SUMMARY
Ubiquitination (Ubiquitylation) is a common protein modification that regulates a multitude of processes within the cell. This modification is typically accomplished through the covalent binding of ubiquitin to a lysine residue onto a target protein and is catalyzed by the presence of three enzymes: an activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-protein ligase (E3). In recent years, ubiquitination has risen as a major signaling regulator of immunity and microbial pathogenesis in the mammalian system. Still, little is known about how ubiquitin relates specifically to vector immunology. Here, we provide a brief overview of ubiquitin biochemistry and describe how ubiquitination regulates immune responses in arthropods of medical relevance. We also discuss scientific gaps in the literature and suggest that, similar to mammals, ubiquitin is a major regulator of immunity in medically-important arthropods.
INTRODUCTION
In the 1930s, novel findings in cell biology indicated that intracellular proteins are in a constant state of synthesis and degradation. The coordination of this process was unknown. In 1978, researchers discovered one of the most important regulatory proteins found in eukaryotes: ubiquitin. This discovery won the Nobel Prize in 2004 and revolutionized our understanding of sophisticated protein networks controlling basic processes inside eukaryotic cells. Ubiquitin, from Latin “ubique”, meaning “everywhere”, is a highly conserved 76-amino acid polypeptide found in nearly every tissue in eukaryotes (Jiang et al., 2011, Collins et al., 2010). Ubiquitin is involved in a multitude of functions, from DNA transcription to protein degradation. Among these, immune response and microbial pathogenesis have emerged as crucial events under the control of the ubiquitination (ubiquitylation) machinery (Jiang et al., 2011, Collins et al., 2010). Little is known about how ubiquitination relates to vector immunology. Mosquitoes, ticks and other arthropod vectors have not been widely studied in the context of ubiquitination. This review aims to discuss information available for ubiquitination and vector immunology.
UBIQUITIN BIOCHEMISTRY AND THE UBIQUITINATION PROCESS
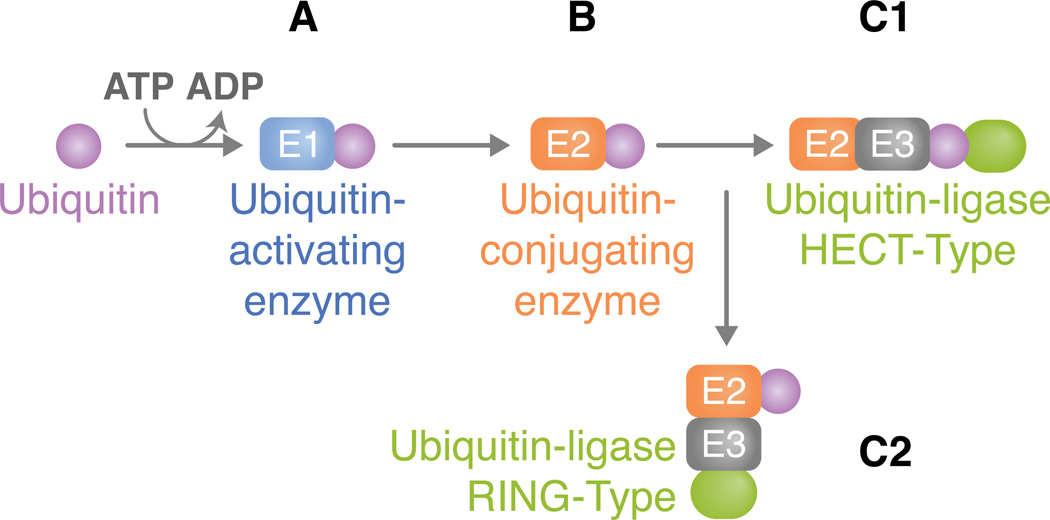
Ubiquitin contains seven lysine (K) residues (K6, K11, K27, K29, K33, K48, and K63), which are responsible for forming linkages with the K of a target protein or with the K of another ubiquitin (Jiang et al., 2011, Collins et al., 2010). Both monoubiquitination and polyubiquitination may occur and this process involves three enzymes: (1) a ubiquitin-activating enzyme (E1), (2) a ubiquitin-conjugating enzyme (E2), and (3) a ubiquitin-protein ligase (E3) (Figure 1). For a ubiquitin to be added to a target lysine, a thiol ester bond between the E1 and ubiquitin is first formed in a two-step ATP-dependent reaction. This is followed by the transfer of the activated ubiquitin to an active site cysteine residue on the E2. Ubiquitin is then attached to a target protein either directly by the E2 or with the E3, which may act as a scaffold and/or bind to ubiquitin and transfer it to the substrate. The ability to interact with the substrate confers the E3 specificity and, for this reason, a great number of E3 enzymes are usually found in a single organism, whereas a smaller number of E2 and very few E1 tend to be observed.
Figure 1. The ubiquitination pathway.
