Abstract
Cystic fibrosis (CF) is an autosomal recessive disease caused by mutations in the gene encoding the CF transmembrane conductance regulator. Disruption of electrolyte homeostasis at mucosal surfaces leads to severe lung, pancreatic, intestinal, hepatic, and reproductive abnormalities. Loss of lung function as a result of chronic lung disease is the primary cause of death from CF. Using high-throughput sequencing to survey microbes in the sputum of 16 CF patients and 9 control individuals, we identified diverse microbial communities in the healthy samples, contravening conventional wisdom that healthy airways are not significantly colonized. Comparing these communities with those from the CF patients revealed significant differences in microbial ecology, including differential representation of uncultivated phylotypes. Despite patient-specific differences, our analysis revealed a focal microbial profile characteristic of CF. The profile differentiated case and control groups even when classically recognized CF pathogens were excluded. As a control, lung explant tissues were also processed from a group of patients with pulmonary disease. The findings in lung tissue corroborated the presence of taxa identified in the sputum samples. Comparing the sequencing results with clinical data indicated that diminished microbial diversity is associated with severity of pulmonary inflammation within our adult CF cohort.
INTRODUCTION
Cystic fibrosis (CF) is a profoundly life-shortening disease (1) whose morbidity centers on its pulmonary manifestations. A chronic, progressive process slowly destroys the airways, leading to severe lung dysfunction and death from respiratory failure (2–4). Treatment of CF pulmonary disease has focused on enhancing mobilization of secretions from the lower airway and the use of long-term antibiotic therapy to suppress bacterial activity at airway surfaces (1, 5). Paradoxically, antibiotic therapy often confers clinically detectable benefit even in the context of continuing infection with antibiotic-resistant organisms or with microbes that have established biofilms with minimum inhibitory concentrations far above the applied dose. This paradox was recognized in the clinical setting and has been difficult to reconcile with laboratory observations. Emerging hypotheses point to nontraditional mechanisms of antibiotic action at sublethal antibiotic concentrations (2, 6–8).
These observations imply that Koch’s postulate, the long-standing paradigm of infectious disease, wherein pathogenesis derives from the presence of a primary and culture-identifiable organism, insufficiently explains pathogenesis in the airway microbial ecology of CF-related lung disease (9). Several years ago, the application of 16S ribosomal RNA (rRNA) gene sequence analysis methods to bronchopulmonary samples proved the use of culture-independent approaches by revealing the presence of microbial species not previously recognized in CF (10–13). Subsequent studies found higher incidences—but incomplete penetrance—of recognized CF pathogens in patients relative to pulmonary disease controls (14, 15), and a lower-resolution terminal restriction fragment length polymorphism (T-RFLP) study found a higher incidence of such pathogens in CF patients compared with healthy controls (14).
High-throughput pyrosequencing of 16S ribosomal DNA (rDNA) has been demonstrated as a means of increasing the sensitivity and robustness of microbial surveys of CF patients (16–18). Measures of microbial diversity within CF patient populations have been compared with patient genotype, clinical status, and prescribed therapies, but the strongest correlation found so far is that diversity within a given patient decreases over time (4, 19). A prejudice that healthy lungs are devoid of microbes, based largely on negative clinical culture results, has impaired testing of the alternative hypothesis that the major feature of pulmonary communities is not the presence of microbes generally but particular perturbations of normal pulmonary microbe communities. Notwithstanding molecular evidence of bacterial populations in the lungs of individuals not diagnosed with lung disease (20–22), few investigations that compare the microbial ecology of healthy and diseased airways have been made. Determination of the microbial communities that are resident in the lungs of both healthy control and disease subjects is necessary to gain insight into the pathogenesis of human pulmonary disease.
To quantitatively examine the CF pulmonary microbiome relative to that in healthy individuals, we undertook a phylogenetic survey of microbes in the sputum of 16 CF patients and 9 healthy control individuals. Using the 454 DNA pyrosequencing platform, we obtained a molecular census of microbes in each individual in the form of thousands of microbial 16S amplicon sequences. Each quality-filtered sequence was classified to a phylogenetic group using the naïve Bayesian rRNA gene sequence classifier from the Ribosomal Database Project (RDP) (23). We discovered a pervasive CF-specific profile and identified, in the CF pulmonary milieu, a subset of organisms (including but not limited to recognized CF pathogens) that associated strongly with CF and either survived or thrived under current paradigms of treatment. As a further control, we sequenced lung tissues explanted from CF and other patients (table S1) who were undergoing lung transplants and validated the rich microbial communities and taxa that constituted the CF microbiome in the sputum samples. The identification of a CF-specific microbial profile prompted us to test for and discover correlations between microbial ecology and clinical parameters.
RESULTS
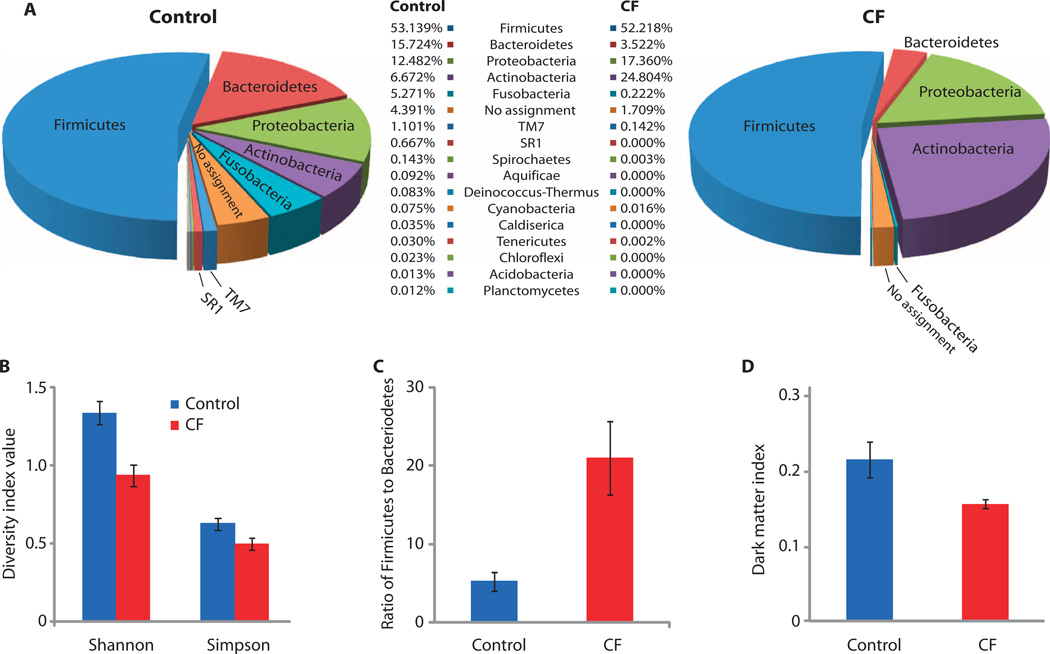
Using the 454 DNA pyrosequencing platform, we analyzed the diversity of microbial consortia in CF and control sputum samples. More than 98% of the 283,470 filtered 16S ribosomal gene sequence reads obtained were classified at the phylum level with at least 95% RDP classification confidence (Fig. 1A). The samples from healthy control subjects were significantly more diverse at the phylum level than were the CF samples—by the Shannon [, where R is richness and pi is the proportion of group i; t test for unequal variances, P = 0.01] and Gini-Simpson (; t test for unequal variances, P = 0.01) indices (Fig. 1B)—with higher richness (R is the number of organism types; t test for unequal variances, P < 0.001) and Pielou evenness (J =H/log(R); t test for unequal variances, P = 0.03). Reduced richness in CF patient samples vis-à-vis healthy and disease control individuals was suggested in the data from two earlier studies (13, 14).
