Abstract
Autophagy is an essential cellular pathway for degrading defective organelles and aggregated proteins. Defects in autophagy have been implicated in the neurodegenerative disorder Huntington's disease (HD), in which polyglutamine-expanded huntingtin (polyQ-htt) is predominantly cleared by autophagy. In neurons, autophagosomes form constitutively at the axon tip and undergo robust retrograde axonal transport toward the cell body, but the factors regulating autophagosome dynamics and autophagosome maturation are not well understood. Here, we show that both huntingtin (htt) and its adaptor protein huntingtin-associated protein-1 (HAP1) copurify and colocalize with autophagosomes in neurons. We use live-cell imaging and RNAi in primary neurons from GFP-LC3 transgenic mice to show that htt and HAP1 control autophagosome dynamics, regulating dynein and kinesin motors to promote processive transport. Expression of polyQ-htt in either primary neurons or striatal cells from HD knock-in mice is sufficient to disrupt the axonal transport of autophagosomes. Htt is not required for autophagosome formation or cargo loading. However, the defective autophagosome transport observed in both htt-depleted neurons and polyQ-htt-expressing neurons is correlated with inefficient degradation of engulfed mitochondrial fragments. Together, these studies identify htt and HAP1 as regulators of autophagosome transport in neurons and suggest that misregulation of autophagosome transport in HD leads to inefficient autophagosome maturation, potentially due to inhibition of autophagosome/lysosome fusion along the axon. The resulting defective clearance of both polyQ-htt aggregates and dysfunctional mitochondria by neuronal autophagosomes may contribute to neurodegeneration and cell death in HD.
Keywords: autophagy, axonal transport, dynein, huntingtin, Huntington's disease, mitophagy
Introduction
Macroautophagy (referred to here as autophagy) is a critical cellular degradation pathway mediated by the autophagosome, a double-membrane vesicle that engulfs organelles and protein cargo (Xie and Klionksy, 2007). Defective autophagy leads to neurodegeneration (Hara et al., 2006; Komatsu et al., 2006) and is implicated in neurodegenerative diseases including Huntington's disease (HD; Martinez-Vicente et al., 2010; Wong and Cuervo, 2010). In neurons, autophagosome formation is a polarized process (Maday et al., 2012). Nascent autophagosomes form and engulf cargo at the axon tip and subsequently undergo robust retrograde axonal transport toward the soma (Lee et al., 2011; Maday et al., 2012). Despite the pronounced unidirectional motility observed during autophagosome movement along the axon, both retrograde and anterograde motor proteins remain robustly associated with autophagosomes (Maday et al., 2012), suggesting that regulatory proteins such as scaffolding proteins may be necessary to promote efficient transport by controlling motor activity.
Huntingtin (htt) regulates the transport dynamics of various organelles (Gunawardena et al., 2003; Gauthier et al., 2004; Caviston et al., 2007; Her and Goldstein, 2008; Power et al., 2012). Htt binds directly to the retrograde motor dynein (Caviston et al., 2007) and interacts with the dynein-activator dynactin and the anterograde motor kinesin-1 via the adaptor protein huntingtin-associated protein-1 (HAP1; Li et al., 1995; Engelender et al., 1997; Li et al., 1998; McGuire et al., 2006; Twelvetrees et al., 2010). Therefore, the formation of a htt-HAP1-motor complex may regulate autophagosome dynamics along the axon.
Polyglutamine expansions in htt (polyQ-htt) cause HD, and both soluble and aggregated polyQ-htt are cleared by autophagy (Ravikumar et al., 2002; Qin et al., 2003). Disruption of autophagy accelerates polyQ-htt aggregate formation and cell death (Ravikumar et al., 2004; Sarkar et al., 2007) and defects in autophagic cargo loading (Martinez-Vicente et al., 2010) have been observed in HD models, further suggesting a link between defects in autophagy and HD.
Here, we find a role for htt and HAP1 as regulators of autophagosome transport. Using live-cell imaging in primary neurons, we show that the htt/HAP1 complex enhances retrograde motility, promoting efficient autophagosome transport toward the soma. Autophagosome transport is disrupted in primary neurons expressing pathogenic polyQ-htt and also in striatal cells from HD knock-in mice. Using RNAi to deplete endogenous htt, we find that htt is not required for the initial steps of autophagosome formation or cargo loading. Rather, the defective autophagosome transport observed in both htt-depleted neurons and polyQ-htt-expressing neurons results in the aberrant accumulation of autophagosomes with engulfed mitochondrial fragments, suggesting that cargo degradation is impaired. Therefore, defects in autophagosome transport and cargo degradation may contribute to the inefficient clearance of dysfunctional mitochondria and the accumulation of polyQ-htt observed in the neurons of patients with HD.
Materials and Methods
Reagents.
GFP-LC3 transgenic mice, strain name B6.Cg-Tg(CAG-EGFP/LC3)53Nmi/NmiRbrc, were developed by N. Mizushima (Tokyo Medical and Dental University, Tokyo, Japan; Mizushima et al., 2004) and deposited into the RIKEN BioResource Center (Japan). Immortalized STHdhQ striatal cell lines from control HdhQ23/Q23 and HD-knock-in Hdh Q111/Q111 mice were developed by M. MacDonald (Richard B. Simches Research Center, Boston, MA; Trettel et al., 2000) and deposited into the Coriell Cell Repositories (Camden, NJ). siRNA to htt (5′-GCAGCUUGUCCAGGUUUAUUU-3′) and HAP1 (5′-GAAGUAUGUCCUCCAGCAAUU-3′) were obtained from Dharmacon (Thermo Scientific). Constructs used were DsRed2-mito (gift from T. Schwarz, Harvard Medical School, Boston, MA), monomeric RFP-Ub (Addgene), LAMP1-RFP (Addgene), hHAP1a (gift from X. J. Li, Emory University, Atlanta, GA), and mCherry-EGFP-LC3 (gift from T. Johansen, University of Tromsø, Tromsø, Norway; Pankiv et al., 2007). Htt-Q23, htt-Q23-Δdyn, htt-Q23-ΔHAP1, htt-Q100, htt-68Q, and HAP1-KBD were gifts from F. Saudou (Institut Curie, Orsay, France; Pardo et al., 2010) and J. Kittler (University College London, London, United Kingdom). pEGFP-LC3 (a gift from T. Yoshimori, Osaka University, Osaka, Japan; Kimura et al., 2007), dynein intermediate chain 1B (DIC1B; a gift from K. Pfister, University of Virginia, Charlottesville, VA), and Kif5C tail (a gift from M. Setou, Hamamatsu University School of Medicine, Shizuoka, Japan) were recloned into pmCherry (Takara). DIC isoforms 1A and 2C (gifts from K. Pfister, University of Virginia, Charlottesville, VA) were recloned into pcDNA3.1mychisA (Invitrogen). Antibodies used were a polyclonal antibody against LC3B (Abcam) and monoclonal antibodies against: htt (MAB2166; Millipore), HAP1 (611302; BD Biosciences), dynein intermediate chain (DIC) (clone 74.1; Millipore), dynactin p150Glued (610474; BD Biosciences), kinesin-1 heavy chain (clone H2; Millipore), GAPDH (ab9484; Abcam), α-tubulin (DM1A; Sigma), HA (MMS-101P; Covance), myc (R950; Invitrogen), and Hsp60 (SPA-806; Enzo Life Sciences).
Cell culture and transfection.
