Summary
Drosha is the main RNase III-like enzyme involved in the process of microRNA (miRNA) biogenesis in the nucleus. Using whole-genome ChIP-on-chip analysis, we demonstrate that, in addition to miRNA sequences, Drosha specifically binds promoter-proximal regions of many human genes in a transcription-dependent manner. This binding is not associated with miRNA production or RNA cleavage. Drosha knockdown in HeLa cells downregulated nascent gene transcription, resulting in a reduction of polyadenylated mRNA produced from these gene regions. Furthermore, we show that this function of Drosha is dependent on its N-terminal protein-interaction domain, which associates with the RNA-binding protein CBP80 and RNA Polymerase II. Consequently, we uncover a previously unsuspected RNA cleavage-independent function of Drosha in the regulation of human gene expression.
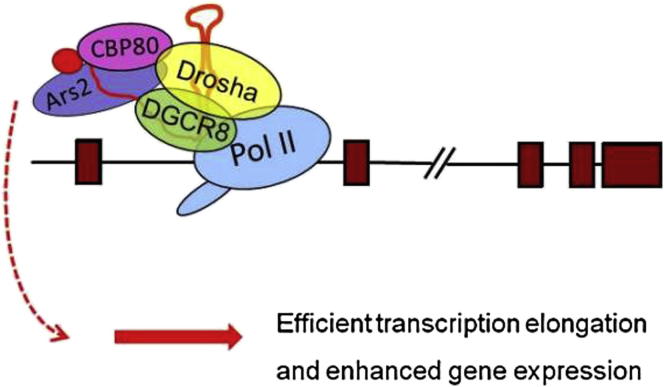
Graphical Abstract
Highlights
-
•
Drosha binds promoter-proximal regions of transcribed human genes
-
•
Drosha binding is not associated with RNA cleavage or miRNA processing
-
•
Drosha regulates nascent gene transcription
-
•
Drosha interacts with CBP80 and RNA Pol II through its N-terminal domain
In higher eukaryotes, the Microprocessor complex (Drosha endonuclease and DGCR8 RNA-binding protein) recognizes and excises RNA hairpins from larger nuclear transcripts. This ultimately leads to cytoplasmic microRNA production or, in some cases, direct downregulation of gene expression through RNA cleavage. In this study, Gromak, Proudfoot, and colleagues show that Drosha-DGCR8 binds numerous gene promoters not to cleave RNA but, rather, to form a molecular interaction surface. This helps recruit additional factors to gene promoters with a consequent increase in gene activity.
Introduction
MicroRNAs (miRNAs) are short noncoding regulatory molecules, involved in diverse biological processes. Biogenesis of miRNAs involve a nuclear phase, where the Microprocessor complex, comprising Drosha, an RNase III-like enzyme and its cofactor DGCR8, process primary miRNAs (pri-miRNAs) into a 70 nt pre-miRNA (Han et al., 2004, Lee et al., 2003, Zeng et al., 2005). This occurs cotranscriptionally from both independently transcribed and intron-encoded miRNAs (Ballarino et al., 2009, Kim and Kim, 2007, Morlando et al., 2008). Following Drosha-mediated RNA cleavage and pre-miRNA release from the nascent RNA, 5′ and 3′ nascent RNA ends are trimmed by 5′-3′ Xrn2 and 3′-5′ exosome (Morlando et al., 2008), and the pre-miRNA precursor is exported to the cytoplasm (Lund et al., 2004, Yi et al., 2003). Here, a second RNase III enzyme, Dicer, further processes the pre-miRNA into the mature miRNA duplex (Bernstein et al., 2001) that targets specific mRNAs for degradation or translational inactivation (reviewed in Bartel, 2009).
MiRNA levels are tightly regulated at the posttranscriptional level by a number of RNA-binding proteins (Siomi and Siomi, 2010). Furthermore, Drosha can directly regulate levels of Microprocessor complex by cleaving hairpin structures in DGCR8 mRNA, thereby decreasing DGCR8 protein levels (Han et al., 2009, Triboulet et al., 2009). Along the same lines, Drosha knockdown in Drosophila leads to upregulation of some mRNAs containing conserved RNA hairpins, potentially recognized by the Microprocessor complex (Kadener et al., 2009). Several recent studies demonstrated the ability of Microprocessor complex to cleave mRNAs, thus regulating their expression. Many Drosha-dependent mRNA cleavage events were identified in mESCs, consistent with Microprocessor regulation of coding mRNAs through direct cleavage (Karginov et al., 2010). Drosha can also cleave the TAR hairpin of the HIV-1 transcript, resulting in premature termination of RNA polymerase II (Pol II) (Wagschal et al., 2012). A recent DGCR8 HITS-CLIP analysis extended these observations and revealed general noncanonical functions of the Microprocessor complex (Macias et al., 2012). Transcriptome and proteome studies of mice lacking Drosha and Dicer suggest that both enzymes have nonredundant functions, as their deficiency can induce different phenotypes (Chong et al., 2010). Although many RNAs were stabilized by Drosha depletion, some were downregulated, consistent with Drosha possessing independent functions to its role in canonical miRNA biogenesis.
In human cells Drosha exists in two distinct multiprotein complexes (Gregory et al., 2004). The smaller complex, containing just Drosha and DGCR8, is necessary and sufficient for miRNA processing. The larger complex, displaying only weak pre-miRNA processing activity in vitro, contains DEAD-box RNA helicases, double-stranded RNA-binding proteins, hnRNP proteins, members of FUS/TLS family of proteins, and the SNIP1 protein, implying additional functions in gene expression. Thus, DEAD box helicases p68/p72 increase Drosha processing efficiency for a subset of miRNAs and at gene-specific promoters interact with transcriptional coactivators and Pol II and regulate alternative splicing (Fuller-Pace and Ali, 2008). Nuclear scaffolding protein hnRNPU and members of FUS/TLS family are also associated with regulation of transcription (Wang et al., 2008). SNIP1, a component of a large SNIP1/SkIP-associated complex, involved in transcriptional regulation and cotranscriptional processing, interacts with Drosha and plays a role in miRNA biogenesis (Fujii et al., 2006, Yu et al., 2008). Ars2 is implicated in RNA silencing that functions in antiviral defense in flies and cell proliferation in mammals (Gruber et al., 2009, Sabin et al., 2009). It interacts with the nuclear cap-binding complex (CBP20/CBP80) and is involved in miRNA biogenesis, suggesting a link between RNA silencing and RNA-processing pathways. CBP20/CBP80 proteins are also implicated in miRNA biogenesis in plants (Kim et al., 2008). Overall, the existence of this large Drosha-complex with only weak miRNA-processing activity suggests that Drosha may play multiple roles in miRNA-independent gene regulation.