Fig. 1.
Phylum-level analysis of 16S rRNA gene sequences from CF patients and healthy control individuals. (A) Representation of microbial phyla in subject sputum. (B) Diversity of microbiota in the sputum of CF and control patients. (C) Ratio of Firmicutes to Bacteroidetes in the sputum of CF and control patients. (D) Representation of uncultivated organisms in the sputum of CF and control patients according to our DMI (see Materials and Methods for definitions).
The microbial profiles obtained varied strongly among individuals within each cohort as well as across the two cohorts. Firmicutes were the most prevalent phylum in both sample groups (Fig. 1A; see fig. S1 and table S2 for detail on phylum-level data), representing about half the sequences in each group. Bacteroidetes were unequally represented, with 15.7% in the control group and only 3.5% in the CF group (t test for unequal variances, P = 0.04). Conversely, Actinobacteria were much more prevalent in the CF samples at 24.8% than in the healthy control samples (6.7%; t test for unequal variances, P = 0.01). A number of microbial phyla with no cultured representative (that is, no reported laboratory isolate) were apparent in the data set, including the phyla TM7 and SR1. These phyla were differentially represented across the control and CF sample groups; for instance, SR1 occurred in 6 of 9 control samples and in 0 of 16 CF samples (z test, P = 0.0001), and the incidence of TM7 was significantly greater in control samples (1.1%) than in CF samples (0.14%) (t test for unequal variances, P = 0.01). Fusobacteria in particular were enriched in controls versus CF samples (t test for unequal variances, P = 0.03) and contributed to phylum-level diversity.
We also found a significant difference in the ratios of Firmicutes to Bacteroidetes (F/B) (Fig. 1C) in the sputum of control individuals (average ratio, 5:1) versus that of CF patients (average ratio, 21:1) (t test for unequal variances, P = 0.002). Higher F/B ratios in the gut microbiota have been correlated with obesity (24, 25), whereas lower gut F/B ratios have been associated with autoimmunity and type 1 diabetes (26).We analyzed the representation of cultured versus uncultured phyla by devising a dark matter index (DMI) (Fig. 1D). The analysis revealed that control samples have a higher proportion of organisms that are poorly represented in culture collections and genome sequence databases compared with the CF samples (t test for unequal variances, P = 0.02).
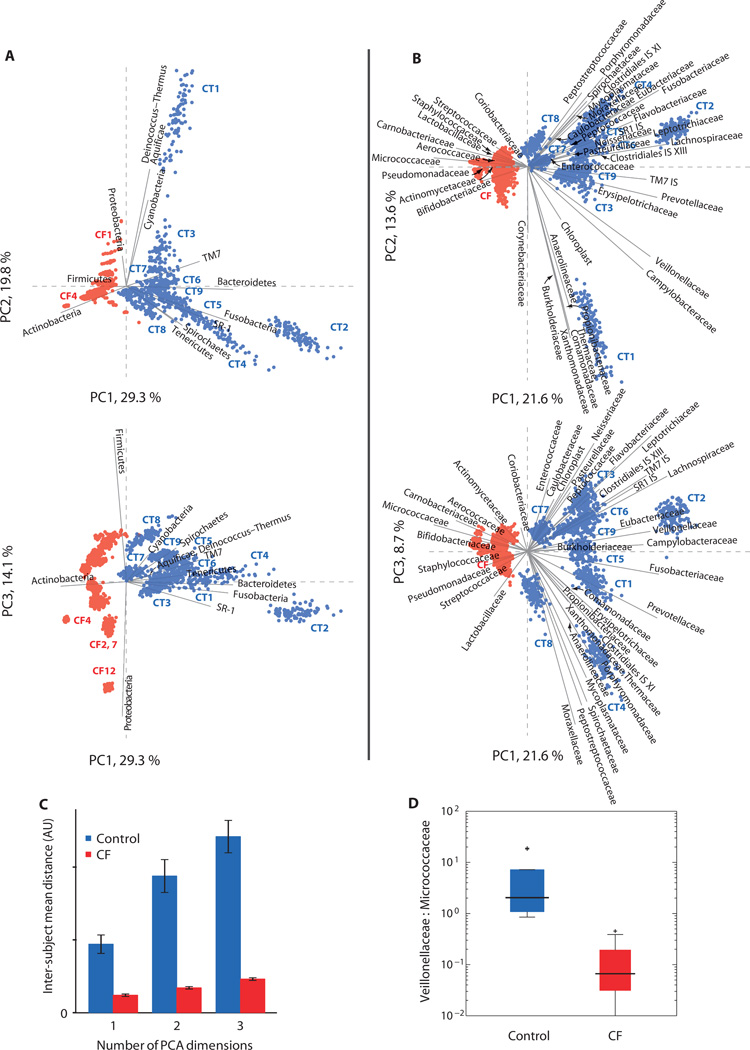
To systematically explore differences among individual subjects and the control and CF groups, we performed a principal components analysis (PCA) of taxon abundances in the sputum samples (Fig. 2). PCA is a mathematical method for transforming data into a new set of coordinates for exploring variation in high-dimensional data sets, displaying the variation in such data sets with lower dimensionality, and finding hidden (composite) variables. The new data coordinates (“principal components”) are linearly independent coordinates each chosen to describe the maximum degree of variation in the original data set, and are ordered by the amount of sample variation described. The first principal component is a coordinate freely chosen to ordinate (explain) the maximum possible variation in the original data set (expressed as a percentage of the total variation); the second coordinate is that which ordinates the most data set variation while being linearly independent from (that is, orthogonal to) all other principal components, and so on. Thus, a significant portion of the variation in a high-dimensional data set such as the abundances of a number of microbial taxa (dimensionality = no. of taxa) can be rendered for print in a small number of dimensions by plotting the data against the first few principal components (Fig. 2 shows two-dimensional plots of the first principal component versus the second and third principal components). The meaning of the principal components is understood by interpretation of their relationship to the plotted data and the original coordinates (microbial taxa in our case).
Fig. 2.
PCA of sequence classification occurrence. (A) Phylum-level and (B) family-level classification data are presented on the same scale in the two representations in each part. Data variance in each populated category (here, 12 phyla or 41 families) is normalized to minimize the influence of large variations in the abundance of one group across samples on the overall analysis. To test the PCA for robustness and dependence on sequencing depth (e.g., counting statistics for rare species), we repetitively rarefied the data, sampling 1000 sequences 100 times (with replacement) from each sample, and performed PCA using all 2500 (25 samples by 100 subsamples) subsamples. Points correspond to individual bootstrap replicate samples, with study subject clusters marked where distinct. The 16 CF samples (red points) cluster separately from controls (blue points). Solid gray lines show the projected original coordinates (corresponding to microbial taxonomic groups). (C) Mean interindividual PCA distance for the two study populations. Mean distances in the family PCA space among the 900 control subsamples (n = 360,099) and 1600 CF subsamples (n = 1,200,099) are plotted in consideration of an increasing number of PCA components (PC1, PC1, and PC2, and PC1, PC2, and PC3). Controls are separated to a much greater degree than are CF samples (t test for unequal variances, P < 10−6). (D) Ratio of Veillonellaceae to Micrococcaceae can be used to segregate subject population by clinical status. Box plots represent data quartiles with outliers indicated.