(A) For an ubiquitin to be added to a target lysine, a thiol ester bond between the E1 and ubiquitin is first formed in a two-step ATP-dependent reaction. Ubiquitin is activated through its combination with MgATP. This, in turn, forms a ubiquitin adenylate intermediate that acts as a ubiquitin donor to the E1 active site, creating an E1 molecule that consists of both an activated thiol ester ubiquitin and an adenylated ubiquitin. (B) The thiol-linked ubiquitin is then transferred to the E2. E2s typically share a core domain of about 150 amino acids and a cysteine residue in the active site to which E1 ubiquitin is transferred. (C) The ubiquitin ligase is typically a protein complex that attaches to both the substrate and the E2. The method in which substrate binding occurs varies between direct interactions or through auxiliary proteins. E3 enzymes can be classified according to the domains they carry. Here, we address two types of ubiquitin ligases: (C1) HECT type or (C2) RING type. (C1) HECT domain ligases have a domain that is approximately 350 residues. They serve a catalytic role in the conjugation of ubiquitin to the target protein (green oval). HECT domain proteins possess a conserved cysteine residue through which the activated E2 ubiquitin can form a thiol ester bond, thus leading to its transfer from E2. After attachment of ubiquitin, a lysine residue of the target protein must be introduced to the active site in order for substrate-ubiquitin ligation to occur. Ubiquitin is ultimately conjugated to a NH2 group of the target protein. The difference in the NH2 terminal domain within various HECT domain proteins is responsible for E3 substrate specificity. (C2) Unlike the HECT domain ligase that serves as a direct intermediate between E2 and substrate ubiquitin transfer, the RING type functions as a molecular scaffold that brings proteins together instead of acting as a chemical catalyst. It is defined by a conserved series of histidine and cysteine residues that is patterned to form a cross-brace structure, which permits the binding of two zinc cations.
E3s are primarily of two types, according to their domains. Homologous to the E6-AP Carboxyl Terminus (HECT) domain ligases are a family of E3s with catalytic roles in the conjugation of ubiquitin to the target protein (Skaug et al., 2009). Unlike the HECT ligases, the Really Interesting New Gene (RING)-type functions as a scaffold that brings proteins together. Certain E3s only require the E2 for ubiquitination, but in some cases, a protein complex that contains the RING finger is mandatory (Skaug et al., 2009). In addition, studies have discovered a protein family that closely resembles the RING finger, but lacks the zinc coordinating residues within the RING domain. This novel protein family, termed U-box, has the ability to elongate ubiquitin chains, and for several U-box proteins, the elongation process is dependent on both E1 and E2, but not on E3 (Skaug et al., 2009). This has led to the categorization of the U-box proteins as a subfamily of E3 enzymes. Deubiquitinases (DUBs) remove ubiquitin chains from target proteins and have the opposite role of E3s. DUBs may be grouped into: 1) enzymes that cleave precursors comprised of multiple ubiquitin proteins or ubiquitin fused to the amino terminus of ribosomal proteins; 2) DUBs that remove ubiquitin from proteins modified post-translationally; and 3) DUBs that edit ubiquitin chains by altering the linkage type (e.g., K63 or K48) (Harhaj et al., 2012).
Depending on which K the ubiquitin attaches, a different outcome may be expected. Typically, modification by K48 polyubiquitination leads to targeting of the substrate for proteasome degradation. K63 linkages, however, have non-proteolytic roles in transcription, DNA repair, protein trafficking and signaling (Skaug et al., 2009). Monoubiquitination has been involved in transcription (Greer et al., 2003), endocytosis and DNA repair (Gregory et al., 2003), while a range of other linkages, such as K6, K11, K27, K29, K33, may also occur but have mostly undetermined roles. Linear ubiquitin chains may also be formed by a ubiquitin ligase complex known as the linear ubiquitin chain assembly complex (LUBAC) - a regulator of nuclear factor (NF)-κB (Tokunaga et al., 2012).
AN INNATE PERSPECTIVE
Recent work revealed that ubiquitination plays a central role in mammalian innate immunity. Ubiquitin is a highly conserved regulatory protein and the pathways underlying innate immunity are also conserved throughout the animal kingdom. Within this context, ubiquitin regulation and pathogen-mediated immunity in Drosophila and arthropod vectors of medical relevance will be discussed.