Dorsal root ganglia (DRGs) were dissected from spinal columns of adult mice of either sex that were <1 year old. To isolate neurons, DRGs were treated with 20 U/ml papain (Worthington), followed by 2 mg/ml collagenase II (Invitrogen) and 2.4 mg/ml dispase II (Roche). Neurons were dissociated in HBSS (Invitrogen) supplemented with 5 mm HEPES and 10 mm d-glucose, pH 7.35, and purified using a 20% Percoll gradient (Sigma) for 8 min at 1000 × g. DRG neurons were transfected with 0.5–2 μg of plasmid DNA, 12–30 pmol of siRNA, or both using the Amaxa Basic Neuron SCN Nucleofector kit (Lonza) and then plated onto coverslips or glass-bottom dishes (FluoroDish; World Precision Instruments) coated with 0.01% poly-l-lysine and 20 μg/ml laminin. Neurons were maintained for 2 DIV in F-12 medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 2 mm l-glutamine, and 100 U/ml penicillin-streptomycin at 37°C in a 5% CO2 incubator. Striatal cells were cultured and differentiated as described previously(Trettel et al., 2000) and imaged 2 DIV after differentiation. Both striatal cells and COS-7 cells were transfected using FuGENE 6 (Promega). All live-cell imaging was performed in low fluorescence nutrient medium (Hibernate A; BrainBits) supplemented with 2% B27 supplement (Invitrogen) and 2 mm GlutaMax (Invitrogen). DRGs were labeled with 100 nm LysoTracker Red (Invitrogen) diluted in F-12 medium for 30 min at 37°C in a 5% CO2 incubator, followed by two washes with Hibernate A medium before imaging. All experiments involving animals were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania.
Biochemistry.
Autophagosome-enriched fractions were prepared from GFP-LC3 transgenic mice brains using a protocol adapted from Morvan et al. (2009) and Strømhaug et al. (1998), as described previously (Maday et al., 2012). Brains were homogenized in 10 ml of 250 mm sucrose in 10 mm HEPES-KOH, pH 7.4, using a 30 ml homogenizer with a round-bottom Teflon pestle. The homogenate was separated using a Nycondenz gradient and volumes of the gradient steps were then scaled proportionately for the SW41 rotor (Beckman Coulter). Equal total protein of low-speed supernatant and the autophagosome-enriched fraction were separated by SDS-PAGE and analyzed by Western blot. For protein analysis of neuronal lysates, DRG neurons were cultured for 2 DIV, washed with Dulbecco's PBS (Invitrogen), pH 7.4, at room temperature, and scraped into ice-cold lysis buffer [0.5 mm DTT, 1 mm leupeptin, 1 mm Pepstatin-A, and 1 mm TAME (Tosyl-l-arginine methyl ester)]. Neuronal lysates were analyzed by SDS-PAGE and Western blot according to standard protocols.
Immunofluorescence.
For immunofluorescence analysis, DRG neurons were plated on coverslips and cultured for 2 DIV. Cells were washed once in PBS (150 mm NaCl in 50 mm NaPO4, pH 7.4) and fixed in 4% paraformaldehyde with 4% sucrose in PBS for 5 min at room temperature. Cells were then washed twice in PBS and blocked and permeabilized in 2% (w/v) BSA and 0.1% (w/v) saponin in PBS for 1 h at room temperature. Fixed cells were incubated in primary antibody for 1 h, washed 3 × 5 min, incubated in secondary antibody for 1 h, washed 3 × 5 min and mounted on glass slides with ProLong gold (Invitrogen).
Immunoprecipitation.
To examine coimmunoprecipitation of htt with dynein isoforms, COS-7 cells were cotransfected for 24 h with myc-tagged DIC isoforms and HA-tagged wild-type htt or polyQ-htt using FuGENE 6 (Promega). Cells were lysed in 80 mm PIPES, 1 mm EGTA, 1 mm MgCl2, 50 mm NaCl, 0.5 mm DTT, 0.5% Triton X-100, and protease inhibitors (1 mm PMSF, 1 mm leupeptin, and 1 mm TAME). Lysates were immunoprecipitated using Protein G-coupled Dynabeads (Invitrogen) incubated in anti-myc antibody, and washed in the above buffer without detergent. Equal total protein levels of lysates and eluates were analyzed by SDS-PAGE and Western blot according to standard protocols. Measured band intensities of htt coimmunoprecipitating with DIC were normalized to account for differential DIC isoform expression and immunoprecipitation, and are expressed as the ratio of the DIC2C:DIC1A interaction for each htt construct.
Microscopy.
Images of GFP-LC3 motility (Figs. 2, 3A, 4, 5), mCherry-EGFP-LC3 motility (Fig. 6E) and polyQ-htt (Fig. 9) were acquired on an inverted epifluorescence microscope (DMI6000B; Leica) with an Apochromat 63× 1.4 numerical aperture (NA) oil-immersion objective (Leica) in a temperature-controlled chamber (37°C). Digital images were acquired with a charge-coupled device camera (ORCA-R2; Hamamatsu Photonics) using LAS-AF software (Leica) at 1 frame every 3 s for a 3 min duration. Dual-color videos were acquired as consecutive green-red images for 3 min duration.
Figure 2.
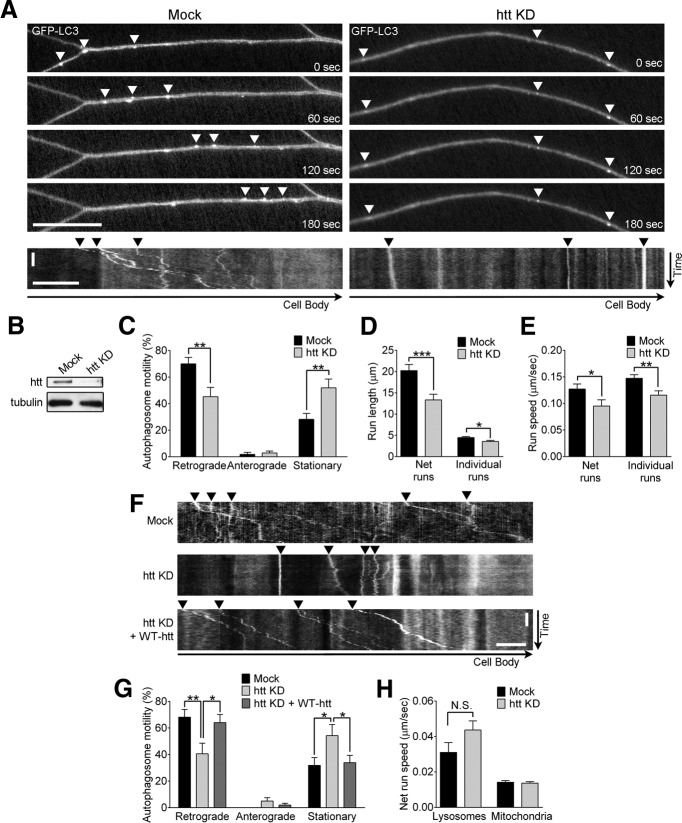
Htt regulates autophagosome transport in neurons. A, Representative time series and corresponding kymographs of GFP-LC3 autophagosome transport in axons of control (mock) and htt siRNA-depleted (htt KD) primary DRG neurons from GFP-LC3-transgenic mice. Autophagosomes in control neurons demonstrate robust retrograde transport (arrowheads), whereas autophagosomes in htt-depleted neurons show disrupted motility (arrowheads). B, Htt protein levels are efficiently depleted by htt siRNA in immunoblot. C, Depleting htt decreases the percentage of retrograde autophagosomes (moving ≥10 μm/3 min in the retrograde direction) and increases the percentage of stationary autophagosomes (moving <10 μm/3 min) in neurons (mock, n = 23; htt KD, n = 23). The percentage of anterograde autophagosomes (moving ≥10 μm/3 min in the anterograde direction) was not altered. D, E, Run lengths (D) and run speeds (E) from autophagosome net runs (total distance traveled over 3 min) and individual runs (distance traveled before changing direction or speed) are reduced by htt depletion in primary neurons (net runs: mock, n = 143; htt KD, n = 162; individual runs: mock, n = 667; htt KD, n = 608). F, Representative kymographs of GFP-LC3 autophagosome transport demonstrate rescue of retrograde autophagosome transport (arrowheads) in htt-siRNA neurons expressing siRNA-resistant wild-type htt Q23 (htt KD + WT-htt) compared with htt siRNA-depleted (htt KD) primary DRG neurons. Rescued neurons have similar autophagosome transport to control neurons (mock). G, Expression of wild-type htt rescues the percentage of retrograde autophagosomes in htt-siRNA neurons (mock, n = 17; htt KD, n = 15; htt KD + WT-htt, n = 12). H, Htt depletion does not disrupt net run speeds of lysosomes or mitochondria in primary DRG neurons (lysosomes: mock, n = 126; htt KD, n = 149; mitochondria: mock, n = 122; htt KD, n = 177). Horizontal scale bars in A and F, 10 μm. Vertical scale bars, 1 min. Values represent means ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; N.S. (not significant, p ≥ 0.05).