Figure 2A shows the PCA of the phylum-level classification data. The first principal component (PC1), which explained about one-third of the total variance in the data sets (inclusive of additional variance introduced by our resampling procedure), efficiently separated the healthy control and CF samples (Fig. 2A, upper plot, view separation of sample groups along PC1 axis in plot of PC1 versus PC2), indicating the presence of a stereotyped distinction between the control and the CF samples at the phylum level. The second PCA component, which represented one-fifth of the data set variance, described differences among the control samples (Fig. 2A, upper plot, view spread of control samples along PC2 axis in plot of PC1 versus PC2). The third component, which represented 14% of the data set variance, stratified both sample categories across exclusive regions of the graph (Fig. 2A, lower plot, view spread of control and CF samples along PC3 axis in separate regions of PC1 versus PC3 plot). By projecting the original data coordinates that corresponded to the microbial phyla as vectors on the PCA plots, we determined which microbial groups drove distinction among PCA-segregated subject groups. Bacteroidetes, Fusobacteria, TM7, SR1, Spirochaetes, and Tenericutes were associated with health, whereas Proteobacteria, Firmicutes, and particularly Actinobacteria were associated with CF. The clustering of all 16 CF samples in one region of the PC1–PC2 plot suggests the existence of a characteristic pulmonary microbiome for CF patients.
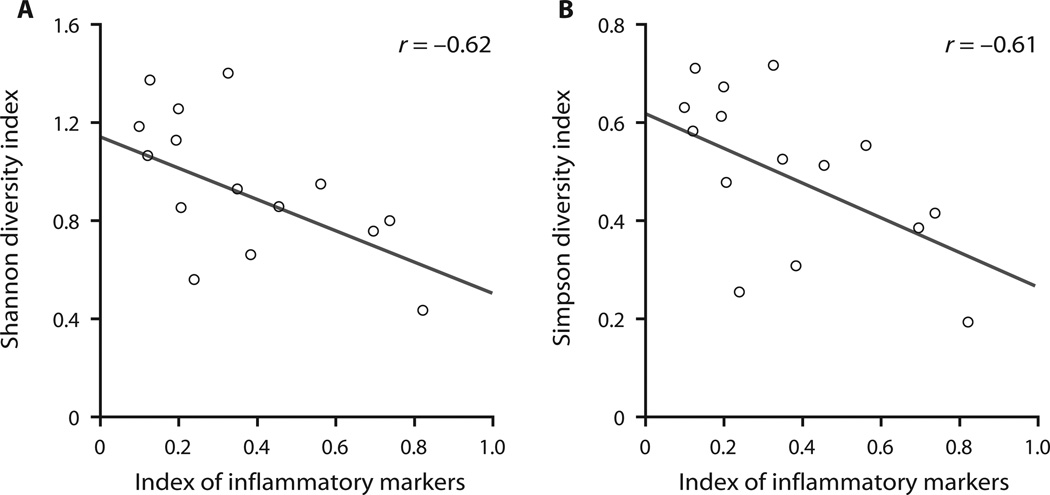
We tested for correlation between microbial diversity within the CF group and clinical measures of patient status at the time of sampling. Because some clinical metrics were strongly correlated with one another (see Materials and Methods and fig. S3), we computed indices that represented two underlying variables: inflammation and pulmonary function. A significant correlation between phylum-level diversity and inflammatory markers was found [r = 0.61 to 0.62, P = 0.02, where P is the probability that random data yield an equal or stronger correlation coefficient than observed in the data under test (|rrandom| ≥ |robserved|); Fig. 3], but diversity did not correlate with pulmonary function in the present cohort. Clinical status is known to decline over a patient’s lifetime and to correlate with reduced microbial diversity (4, 19). Our cohort was relatively uniform in age, such that the major differentiator was clinical status (or rate of clinical decline), not patient age per se. Nonetheless, we tested for correlation of age with microbial diversity and found positive but statistically insignificant values. Taxonomic diversity at the family level correlated less strongly with inflammation than did phylum-level diversity. This may indicate that functional diversity among microbes is the key variable related to patient clinical status rather than phylogenetic diversity.
Fig. 3.
Phylum-level correlations with clinical parameters. (A and B) Scatter plots reveal correlation of the Shannon diversity index (A) and the Simpson diversity index (B) with the inflammatory severity index.
We also analyzed RDP classifications at the family level. Of the filtered reads, 96% could be classified with a microbial family at 80% or better confidence; the high fraction of classified sequences indicates an overall sequence quality that justifies classification at this level of resolution. These data revealed a diverse (more so in the healthy control samples) microbial community that consists of many taxa, including aerobes, obligate anaerobes, and uncultured organisms (see fig. S2 and table S3 for family-level data).
Figure 2B shows the results of the PCA on family-level abundance data for 41 microbial families. In this analysis, the samples from control subjects formed dispersed foci and remained distinct from one another, indicating that personalized pulmonary microbiomes exist in the healthy cohort. Markedly, all 1600 data points derived from the 16 CF patient samples collapsed within a confined region of the graph that was separate from the healthy control data points and exhibited an overall range similar to that found for individual control subjects. The cluster of CF data held even when the first three principal components, representing almost half the total data set variance (which includes intersubject biological variability and technical variability from the resampling procedure), were considered together (Fig. 2C).
As noted above, analysis of the CF samples revealed microbial communities with lower diversity than those of individual control samples. The collapse of the CF cluster demonstrates that the CF samples were far less divergent from one another than were the control samples. This effect is represented in Fig. 2C, which indicates that the inter-CF sample distance was significantly smaller than the inter-control sample distance, corroborating the visual collapse of the CF samples in Fig. 3B.
The original data coordinates also appear in Fig. 3B and allow identification of families that drive segregation of the control and CF samples in the PCA plot. Families that contain the most-recognized CF pathogens associate with the CF group, including Pseudomonadaceae, Streptococcaceae, and Staphylococcaceae. Notably, the families Burkholderiaceae and Pasteurellaceae, which include species whose presence is known to correlate with poor clinical outcomes in CF, do not associate with the CF group. Only a small number of Burkholderiaceae sequences were found in total, and these sequences were inconsistent with the presence of Burkholderia cepacia complex (BCC); BCC infection has been associated with morbidity and mortality in CF patients. Pasteurellaceae family members were more consistently represented in the control group than in the CF group, with the exception of the genus Haemophilus, which was found in both control and patient samples. Although the occurrence in control subjects of taxonomic families (such as Pasteurellaceae) that encompass known pulmonary pathogens is surprising, the Pasteurellaceae family includes not only pathogens such as Haemophilus influenzae but also commensal organisms such as Actinobacillus indolicus, Lonepinella koalarum, and the capnophile (CO2-loving) Mannheimia succiniciproducens (27).
Many actinobacterial families—Actinomycetaceae, Micrococcaceae, and Bifidobacteriaceae—associate with CF, but not all do (for example, Corynebacterineae and Propionibacterineae). The Micrococcaceae have the strongest disease association overall, with most of these sequences classified as Rothia and making strong hits to the sequenced genomes of two species, Rothia dentocariosa and Rothia mucilaginosa. Rothia spp. have been previously isolated from CF sputum samples (28) but are not generally considered to be pathogens characteristic of CF. The Micrococcaceae, which make up about 20% of the sequences we recovered from CF patients, constitute high-penetrance organisms that may worsen the pulmonary status of patients with CF. The abundance of these organisms is strongly and inversely correlated with the abundance of Pseudomonas spp. in our cohort [r = −0.56, P = 0.02, where P is the probability that random data yield an equal or stronger correlation coefficient than observed in the data under test (|rrandom| ≥ |robserved|)]. Carnobacteriaceae, Aerococcaceae, and Lactobacillaceae also show strong associations with CF and should be investigated as agents of disease, as organisms whose growth may be favored by current paradigms of intervention in CF, and as possible targets for new therapies.