Using genome-wide and gene-specific approaches we now show that Drosha binds to the promoter-proximal regions of many human genes in a transcription-dependent manner. Similarly, DGCR8 binds promoter-proximal regions of many human genes, suggesting that the whole Microprocessor is recruited at promoter regions. We also find that Drosha interacts with Pol II and its depletion from human cells causes transcriptional downregulation with a concomitant decrease in nascent and mature mRNA levels. This positive function of Drosha in gene expression is mediated through its interaction with the RNA-binding protein CBP80 and dependent on the N-terminal protein-interaction domain of Drosha. Thus, results presented in this paper demonstrate an miRNA- and cleavage-independent function of Drosha.
Results
Drosha Binds 5′ Ends of Human Genes in a Transcription-Dependent Manner
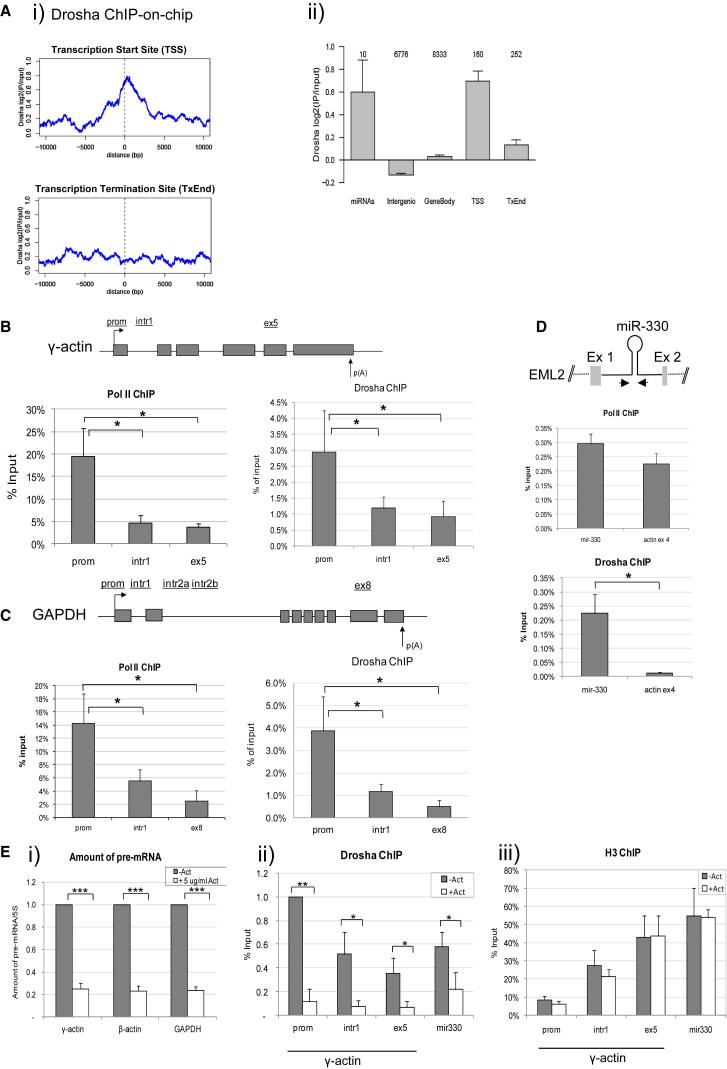
Pre-miRNA processing occurs cotranscriptionally. Consequently binding of Drosha to nascent miRNA sequences can be detected using chromatin immunoprecipitation (ChIP) analysis (Morlando et al., 2008). To investigate genome-wide binding of Drosha, we employed a ChIP-on-chip approach using human 5.1.1 ENCODE array. Our results show that, in addition to miRNA regions, Drosha binds many human genes. This binding is enriched at the transcriptional start sites (TSSs), compared to the ends of transcripts (TxEnd), other gene body, or intergenic regions (Figure 1A; full gene list in Table S1). In particular, we observed that, out of 160 TSS-proximal probes, 55 (34%) had more than 2-fold enrichment of Drosha-ChIP signal over the input signal. This is significantly more than 1,669 (10%) out of 15,998 total probes or 953 (11%) out of 8,333 probes in the gene bodies (p < 0.0001, Fisher’s exact test). The same level of enrichment was observed for four (40%) out of ten probes overlapping with annotated miRNA regions (Figure 1Aii). By way of illustration, we present SELENPB1 and EEF1A1 genes, which scored positively in our genomic analysis, where Drosha selectively binds to 5′-proximal gene regions (Figure S1). Genes that are not expressed in HeLa cells such as IL4 and TARM1 demonstrated only background Drosha signal (Figure S1). We also identified genes, such as G6PD, ZNF687, and FUNDC2, which are expressed in HeLa cells but did not demonstrate any significant Drosha enrichment (Figure S3A). We further confirmed these array data using gene-specific primers in ChIP experiments. In particular, we observed an enrichment of Drosha binding to promoter-proximal region of SELENBP1 gene, colocalizing with Pol II peak (Figure S2A). We did not detect any significant binding of Drosha or Pol II to IL4 and TARM1 genes, further confirming ChIP-on-chip results (Figures S2B and S2C). We also did not detect any significant Drosha binding over promoter-proximal region, enriched for Pol II, on G6PD gene (Figure S3C).
Figure 1.
Drosha Binds 5′ Ends of Human Genes in a Transcription-Dependent Manner
(A) (i) Metagene profiles of Drosha binding around transcriptional start sites (TSS, top) and transcriptional termination sites (TxEnd, bottom) based on Drosha ChIP-on-chip analysis in HeLa cells using ENCODE array. (ii) Average Drosha ChIP enrichment over input (log2) for probes overlapping miRNAs, intergenic regions, gene bodies, TSSs, and TxEnds. Error bars represent SEM. Number of probes covering each region is indicated above the bars. TSS signal is significantly enriched (p < 0.001, t test) compared to all intergenic gene bodies or TxEnd regions.
(B–D) Pol II and Drosha ChIP analyses on γ-actin (B) and GAPDH (C) genes and the miR-330 genomic locus, compared to β-actin ex 4 (D) in HeLa cells. ChIP amplicons are shown above the gene diagrams.
(E) (i) qRT-PCR analysis of γ-actin, β-actin and GAPDH pre-mRNAs from HeLa cells mock-treated or treated with 5 μg/ml actinomycin D for 6 hr, normalized to 5S rRNA. The amount of RNA in the untreated cells was taken as 1. Drosha (ii) and H3 (iii) ChIPs on γ-actin gene and miR-330 gene locus in HeLa cells, mock-treated or treated with 5 μg/ml of actinomycin D for 6 hr.
Bars in (B)–(E) represent the average values from at least three biological experiments ±SD. See Figures S1–S5.