Figure 3.
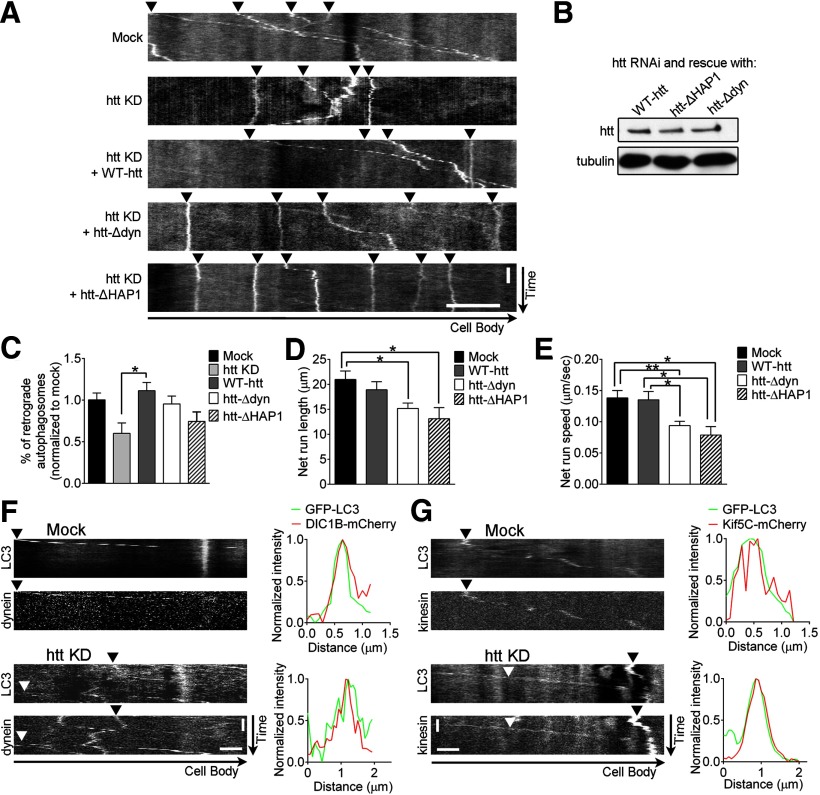
Htt regulates autophagosome transport by binding to dynein and HAP1. A, Representative kymographs of GPF-LC3 autophagosome transport demonstrate that siRNA-resistant dominant-negative htt constructs that cannot bind dynein (htt KD + htt-Δdyn) or HAP1 (htt KD + htt-ΔHAP1) are unable to rescue autophagosome motility (arrowheads) in htt siRNA-depleted primary DRG neurons, compared with normal full-length htt (htt KD + WT-htt). B, WT-htt, htt-Δdyn, and htt-ΔHAP1 are all efficiently expressed in neurons by immunoblot. C, WT-htt rescues the percentage of retrograde autophagosomes to control levels, whereas htt-Δdyn and htt-ΔHAP1 are unable to do so (mock, n = 41; htt KD, n = 14; WT-htt, n = 37; htt-Δdyn, n = 25; htt-ΔHAP1, n = 23). D, E, Run lengths (D) and run speeds (E) from autophagosome net runs (total distance traveled over 3 min) are not rescued by htt-Δdyn or htt-ΔHAP1 compared with WT-htt (mock, n = 184; WT-htt, n = 135; htt-Δdyn, n = 207; htt-ΔHAP1, n = 67). F, Representative kymographs and corresponding line scans show colocalization and cotransport (arrowheads) of a neuronal-specific isoform of the retrograde motor dynein (DIC1B-mCherry) with GFP-LC3 autophagosomes in both control neurons (mock) and htt-depleted (htt KD) primary DRG neurons. G, Representative kymographs and corresponding line scans show colocalization and cotransport (arrowheads) of the anterograde motor kinesin (Kif5C-mCherry) with GFP-LC3 autophagosomes in both control neurons (mock) and htt-depleted neurons (htt KD). Line scan intensities are normalized per marker and per condition. Horizontal scale bars: A, 10 μm; F, G, 2 μm. Vertical scale bars, 1 min. Values represent means ± SEM. *p < 0.05; **p < 0.01.
Figure 4.
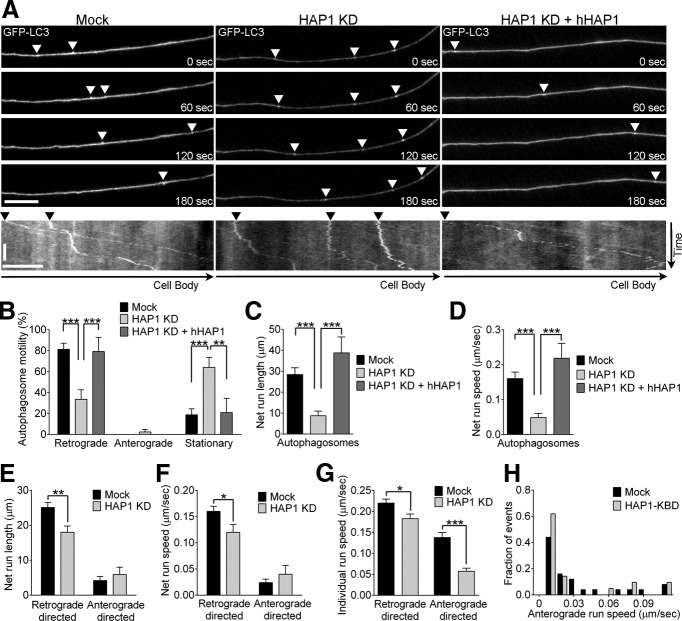
HAP1 depletion inhibits autophagosome transport in neurons. A, Representative time series and corresponding kymographs of GFP-LC3 autophagosome transport in axons of primary DRG neurons demonstrate that autophagosome transport is disrupted in HAP1 siRNA-depleted neurons (HAP1 KD) compared with control neurons (mock) and is rescued by expression of siRNA-resistant human HAP1 (HAP1 KD + hHAP1; arrowheads). Two autophagosomes are shown moving in control neurons (Mock) with one leaving the frame of view by 180 s (see corresponding kymograph). B, Depleting HAP1 decreases the percentage of retrograde autophagosomes and increases the percentage of stationary autophagosomes in neurons; these defects are rescued by expression of siRNA-resistant human HAP1 (HAP1 KD + hHAP1) (mock, n = 10; HAP1 KD, n = 14; HAP1 KD + hHAP1, n = 6). C, D, HAP1 depletion reduces net run lengths (C) and net run speeds (D) of autophagosomes; these defects are rescued by hHAP1 expression (mock, n = 52; HAP1 KD, n = 39; HAP1 KD + hHAP1, n = 17). E, F, HAP1 depletion reduces the run lengths (E) and run speeds (F) of autophagosome net runs (total distance traveled over 3 min) in the retrograde direction, but not in the anterograde direction (retrograde-directed: mock, n = 180; HAP1 KD, n = 107; anterograde-directed: mock, n = 30; HAP1 KD, n = 30). G, HAP1 depletion reduces individual run speeds in both the retrograde and anterograde direction (retrograde-directed: mock, n = 709; HAP1 KD, n = 336; anterograde-directed: mock, n = 269; HAP1 KD, n = 175). Retrograde directed refers to runs of any length in the retrograde direction; anterograde directed refers to runs of any length in the anterograde direction. H, Expression of HAP1 lacking the kinesin-binding domain (HAP1-KBD) results in an increase in the fraction of autophagosomes expressing slower anterograde run speeds (mock, n = 25; HAP1-KBD, n = 21). Horizontal scale bar, 10 μm. Vertical scale bar, 1 min. Values represent means ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 5.