Drosophila: the strength of a model
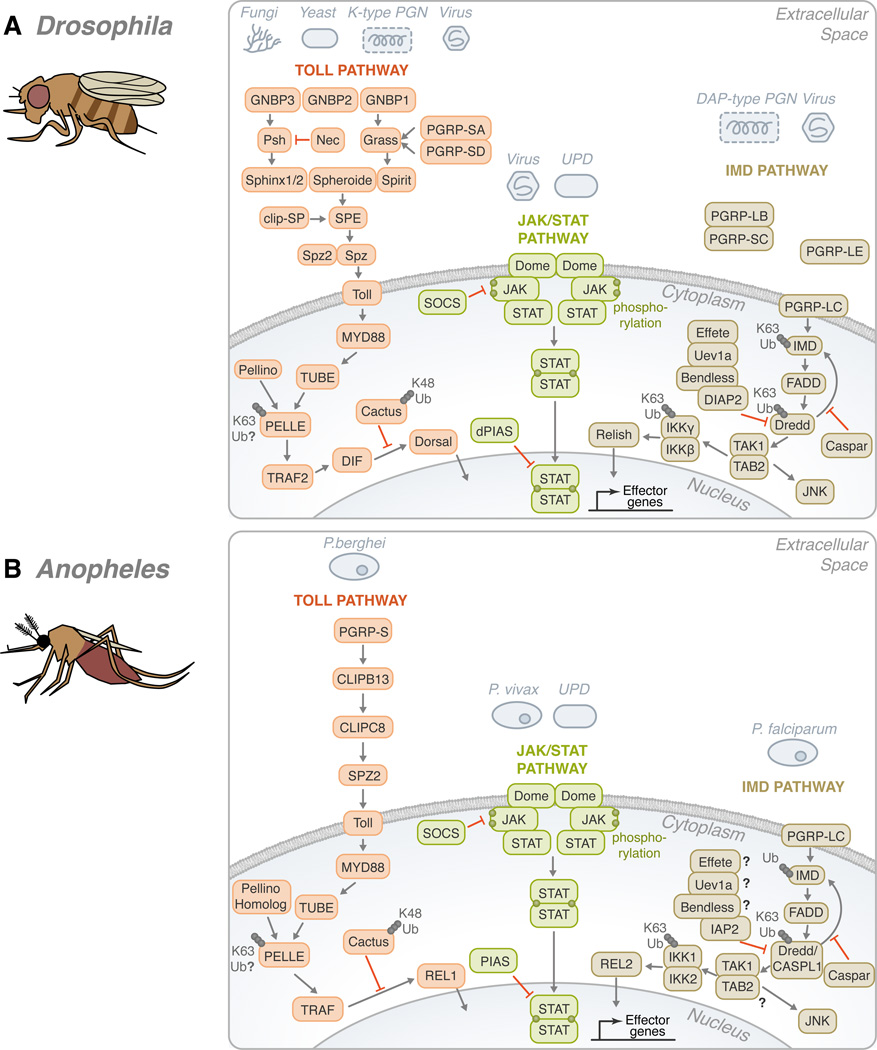
The Toll pathway is an evolutionarily conserved cascade involved in the establishment of the dorsal-ventral axis in Drosophila (Anderson et al., 1985). It also shares similarities with the mammalian myeloid differentiation primary response protein 88 (MyD88)-dependent pathway, which involves NF-κB activation and direct Toll-like receptor recognition. In Drosophila, this cascade is activated mostly in the presence of lysine-type peptidoglycans, fungi, viruses and yaest (Figure 2A) (Valanne et al., 2011). Peptidoglycan recognition proteins (PGRPs) and Gram-negative binding proteins (GNBPs) play the main role in recognition upstream of Toll and lead to processing and activation of Spätzle, followed by binding of this cytokine to the Toll receptor. Downstream, the cascade involves phosphorylation and ubiquitin regulation. First, Pelle - homologous to the mammalian IL-1 receptor-associated kinases (IRAK) family of kinases - is recruited. Then, phosphorylation and proteasomal-mediated degradation of Cactus, the Drosophila inhibitor of κB homolog, takes place due to K48 linkages. Recently, another highly conserved protein named Pellino was suggested to interact with Pelle/IRAK, most likely via ubiquitination of Pelle (Moynagh, 2009).
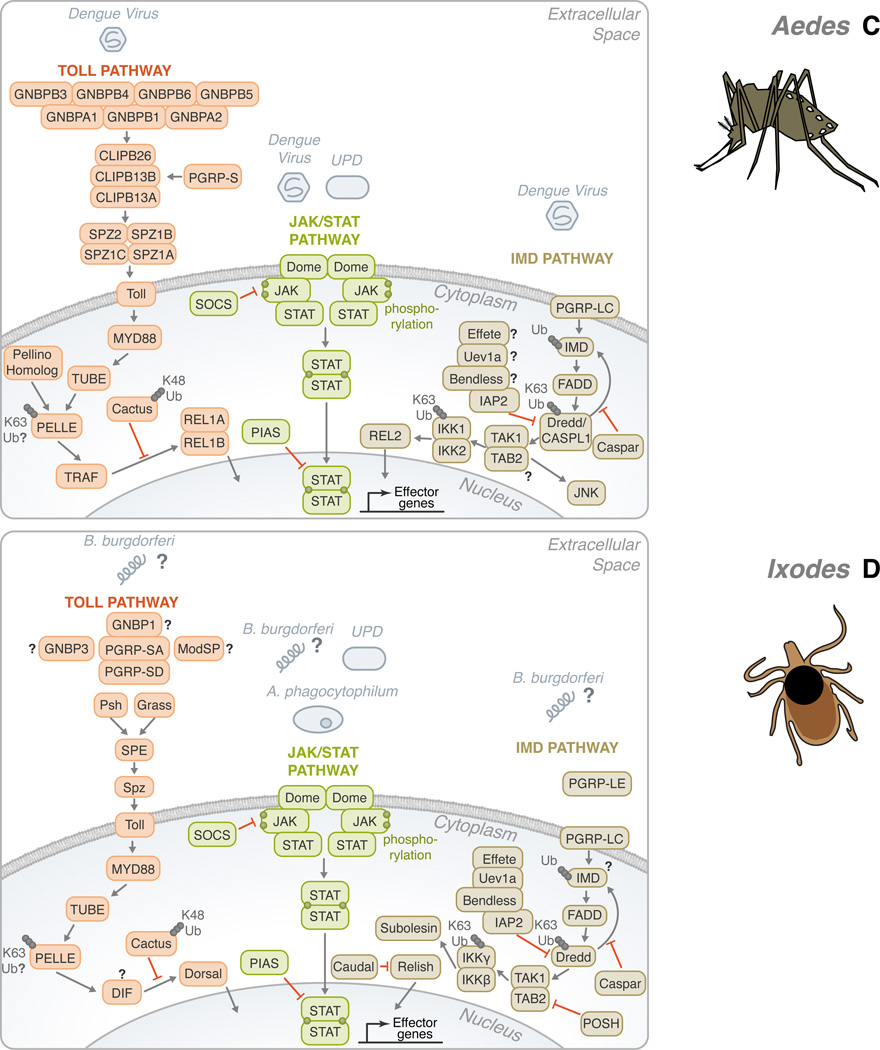
Figure 2. Conservation of the Toll, IMD, and JAK/STAT pathways in Drosophila melanogaster, Anopheles gambiae, Aedes aegypti and Ixodes scapularis.