Other families, including Fusobacteriaceae, Lachnospiraceae, Leptotrichiaceae, Prevotellaceae, and Veillonellaceae, showed strong correlations with varied states of health in the control group. Fusobacteriaceae and Leptotrichiaceae are known components of the human oral microbiota, but the sequences we sampled in sputum are most likely of pulmonary origin for the following reasons. First, it is known that the mixing of saliva with viscous sputum is limited (29), and induced sputum is an accepted method for pulmonary sampling (22, 29, 30). Second, compared to the control group, the prevalence of these microbial families is markedly diminished and, in many cases, entirely absent from the CF group, despite the use of identical sampling methods. Third, the quantities of microbial DNA recovered from the CF and control sputum samples were statistically indistinguishable (see Materials and Methods for details and table S4) and in line with quantities determined by real-time quantitative polymerase chain reaction (qPCR) in explanted lung tissue samples (table S4). Fourth, the sputum samples from control and CF individuals shared a larger fraction of genera with each other than with saliva samples from two independent studies that used comparable methodology (fig. S4) (31, 32). The co-occurrence of 30 to 40 core genera between the oral and pulmonary samples was not surprising because colonization of the airway by oral taxa was observed previously (29). Fifth, a recent study demonstrated marked homogeneity of microbial communities found along the length of the respiratory tract, indicating that contamination of deep-origin sputum by transit through the upper respiratory tract is of minimal consequence (22). Finally, sequencing of 21 explanted lung samples from seven individuals corroborated both the richness of microbial taxa evidenced in the sputum samples and the correspondence between the microbial taxa found in sputum samples and the microbes present in the lungs themselves. In a concordance analysis for the presence and absence of microbial families, more than 85% concordance was found in the flora present in CF lung tissue and the sputum of CF patients and more than 82% concordance was found between microbial families scored in the control sputum samples and those scored in the lung tissue samples in our study (table S5).
DISCUSSION
We have demonstrated the existence of an endemic pulmonary microbiome in healthy individuals and shown that these communities are in fact significantly more diverse and endowed with more uncultured microbial content than those found in CF patients. The community profiles in both subject groups were highly variable from one individual to another, although greater intersubject variability was found in the control cohort. Some of the most marked findings in this study are commonalities identified among the CF sputum samples, best visualized in the family-level PCA (Fig. 2, B and C). In this analysis, the cluster containing 16 CF samples is highly focused in one region of the graph, with all the samples overlapped with one another (indicating statistical congruence), even when considering the first three principal components (Fig. 2C), which together describe nearly half of the total variance in the analysis.
This is not to say that all CF patients harbor the same microbiome, but rather, that there exists a signature in the microbial profile of CF sputum that is well characterized by the first PCA component that describes the family-level data (Fig. 2B). At the phylum level, the third principal component stratified CF patients along a Firmicutes-Proteobacteria axis (Fig. 2A); such differences in microbial communities between patients are likely to have clinical significance. From the PCA, it is also clear that the CF signature is not determined by one or two pathogenic species. Instead, the signature consists of the presence or increased abundance of more than half a dozen microbial families and the absence or decreased abundance of many other microbial families compared with healthy controls. Thus, the signature is that of a defined but complex microbial community characteristic of CF patients, including groups not routinely detected or analyzed in clinical culture.
Many of the taxa that drove segregation of the sample groups in the PCA are not known to contain pathogenic species. For instance, the ratio of Veillonellaceae to Micrococcaceae was much higher in controls than in CF cases, even though neither family contains widely recognized CF pathogens; the CF and control groups were completely separated on the basis of this metric, with values roughly greater than 1 characterizing the control group and values less than 1 characterizing the CF group (Fig. 2D). The ability to stratify the disease and control groups based on these taxa could form the basis of new molecular diagnostic tests. The ability to do so based on taxa not routinely monitored in the clinic suggests that there is much to learn about the ecology of pulmonary microbial communities and that this knowledge is likely to inform treatment of CF patients. On the other hand, pathogenic taxa that are unmonitored in the clinic might resist nonspecific antimicrobial therapies, exhibit virulence, and contribute to disease. The lungs of CF patients show infection and inflammation; thus, it is also possible that taxa absent from the CF samples and present in the control samples have the job of protecting the pulmonary system, similar to the role played by protective microbial species identified in the gut (33–36) wherein commensal organisms inhibit pathogen outgrowth, expression of virulence genes, or inflammation. Such protective communities could be displaced by pathogens in the CF airways or selected against as a result of prolonged antibiotic therapy. Negative correlations between microbial community diversity and pathogen growth are known in model CF communities (37).
In the gut, researchers have observed marked (if largely reversible) changes in microbial consortia and in the state of the host epithelium upon antibiotic treatment (38–42), which also has been connected to dysbiosis of the gut consortia, chronic inflammatory states (43, 44), and improper immune development (45, 46). Although it is possible that antibiotic treatment accounts for the lower diversity we observed in the lungs of CF patients, there are several reasons to consider this hypothesis unlikely. On the basis of the findings reported in the literature, patterns of response to prolonged antibiotic treatment in the lung appear to differ significantly compared with the responses in the gut to episodic antibiotic administration (38–42, 44). Because many CF patients are treated on an ongoing basis with multiple antibiotics that often have long half-lives and accumulate to high concentrations in lung tissue, changes to microbial community structure in response to additional antibiotic treatments may be muted. Evidence suggests that the clinical benefits that arise from subsequent additional antibiotic treatments do not result from the elimination of pathogenic bacterial populations but rather from noncanonical modes of action against pathogens such as suppression of virulence (47–49) or direct anti-inflammatory activity of the antibiotic compounds (50–52).
The CF subjects in our cohort were treated with zero to three antibiotics from six different classes. The correlations between microbial diversity and antibiotic administration or the number of administered antibiotics were consistent with random data, indicating no significant relationship between these parameters in our cohort. The three samples from CF patients not treated with antibiotics in the 6 months before sampling fell into the CF cluster, supporting the idea that the stereotyped CF profile we identified is characteristic of the disease and is not an epiphenomenon resulting from antibiotic treatment. Another fact supporting this conclusion is that the association we observe between microbial diversity and clinical measures of inflammation is observed within the CF group, not between the CF group and the untreated control group.
The strong connection between microbial diversity and inflammation in the lung directly connects clinical parameters with quantitative details of microbial ecology and suggests that the pattern of diversity contributes more to clinical presentation than the load of any particular pathogen. This finding and that of an endemic pulmonary microbiome in healthy individuals have the potential to inform current treatment paradigms for lung diseases characterized by infection or inflammation, including chronic obstructive pulmonary disease, diffuse bronchiolitis, bronchiectasis, and asthma (53, 54). Rather than aggressively prescribing broad-spectrum antibiotics, clinicians might introduce targeted antimicrobials and probiotic therapies (55, 56) intended to regulate pathogen activity and enhance the efficacy of natural immune mechanisms with reduced long-term toxicity to the patient and the healthy microbiome (57).
MATERIALS AND METHODS
Methods summary
We extracted DNA from subject samples and preparatively amplified the 16S ribosomal gene sequences using bar-coded PCR universal fusion primers, where the 16S complementary portions were 515F/1391R (58) for sputum samples and 515F/907R for lung explant samples. This PCR product was quantified with real-time and digital PCR (59) and sequenced on the 454 platform (sputum samples) or Ion Torrent platform (lung explant samples).
Subject recruitment
Human subject research approval was obtained for sample collection, and subjects signed informed consent that allowed the banking of their specimens for later use. For this study, we included sputum samples from a cohort of adults with significant CF disease; Table 1 presents the characteristics of the CF subjects from whom sputum samples were obtained. All these subjects had a history of chronic Pseudomonas aeruginosa infection and had a sputum culture within a month of study participation that grew predominantly P. aeruginosa (mucoid and nonmucoid species). Sputum samples were selected at time points for which clinical status and medication (including antibiotics) had been stable for 6 months or more. Control individuals were adults with no history of pulmonary disease and no systemic antibiotic use 6 months before sampling. All of the CF patients were at a stable clinical baseline at the time of sampling (that is, were not experiencing pulmonary exacerbation). Many of the patients were receiving antibiotic treatment at this time, although three were not. Because six different antibiotics were prescribed variously to subsets of individuals, the statistical power of this study to determine antibiotic treatment–microbiome correlations was limited.