To investigate the function of Drosha at the beginning of human genes in more detail, we compared Pol II and Drosha binding profiles for specific human genes. Both Pol II and Drosha were enriched over the promoter-proximal regions of human γ-actin, GAPDH, PTB, β-actin, and intronless TAF7 genes (Figures 1B, 1C, and S4A–S4C). The enrichment of Drosha observed over these genes is higher than over the miR-330 locus within the EML2 gene, used as a positive control for Drosha binding (Morlando et al., 2008) (Figure 1D). As a negative control, we used ChIP for histone H3, which was found to be depleted from promoter-proximal regions and enriched in the body of these genes (Figure 1Eiii). To test if Drosha binding to these genes is transcription dependent, we treated HeLa cells with Pol II transcriptional inhibitor actinomycin D. Following this treatment, we observed ∼80% reduction in pre-mRNA levels and a substantial reduction in Drosha binding to miR-330 locus, γ-actin, and β-actin genes (Figures 1E and S4C). In contrast, we saw no effect on histone H3 binding (Figure 1Eiii). These results indicate that, in addition to miRNA sequences, Drosha binds 5′ end regions of human protein-coding genes in a transcription-dependent manner. Furthermore, we also searched for potential correlation between Drosha recruitment using our Drosha ChIP-array data and level of Pol II signal determined from available chromatin immunoprecipitation sequencing (ChIP-seq) experiments in UCSC depository. We have calculated Pol II signal in 1 kb region around the TSS for Refseq transcripts covered by probes in the Drosha array. Following this analysis, we identified a weak (Pearson R = 0.41) but significant (p = 1e-6) correlation between the Drosha probe signal at TSS and Pol II ChIP density (Figure S5), indicating that Drosha is preferentially recruited to the Pol II-rich regions. Overall, these results suggest that Drosha binding may be required for enhanced gene expression.
DGCR8 Binds Promoter-Proximal Regions of Human Genes
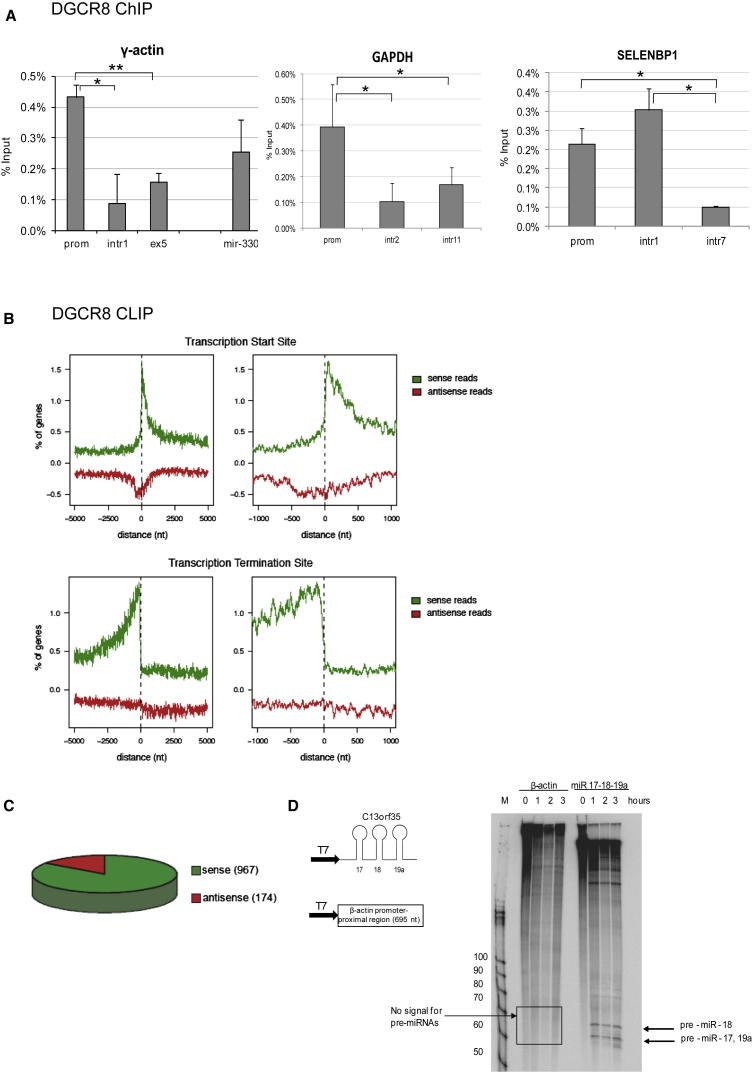
Drosha is known to bind miRNA sequences as part of the Microprocessor complex, which also contains DGCR8, a double-stranded RNA-binding protein that is deleted in the DiGeorge syndrome (Landthaler et al., 2004). To investigate whether Drosha binds 5′ regions of human genes together with DGCR8, we carried out ChIP experiments in HeLa cells (Figure 2A). DGCR8 was found to be enriched over the miR-330 locus, γ-actin, GAPDH, and SELENBP1 promoter-proximal gene regions (Figure 2A). These regions overlap Drosha binding regions (Figures 1, S2, and S4), suggesting that Drosha and DGCR8 may form a complex at promoter-proximal regions of human genes.
Figure 2.
DGCR8 Binds Promoter-Proximal Regions of Human Genes
(A) DGCR8 ChIP analysis on γ-actin, GAPDH, and SELENBP1 genes and the miR-330 genomic locus in HeLa cells. SELENBP1 gene diagram is in Figure S2A. Bars represent the average values from at least three biological experiments ±SD.
(B) DGCR8 HITS-CLIP. Distribution of significant sense (green) and antisense (red) reads mapping around transcription start sites (TSSs; top panels) and transcription termination regions (TTSs; bottom panels) of protein coding genes, as determined by CEAS software, using FDR < 0.01. The y axis shows the number of genes detected as a percentage, whereas the x axis represents the location in relation to the position of TSS and TTS.
(C) Distribution of sense and antisense DRCG8 CLIP binding sites. 1,101 DGCR8 binding sites in the sense orientation correspond to 967 genes, and 196 DGCR8 antisense binding sites correspond to 174 genes.
(D) Drosha in vitro processing reactions in HeLa nuclear extracts with β-actin promoter-proximal region and miR17-18-19a sequences. Arrows indicate the positions of pre-miRNAs.
See also Figures S6 and S7.