(A) In Drosophila, gram-negative bacteria binding proteins (GNBPs) and peptidoglycan recognition proteins (PGRPs) have been shown to activate the Toll pathway in the presence of stimulants, such as fungi, yeast, lysine-type peptidoglycan (K-type PGN) and viruses. These recognition proteins signal downstream to Persephone (psh) and Grass. CLIP domain serine proteases (clip-SP) modulate the signaling after recognition as well. Sphinx1/2, Spheroide, and Spirit initiate the activation of Spätzle through the Spätzle processing enzyme (SPE). Spätzle binds to Toll which recruits three Death domain-containing molecules: MyD88, Tube, and Pelle. Pellino/Pellino homolog, perhaps, acts as a positive regulator of immunity by ubiquitinating Pelle. After which, TNF-receptor-associated factor (TRAF) signals to Dorsal-immune related factor (DIF), followed by signaling to Dorsal. The activation is facilitated by the degradation of Cactus through K48 ubiquitination. The transcription factor translocates to the nucleus in order to upregulate immune genes. The Toll pathway is highly conserved in: (B) Anopheles, (C) Aedes, and (D) Ixodes. The Toll pathway has been demonstrated to recognize Plasmodium berghei in (B) Anopheles and the Dengue virus in (C) Aedes. TRAF signaling initiates REL1 and REL1A /1B activity in (B) Anopheles and (C) Aedes, respectively. (A) On the other hand, the Drosophila IMD pathway recognizes primarily mono-diaminopimelic acid-type peptidoglycans (DAP-type PGN). Fas-associated protein with death domain (FADD) is recruited to IMD. FADD binds to the Death related ced-3/Nedd2-like caspase (Dredd)/CASPL1. Dredd can cleave IMD. Inhibitor of apoptosis (IAP) can also associate with Dredd/CASPL1. Effete, Uev1a, and Bendless play a role in the regulation of this step and caspar may also inhibit the activity of IMD-dependent transcription factors. TGF-β activated kinase (TAK1), TAK1 binding protein 2 (TAB2) complex forms as signaling continues. Two avenues may result from the IMD pathway: JNK or NF-κB. For NF-κB activation, Relish translocates to the nucleus to activate effector genes. There are several potential sites of ubiquitination throughout the IMD pathway: IMD, Dredd/CASPL1, and the IKK complex. Like the Toll pathway, the IMD pathway is found in many species: (B) Anopheles, (C) Aedes, and (D) Ixodes. P. falciparum and the dengue virus can trigger the IMD pathway in (B) Anopheles and (C) Aedes, respectively. While Relish is regulated by the IMD pathway in (A) Drosophila and (D) Ixodes, REL2, the homolog of Relish, acts as the transcription factor in (B) Anopheles and (C) Aedes. The third pathway is the JAK/STAT pathway. (A) A ligand derived from the unpaired (UPD) gene activates the pathway by binding to Domeless (Dome). Phosphorylated JAK promotes the dimerization of STAT. Dimerized STAT can proceed to the nucleus. Countering the activation, both SOCS and PIAS negatively regulate the JAK/STAT pathway. Though the JAK/STAT pathway is evolutionarily conserved across the organisms discussed, various pathogens have demonstrated the ability to activate the JAK-STAT pathway, such as: (B) Plasmodium vivax, (C) dengue virus and (D) A. phagocytophilum. For the Toll, IMD and JAK/STAT pathways, B. burgdorferi recognition in I. scapularis remains mostly elusive. The information illustrated here was obtained from gene sequences available at vectorbase (www.vectorbase.org) and the following articles: (Waterhouse et al., 2007, Liu et al., 2012, Souza-Neto et al., 2009, Cirimotich et al., 2010, Xi et al., 2008, Hoffmann, 2003).
