and neck squamous cell carcinoma: a clinicopathological study. Oral Oncol. 2004;40:688–696. doi: 10.1016/j.oraloncology.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Kumar-Sinha C, Ignatoski KW, Lippman ME, Ethier SP, Chinnaiyan AM. Transcriptome analysis of HER2 reveals a molecular connection to fatty acid synthesis. Cancer Res. 2003;63:132–139. [PubMed] [Google Scholar]
- Lee JJ, Drakaki A, Iliopoulos D, Struhl K. MiR-27b targets PPARgamma to inhibit growth, tumor progression and the inflammatory response in neuroblastoma cells. Oncogene. 2012;31:3818–3825. doi: 10.1038/onc.2011.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen JV, Brusselmans K, Verhoeven G. Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care. 2006;9:358–365. doi: 10.1097/01.mco.0000232894.28674.30. [DOI] [PubMed] [Google Scholar]
- Robbins GT, Nie D. PPAR gamma, bioactive lipids, and cancer progression. Front Biosci. 2012;17:1816–1834. doi: 10.2741/4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta RG, Williamson E, Patel MK, Koeffler HP. A ligand of peroxisome proliferator-activated receptor gamma, retinoids, and prevention of preneoplastic mammary lesions. J Natl Cancer Inst. 2000;92:418–423. doi: 10.1093/jnci/92.5.418. [DOI] [PubMed] [Google Scholar]
- Liu H, Zang C, Fenner MH, Possinger K, Elstner E. PPARgamma ligands and ATRA inhibit the invasion of human breast cancer cells in vitro. Breast Cancer Res Treat. 2003;79:63–74. doi: 10.1023/a:1023366117157. [DOI] [PubMed] [Google Scholar]
- Burton JD, Goldenberg DM, Blumenthal RD. Potential of peroxisome proliferator-activated receptor gamma antagonist compounds as therapeutic agents for a wide range of cancer types. PPAR Res. 2008;2008:494161. doi: 10.1155/2008/494161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rageul J, Mottier S, Jarry A, Shah Y, Theoleyre S, Masson D, et al. KLF4-dependent, PPARgamma-induced expression of GPA33 in colon cancer cell lines. Int J Cancer. 2009;125:2802–2809. doi: 10.1002/ijc.24683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Yuan H, Kopelovich L, Yin Y, Lu J, Glazer RI. Drug-targeted inhibition of peroxisome proliferator-activated receptor-gamma enhances the chemopreventive effect of anti-estrogen therapy. Oncotarget. 2012;3:345–356. doi: 10.18632/oncotarget.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton-McDowall ML, Feil D, Robker RL, Thompson JG, Dunning KR. Utilization of endogenous fatty acid stores for energy production in bovine preimplantation embryos. Theriogenology. 2012;77:1632–1641. doi: 10.1016/j.theriogenology.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Ferguson EM, Leese HJ. A potential role for triglyceride as an energy source during bovine oocyte maturation and early embryo development. Mol Reprod Dev. 2006;73:1195–1201. doi: 10.1002/mrd.20494. [DOI] [PubMed] [Google Scholar]
- Zu XY, Zhang QH, Liu JH, Cao RX, Zhong J, Yi GH, et al. ATP citrate lyase inhibitors as novel cancer therapeutic agents. Recent Pat Anticancer Drug Discov. 2012;7:154–167. doi: 10.2174/157489212799972954. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Hauser S, Adelmant G, Sarraf P, Wright HM, Mueller E, Spiegelman BM. Degradation of the peroxisome proliferator-activated receptor gamma is linked to ligand-dependent activation. J Biol Chem. 2000;275:18527–18533. doi: 10.1074/jbc.M001297200. [DOI] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- Christgen M, Ballmaier M, Lehmann U, Kreipe H. Detection of putative cancer stem cells of the side population phenotype in human tumor cell cultures. Methods Mol Biol. 2012;878:201–215. doi: 10.1007/978-1-61779-854-2_13. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu H, Urvalek AM, Li T, Yu L, Lamar J, et al. KLF8 promotes human breast cancer cell invasion and metastasis by transcriptional activation of MMP9. Oncogene. 2011;30:1901–1911. doi: 10.1038/onc.2010.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.