To substantiate these findings in a genome-wide fashion, we analyzed a DGCR8 HITS-CLIP experiment that was performed with endogenous and overexpressed DGCR8 protein in HEK293T cells to identify DGCR8 endogenous RNA targets (Macias et al., 2012). In particular, we observed 1,101 DGCR8 clusters, corresponding to 967 genes, in the sense orientation (see supplemental material in Macias et al., 2012) and 196 DGCR8 clusters, corresponding to 174 genes, in antisense orientation (Figure 2C; Table S2 for gene identity). Interestingly, significant clusters of DGCR8 binding were seen at −1,000/+200 bp from the TSSs and just upstream of the transcription termination regions (TTSs) (false discovery rate [FDR] < 0.01) (Figure 2B). In particular, we observed significant enrichment of DGCR8 binding in the region within 200 nt downstream of the TSS (sense versus antisense ks test p < 2.2e-16; Figure 2B). DGCR8 binding in this region is most pronounced in genes expressed at medium and high levels when compared to genes expressed at lower levels (ks test p < 2.2e-16 in both cases) (Figure S7A). These results clearly indicate that DGCR8, similar to Drosha, binds to promoter-proximal regions of human genes. However, it should be noted that when we analyzed DGCR8 binding to spliced mRNA transcripts (cDNAs) (Figure S6), we also see an enrichment over exonic sequences immediately downstream of the TSS. Again, this is seen in genes with medium and high expression (ks test p < 2.2e-16 in both cases) (Figure S7B). This may be related to the reported binding of DGCR8 to multiple exonic regions (Macias et al., 2012), potentially reflecting an additional function.
Promoter-Proximal Binding of Drosha Is Not Related to miRNA Synthesis or RNA Cleavage
Binding of Drosha to promoter-proximal regions of human genes may be related to its potential role in processing miRNAs from these genomic regions, even though such elements are not annotated in the human genome databases. To test if Drosha binds and cleaves miRNAs present in promoter-proximal regions, we performed in vitro pri-miRNA cleavage assays in HeLa nuclear extracts (Guil and Cáceres, 2007). Using in vitro transcription reactions, we generated radiolabeled RNAs from gene regions, identified by Drosha ChIP. We employed miR-17-92 cluster within the C13orf25 human genomic region as a positive control for miRNA production. Following in vitro cleavage assay in HeLa extracts, miRNAs were generated from the control miR-17-19a sequences but not from the promoter-proximal region of the β-actin gene (Figure 2D). We also failed to detect pre-miRNAs or RNA cleavage products in vitro for promoter-proximal regions of other genes, identified in Drosha ChIP assays (data not shown). These findings suggest that Drosha binding over promoter-proximal regions is not associated with miRNA synthesis and RNA cleavage.
Drosha Knockdown Causes Transcriptional Downregulation and a Decrease in Pre-mRNA and Poly(A)+ RNA
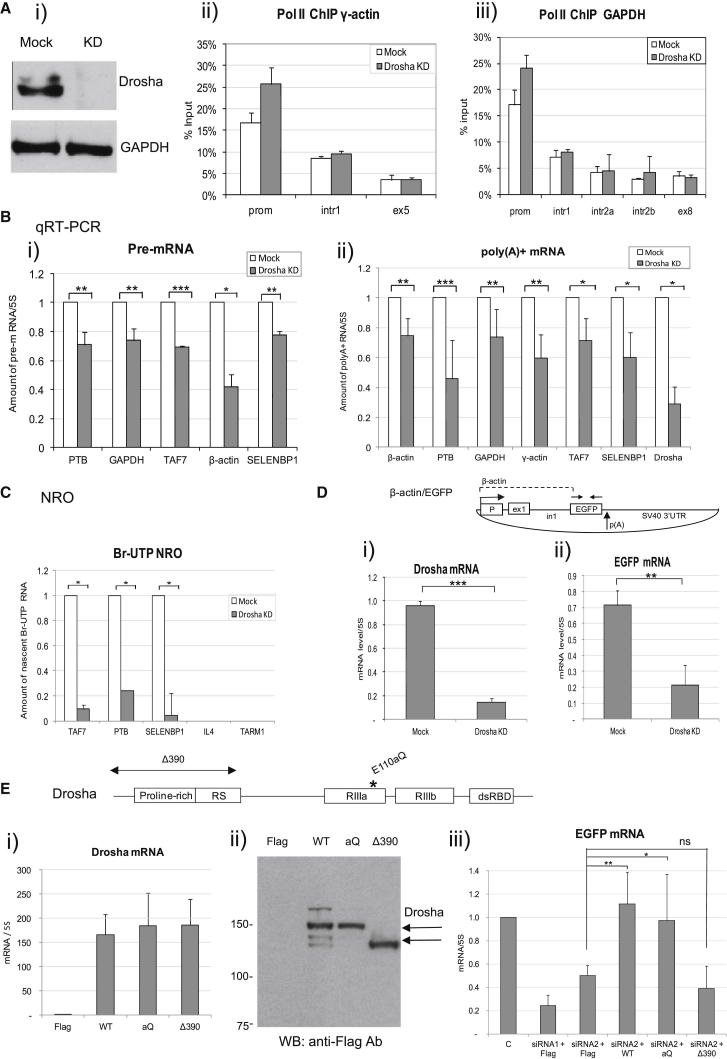
To investigate the promoter-associated function of Drosha, we performed RNAi-induced Drosha knockdown in HeLa cells (Figure 3A). Following Drosha depletion, we observed a small increase in Pol II binding over the promoters of γ-actin and GAPDH genes, using ChIP analysis (Figure 3A). We also observed an ∼20%–40% decrease in pre-mRNA levels and ∼30%–60% decrease in poly(A)+ RNA levels in HeLa cells depleted for Drosha (Figure 3B). In contrast, we did not detect any significant change in the levels of poly(A)+ and pre-mRNA corresponding to G6PD, ZNF687, and FUNDC2 genes, for which we observed lack of promoter-proximal Drosha binding (Figures S3B and S3C). This suggests the specificity of observed transcriptional effects, following Drosha depletion.
Figure 3.
Drosha Depletion Causes Transcriptional Downregulation and Decreases Pre-mRNA and Poly(A)+ RNA Levels
(A) Western blot analysis of protein extracts (50 μg) from mock-treated and Drosha siRNA-treated HeLa cells, probed with Drosha and GAPDH antibodies (i). Pol II ChIP across γ-actin (ii) and GAPDH (iii) genes in mock-treated and Drosha-depleted HeLa cells.
(B) qRT-PCR analysis of pre-mRNA (i) and poly(A)+ RNA (ii) in mock-treated and Drosha-depleted cells. The level of RNA in mock-treated samples was taken as 1.
(C) Br-UTP NRO analysis carried out in mock-treated and Drosha-depleted cells. Amount of nascent Br-UTP RNA was calculated by subtracting the background of U-RNA produced over a specific gene probe. The level of Br-UTP RNA in mock-treated samples was taken as 1.