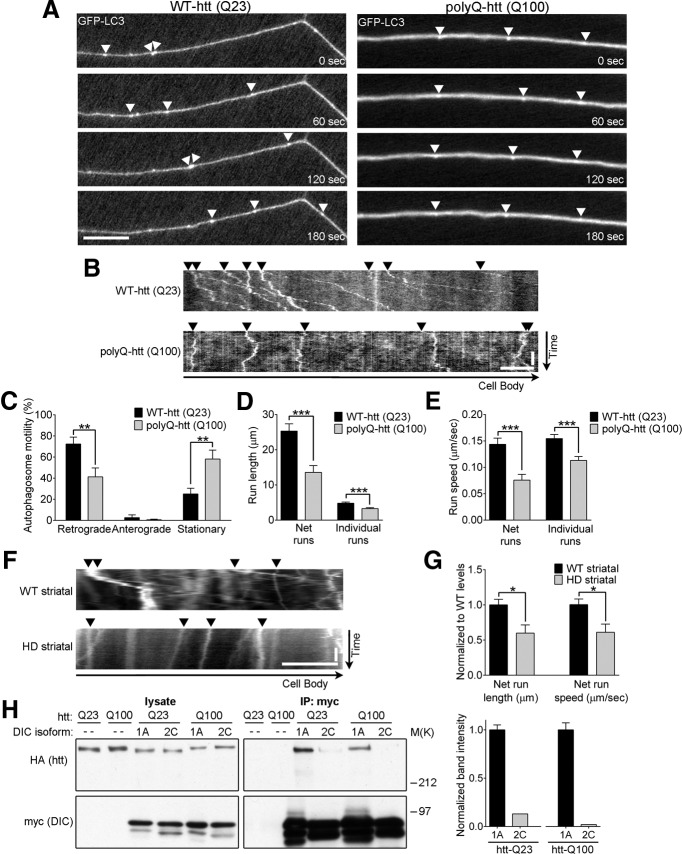
Pathogenic polyQ-htt disrupts autophagosome dynamics in neurons. A, B, Representative time series (A) and kymographs (B) of GPF-LC3 show that expression of siRNA-resistant polyQ-htt (Q100) in siRNA-htt-depleted primary DRG neurons disrupts autophagosome transport (arrowheads) compared with expression of wild-type htt (WT-htt, Q23). C, PolyQ-htt decreases the percentage of retrograde autophagosomes and increases the percentage of stationary autophagosomes in neurons (WT-htt, n = 19; polyQ-htt, n = 17). D, E, Run lengths (D) and run speeds (E) from autophagosome net runs (total distance traveled over 3 min) and individual runs (distance traveled before changing direction or speed) are reduced by polyQ-htt compared with wild-type htt in primary neurons (net runs: WT-htt, n = 97; polyQ-htt, n = 77; individual runs: WT-htt, n = 526; polyQ-htt, n = 393). F, Representative kymographs of mCherry-LC3 show reduced autophagosome motility in neurites of differentiated striatal cells from HD homozygous knock-in mice (HdhQ111/Q111) compared with striatal cells from wild-type mice (HdhQ7/Q7). G, Autophagosome net run lengths and net run speeds are reduced in HdhQ111/Q111 striatal cells (WT, n = 163; HD, n = 50). H, Coimmunoprecipitation experiments show that both wild-type htt (Q23) and polyQ-htt (Q100) preferentially bind to neuronal-specific isoform DIC1A compared with the ubiquitously expressed isoform DIC2C. Band intensities of coimmunoprecipitated htt were normalized for efficiency of DIC expression and immunoprecipitation and expressed as the relative ratio of DIC2C: DIC1A interaction for each htt construct. Horizontal scale bars, 10 μm. Vertical scale bars, 1 min. Values represent means ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 6.
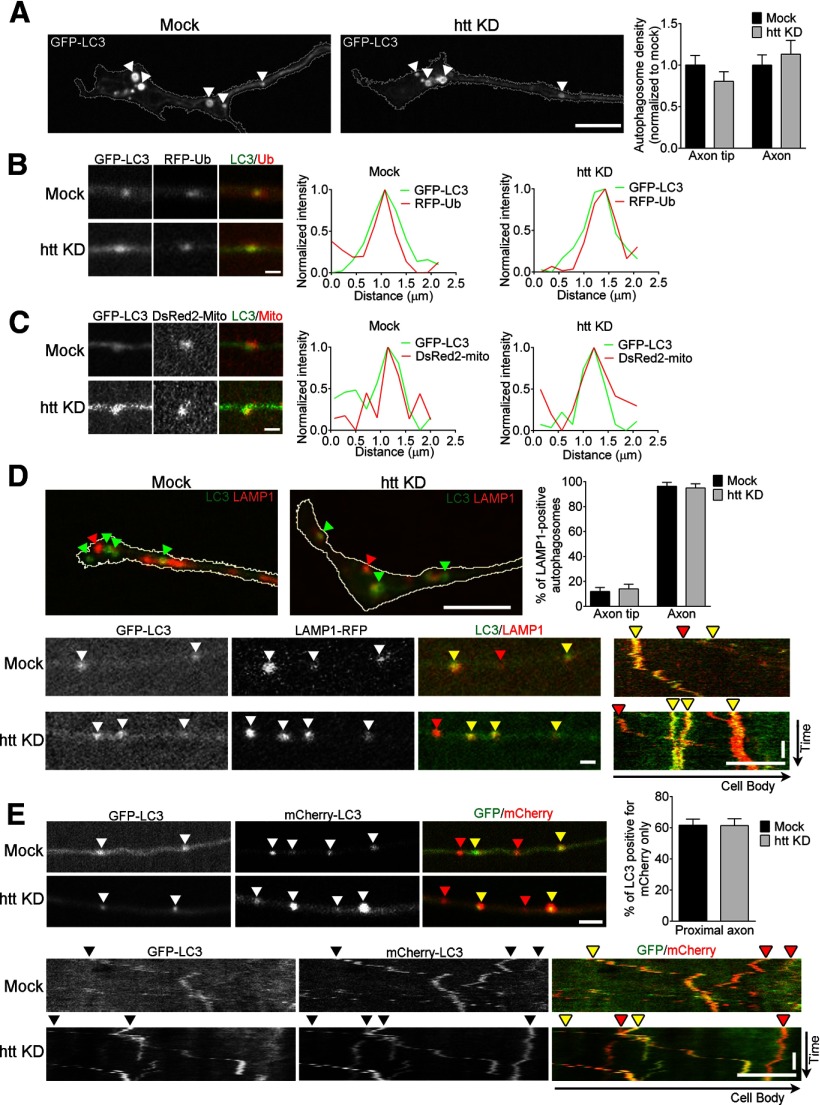
Htt is not required for initial steps of autophagosome formation, cargo loading, and initial maturation. A, Representative confocal live-cell images of GFP-LC3 autophagosomes demonstrate autophagosomes forming (arrowheads) in the distal tip of the neurite from puncta into ring-like structures in both control (mock) and htt-depleted (htt KD) primary DRG neurons. Autophagosome density at the axon tip and along the axon is not disrupted by htt depletion (axon tip: mock, n = 10; htt KD, n = 13; axon: mock, n = 23; htt KD, n = 23). B, C, Representative images and corresponding line scans of GFP-LC3 autophagosomes colocalized with RFP-Ub ubiquitinated protein cargo (B) and DsRed2-mito mitochondrial fragments (C) in axons of both control (mock) and htt-depleted (htt KD) neurons. Line scan intensities are normalized per marker and per condition. D, Representative images of GFP-LC3 and LAMP1-RFP at the axon tip (top) and representative images and corresponding kymograph of GFP-LC3 autophagosomes positive for LAMP1-RFP (yellow arrowheads) along the axon (bottom) in both control (mock) and htt-depleted (htt KD) neurons (axon tip: mock, n = 19; htt KD, n = 22; axon: mock, n = 22; htt KD, n = 20). E, Representative images (top) and kymographs (bottom) of acidification of autophagosomes (red arrowheads) in neurons expressing mCherry-EGFP-LC3 in both control (mock) and htt-depleted (htt KD) primary DRG neurons. In acidic environments, the GFP moiety is preferentially quenched with persistent mCherry fluorescence (mock, n = 16; htt KD, n = 13). Horizontal scale bars: A, D, E (kymographs), 10 μm; B–E (images), 1 μm. Vertical scale bars, 1 min.