Another important pathway in immunity is the IMD (immune deficiency) pathway. IMD is a death domain-containing protein that shares homology with the receptor interacting protein (RIP) of the mammalian tumor necrosis factor receptor pathway. This cascade is stimulated mostly by diaminopimelic acid (DAP)-type peptidoglycans. Drosophila inhibitor of apoptosis protein 2 (DIAP2) was identified as an E3 that targets the Dredd caspase for K63 polyubiquitination (Meinander et al., 2012). Dredd cleaves IMD and allows for its association with DIAP2 (Paquette et al., 2010) and further conjugation of IMD and K63 polyubiquitin chains. The E2 ligases Effete, Bendless and Uev1a also mediate this process and polyubiquitination seems to be crucial for the activation of TGF-β activated kinase (TAK1) and the Drosophila homolog of the mammalian I-κB kinase (IKK) complex (Paquette et al., 2010). This kinase forms a complex with the adaptor molecule TAB2, and likely binds K63 polyubiquitin chains (Paquette et al., 2010). This leads to the induction of the two branches of the IMD pathway: c-Jun N-terminal kinases (JNK) and NF-κB (Zhuang et al., 2006).
The Janus Kinase and Signal Transducers and Activators of Transcription (JAK/STAT) pathway is also present in D. melanogaster and has been associated with cell proliferation, differentiation, morphogenesis, heterochromatin stability and immunity (Arbouzova et al., 2006). This pathway involves the secreted protein ligand Unpaired, the transmembrane receptor Domeless (Dome), the JAK Hopscotch, the STAT transcription factor, as well as the negative regulator protein inhibitors of activated STAT (PIAS) and the suppressor of cytokine signaling (SOCS). Phosphorylation of Dome and STAT are known to regulate this cascade in Drosophila (reviewed in (Agaisse et al., 2004)). Proteasomal-dependent regulation of JAK (Ungureanu et al., 2002) and STAT ubiquitination and degradation have been demonstrated in mammals (Ungureanu et al., 2005). The PIAS protein family also shows small ubiquitin-related modifier (SUMO) E3 activity (Sharrocks, 2006). Though this pathway seems to be conserved, the regulatory role of ubiquitin is yet to be confirmed in Drosophila.
VECTOR-BORNE DISEASES: THE UBIQUITOUS NIGHTMARE
Vector-borne diseases such as malaria, dengue and West Nile encephalitis are caused by the transfer of pathogenic microorganisms from a vector (e.g., a mosquito) to a mammalian host. They constitute a large portion of the neglected diseases that affect the health of billions of people worldwide, and vector-borne pathogens are estimated to infect half of the world’s population (Table S1) (Hotez et al., 2009). The role of ubiquitination during pathogen colonization of arthropod vectors remains mostly elusive.
Ubiquitination and mosquito-borne pathogens: are we there yet?
Mosquitoes are the most important vectors of human diseases in the world. The immunity of Anopheles gambiae, the primary vector of Plasmodium falciparum in sub-Saharan Africa, has been the focus of several studies aiming to understand the parasite-vector interaction. The Anopheles genome codifies two NF-κB transcription factors, Rel1 and Rel2, orthologous to Drosophila Dorsal and Relish, respectively (Figure 2B). The knockdown of Rel2 and its regulator, Caspar - which carries ubiquitin-associated and ubiquitin-like domains - showed that Rel2 controls A. gambiae resistance to P. falciparum (Garver et al., 2009). This Rel1/Rel2-dependent response regulates basal expression of the major anti-parasitic factors, such as thioester containing protein 1 (TEP1) (Garver et al., 2012). Serine protease inhibitors (serpins), as well as CLIP domain proteases, implicated in antimicrobial synthesis in Drosophila, have been identified as regulators of proteolytic cascades in A. gambiae (Volz et al., 2005). One can expect that ubiquitination regulates intracellular kinase activation of NF-κB-dependent events in Anopheles - similar to what is observed in humans and Drosophila.
Aedes aegypti is another important disease vector. It transmits yellow fever, dengue fever and filariasis. Dengue is the most common arthropod-borne viral disease in humans, and half of the world’s population is at risk (Wilder-Smith et al., 2012). This mosquito presents two isoforms of Rel1 that seem to cooperate in enhancing gene expression (Waterhouse et al., 2007) (Figure 2C). Both the RNA interference (RNAi) and the JAK/STAT pathways modulate dengue infection in mosquitoes but in depth studies are needed to address regulation by ubiquitination. RNAi screen approaches have identified ubiquitin-related proteins as host factors that promote viral replication (Krishnan et al., 2008, Sessions et al., 2009). Dipteran and mammalian hosts have been found to share ubiquitin-related host factors associated with dengue virus infection (Sessions et al., 2009). In humans, many components of the ubiquitin machinery have their expression altered upon dengue virus infection (Fink et al., 2007).
West Nile virus (WNV) is a RNA virus of re-emerging importance. It is transmitted by Culex spp. mosquitoes, and there is very little known about the West Nile-mosquito interactions. Culex quinquefasciatus gene family members share large similarities with A. aegypti (Bartholomay et al., 2010) suggesting that common antiviral mechanisms exist in the WNV-Culex system. Interestingly, WNV infection seems to be reduced upon silencing of the endocytosis-related ubiquitin ligase CBLL1 in human HeLa cells. WNV virions were retained in the plasma membrane and treatment with a proteasomal inhibitor further abolished WNV infection (Krishnan et al., 2008). Furthermore, a WNV envelope protein named WNV-E inhibited dsRNA-induced cytokine production in murine macrophages by altering the pattern of RIP1 ubiquitination and NF-κB activation (Arjona et al., 2007).