(D) Diagram of β-actin/EGFP plasmid. Promoter, exon 1, and intron 1 sequences are derived from the human β-actin gene, indicated by dashed line. qRT-PCR primers are shown above the diagram. SV40 3′ UTR contains a poly(A) signal, indicated with an arrow. qRT-PCR analysis of Drosha (i) and EGFP (ii) mRNAs in mock-treated and Drosha-depleted HeLa cells.
(E) Top, domain structure of Drosha protein. Double-stranded RNA-binding (dsRBD), RNaseIII (RIIIa, RIIIb), proline-rich, and serine-arginine (RS) domains are shown. Positions of catalytic E110aQ (aQ) mutation and Δ390 deletion of the N-terminal proline-rich and RS-rich domains are indicated above the diagram. Bottom, qRT-PCR analysis of Drosha mRNA (i) and EGFP mRNA (iii) in Drosha-depleted 293T cells, overexpressed with RNAi-resistant Flag, Drosha WT, aQ, and Δ390 constructs. The level of Drosha (i) or EGFP (iii) mRNA in cells, overexpressed with Flag was taken as 1. siRNA2, targeting 3′ UTR of endogenous Drosha mRNA, was used for Drosha depletion in overexpression experiments. (ii) Western blot of total protein extracts (50 μg) from 293T cells, overexpressed with Flag, Drosha WT, aQ, and Δ390 plasmids, probed with Flag antibody. The positions of the marker are indicated on the left.
Bars in (A)–(E) represent average values from at least three independent experiments ±SD. In (B), (D), and (E), the levels of RNA were normalized to 5S rRNA.
To evaluate if Drosha has a direct effect on transcription, we employed nuclear run-on (NRO) using Br-UTP as the labeled nucleotide (Core and Lis, 2008, Lin et al., 2008). The advantage of Br-UTP NRO over Pol II ChIP is the ability of Br-UTP NRO to detect actively transcribing polymerases, as opposed to total Pol II levels detected by ChIP. As demonstrated in Figure 3C, we observed a substantial decrease in the level of nascent transcription for TAF7, PTB, and SELENBP1 genes in Drosha-knocked-down cells. As a negative control for this experiment, we used IL4 and TARM1 genes. These genes are not expressed in HeLa cells; hence, we did not observe any detectable transcription signal in Br-UTP NRO reactions. These results suggest that Drosha has a direct positive effect on transcription, consistent with the genome-wide expression data, where highly expressed genes correlated with increased Drosha binding on TSSs (Figure 1A). Furthermore, the antisense transcripts detected over the promoter-proximal region of the β-actin gene were also decreased upon Drosha depletion (Figure S4D). These results further indicate that Drosha is not involved in the cleavage of nascent sense or antisense RNA transcripts, pointing toward a cleavage-independent function of Drosha in human gene expression.
Drosha Regulates the Expression of a Heterologous Reporter
We next employed a β-actin/EGFP reporter plasmid, where transcription of EGFP is driven by the β-actin promoter, fused to β-actin exon 1 and intron 1 regions (Qin and Gunning, 1997) (Figure 3D). As described above, Drosha binds the promoter-proximal region of β-actin gene (Figure S4C). Interestingly, in HeLa cells depleted for Drosha we observed ∼75% reduction in expression of EGFP mRNA from the plasmid (Figure 3D), recapitulating downregulation of endogenous β-actin mature and pre-mRNAs (Figure 3B). These results suggest that Drosha plays a positive role in the regulation of β-actin/EGFP expression.
Next, we cotransfected β-actin/EGFP reporter plasmid with RNAi-resistant Flag-tagged Drosha (WT), a catalytic mutant E110aQ (aQ), unable to process miRNAs, and Δ390 construct into 293 cells, depleted for Drosha, using siRNA2 (Han et al., 2004). The Δ390 Drosha construct is active in miRNA processing but lacks the N-terminal proline-rich and RS-rich domains, proposed to be important for Drosha protein-protein interactions (Han et al., 2004) (Figure 3E). Both mRNAs and proteins corresponding to WT, aQ, and Δ390 Drosha were expressed at a high level in 293T cells, depleted for endogenous Drosha (Figure 3Ei and 3Eii). Overexpression of both WT and aQ catalytic Drosha mutant resulted in the increase of EGFP mRNA (Figure 3Eiii). In contrast, overexpression of Δ390 construct had no significant effect on the expression of EGFP mRNA. These results suggest that Drosha plays a positive role in the regulation of the β-actin promoter. Importantly, this Drosha function is independent of its ability to cleave RNA but does require the N-terminal protein-protein interaction domain.
Drosha Regulates Human Gene Expression through Interaction with CBP80 and RNA Polymerase II
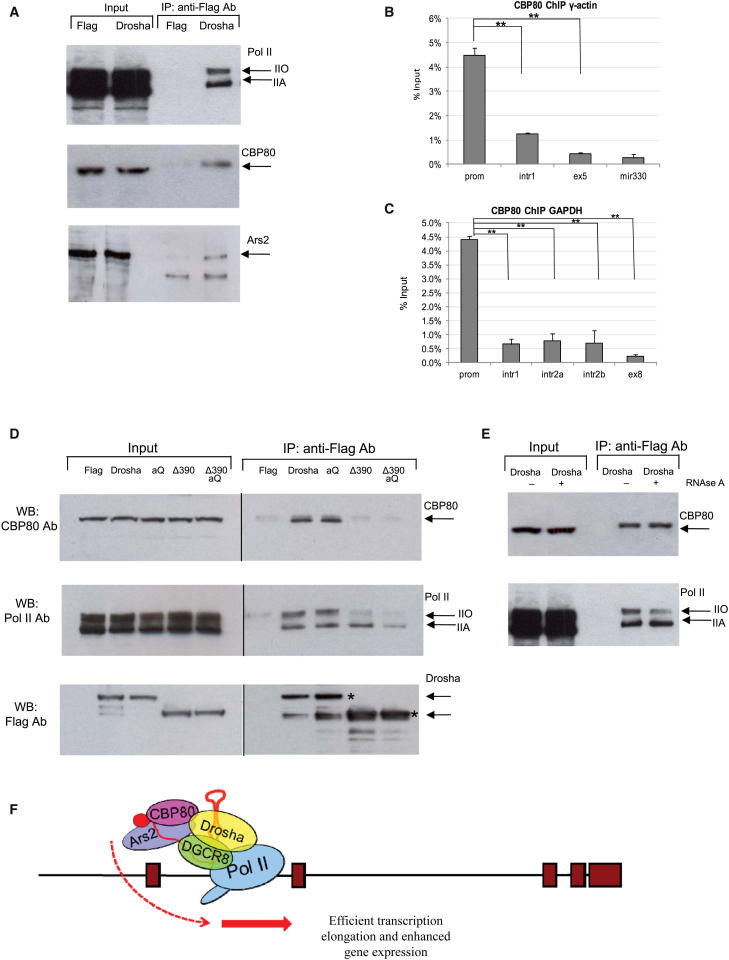
We hypothesize that the cleavage-independent stimulatory role of Drosha on gene expression may be mediated through its association with protein cofactors. We therefore performed coimmunoprecipitation experiments and found that transiently overexpressed Flag-tagged Drosha interacts with both unphosphorylated (IIA) and phosphorylated (IIO) forms of Pol II in 293T cells, supporting its role in transcriptional regulation. Drosha also interacts with CBP80 and Ars2 proteins, involved in miRNA biogenesis and RNA processing pathways, respectively (Gruber et al., 2009, Sabin et al., 2009) (Figure 4A). Furthermore, by ChIP analysis we found that CBP80 protein binds promoter-proximal regions of γ-actin and GAPDH genes, colocalizing with Drosha binding (Figures 4B and 4C). We next observed that Drosha-CBP80 interaction is not affected by the aQ mutation in the catalytic domain of Drosha but is significantly reduced when the N-terminal domain of Drosha is deleted in Δ390 construct or when both Δ390 and aQ mutations are combined (Figure 4D, top panel). These results strongly support the view that Drosha interacts with CBP80 through its N-terminal RS domain. We also studied the interaction of Drosha with Pol II (Figure 4D, middle). Similar to CBP80, a catalytic mutation in Drosha (aQ) does not affect its interaction with Pol II. However, the Drosha Δ390 construct showed reduced interaction with Pol II, and specifically with its phosphorylated form (IIO). A similar effect was observed with Δ390 aQ construct. This suggests that the N-terminal domain of Drosha is particularly important for Drosha-mediated transcriptional effects in human cells because it mediates interaction of Drosha with CBP80 and Pol II. We also observed that interaction of Drosha with CBP80 and Pol II is RNA independent (Figure 4E). Consequently, this result provides a molecular explanation for the ability of Drosha to regulate gene expression in a miRNA- and cleavage-independent manner.