Figure 9.
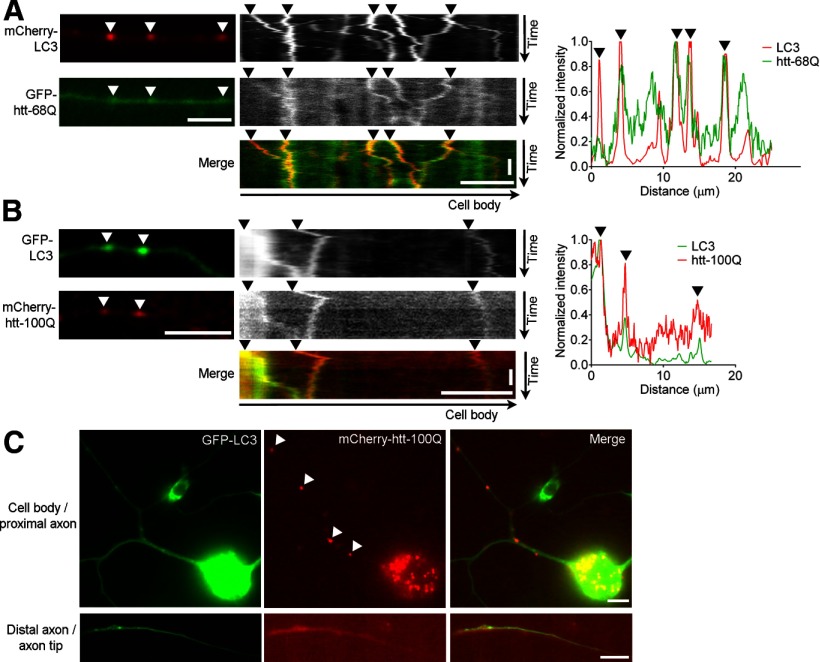
Disrupted autophagosome dynamics lead to inefficient clearance of pathogenic polyQ-htt from the distal axon. A, B, Representative images and kymographs and corresponding linescans show disease-associated cleaved N-terminal fragment of GFP-polyQ-htt (Q68; A) and full-length mCherry-polyQ-htt (Q100; B) colocalized and cotransporting as autophagic cargo with retrogradely moving autophagosomes (arrowheads) in axons of primary neurons. C, Representative image of ineffective clearance and aggregate formation of mCherry-polyQ-htt (Q100) in the cell body, axon (arrowheads), and distal axon tip of primary neurons. Horizontal scale bars, 10 μm. Vertical scale bars, 1 min.
Images of lysosome and mitochondrial motility (Fig. 2H), cotransport of GFP-LC3 with motors (Fig. 3F,G), autophagosome biogenesis, GFP-LC3 cotransport with cargo, and maturation and fusion with lysosomes (Figs. 6A–D, 7, 8) were acquired on a spinning-disk confocal (UltraVIEW VoX; PerkinElmer) on a Nikon Eclipse Ti microscope using an Apochromat 100× 1.49 NA oil-immersion objective (Nikon) in a temperature-controlled chamber (37°C). Digital images were acquired with an EM charge-coupled device camera (C9100; Hamamatsu Photonics) using Volocity software (PerkinElmer) at 1 frame every 3–5 s for 3–10 min duration. Dual-color videos were acquired as consecutive green-red images for colocalization/cotransport experiments. Videos of mitochondria cotransported with autophagosomes were acquired using longer exposure times to allow visualization of mitochondrial fragments within autophagosomes. Neurons fixed for immunofluorescence experiments (Fig. 1B–D) were also imaged using the confocal microscope described above. All images were assembled using ImageJ and Adobe Photoshop.
Figure 7.
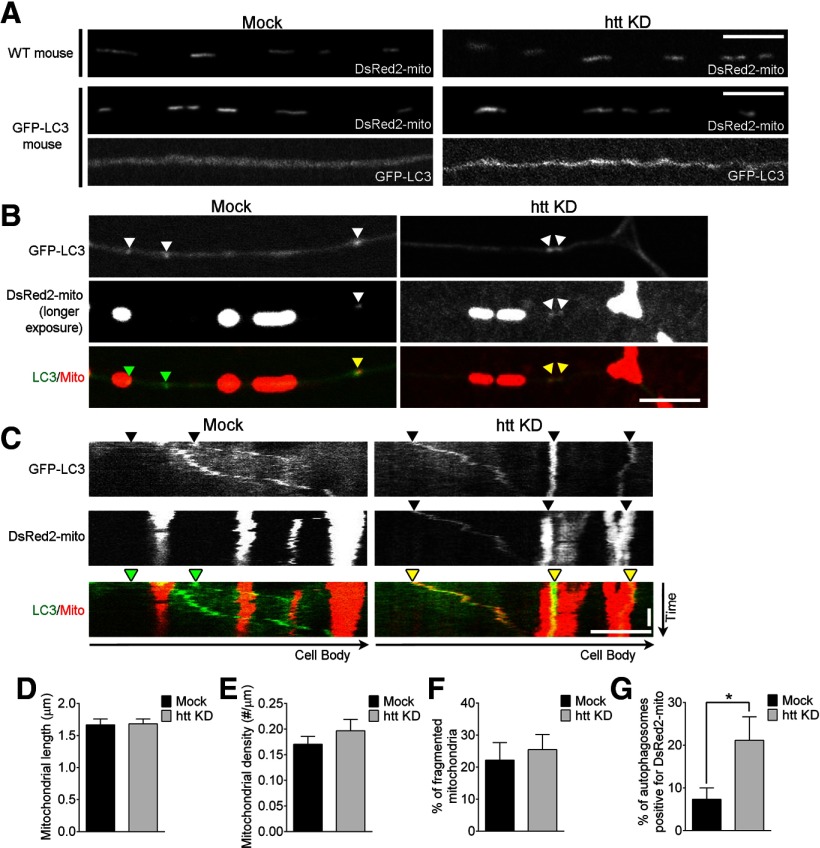
Depletion of htt leads to inefficient mitochondrial cargo degradation in autophagosomes. A, Htt depletion does not disrupt mitochondria morphology in either wild-type mice (WT) or GFP-LC3-transgenic mice. B, C, Representative images (B) and kymographs (C) of increased DsRed2-mito mitochondrial fragments colocalized and cotransporting with GFP-LC3 autophagosomes (yellow arrowheads) in axons of htt-depleted neurons. Mitochondria were imaged at longer exposures to allow for visible cotransport of mitochondrial fragments with autophagosomes. Images and kymographs are taken from different neurons. D–F, Htt depletion did not disrupt mitochondrial length (D; mock, n = 122; htt KD, n = 177), density along the axon (E), or percentage of fragmented mitochondria (length < 1 μm; F) in primary DRG neurons (mock, n = 9; htt KD, n = 12). G, Htt depletion increased the percentage of GFP-LC3 autophagosomes containing DsRed2-mito mitochondria per neurite (mock, n = 32; htt KD, n = 38). Horizontal scale bars, 10 μm. Vertical scale bar, 1 min. Values represent means ± SEM. *p < 0.05.