Table 1.
Summary of CF subject characteristics.
| Age in years (mean ± SD) | 28.14 ± 7.23 |
| Gender | 10 male, 6 female |
| FVC (mean ± SD), liters | 3.88 ± 0.60 |
| % – Predicted | 85.65 ± 8.9 |
| FEV1 (mean ± SD), liters | 2.72 ± 0.54 |
| % – Predicted | 71.89 ± 12.70 |
| CFTR genotype | |
| p.Phe508del/p.Phe508del | 7 |
| p.Phe508del/other | 4 |
| Other/other | 2 |
| Sweat chloride (mM) | 97.40 ± 29.29 |
| Number of patients on chronic antibiotic therapy (not mutually exclusive) | |
| Oral azithromycin | 9/15 |
| Inhaled tobramycin | 8/15 |
| Inhaled colistin | 5/15 |
| Oral dicloxacillin | 1/15 |
| Oral levofloxacin | 1/15 |
FVC, full vital capacity; FEV1, forced expiratory volume, 1 s; CFTR, gene encoding the cystic fibrosis transmembrane conductance regulator.
Explanted lung tissue samples were collected from normally scheduled transplant surgeries at the Stanford Medical Center in 2011. Recipients had been diagnosed with either CF (three), idiopathic pulmonary fibrosis (one), interstitial lung disease (two), or chronic obstructive pulmonary disease (one). Table S1 presents data on the lung transplant patients from whom explanted tissues were obtained.
Sample collection and preservation
The methodology to obtain lower airway secretions has been well standardized by our group. In brief, subjects were asked to inhale nebulized (high-output nebulizer) 3% hypertonic saline solution for 3 min. The subjects then are asked to take a deep inhalation, clear their mouth of saliva, forcefully cough three times, and expectorate into a sterile container. This maneuver is repeated four more times, and all the sputum collected was preserved by mixing with RNA later (Ambion), pooled into a single specimen, immediately snap-frozen in liquid nitrogen for later batch processing, and stored at −80°C.
Induced sputum is an accepted method of sampling the lungs in CF studies that, despite limitations, compares favorably with alternative methods (29, 30, 60–62). Sputum samples transit the upper respiratory system during collection; however, the high viscosity of sputum minimizes its mixing with fluids in the upper respiratory tract (for example, saliva) (29). On the other hand, the upper respiratory system is contiguous with the lung, and its microbial consortia possibly affects the etiology of CF. Temporal variation in mucus properties and the geography of production inside the lung could affect the geography and efficiency (cells per milliliter of sputum vis-à-vis cells per gram of lung tissue) of sampling.
The explanted lung tissue was refrigerated and processed within 24 hours of the transplant procedure. Samples up to 0.5 g were excised from the explants and frozen at −80°C until they were processed for DNA extraction (90 samples were obtained in total).
DNA extraction
DNA was extracted from each sputum or explant sample with the Qiagen DNeasy Blood & Tissue Kit, generally following the pretreatment steps for Gram-positive bacteria, including the optionally indicated mechanical disruption step. In detail, 50 µl of sputum or 50 mg of explant tissue was washed twice with phosphate-buffered saline (tissue samples were not washed) and then combined in 2-ml microcentrifuge tubes with 50 mg of acid-washed glass beads (equal mass portions: 212 to 300 µm, Sigma G1277; 150 to 212 µm, Sigma G1145; and 425 to 600 µm, Sigma G8772) and 300 µl of 20 mM tris, 2 mM EDTA, and lysozyme (20 mg/ml). Disruption was carried out for 5 min at 30 Hz in the TissueLyser II instrument (Qiagen). Fifty percent Tween 20 (9 µl) was added to the sample, which was then mixed and allowed to incubate for 30 min at 37°C. DNeasy buffer AL (300 µl) and proteinase K (15 mg/ml) (38 µl) were then added, and a 30-min incubation at 56°C was carried out. Two hundred proof ethanol (300 µl) was added to the samples, which were mixed and applied to DNeasy spin columns. The columns were washed with 500 µl of DNeasy buffer AW1 and then with 500 µl of the DNeasy buffer AW2. The columns were dried before elution of the extracted DNA with DNeasy buffer AE (200 µl).
Quantification of the extracted DNA
The extracted product was initially quantified by ultraviolet light absorption at 260 nm on a NanoDrop spectrophotometer (Thermo Scientific). The results indicated that the quantity of extracted DNA was somewhat lower in control sputum samples (2.1 ± 0.7 ng/µl) compared with the CF sputum samples (2.3 ± 1.2 ng/µl). 16S gene copy numbers were estimated by qPCR with universal small subunit rRNA (SSU rRNA) gene primers (58) and by the universal TaqMan scheme (59, 63) with the locked nucleic acid FAM probe 149 (Roche). The primers, with sequences 515F GTGCCAGCMGCCGCGGTAA, 515F-UPL GGCGGCGAGTGCCAGCMGCCGCGGTAA, and 1391R GACGGGCGGTGWGTRCA (sputum samples) or 907R CCTCCGTCAATTCCTTTRAGTTT (explant samples), were obtained as desalted synthesis products from Integrated DNA Technologies (IDT). A standard hot-start (10 min, 95°C), two-step (95°C, 30 s; 60°C, 60 s) PCR program was used on an MX3005-P thermocycler (Stratagene) with Applied Biosystems TaqMan Gene Expression Master Mix. Escherichia coli genomic DNA was serially diluted to generate a standard curve. The qPCR-determined SSU rRNA gene concentration values for the sputum and tissue sample types were comparable.
After the removal of human DNA sequences (see Sequence quality filtering section), the control samples again showed a slightly lower quantity of DNA than was found in the CF samples (table S4). We interpreted these values as amplifiable rDNA units and found that the difference in the average DNA quantity between the cohorts was not significant (P = 0.37).
Amplification and library generation
Between 100,000 and 1,000,000 amplifiable rDNA units, determined by qPCR quantification of the extracted DNA, were used as the template for a preparative PCR to generate adapted and bar-coded 16S amplicons for 454 DNA pyrosequencing with titanium chemistry. For sputum samples, the primer sequences were Ti-A-MID-515F (5′-CATCCCTGCGTGTCTCCGAC-TCAG-XXXXXXXXXX-GTGCCAGCMGCCGCGGTAA-3′) and Ti-B-1391R (5′-CCCTGT-GTGCCTTGGCAGTC-TCAG-CA-GACGGGCGGTGWGTRCA-3′), where XXXXXXXXXX represents one of 20 standard Roche 10-base bar code sequences used in this study. These primers were obtained polyacrylamide gel electrophoresis–purified from IDT.
The reverse primer 1391 was found to be insufficiently selective for microbial DNA against the stronger human signal in tissue samples in 454 sequencing. For this reason, the 907R reverse primer was used to generate indexed libraries that could be deep-sequenced on Illumina MISeq instrument. After failed sequence runs and Illumina primer redesign, the highest-quality Illumina libraries were converted for fast-turnaround sequencing on the Ion Torrent Personal Genome Machine with the primers PGM-B-515F (5′-CCTCTCTATGGGCAGTCGGT-GATCAGTGCCAGCMGCCGCGGTAA-3′) and 5′-PGM-A-P7 (CCATCTCATCCCTGCGTGTCTCCGACTCAGGGCAAGCAGAA-GACGGCATACGAGAT-3′). The thermal program—94°C, 5 min; then 6 to 10 cycles of 92°C, 20 s; 50°C, 30 s; 65°C, 60 s; 75°C, 60 s; 70°C, 60 s—was applied to the samples with an additional 10-min 70°C final extension step. The PCRs were carried out in a 50-µl volume with Platinum HiFi Master Mix (Invitrogen) on an MJ PTC-100 thermocycler. Reaction products were purified by the QIAQuick method (Qiagen), followed by a solid-phase reversible immobilization purification and size-selection step on calibrated AMPure magnetic beads (Agentcourt) according to the Roche/454 method.