Figure 4.
Drosha Regulates Human Gene Expression through Its Interaction with CBP80 and Pol II
(A) Pull-down experiments performed with anti-Flag in 293 cells, overexpressed with Flag and Drosha WT-Flag constructs. Western blots were probed with Pol II, Ars2, and CBP80 antibodies. Input represents 1% of material added to IP.
(B and C) CBP80 ChIP analyses across γ-actin (B) and GAPDH (C) genes and miR-330 gene locus in HeLa cells. Bars represent average values from at least three independent experiments ±SD.
(D) Pull-down experiments performed with anti-Flag in 293 cells, overexpressed with Flag, Drosha WT, aQ, Δ390, and Δ390 aQ constructs. Input represents 1% of material added to IP. Western blot was probed with CBP80 (top panel) and Pol II (middle panel) and FLAG (bottom panel) antibodies. Asterisks indicate full-length proteins.
(E) Pull-down experiments performed with anti-Flag in 293 cells, overexpressed with Drosha WT construct. Cell extracts were nontreated (−) or treated (+) with 50 μg/ml of RNase A. Input represents 1% of material added to IP. Western blot was probed with CBP80 (top panel) and Pol II (bottom panel) antibodies.
(F) Model for the function of Drosha in the regulation of human gene expression. Following transcription of nascent RNA by Pol II, Drosha, most probably in a complex with DGCR8, binds GC-rich hairpins at the promoter-proximal regions of human genes. Drosha does not cleave such hairpins, but it interacts with Pol II and CBP80 through its N-terminal proline-rich domain. Drosha can enhance gene expression, independently of its RNA cleavage function. Depletion of Drosha results in decrease of nascent transcription and subsequent decrease in the amount of nascent and poly(A)+ mRNA.
Discussion
We demonstrate the existence of a miRNA- and cleavage-independent function of Drosha in the regulation of human gene expression. Combining whole-genome analysis with gene-specific approaches, we demonstrate that Drosha binds to the 5′ ends of human genes in a transcription-dependent manner (Figure 1). This binding is not associated with miRNA biogenesis or RNA cleavage but correlates with the level of gene expression (Figures 1 and 2). Depletion of Drosha from HeLa cells led to direct downregulation of transcription based on nuclear run-on and Pol II accumulation at the promoters of these genes, resulting in reduced levels of pre-mRNA and poly(A)+ mRNA (Figure 3). This suggests a positive effect of Drosha on gene transcription. ChIP and HITS-CLIP analysis of DGCR8 also revealed binding sites at promoter-proximal regions, enriched at genes expressed at a higher level, suggesting that Drosha and DGCR8 may bind to promoter regions as a complex (Figures 2 and S7). A positive function of Drosha in the regulation of gene expression was further confirmed using a heterologous reporter construct, where Drosha overexpression influences reporter expression (Figure 3). This function of Drosha is independent of its catalytic activity and mediated through its N-terminal domain, which interacts with the RNA-binding protein CBP80 and Pol II, which also bind to the 5′ end of human genes (Figures 3 and 4). Taken together, these results suggest a model whereby Drosha promotes gene expression in a cleavage-independent manner by binding to RNA hairpins formed at the 5′ ends of the nascent RNAs and interacting with CBP80 and Pol II, through its N-terminal RS domain (Figure 4F).
Initially, we hypothesized that this positive function of Drosha may be associated with generation/regulation of short promoter-associated transcripts (PROMPTs), originally predicted to regulate transcription through changes in chromatin structure, promoter methylation, or Pol II recycling (Preker et al., 2008). PROMPTs may direct Drosha to the beginning of the gene through interaction with its binding factors. In our studies, both sense and antisense transcripts, detected over the promoter-proximal regions, were destabilized in Drosha-depleted cells (Figure S4D), suggesting that Drosha does not cleave these transcripts and its function is not mediated through PROMPTs.
Drosha binding at the beginning of human genes may be also related to Pol II release from promoter-proximal pausing events and be required for the recruitment of transcription/RNA processing factors. Interestingly, Drosha binding coincides with the binding site of the negative elongation factor NELF, which defines the “late” elongation checkpoint, ensuring conversion to productive elongation mediated by phosphorylation of Pol II CTD, NELF, and DSIF by the positive transcription elongation factor b (P-TEFb) (Egloff et al., 2009). As both nucleosome positioning and chromatin modifications play an important role in the elongation functions of P-TEFb, we also checked whether Drosha knockdown causes a change in chromatin modifications. However, we did not observe any significant changes (data not shown). This suggests that Drosha regulates gene expression through a different mechanism. GC-rich sequences are known to be enriched in the promoter regions of human genes (Calistri et al., 2011). Transcripts for these sequences are likely to form RNA hairpin structures, predicted to be suitable for Microprocessor binding mediated by DGCR8 (Macias et al., 2012; Table S1). If Drosha were to cleave such RNAs, the consequences of Drosha cleavage across the whole human genome would be detrimental to cellular survival. We hypothesize that Drosha does not cleave promoter-associated RNA but is instead utilized as a positive regulator of gene expression. We further analyzed the structures found in promoter transcripts and compared them to miRNA, snoRNAs, and protein-coding region structures, found to be bound by DGCR8 in CLIP analysis. Our bioinformatic analysis confirmed that structures in promoter regions have shorter stems, higher minimum free energy, and lower base-pairing probability (Figure S8). In addition, promoter transcripts also lack motifs important for the processing of the stem, such as TG dinucleotide at the base of the stem and CNNC motif downstream of the stem, as described in Auyeung et al. (2013). Taken together, this analysis suggests that promoter structures may be less stable, and their recognition and processing by the Microprocessor complex may be less efficient compared to miRNAs or other sequences cleaved by Drosha.