Figure 8.
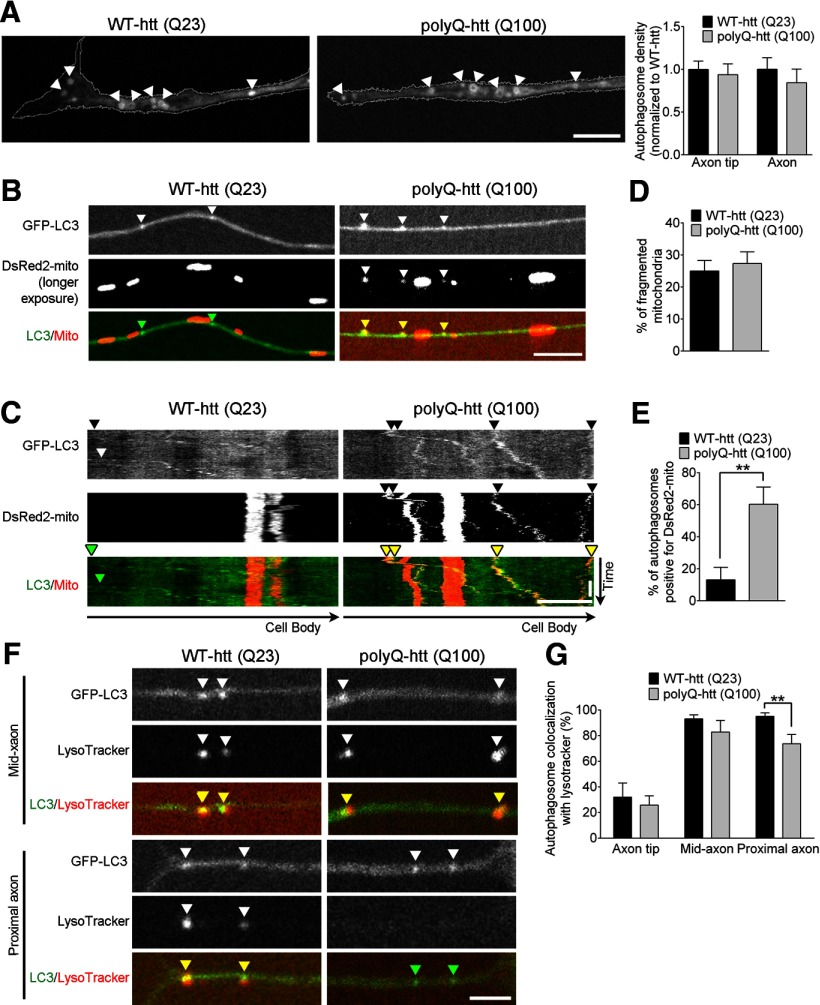
Pathogenic polyQ-htt causes inefficient mitochondrial cargo degradation in neurons. A, Confocal live-cell images showing GFP-LC3 autophagosomes forming (arrowheads) in the distal tip of the neurite from puncta into ring-like structures in htt siRNA-depleted primary DRG neurons from GFP-LC3 transgenic mice expressing either siRNA-resistant wild-type htt (Q23) or polyQ-htt (Q100). Autophagosome density at the axon tip and along the axon is not altered by expression of polyQ-htt (Q100) in primary DRG neurons (axon tip: WT-htt, n = 12; polyQ-htt, n = 11; axon: WT-htt, n = 14; polyQ-htt, n = 17). B, C, Representative images (B) and kymographs (C) of increased DsRed2-mito mitochondrial fragments colocalized and cotransporting with GFP-LC3 autophagosomes (yellow arrowheads) in axons of polyQ-htt (Q100)-expressing neurons. Mitochondria were imaged at longer exposures to allow for visible cotransport of mitochondrial fragments with autophagosomes. D, PolyQ-htt expression did not disrupt percentage of fragmented mitochondria (length < 1 μm) in primary DRG neurons (WT-htt, n = 12; polyQ-htt, n = 17). E, PolyQ-htt expression increased the percentage of GFP-LC3 autophagosomes containing DsRed2-mito mitochondria per neurite (WT-htt, n = 12; polyQ-htt, n = 17). F, G, Representative images (F) and quantification (G) showing similar levels of LysoTracker Red-positive autophagosomes at the axon tip and mid-axon, but reduced LysoTracker Red-positive autophagosomes in the proximal axon of neurons expressing polyQ-htt (Q100) compared with neurons expressing WT-htt (Q23) (axon tip: WT-htt, n = 20; polyQ-htt, n = 18; mid-axon: WT-htt, n = 16; polyQ-htt, n = 15; proximal-axon: WT-htt, n = 14; polyQ-htt, n = 14). Horizontal scale bars: A–C, 10 μm; F, 5 μm. Vertical scale bar, 1 min. Values represent means ± SEM. **p < 0.01.
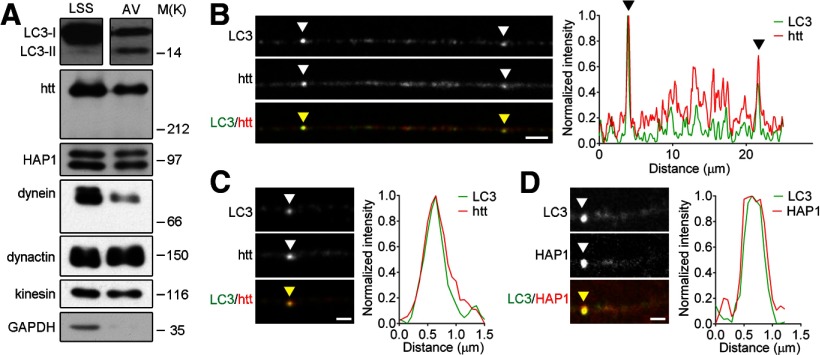
Figure 1.
Htt associates with autophagosomes in neurons. A, Autophagosome-enriched fraction (AV) prepared from mouse brain containing LC3-II, the lipidated form of LC3, is positive for htt, HAP1, the retrograde motor complex proteins dynein and dynactin, and the anterograde motor protein kinesin, but not for the cytoplasmic protein GAPDH. Equal total protein was loaded from the low speed supernatant fraction (LSS) obtained before purification for comparison by immunoblot. B–D, Representative images with corresponding linescans of immunostaining of endogenous LC3 with htt (B,C) and HAP1 (D) in axons from primary DRG neurons demonstrate colocalization of htt and HAP1 with autophagosomes. Arrowheads highlight areas of colocalization. Horizontal scale bars in B–D, 1 μm.
Image analysis.
Kymographs were created using MetaMorph (Molecular Devices) for time-lapse epifluorescent images and the Multiple Kymograph plugin (submitted by J. Rietdorf and A. Seitz, European Molecular Biology Laboratory, Heidelberg, Germany) in ImageJ for time-lapse confocal images. From these kymographs, GFP-LC3 puncta were classified as retrograde (moved ≥10 μm in the retrograde direction), anterograde (moved ≥10 μm in the anterograde direction), or stationary (moved <10 μm during the duration of the 3 min video). The percent motility of autophagosomes along the axon (retrograde, anterograde, or stationary) was calculated as a percentage of the total number of autophagosomes imaged per neuron. Run lengths and run speeds of organelles were manually measured for net runs (the total distance a puncta traveled during the 3 min video) and individual runs (the distance a puncta traveled before switching direction or speed) by drawing a slope from the beginning to the end of the run on the kymograph. Neurons from htt-depleted or htt rescue cultures were analyzed for the percentage of retrograde autophagosomes and then normalized to the percentage of retrograde autophagosomes from control neurons of corresponding experiments performed in parallel (Fig. 3C). For HAP1 depletion experiments, net runs and individual runs were further classified as retrograde directed (moved any distance in the retrograde direction) or anterograde directed (moved any distance in the anterograde direction).