Tricky ticks and the track of ubiquitin
On a global basis, ticks are second only to mosquitoes as vectors of disease-causing agents. For example, they transmit Anaplasma phagocytophilum, which causes human granulocytic anaplasmosis, and Borrelia burgdorferi, the agent of Lyme disease, the most prevalent vector-borne disease in the United States. This arthropod has a well-developed innate immune system, but only very little has been recognized about tick immunology, especially when compared to other vectors, such as mosquitoes (Kopacek et al., 2011). Comparative genomics approaches have illustrated that the core set of genes involved with the Toll, IMD, JAK/STAT and RNAi pathways exist in ticks (Figure 2D). A few articles have also addressed antimicrobial expression during pathogen infection in ticks (Liu et al., 2012). In fact, differential expression of ubiquitin-related proteins has been described in tick cells upon infection with A. marginale (de la Fuente et al., 2007), which causes bovine anaplasmosis. Vaccination studies using an ubiquitin peptide have been performed for the control of cattle tick (Almazan et al., 2010). Mechanistic approaches, however, are still needed to elucidate the regulatory cascades underlying ubiquitination and tick immunology.
Kissing bugs, sandflies and fleas
Kissing bugs, especially Rhodnius prolixus, constitute perhaps the best understood vectors other than mosquitoes and ticks. Studies in immunology in triatomines have been reported not only against Trypanosoma cruzi, the agent of Chagas disease, but also T. rangeli (Borges et al., 2008, Garcia et al., 2010). Interestingly, T. cruzi seems to secrete a RING domain protein directly into the nucleus of HeLa cells. This protein showed E3 activity and a list of putative partners were identified (Hashimoto et al., 2010). Mechanistic studies should provide insight on how this pathogen uses ubiquitination to colonize humans, and perhaps, arthropod vectors. A few genes implicated in the immunity of the key vector of Leishmania in the Americas, the sand fly Lutzomyia longipalpis, have been identified by expressed sequence tagging sequencing. An ubiquitin conjugation enzyme has also been described in the L. longipalpis cDNA library (Ramalho-Ortigao et al., 2001). Defensin, a glycine-rich protein, PGRP and serpin were also found in the genome of this phlebotomine. Although they are believed to be upregulated after a blood meal, their precise relationship with Leishmania is unclear.
Although flea gene expression has been investigated by a few groups (Dreher-Lesnick et al., 2010, Zhou et al., 2012), flea immunology remains overlooked. Yersinia pestis, the agent of plague, regulates gene expression both in fleas and mammals (Vadyvaloo et al., 2010). However, how fleas respond to the presence of this pathogen i s poorly known. A putative E3 catalytic site was recently identified using modeling tools on the Yersinia outer protein M effector. This novel E3 domain was also identified in several other bacteria and described as a potential immunomodulatory molecule in both mammalian and arthropod immunity (Soundararajan et al., 2011).
CONCLUSIONS AND FUTURE DIRECTIONS
Information concerning the role of ubiquitination during pathogen colonization of arthropod-disease vectors is lacking. We speculate that comparative genomics approaches and the development of tools to be used across different arthropod systems should reveal ubiquitin machinery as an essential regulatory mechanism of arthropod-borne infection. The use of cell lines has greatly contributed to the understanding of the enzymatic activity of many ubiquitin-related proteins; however, the biochemical functions of ubiquitin-related proteins must be analyzed in vivo. Ubiquitin is involved in a range of cellular processes and targets a multiplicity of substrates. Therefore, careful interpretation of experimental approaches analyzing the physiological roles of ubiquitin in parallel with immune-related function is a crucial step for further development. These findings may in turn be used to enhance the immune response to pathogens and manage vectorial competence. Attenuated pathogen strains could perhaps be developed; and vector factors necessary for pathogen propagation could be blocked in order to disrupt the pathogen or parasite life cycle. Considerable effort should also be placed towards investigating the mechanistic basis of vector colonization, taking into consideration genotypic and phenotypic variations.
One could argue that the manipulation of the arthropod ubiquitination machinery during pathogen transmission may provide novel strategies to tackle the problem of vector-borne diseases. The “ubiquitin” approach for therapeutic development was adopted in mammals and drugs targeting various signaling pathways have been approved by the Food and Drug Administration for use in the clinic. For example, the proteasome inhibitor Bortezomib (Millenium Pharmaceuticals) has been very effective against myeloma and mantle cell lymphoma, whereas Nutlin (Roche) has been used against acute myeloid leukemia and multiple myeloma (Eldridge et al., 2010). Currently, a plethora of small molecules from several corporations and other institutions have also entered clinical trials (www.clinicaltrials.gov). Chemical screening assays with the intent of inhibiting the activity of E3 ubiquitin ligases or deubiquitinases in arthropod vectors could be applied. This type of approach has been successfully used to uncover effector molecules that regulate caspase-dependent cell death in Drosophila (Chew et al., 2009). Thus, the argument that a therapeutic can manipulate ubiquitin regulation in vector signaling pathways may not be entirely far-fetched. Nonetheless, it should be recognized that developing prophylactic measures for vector-borne diseases may be considered inherently more complex when compared to other human illnesses. Vectors themselves draw another layer of complexity for therapeutic or vaccine development.