Evidence that components of the miRNA machinery may have additional functions comes from several previous studies. In early-stage thymocytes, Drosha recognizes and cleaves mRNAs harboring secondary stem-loop structures (Chong et al., 2010). Drosha can also regulate viral gene expression of Kaposi’s Sarcoma-associated Herpesvirus (KSHV) (Lin and Sullivan, 2011). DGCR8 together with Drosha controls the abundance of many cellular RNAs, including noncoding RNAs, mRNAs, and alternatively spliced isoforms (Macias et al., 2012). Finally, Microprocessor can regulate transcription of some cellular mRNA and HIV provirus, causing Pol II premature termination through the cleavage of the TAR stem-loop structure (Wagschal et al., 2012). These regulatory activities of Drosha each involve recognition and cleavage of target RNAs in an analogous manner to its function in miRNA biogenesis in contrast to the positive function of Drosha on transcription described here. Possibly this positive function of Drosha is related to the action of RNA-binding factors, known to modulate the function of Drosha in human cells. Such factors are likely to interact with the N-terminal proline-rich and RS domains, proposed to be the main sites of Drosha protein-protein interactions. We confirmed the previously described interaction between Drosha and CBP80 and Ars2 proteins (Gruber et al., 2009, Sabin et al., 2009). Furthermore, our results also show that Drosha interacts with the C-terminal domain (CTD) of Pol II (Figure 4). This interaction may be direct or mediated via proteins, containing RS domains and interacting with the CTD, potentially modulating Drosha’s cleavage-independent function. Interestingly, in Drosophila the key RNAi components Dicer 2 and Argonaute 2 associate with chromatin and interact with the core transcription machinery, affecting Pol II dynamics (Cernilogar et al., 2011), further supporting the role of RNAi machinery in transcriptional regulation. Because both Drosha and Dicer may have additional roles to miRNA biogenesis future studies will be necessary to evaluate the full spectrum of molecular functions that these RNase-III-like enzymes play in human cells. This will represent an exciting avenue for future research in gene expression.
Experimental Procedures
Plasmids
pCK-Flag WT, aQ, and Δ390 Drosha constructs (Han et al., 2004) and β-actin/EGFP plasmid (Qin and Gunning, 1997) were described previously. pflag-CMV-2 (Flag) was purchased from Sigma-Aldrich (E7398).
RNA Analysis
Total RNA was harvested using TRIZOL reagent (Invitrogen) followed by DNase I treatment (Roche). Total RNA (2 μg) was reverse-transcribed using SuperScript Reverse Transcriptase (Invitrogen) and random hexamer primers (Invitrogen) or gene-specific primers, as described in the Supplemental Experimental Procedures and Table S3, followed by qPCR. Gene-specific primers were used in all figures, apart from Figure 3Eiii, where random hexamers were used for quantitative RT-PCR (qRT-PCR), following the manufacturer’s description (Invitrogen).
In Vitro Processing Assays
MiR-17-19 and γ-actin gene regions were amplified from genomic DNA using T7+β-actin (F/R) and T7+miR-17-19 (F/R) primers accordingly. In vitro processing was performed as in Guil and Cáceres (2007).
RNAi and Protein Analysis
The RNAi was carried out as described (Wollerton et al., 2004). mRNA target sequence for Drosha small interfering RNA (siRNA) duplex was 5′-CGAGUAGGCUUCGUGACUU-3′ (siRNA1) and 5′-GAGUAUUUACUUGCUCAG UAA-3′ (siRNA2). siRNA1 was used in all figures apart from Figure 3E, where siRNA2 was used. Pull-downs were carried out in RNase A nontreated extracts, except Figure 4E where 50 μg/ml RNase A was used to treat cell extracts during the pull-down procedure as described in Gruber et al. (2009). The immunoprecipitated proteins were detected by western blotting. Western blots were probed with Drosha (AbCam), actin (Sigma-Aldrich), GAPDH (Sigma-Aldrich), Pol II (Covance), and CBP80 (Sigma) antibodies.
ChIP and HITS-CLIP Analyses
ChIP analysis was carried out as previously described (West et al., 2004). Five micrograms of each antibody was used per ChIP. The immunoprecipitated DNAs were used as templates for qPCR. Drosha Chip-on-chip experiments were carried out as described in De Gobbi et al. (2007) and in the Supplemental Information. HITS-CLIP for DGCR8 was based on a published protocol (Wang et al., 2009) with minor modifications, as described in Macias et al. (2012) and the Supplemental Information.
Br-UTP Nuclear Run-on Analysis
The Br-UTP NRO was carried out largely as described (Lin et al., 2008, Skourti-Stathaki et al., 2011) with some modifications. Nuclear pellets were resuspended in transcription buffer (40 mM Tris-HCl [pH 7.9], 300 mM KCl, 10 mM MgCl2, 40% glycerol, 2 mM DTT) and 10 mM mix of rATP, rCTP, rGTP, and Br-UTP or rUTP (in the control samples). The NRO reaction was performed at 30°C for 30 min. Total RNA was isolated using TRIzol reagent (Invitrogen) according to manufacturer’s instructions and treated with RNase-free DNase I (Roche). Two microliters of anti-BrU antibody (Sigma-Aldrich) was preincubated with 30 μl of Protein G Dynabeads (Upstate) and 10 μg tRNA per sample for 1 hr at 4°C. The beads were washed three times with RSB-100 buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 2.5 mM MgCl2, 0.4% Triton X-100) and resuspended in 150 μl RSB-100 with 40 U RNase-OUT (Invitrogen) and 5 μg of glycogen. Total RNA was added to the beads and incubated for additional 1 hr at 4°C. The beads were washed three times with RSB-100 buffer. RNA bound to the beads was extracted with TRIzol reagent followed by DNase I treatment. The RT reaction was performed using SuperScript III Reverse Transcriptase (Invitrogen) following the manufacturers’ instructions. The real-time quantitative PCR was performed using a Corbett Research Rotor-Gene GG-3000 machine. The PCR mixture contained QuantiTest SYBR green PCR master mix (Qiagen), 2 μl of template cDNA, and primers from Table S3. Cycling parameters were 95°C for 15 min, followed by 45 cycles of 95°C for 15 s, 58°C for 20 s, and 72°C for 20 s. Fluorescence intensities were plotted against the number of cycles by using an algorithm provided by the manufacturer. Amount of nascent Br-UTP RNA was calculated by subtracting the background of U-RNA produced over a specific gene probe.