Sequence library quantification
The purified libraries were quantified with the previously described digital PCR method (59), except that the 48.770 Digital Array (Fluidigm) was used for the microfluidic digital PCR step and amplification primers complementary to the Titanium adaptor sequences were used. Briefly, serial dilutions of the sequencing libraries were made in 20 mM tris buffer with 0.02% Tween 20. The quantitated libraries were then diluted to 105 molecules/µl in 20 mM tris with 0.02% Tween 20 and aliquoted for storage at −60°C.
Sequencing of 16S amplicons
We carried out 16S amplicon sequencing on the 454 platform using “Titanium” chemistry for the sputum samples. Emulsion PCR was carried out with DNA/bead ratios between 0.08:1 and 0.3:1. We chose sequencing of the V4/V5 region of the 16S gene on the 454 Titanium platform over alternative methods including 16S gene clone sequencing (12, 14, 15), temporal temperature gradient gel electrophoresis (11), length heterogeneity PCR (10), T-RFLP analysis (12, 13, 62), and microarray approaches such as the Phylochip (19) for a variety of reasons. Although full-length 16S gene sequences are routinely obtained when sequencing clones and aid phylogenetic assignment at high resolution, V4/V5 sequences can be classified with very high confidence at lower levels of phylogenetic resolution (64, 65). All of these methods are subject to biases introduced at early steps including DNA extraction and PCR amplification and during the preparation of sequencing libraries and the sequencing procedures. Specific procedures can be established to minimize these biases and should be regularized across samples such that intersample analyses detect sample-dependent rather than procedural variation (the differences in primers and sequencing platform between sputum and lung explant samples are the reasons we limit the comparison of the disparate sample types to presence/absence).Molecular bar coding and large sample sets are necessary to reduce per-sample costs in high-throughput screening, and accurate library quantification (59) is critical to obtain adequate representation of all the samples.
We obtained between 1971 and 62,807 raw 16S sequence reads from each sputum sample (387,309 total reads). Between 3 and 80% of the sequences obtained from each sample using the degenerate primers were identified as host-derived. The fraction of human DNA was 80% higher in patient samples (average, 32%) than in controls (average, 18%), and this difference was significant (t test for unequal variances, P = 0.002). After eliminating human sequences and reads less than 150 base pairs (bp) long, the number of high-quality microbial reads per sample ranged from 1389 to 48,525 (219,804 in all). Using the RDP classifier, we assigned a microbial phylum to 219,728 reads. A total of 862,806 reads were obtained from 21 explant samples, a negligible fraction of which originated from host cells. Four hundred fifty-four sequences were demultiplexed with the Roche-supplied SFF file software tool with standard parameters, whereas PGM sequences were demultiplexed in the MOTHUR environment (66).
Sequence quality filtering
Human sequences were filtered from all sequence data on a local server by running the basic local alignment search tool plus (blast+) (http://blast.ncbi.nlm.nih.gov) with the following command: blastn -query [query file] -db [human genome database] -num_descriptions 20 -culling_limit 2 -max_target_seqs 10 -out [output filename] -task megablast -evalue 20 -outfmt 6 -num_threads 6. Sequence reads with hits were omitted from subsequent analyses. The results were insensitive to moderate changes in the parameter values. Custom code that was run in the Matlab (MathWorks) environment was used to check for and trim primer sequences and eliminate 454 reads shorter than 150 bases. To compare with a more stringent filtering procedure, we alternatively filtered 454 reads from selected samples in the MOTHUR environment for quality, alignment frame (perfect start required against the curated Silva bacterial database), maximum homopolymer size (no greater than 8), the absence of ambiguous bases, and chimeras (chimera slayer algorithm) after removal of human sequences. Although some additional sequences were removed, the resulting distribution of sequence classifications was not significantly altered, indicating the use of classification confidence filtering (indicated as Classification, below) in rejecting problematic sequences from the classification results. PGM reads representing the explant samples were filtered similarly in the MOTHUR environment, but with a minimum length requirement of 125 bp rather than 150 bp, and without the chimera removal step. PGM reads (317,081) remained after these filtering steps, with the 125-bp requirement dominating the attrition of reads.
Classification
Phylogenetic classification was carried out with the RDP MultiClassifier (August 2010 release) (23). More than 98% of the filtered 454 reads were assigned to one of the RDP phyla at the 95% confidence level. For classification of 454 reads at each taxonomic level, we required 95% confidence at the phylum level and 80% confidence at every descending taxonomic level. The results were relatively insensitive to variation of the “confidence level” parameter between 50 and 80% at these levels of phylogenetic resolution. Most of the filtered 454 sequences (96%) were successfully classified down to the family level with greater than 80% confidence. High-confidence genus-level classifications corresponding to candidate phyla were back-propagated to intermediate taxonomic levels. The PGM reads were similarly classified, except that 50% classification confidence was accepted for these shorter sequences.
Principal components analysis
The classification data were analyzed with a custom code in the Matlab environment. These bootstrap samples were mean-centered and variance-normalized with a custom Matlab code to even out the contribution of each classification category. This makes PCA less sensitive to the outgrowth of a particular pathogenic strain, for instance, and allows variation in less abundant (but still adequately sampled) groups to contribute to the analysis. Taxa with overall occurrence greater than 0.03% or representation of at least 0.01% in three or more samples were included (12 phyla, 41 families). For each sample, 1000 read classifications were sampled 100 times with replacement. This type of subsampling is commonly used to empirically describe the statistics of a given data set. The approach is referred to as “the bootstrap” in statistics (67) and has several advantages over classical methods, including accuracy for non-Gaussian error distributions and faster convergence than regression analysis.
PCA was carried out on the entire collection of bootstrap samples. The distances among CF samples and those among healthy control samples in the PCA space were calculated with a custom Matlab code. We tabulated Euclidian distances between all combinations of intracategory bootstrap replicates (excluding bootstrap replicate comparison within a given subject sample) in the PCA space with the indicated dimensionality (Fig. 2C) and calculated the mean and SD of the distance value distributions.
DMI and clinical indices
The DMI of a phylogenetic category, DMIcategory, is defined as follows: If A = 0, DMIcategory = 1, else, DMIcategory = 0.8 × [log(A) − log(B)]/log(B), where A is the number of Genomes OnLine Database (GOLD) genomes and genome projects in the phylogenetic category, and B is the number of GOLD genomes and genome projects in the phylogenetic domain. The DMI of a given sample (DMIsample) is defined as the dot product of the phylum fractional abundance vector and the calculated DMIcategory vector.
Groups of correlated clinical variables were variance-normalized and averaged to form indices in the following combinations: The pulmonary severity index was determined from the functional vital capacity percentage predicted (FVC%) and the forced expiratory volume in 1 s, percentage predicted (FEV1%). The inflammatory severity index was determined from measurements of sputum elastase, sputum T protein, sputum tumor necrosis factor–α, sputum interleukin-6, and sputum interleukin-8 concentrations. The two spirometry parameters (FVC% and FEV1%) were correlated with r = 0.77, and the correlation coefficients of the 10 pairs of inflammatory metrics averaged r = 0.49 with an SD of 0.24. Figure S3 presents a histogram of these values.