To assess colocalization of LC3 with motors or cargo, line scans were generated using ImageJ and normalized per protein and per condition. Autophagosome density at the axon tip was calculated using the total number of autophagosomes observed at the beginning of a video in a given axon tip (total area < ∼50 μm from the end of a neurite). Autophagosome density along the axon was calculated from kymographs using the total number of autophagosomes observed divided by the length of the kymograph. The percentage of LAMP-1-positive GFP-LC3 puncta was calculated using the total number of autophagosomes observed at the axon tip (total area < ∼50 μm from the end of a neurite) or along the axon (>100 μm from the end of a neurite) at a given time point. The percentage of mCherry-EGFP-LC3 puncta that had mCherry or GFP fluorescence was calculated as a fraction of the total number of LC3-fluorescent puncta at the proximal axon (<100 μm from the cell body). The percentage of DsRed2-mito-positive GFP-LC3 puncta was calculated using the total number of autophagosomes observed in a kymograph along the axon (>100 μm from the end of a neurite). Mitophagic flux was determined at the axon tip as the percentage of DsRed2-mito-positive GFP-LC3 puncta per micrometer per minute observed at the distal axon tip (45–60 μm from the end of the neurite). The percentage of LysoTracker Red-positive GFP-LC3 puncta was calculated at the axon tip (<∼50 μm from the end of a neurite), the mid-axon (>100 μm from the end of a neurite) or the proximal axon (<100 μm from the cell body) at a given time point. Axon tips were outlined using threshold analysis in ImageJ.
Statistical analysis.
Statistics and graphing were performed using Prism (GraphPad) software. Comparisons of two datasets were performed using unpaired two-tailed Student's t test. Comparisons of percentage motility of autophagosomes for multiple datasets were performed using two-way ANOVA with Tukey's post hoc test. All other comparisons of multiple datasets were performed using one-way ANOVA with Tukey's post hoc test.
Results
Htt regulates autophagosome transport in neurons
Htt regulates the transport of organelles including brain-derived neurotrophic factor (BDNF)-containing vesicles and recycling endosomes (Gunawardena et al., 2003; Gauthier et al., 2004; Caviston et al., 2007; Her and Goldstein, 2008; Power et al., 2012). Here, we investigated whether htt and its adaptor protein HAP1 regulate autophagosome dynamics. Htt has been localized previously to the outer membrane of liver-isolated autophagosomes (Atwal et al., 2007; Martinez-Vicente et al., 2010). To determine whether htt and HAP1 are associated with neuronal autophagosomes, we isolated autophagosomes from mouse brain (Strømhaug et al., 1998; Morvan et al., 2009), enriching for LC3-II, the membrane-associated lipidated form of the autophagosome marker LC3 (Kabeya et al., 2000; Bampton et al., 2005; Klionsky et al., 2012). Htt and the HAP1 isoforms HAP1a and HAP1b copurified in LC3-II-enriched fractions (Fig. 1A), along with dynein, dynactin, and kinesin-1, as shown previously (Maday et al., 2012). Immunofluorescent staining of LC3 in axons from primary neurons showed ∼65% of LC3 puncta colocalized with endogenous htt (Fig. 1B,C). Endogenous HAP1 also showed >50% colocalization with LC3 puncta in axons of primary neurons (Fig. 1D).
To study the regulation of autophagosome dynamics in primary neurons, we isolated DRG neurons from GFP-LC3 transgenic mice (Mizushima et al., 2004) and analyzed GFP-LC3 labeled motility along neurites using live-cell imaging. DRG neurites extend up to 1000 μm from the soma (Perlson et al., 2009); microtubules in these neurites are uniformly polarized (Maday et al., 2012). Using this model system, we have found that autophagosomes form constitutively at neurite tips and undergo robust unidirectional transport toward the soma (Maday et al., 2012) driven by dynein and dynactin (Ravikumar et al., 2005; Jahreiss et al., 2008; Kimura et al., 2008; Lee et al., 2011; Maday et al., 2012).
We depleted htt from DRGs using fluorescently labeled siRNA; ∼80% of neurons were positive for the htt-siRNA, leading to reduced htt levels on immunoblot (Fig. 2B). Htt depletion did not significantly decrease neurite length under our conditions (mock, 507 ± 19.1 μm vs htt KD, 475 ± 15 μm; p = 0.19). Autophagosomes in control neurons showed robust motility along the neurite, with the majority (70 ± 4.8%) moving in the retrograde direction (≥10 μm/3 min); few (28 ± 4.5%) were stationary (<10 μm/3 min), as described previously (Maday et al., 2012). In contrast, htt-depleted neurons had a significantly decreased percentage of retrograde autophagosomes (45 ± 6.9%; p < 0.01) and a significant increase in bidirectional or stationary autophagosomes (52 ± 6.6%; p < 0.01; Fig. 2A,C). Further, autophagosomes in htt-depleted neurons exhibited reduced run lengths and run speeds and both net runs and individual runs were decreased (Fig. 2D,E).
To confirm specificity of knockdown, we rescued neurons depleted of endogenous htt with a full-length human htt-Q23 construct (WT-htt) resistant to the oligos used for siRNA. Rescue with WT-htt significantly increased the percentage of retrograde autophagosomes to levels similar to control neurons (Fig. 2F,G) and significantly increased autophagosome net run lengths (htt KD, 17.2 ± 2.03 μm vs htt KD + WT-htt, 25.2 ± 2.41 μm; p < 0.05) and net run speeds (htt KD, 0.10 ± 0.012 μm/s vs htt KD + WT-htt, 0.15 ± 0.015 μm/s; p < 0.05). Importantly, these effects were specific to autophagosomes because neither lysosomal nor mitochondrial velocities were reduced by htt depletion (Fig. 2H).
Htt binds dynein directly and binds both dynactin and kinesin-1 indirectly via the adaptor HAP1 (Li et al., 1995; Engelender et al., 1997; Li et al., 1998; McGuire et al., 2006; Caviston et al., 2007; Twelvetrees et al., 2010). Therefore, htt might regulate autophagosome transport either by interacting with bound motor proteins via a scaffolding activity or, alternatively, by recruiting motor proteins to autophagosomes. To test the first hypothesis that htt acts as a scaffold to regulate efficient motor function, we depleted endogenous htt from GFP-LC3 primary DRG neurons and rescued with an siRNA-resistant wild-type htt construct (WT-htt) or dominant-negative htt constructs unable to bind to either dynein (htt-Δdyn) or HAP1 (htt-ΔHAP1; Pardo et al., 2010; Fig. 3B). Although WT-htt could fully rescue defects in retrograde motility induced by htt depletion, autophagosomes in neurons expressing either htt-Δdyn or htt-ΔHAP1 demonstrated significantly disrupted autophagosome transport (Fig. 3A,C). Moreover, both htt-Δdyn and htt-ΔHAP1 were unable to rescue autophagosome net run length or speed (Fig. 3D,E). These results are consistent with the hypothesis that the interactions of htt and HAP1 with microtubule motors regulate their activity to promote efficient unidirectional transport.
We next tested the hypothesis that htt recruits motor proteins to autophagosomes. We depleted htt in DRG neurons and used live-cell imaging to determine that both dynein (Fig. 3F) and kinesin-1 (Fig. 3G) were still cotransported with GFP-LC3-labeled autophagosomes, as shown in representative images and corresponding line scans from single neurons. This suggests that motor proteins are still recruited to autophagosomes in the absence of htt, potentially by proteins such as FYCO1 (Pankiv et al., 2010). However, because we could not assess the number of bound motors quantitatively under each condition, we cannot exclude the possibility that htt depletion may decrease the number of motors recruited to autophagosomes.