Finally, we suggest that ubiquitin regulatory processes are likely to interfere with blood meal digestion. The interface between blood digestion and nutrient absorption may, in fact, correspond to a process regulated by ubiquitin because protein degradation and amino acid availability are largely controlled by the ubiquitin-proteasome system (Driscoll et al., 2012). This may, in turn, affect the immune response in the gut lining during vector engorgement and pathogen acquisition. These are only a few of the vast number of roles that ubiquitin plays in pathogen-vector interactions. In summary, this review aimed to provide information available in ubiquitination and vector immunology. A profound mechanistic understanding of ubiquitin signaling would be extremely valuable for the future of the field of vector-borne diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank invaluable colleagues for their thoughtful intellectual input and editorial comments on the manuscript. Work in the Pedra laboratory is supported by public health service grant R01 AI093653.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev. 2004;198:72–82. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- Almazan C, Lagunes R, Villar M, Canales M, Rosario-Cruz R, Jongejan F, de la Fuente J. Identification and characterization of Rhipicephalus (Boophilus) microplus candidate protective antigens for the control of cattle tick infestations. Parasitology Res. 2010;106:471–479. doi: 10.1007/s00436-009-1689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KV, Jurgens G, Nusslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell. 1985;42:779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- Arjona A, Ledizet M, Anthony K, Bonafe N, Modis Y, Town T, Fikrig E. West Nile virus envelope protein inhibits dsRNA-induced innate immune responses. J Immunol. 2007;179:8403–8409. doi: 10.4049/jimmunol.179.12.8403. [DOI] [PubMed] [Google Scholar]
- Bartholomay LC, Waterhouse RM, Mayhew GF, Campbell CL, Michel K, Zou Z, et al. Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science. 2010;330:88–90. doi: 10.1126/science.1193162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges AR, Santos PN, Furtado AF, Figueiredo RC. Phagocytosis of latex beads and bacteria by hemocytes of the triatomine bug Rhodnius prolixus (Hemiptera: Reduvidae) Micron. 2008;39:486–494. doi: 10.1016/j.micron.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Chew SK, Chen P, Link N, Galindo KA, Pogue K, Abrams JM. Genome-wide silencing in Drosophila captures conserved apoptotic effectors. Nature. 2009;460:123–127. doi: 10.1038/nature08087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol. 2010;34:387–395. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Brown EJ. Cytosol as battleground: ubiquitin as a weapon for both host and pathogen. Trends Cell Biol. 2010;20:205–213. doi: 10.1016/j.tcb.2010.01.002. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Blouin EF, Manzano-Roman R, Naranjo V, Almazan C, Perez de la Lastra JM, et al. Functional genomic studies of tick cells in response to infection with the cattle pathogen, Anaplasma marginale. Genomics. 2007;90:712–722. doi: 10.1016/j.ygeno.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Dreher-Lesnick SM, Ceraul SM, Lesnick SC, Gillespie JJ, Anderson JM, Jochim RC, et al. Analysis of Rickettsia typhi-infected and uninfected cat flea (Ctenocephalides felis) midgut cDNA libraries: deciphering molecular pathways involved in host response to R. typhi infection. Insect Mol Biol. 2010;19:229–241. doi: 10.1111/j.1365-2583.2009.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll JJ, Chowdhury RD. Molecular crosstalk between the proteasome, aggresomes and autophagy: translational potential and clinical implications. Cancer Letters. 2012;325:147–154. doi: 10.1016/j.canlet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Eldridge AG, O'Brien T. Therapeutic strategies within the ubiquitin proteasome system. Cell Death Differentiation. 2010;17:4–13. doi: 10.1038/cdd.2009.82. [DOI] [PubMed] [Google Scholar]
- Fink J, Gu F, Ling L, Tolfvenstam T, Olfat F, Chin KC, et al. Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl Trop Dis. 2007;1:e86. doi: 10.1371/journal.pntd.0000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia ES, Castro DP, Figueiredo MB, Azambuja P. Immune homeostasis to microorganisms in the guts of triatomines (Reduviidae) Mem Instituto Oswaldo Cruz. 2010;105:605–610. doi: 10.1590/s0074-02762010000500001. [DOI] [PubMed] [Google Scholar]
- Garver LS, Bahia AC, Das S, Souza-Neto JA, Shiao J, Dong Y, Dimopoulos G. Anopheles Imd pathway factors and effectors in infection intensity-dependent anti-Plasmodium action. PLoS Pathogens. 2012;8:e1002737. doi: 10.1371/journal.ppat.1002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver LS, Dong Y, Dimopoulos G. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog. 2009;5:e1000335. doi: 10.1371/journal.ppat.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer SF, Zika E, Conti B, Zhu XS, Ting JP. Enhancement of CIITA transcriptional function by ubiquitin. Nat Immunol. 2003;4:1074–1082. doi: 10.1038/ni985. [DOI] [PubMed] [Google Scholar]