Diversity estimates
The diversity of each sample was estimated at the phylum and family levels on the basis of the Shannon and Gini-Simpson measures as calculated on bootstrap subsamples (100 sets of 1000 read classifications for each sample) with a custom Matlab code.
Comparison of taxon occurrence
Comparisons of the occurrence of taxa in the CF and control sputum samples with that in the explant samples were made at the phylum and family levels. Each taxon was scored as a match in pairwise comparisons if so-classified reads were present in each sample or if reads were present in both samples at less than 0.1%. The definition of absence was relaxed slightly to effectively exclude rare taxa that may not have a clinical impact or be reliably detectable in our approach. The coverage of taxa scored in one sample by another is tabulated as the percentage of matches in the total number of comparisons made. At the family level, >82% of classifications in the control sputum samples were matched by the lung explant sample classifications, whereas >85% of classifications in the CF sputum samples were matched by the lung explant sample classifications (see table S5).
Statistics
Pearson product-moment correlation coefficients among microbiological and clinical variables were calculated in the Matlab programming environment. Significance testing of various metrics was carried out with the Student’s t test for unequal variances (68). P values were determined by integration of Student’s t distribution with appropriately determined degree-of-freedom parameters, except where a z test (69) was indicated for categorical data. P values that report representation differences among microbial taxa were Bonferroni-corrected (70) for multiple testing on the basis of the number of comparisons made (12 comparisons for phyla, 41 comparisons for families). P values for ratios of taxonomic groups were not corrected because specific taxa were chosen for significance testing on the basis of the literature and our PCA results. Error bars represent SEM. In Fig. 2C, the sample number used for calculations of SEM and P values is the number of intersubject comparisons: 36 for the control set and 120 for the CF set.
Supplementary Material
Acknowledgments
We thank the anonymous study subjects for their willingness to participate in this clinical research project, L. Dethlefsen for consulting on primer design and PCR conditions, J. Tsai and A. Potanina for assistance with sequencing library preparation and quantification, and L. Penland for operating the 454 DNA pyrosequencer to sequence these samples. The sequence data sets published in this paper can be found in the Short Read Archive, accession no. SRA057249.
Funding: This work was funded by an NIH Director’s Pioneer Award and NIH 5R01HG004863-02 (to S.R.Q.).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/4/153/153ra130/DC1
Table S1. Characteristics of lung explant samples.
Table S2. Phylum-level taxon occurrence by study subject (taxa with >10 total reads).
Table S3. Family-level taxon occurrence by study subject (taxa with >10 total reads).
Table S4. Amplifiable microbial rDNA abundance in subject samples assessed by qPCR.
Table S5. List of microbial families represented as matching in “presence” or “absence” status in comparisons between sputum cohorts and explant cohort.
Fig. S1. Plots of phylum abundances by cohort for the 12 phyla analyzed in Fig. 2.
Fig. S2. Plots of family abundances by cohort for the 41 phyla analyzed in Fig. 2.
Fig. S3. Histogram of correlation coefficients among the clinical variables making up the index of inflammatory markers.
Fig. S4. Taxon occurrence in the sputum of the control and CF cohorts in comparison with saliva.
Author contributions: P.C.B., D.N.C., C.E.M., and S.R.Q. designed the study. P.C.B., D.N.C., and C.E.M. carried out the experiments. P.C.B., D.N.C., C.E.M., and S.R.Q. analyzed the data and wrote the paper. S.R.Q. and P.C.B. performed the statistical analyses and produced the figures.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 2.Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology. 2007;153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 3.Jelsbak L, Johansen HK, Frost AL, Thøgersen R, Thomsen LE, Ciofu O, Yang L, Haagensen JA, Høiby N, Molin S. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect. Immun. 2007;75:2214–2224. doi: 10.1128/IAI.01282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Andersen GL, Brown R, Fujimura KE, Wu B, Tran D, Koff J, Kleinhenz ME, Nielson D, Brodie EL, Lynch SV. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010;5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.di Sant’agnese PA, Davis PB. Cystic fibrosis in adults, 75 cases and a review of 232 cases in the literature. Am. J. Med. 1979;66:121–132. doi: 10.1016/0002-9343(79)90491-1. [DOI] [PubMed] [Google Scholar]
- 6.Rogers GB, Carroll MP, Bruce KD. Studying bacterial infections through culture-independent approaches. J. Med. Microbiol. 2009;58:1401–1418. doi: 10.1099/jmm.0.013334-0. [DOI] [PubMed] [Google Scholar]
- 7.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 2010;23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers GB, Stressmann FA, Walker AW, Carroll MP, Bruce KD. Lung infections in cystic fibrosis: Deriving clinical insight from microbial complexity. Expert Rev. Mol. Diagn. 2010;10:187–196. doi: 10.1586/erm.09.81. [DOI] [PubMed] [Google Scholar]
- 9.Nelson A, De Soyza A, Perry JD, Sutcliffe IC, Cummings SP. Polymicrobial challenges to Koch’s postulates: Ecological lessons from the bacterial vaginosis and cystic fibrosis microbiomes. Innate Immun. 2012 doi: 10.1177/1753425912439910. [DOI] [PubMed] [Google Scholar]
- 10.Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, Bruce KD. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J Clin. Microbiol. 2003;41:3548–3558. doi: 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolak M, Karpati F, Monstein HJ, Jonasson J. Molecular typing of the bacterial flora in sputum of cystic fibrosis patients. Int. J. Med. Microbiol. 2003;293:309–317. doi: 10.1078/1438-4221-00265. [DOI] [PubMed] [Google Scholar]
- 12.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 2004;42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Kehagia V, Jones GR, Bruce KD. Bacterial activity in cystic fibrosis lung infections. Respir. Res. 2005;6:49. doi: 10.1186/1465-9921-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, Pace NR. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bittar F, Richet H, Dubus JC, Reynaud-Gaubert M, Stremler N, Sarles J, Raoult D, Rolain JM. Molecular detection of multiple emerging pathogens in sputa from cystic fibrosis patients. PLoS One. 2008;3:e2908. doi: 10.1371/journal.pone.0002908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armougom F, Bittar F, Stremler N, Rolain JM, Robert C, Dubus JC, Sarles J, Raoult D, La Scola B. Microbial diversity in the sputum of a cystic fibrosis patient studied with 16S rDNA pyrosequencing. Eur. J. Clin. Microbiol. Infect. Dis. 2009;28:1151–1154. doi: 10.1007/s10096-009-0749-x. [DOI] [PubMed] [Google Scholar]
- 18.Guss AM, Roeselers G, Newton IL, Young CR, Klepac-Ceraj V, Lory S, Cavanaugh CM. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 2011;5:20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klepac-Ceraj V, Lemon KP, Martin TR, Allgaier M, Kembel SW, Knapp AA, Lory S, Brodie EL, Lynch SV, Bohannan BJ, Green JL, Maurer BA, Kolter R. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ. Microbiol. 2010;12:1293–1303. doi: 10.1111/j.1462-2920.2010.02173.x. [DOI] [PubMed] [Google Scholar]
- 20.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WO. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, Martinez FJ, Huffnagle GB. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 26.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyöty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhnert P, Christensen H. Pasteurellaceae: Biology, Genomics and Molecular Aspects. Norfolk, UK: Caister Academic Press; 2008. [Google Scholar]