Statistical Analysis
Unless otherwise stated, results are shown as the average values from at least three independent biological experiments ±SD. The asterisk indicates statistical significance (∗p < 0.05; ∗∗p < 0.05; ∗∗∗p < 0.05), on the basis of an unpaired, two-tailed distribution determined with a Student’s t test.
Author Contributions
N.G. and N.J.P. conceived the study. N.G. performed all experiments and analyzed the data. M.D. performed bioinformatic analysis of Drosha ChIP-on-chip data. S.M., E.E., and M.P. analyzed DGCR8 binding at promoter regions by HITS-CLIP and carried analysis in Figures S6–S8. N.G., J.F.C., and N.J.P. wrote the paper.
Acknowledgments
We thank N. Kim for providing pCK-Flag WT, E110aQ, and ΔN390 Drosha constructs, M. de Gobbi and D. Higgs for the help with ChIP-on-chip experiments, and Natalie Braun for the help with Figure S3C. The work was supported by a Wellcome Trust Programme Grant to N.J.P. and Royal Society University Research fellowship and MRC NIRG MR/J007870/1 to N.G. J.F.C. and S.M. were supported by the Medical Research Council and Wellcome Trust (grant 095518/Z/11/Z). E.E. was supported by grants from the Spanish Ministry of Science and by the Sandra Ibarra Foundation (BIO2008-01091, BIO2011-23920, and CSD2009-00080). M.P. is supported by the Novo Nordisk Foundation.
Published: December 19, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes Supplemental Experimental Procedures, eight figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.11.032.
Contributor Information
Natalia Gromak, Email: natalia.gromak@path.ox.ac.uk.
Nicholas J. Proudfoot, Email: nicholas.proudfoot@path.ox.ac.uk.
Accession Numbers
The NCBI Gene Expression Omnibus accession number for the sequencing raw data for endogenous and overexpressed DGCR8 HITS-CLIP reported in this paper is GSE39086.
Supplemental Information
References
- Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballarino M., Pagano F., Girardi E., Morlando M., Cacchiarelli D., Marchioni M., Proudfoot N.J., Bozzoni I. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell. Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Calistri E., Livi R., Buiatti M. Evolutionary trends of GC/AT distribution patterns in promoters. Mol. Phylogenet. Evol. 2011;60:228–235. doi: 10.1016/j.ympev.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Cernilogar F.M., Onorati M.C., Kothe G.O., Burroughs A.M., Parsi K.M., Breiling A., Lo Sardo F., Saxena A., Miyoshi K., Siomi H. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong M.M., Zhang G., Cheloufi S., Neubert T.A., Hannon G.J., Littman D.R. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Lis J.T. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gobbi M., Anguita E., Hughes J., Sloane-Stanley J.A., Sharpe J.A., Koch C.M., Dunham I., Gibbons R.J., Wood W.G., Higgs D.R. Tissue-specific histone modification and transcription factor binding in alpha globin gene expression. Blood. 2007;110:4503–4510. doi: 10.1182/blood-2007-06-097964. [DOI] [PubMed] [Google Scholar]
- Egloff S., Al-Rawaf H., O’Reilly D., Murphy S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol. Cell. Biol. 2009;29:4002–4013. doi: 10.1128/MCB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Lyakh L.A., Bracken C.P., Fukuoka J., Hayakawa M., Tsukiyama T., Soll S.J., Harris M., Rocha S., Roche K.C. SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol. Cell. 2006;24:771–783. doi: 10.1016/j.molcel.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F.V., Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem. Soc. Trans. 2008;36:609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Gruber J.J., Zatechka D.S., Sabin L.R., Yong J., Lum J.J., Kong M., Zong W.X., Zhang Z., Lau C.K., Rawlings J. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S., Cáceres J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Pedersen J.S., Kwon S.C., Belair C.D., Kim Y.K., Yeom K.H., Yang W.Y., Haussler D., Blelloch R., Kim V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S., Rodriguez J., Abruzzi K.C., Khodor Y.L., Sugino K., Marr M.T., 2nd, Nelson S., Rosbash M. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov F.V., Cheloufi S., Chong M.M., Stark A., Smith A.D., Hannon G.J. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Kim V.N. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Yang J.Y., Xu J., Jang I.C., Prigge M.J., Chua N.H. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol. 2008;49:1634–1644. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M., Yalcin A., Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lin Y.T., Sullivan C.S. Expanding the role of Drosha to the regulation of viral gene expression. Proc. Natl. Acad. Sci. USA. 2011;108:11229–11234. doi: 10.1073/pnas.1105799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Coutinho-Mansfield G., Wang D., Pandit S., Fu X.D. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Macias S., Plass M., Stajuda A., Michlewski G., Eyras E., Cáceres J.F. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat. Struct. Mol. Biol. 2012;19:760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M., Ballarino M., Gromak N., Pagano F., Bozzoni I., Proudfoot N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M.S., Mapendano C.K., Schierup M.H., Jensen T.H. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Qin H., Gunning P. The 3′-end of the human beta-actin gene enhances activity of the beta-actin expression vector system: construction of improved vectors. J. Biochem. Biophys. Methods. 1997;36:63–72. doi: 10.1016/s0165-022x(97)00045-6. [DOI] [PubMed] [Google Scholar]
- Sabin L.R., Zhou R., Gruber J.J., Lukinova N., Bambina S., Berman A., Lau C.K., Thompson C.B., Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Siomi M.C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K., Proudfoot N.J., Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R., Chang H.M., Lapierre R.J., Gregory R.I. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A., Rousset E., Basavarajaiah P., Contreras X., Harwig A., Laurent-Chabalier S., Nakamura M., Chen X., Zhang K., Meziane O. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150:1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Tollervey J., Briese M., Turner D., Ule J. CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods. 2009;48:287–293. doi: 10.1016/j.ymeth.2009.02.021. [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J. Human 5′ —> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Wollerton M.C., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Bi L., Zheng B., Ji L., Chevalier D., Agarwal M., Ramachandran V., Li W., Lagrange T., Walker J.C., Chen X. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Yi R., Cullen B.R. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.