- Gregory RC, Taniguchi T, D'Andrea AD. Regulation of the Fanconi anemia pathway by monoubiquitination. Semin Cancer Biol. 2003;13:77–82. doi: 10.1016/s1044-579x(02)00102-5. [DOI] [PubMed] [Google Scholar]
- Harhaj EW, Dixit VM. Regulation of NF-κB by deubiquitinases. Immunol Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Murata E, Aoki T. Secretory protein with RING finger domain (SPRING) specific to Trypanosoma cruzi is directed, as a ubiquitin ligase related protein, to the nucleus of host cells. Cell Microbiol. 2010;12:19–30. doi: 10.1111/j.1462-5822.2009.01375.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Hotez PJ, Fenwick A, Savioli L, Molyneux DH. Rescuing the bottom billion through control of neglected tropical diseases. Lancet. 2009;373:1570–1575. doi: 10.1016/S0140-6736(09)60233-6. [DOI] [PubMed] [Google Scholar]
- Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2011;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopacek P, Hajdusek O, Buresova V, Daffre S. Tick innate immunity. Adv Exp Med Biol. 2011;708:137–162. [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Dai J, Zhao YO, Narasimhan S, Yang Y, Zhang L, Fikrig E. Ixodes scapularis JAK-STAT pathway regulates tick antimicrobial peptides, thereby controlling the agent of human granulocytic anaplasmosis. J Infect Dis. 2012;206:1233–1241. doi: 10.1093/infdis/jis484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinander A, Runchel C, Tenev T, Chen L, Kim CH, Ribeiro PS, et al. Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 2012;31:2770–2783. doi: 10.1038/emboj.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynagh PN. The Pellino family: IRAK E3 ligases with emerging roles in innate immune signalling. Trends Immunol. 2009;30:33–42. doi: 10.1016/j.it.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Paquette N, Broemer M, Aggarwal K, Chen L, Husson M, Erturk-Hasdemir D, et al. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-κB signaling. Mol Cell. 2010;37:172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Ortigao JM, Temporal P, de Oliveira SM, Barbosa AF, Vilela ML, Rangel EF, et al. Characterization of constitutive and putative differentially expressed mRNAs by means of expressed sequence tags, differential display reverse transcriptase-PCR and randomly amplified polymorphic DNA-PCR from the sand fly vector Lutzomyia longipalpis. Mem Inst Oswaldo Cruz. 2001;96:105–111. doi: 10.1590/s0074-02762001000100012. [DOI] [PubMed] [Google Scholar]
- Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD. PIAS proteins and transcriptional regulation - more than just SUMO E3 ligases? Genes Dev. 2006;20:754–758. doi: 10.1101/gad.1421006. [DOI] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Ann Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- Soundararajan V, Patel N, Subramanian V, Sasisekharan V, Sasisekharan R. The many faces of the YopM effector from plague causative bacterium Yersinia pestis and its implications for host immune modulation. Innate Immunity. 2011;17:548–557. doi: 10.1177/1753425910377099. [DOI] [PubMed] [Google Scholar]
- Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl Acad Sci USA. 2009;106:17841–17846. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Iwai K. Linear ubiquitination: A novel NF-κB regulatory mechanism for inflammatory and immune responses by the LUBAC ubiquitin ligase complex. Endocrine J. 2012;59:641–652. doi: 10.1507/endocrj.ej12-0148. [DOI] [PubMed] [Google Scholar]
- Ungureanu D, Saharinen P, Junttila I, Hilton DJ, Silvennoinen O. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Mol Cell Biol. 2002;22:3316–3326. doi: 10.1128/MCB.22.10.3316-3326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungureanu D, Silvennoinen O. SLIM trims STATs: ubiquitin E3 ligases provide insights for specificity in the regulation of cytokine signaling. Sci STKE. 2005;2005:pe49. doi: 10.1126/stke.3042005pe49. [DOI] [PubMed] [Google Scholar]
- Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathogens. 2010;6:e1000783. doi: 10.1371/journal.ppat.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valanne S, Wang JH, Ramet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- Volz J, Osta MA, Kafatos FC, Muller HM. The roles of two clip domain serine proteases in innate immune responses of the malaria vector Anopheles gambiae. J Biol Chem. 2005;280:40161–40168. doi: 10.1074/jbc.M506191200. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder-Smith A, Renhorn KE, Tissera H, Abu Bakar S, Alphey L, Kittayapong P, et al. DengueTools: innovative tools and strategies for the surveillance and control of dengue. Glob Health Action. 2012;5 doi: 10.3402/gha.v5i0.17273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathogens. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Russell CW, Johnson KL, Mortensen RD, Erickson DL. Gene expression analysis of Xenopsylla cheopis (Siphonaptera: Pulicidae) suggests a role for reactive oxygen species in response to Yersinia pestis infection. J Med Entomol. 2012;49:364–370. doi: 10.1603/me11172. [DOI] [PubMed] [Google Scholar]
- Zhuang ZH, Sun L, Kong L, Hu JH, Yu MC, Reinach P, et al. Drosophila TAB2 is required for the immune activation of JNK and NF-κB. Cell Signal. 2006;18:964–970. doi: 10.1016/j.cellsig.2005.08.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.