- 28.Tunney MM, Field TR, Moriarty TF, Patrick S, Doering G, Muhlebach MS, Wolfgang MC, Boucher R, Gilpin DF, McDowell A, Elborn JS. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 2008;177:995–1001. doi: 10.1164/rccm.200708-1151OC. [DOI] [PubMed] [Google Scholar]
- 29.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Kehagia V, Connett GJ, Bruce KD. Use of 16S rRNA gene profiling by terminal restriction fragment length polymorphism analysis to compare bacterial communities in sputum and mouthwash samples from patients with cystic fibrosis. J. Clin. Microbiol. 2006;44:2601–2604. doi: 10.1128/JCM.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henig NR, Tonelli MR, Pier MV, Burns JL, Aitken ML. Sputum induction as a research tool for sampling the airways of subjects with cystic fibrosis. Thorax. 2001;56:306–311. doi: 10.1136/thorax.56.4.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazarevic V, Whiteson K, Hernandez D, François P, Schrenzel J. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics. 2010;11:523. doi: 10.1186/1471-2164-11-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He F, Morita H, Ouwehand AC, Hosoda M, Hiramatsu M, Kurisaki J, Isolauri E, Benno Y, Salminen S. Stimulation of the secretion of pro-inflammatory cytokines by Bifidobacterium strains. Microbiol. Immunol. 2002;46:781–785. doi: 10.1111/j.1348-0421.2002.tb02765.x. [DOI] [PubMed] [Google Scholar]
- 34.Macdonald TT, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005;307:1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- 35.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujimura KE, Slusher NA, Cabana MD, Lynch SV. Role of the gut microbiota in defining human health. Expert Rev. Anti Infect. Ther. 2010;8:435–454. doi: 10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spasenovski T, Carroll MP, Lilley AK, Payne MS, Bruce KD. Modelling the bacterial communities associated with cystic fibrosis lung infections. Eur. J. Clin. Microbiol. Infect. Dis. 2010;29:319–328. doi: 10.1007/s10096-009-0861-y. [DOI] [PubMed] [Google Scholar]
- 38.Schumann A, Nutten S, Donnicola D, Comelli EM, Mansourian R, Cherbut C, Corthesy-Theulaz I, Garcia-Rodenas C. Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol. Genomics. 2005;23:235–245. doi: 10.1152/physiolgenomics.00057.2005. [DOI] [PubMed] [Google Scholar]
- 39.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl. 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins S, Verdu E, Denou E, Bercik P. The role of pathogenic microbes and commensal bacteria in irritable bowel syndrome. Dig. Dis. 2009;27(Suppl. 1):85–89. doi: 10.1159/000268126. [DOI] [PubMed] [Google Scholar]
- 44.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 46.Alm B, Erdes L, Möllborg P, Pettersson R, Norvenius SG, Aberg N, Wennergren G. Neonatal antibiotic treatment is a risk factor for early wheezing. Pediatrics. 2008;121:697–702. doi: 10.1542/peds.2007-1232. [DOI] [PubMed] [Google Scholar]
- 47.Kita E, Sawaki M, Oku D, Hamuro A, Mikasa K, Konishi M, Emoto M, Takeuchi S, Narita N, Kashiba S. Suppression of virulence factors of Pseudomonas aeruginosa by erythromycin. J. Antimicrob. Chemother. 1991;27:273–284. doi: 10.1093/jac/27.3.273. [DOI] [PubMed] [Google Scholar]
- 48.Gemmell CG. Antibiotics and the expression of staphylococcal virulence. J. Antimicrob. Chemother. 1995;36:283–291. doi: 10.1093/jac/36.2.283. [DOI] [PubMed] [Google Scholar]
- 49.Tateda K, Ishii Y, Matsumoto T, Kobayashi T, Miyazaki S, Yamaguchi K. Potential of macrolide antibiotics to inhibit protein synthesis of Pseudomonas aeruginosa: Suppression of virulence factors and stress response. J. Infect. Chemother. 2000;6:1–7. doi: 10.1007/s101560050042. [DOI] [PubMed] [Google Scholar]
- 50.Jaffé A, Bush A. Anti-inflammatory effects of macrolides in lung disease. Pediatr. Pulmonol. 2001;31:464–473. doi: 10.1002/ppul.1076. [DOI] [PubMed] [Google Scholar]
- 51.Equi A, Balfour-Lynn IM, Bush A, Rosenthal M. Long term azithromycin in children with cystic fibrosis: A randomised, placebo-controlled crossover trial. Lancet. 2002;360:978–984. doi: 10.1016/s0140-6736(02)11081-6. [DOI] [PubMed] [Google Scholar]
- 52.Kabra SK, Pawaiya R, Lodha R, Kapil A, Kabra M, Vani AS, Agarwal G, Shastri SS. Long-term daily high and low doses of azithromycin in children with cystic fibrosis: A randomized controlled trial. J. Cyst. Fibros. 2010;9:17–23. doi: 10.1016/j.jcf.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 53.Jaffé A, Francis J, Rosenthal M, Bush A. Long-term azithromycin may improve lung function in children with cystic fibrosis. Lancet. 1998;351:420. doi: 10.1016/S0140-6736(05)78360-4. [DOI] [PubMed] [Google Scholar]
- 54.Rollins DR, Beuther DA, Martin RJ. Update on infection and antibiotics in asthma. Curr. Allergy Asthma Rep. 2010;10:67–73. doi: 10.1007/s11882-009-0086-2. [DOI] [PubMed] [Google Scholar]
- 55.Bruzzese E, Raia V, Gaudiello G, Polito G, Buccigrossi V, Formicola V, Guarino A. Intestinal inflammation is a frequent feature of cystic fibrosis and is reduced by probiotic administration. Aliment. Pharmacol. Ther. 2004;20:813–819. doi: 10.1111/j.1365-2036.2004.02174.x. [DOI] [PubMed] [Google Scholar]
- 56.Bruzzese E, Raia V, Spagnuolo MI, Volpicelli M, De Marco G, Maiuri L, Guarino A. Effect of Lactobacillus GG supplementation on pulmonary exacerbations in patients with cystic fibrosis: A pilot study. Clin. Nutr. 2007;26:322–328. doi: 10.1016/j.clnu.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 57.Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: An ecological perspective. Sci. Transl. Med. 2012;4:137rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U.S.A. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White RA, III, Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radhakrishnan DK, Corey M, Dell SD. Realities of expectorated sputum collection in the pediatric cystic fibrosis clinic. Arch. Pediatr. Adolesc. Med. 2007;161:603–606. doi: 10.1001/archpedi.161.6.603. [DOI] [PubMed] [Google Scholar]
- 61.Al-Saleh S, Dell SD, Grasemann H, Yau YC, Waters V, Martin S, Ratjen F. Sputum induction in routine clinical care of children with cystic fibrosis. J. Pediatr. 2010;157:1006.e1–1011.e1. doi: 10.1016/j.jpeds.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Rogers GB, Skelton S, Serisier DJ, van der Gast CJ, Bruce KD. Determining cystic fibrosis-affected lung microbiology: Comparison of spontaneous and serially induced sputum samples by use of terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 2010;48:78–86. doi: 10.1128/JCM.01324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Zhang D, Li W, Chen J, Peng Y, Cao W. A novel real-time quantitative PCR method using attached universal template probe. Nucleic Acids Res. 2003;31:e123. doi: 10.1093/nar/gng123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sundquist A, Bigdeli S, Jalili R, Druzin ML, Waller S, Pullen KM, El-Sayed YY, Taslimi MM, Batzoglou S, Ronaghi M. Bacterial flora-typing with targeted, chip-based Pyrosequencing. BMC Microbiol. 2007;7:108. doi: 10.1186/1471-2180-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Claesson MJ, Wang Q, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O’Toole PW. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Efron B, Tibshirani R, Tibshirani R. An Introduction to the Bootstrap. Boca Raton: Chapman & Hall/CRC; 1993. [Google Scholar]
- 68.Welch BL. The generalisation of student’s problems when several different population variances are involved. Biometrika. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 69.Sprinthall RC. Basic Statistical Analysis. Boston: A and B Publishing; 2003. [Google Scholar]
- 70.Dunn OJ. Multiple comparisons among means. J. Am. Stat. Assoc. 1961;56:52–64. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.