Loss of HAP1 disrupts autophagosome transport
To further investigate the role of htt and HAP1 in regulating autophagosome dynamics, we depleted HAP1 in GFP-LC3-expressing DRG neurons using a fluorescently labeled siRNA. HAP1-depleted neurons demonstrated significant disruption of autophagosome transport (Fig. 4A), with an increased percentage of stationary autophagosomes (mock, 19 ± 5.8% vs HAP1 KD, 64 ± 9.5%; p < 0.001) and significantly fewer autophagosomes moving in the retrograde direction (mock, 81 ± 5.8% vs HAP1 KD, 34 ± 9.2%; p < 0.001; Fig. 4B). To confirm specificity of knockdown, we expressed an siRNA-resistant human HAP1 (hHAP1) construct, which rescued autophagosome motility to levels seen in control neurons (Fig. 4A,B). Net run speeds and net run lengths of autophagosomes were also significantly decreased by HAP1 depletion and were rescued by hHAP1 expression (Fig. 4C,D). HAP1 was shown previously to regulate neurite length in PC12 cells (Li et al., 2000; Rong et al., 2006), which have much shorter neurites than the DRGs studied here. However, we found that neurite length in primary DRG neurons at 2 DIV was not affected by HAP1 depletion (mock, 424 ± 12.3 μm vs HAP1 KD, 466 ± 23.2 μm), suggesting that the disruption of autophagosome transport induced by HAP1 depletion is not the result of reduced neurite outgrowth. Because HAP1 binds both dynactin and kinesin, we next investigated whether depleting HAP1 affected either retrograde or anterograde motor protein activity on autophagosomes.
We found that depleting HAP1 specifically decreased the net run lengths and run speeds of retrograde-directed autophagosomes (Fig. 4E,F), suggesting that the predominantly retrograde net runs of an autophagosome are regulated by HAP1. When we examined the speeds of individual runs, we found that HAP1 depletion decreased individual run speeds in both directions (Fig. 4G), suggesting that HAP1 interacts with motor proteins to enhance motor activity for both retrograde and anterograde motors. This was further supported by expressing a dominant-negative HAP1 inhibitor that blocks the HAP1-kinesin interaction (HAP1-KBD; Twelvetrees et al., 2010). HAP1-KBD expression increased the percentage of slow-moving (<0.01 μm/s) anterograde runs (Fig. 4H), whereas depleting HAP1 increased the length of individual anterograde runs (mock, 1.16 ± 0.089 μm vs HAP1 KD, 1.90 ± 0.383 μm; p < 0.05). Therefore, disruption of HAP1 function in primary neurons induces prolonged anterograde runs but at slower speeds. Together, these results support a role for HAP1 in regulating autophagosome motility by promoting efficient retrograde-directed transport.
Pathogenic polyQ-htt disrupts autophagosome dynamics
PolyQ expansions in htt cause HD, an autosomal-dominant neurodegenerative disorder in which autophagy has been found to be critical in clearing both soluble and aggregated forms of pathogenic polyQ-htt (Ravikumar et al., 2002; Qin et al., 2003). To determine whether autophagosome transport is affected by polyQ-htt, we expressed siRNA-resistant wild-type htt (Q23) and polyQ-htt (Q100) in primary neurons depleted of endogenous htt and found that autophagosome motility was inhibited by polyQ-htt expression (Fig. 5A,B). Neurons expressing polyQ-htt had a significant increase in the percentage of stationary autophagosomes (58 ± 8.5%) compared with neurons expressing wild-type htt (25 ± 5.4%; p < 0.01) and exhibited decreased run lengths and speeds (Fig. 5C–E).
Striatal neurons are among the cells selectively affected in HD, so we compared autophagosome motility in striatal cells from wild-type mice (HdhQ7/Q7) and from HD-knock-in mice (HdhQ111/Q111; Trettel et al., 2000) using live-cell imaging of mCherry-LC3. Striatal cells from HD mice demonstrated significant disruption of autophagosome dynamics with decreased net run lengths (60 ± 11.6% of wild-type levels; p < 0.05) and decreased net run speeds (61 ± 11.7% of wild-type levels; p < 0.05; Fig. 5F,G). These results further support the hypothesis that expression of polyQ-htt leads to misregulated autophagosome transport in neurons.
HD causes selective neurodegeneration, although htt is ubiquitously expressed throughout the body (Gusella and MacDonald, 2006). The cell-type specificity observed in HD has been proposed to be mediated by htt's interactions with neuronal-specific proteins (Subramaniam et al., 2009). Therefore, we investigated the ability of htt to bind to differentially expressed isoforms of dynein intermediate chain (DIC), the major cargo-binding subunit of the dynein motor (Kuta et al., 2010). The DIC1 gene (DYNC1I1) is expressed in a neuron-specific manner to produce isoforms DIC1A, DIC1B, and DIC1C, whereas the DIC2C isoform is a product of the DIC2 gene (DYNC1I2) and is expressed ubiquitously (Pfister et al., 1996a, 1996b; Myers et al., 2007; Kuta et al., 2010; Zhang et al., 2013). Here, we investigated the ability of wild-type htt and polyQ-htt to bind to the neuronal-specific isoform DIC1A compared with the ubiquitously expressed isoform DIC2C. We coexpressed myc-tagged DIC isoforms with HA-tagged htt in COS7 cells and immunoprecipitated using anti-myc antibody. Both wild-type htt and polyQ-htt bound strongly to neuronal-specific DIC1A, but much less binding to the ubiquitously expressed DIC2C isoform was observed (Fig. 5H). This preferential association of both wild-type and pathogenic htt with the neuronal-specific dynein isoform predicts that dynein-based transport of autophagosomes may be selectively impaired in neurons and this selectivity may contribute to the neuronal-selective degeneration seen in HD.
Htt is not required for autophagosome formation or cargo loading in neurons
To determine whether htt regulates other aspects of autophagy in neurons, we examined three initial steps that occur before autophagosome transport down the axon: autophagosome formation, cargo loading, and initial fusion with late endosomes. In htt-depleted primary neurons, we observed that autophagosomes continue to form, developing from small puncta into ring-like structures at the axon tip (Fig. 6A). Depleting htt did not affect autophagosome density at the axon tip, the site of constitutive autophagosome formation, nor did htt depletion affect autophagosome density along the axon (Fig. 6A).
Next, we examined cargo loading, focusing on ubiquitinated proteins and mitochondria. Autophagosomes in htt-depleted neurons still engulfed ubiquitinated proteins labeled by RFP-Ub (Fig. 6B), with all autophagosomes positive for RFP-Ub in both mock and htt-depleted neurons (mock, n = 41/41 autophagosomes vs htt-depleted, n = 30/30 autophagosomes). Autophagosomes also effectively engulfed DsRed2-mito-labeled mitochondrial fragments (Maday et al., 2012) in both mock and htt-depleted neurons (Fig. 6C).
Next, we investigated the initial fusion of autophagosomes with late endosomes, an initial step in compartment maturation (Eskelinen, 2008). After formation at the axon tip, autophagosomes acquire the late endosomal marker LAMP1 as they initiate their transit along the axon (Maday et al., 2012). We examined the colocalization of GFP-LC3 and LAMP1-RFP at both the axon tip and along the axon in control and htt-depleted neurons and saw no differences (Fig. 6D); in both cases, few autophagosomes were LAMP1 positive at the axon tip, whereas the majority of autophagosomes along the axon were LAMP1-positive, consistent with previous observations (Lee et al., 2011; Maday et al., 2012).
We next investigated whether autophagosomes in htt-depleted neurons are able to mature into autolysosomes. We used a dual-color mCherry-EGFP-LC3 reporter; the GFP moiety is preferentially quenched as the compartment matures and acidifies (Kimura et al., 2007; Pankiv et al., 2007), leading to an increase in red-only autophagosomes more proximal to the soma (Maday et al., 2012). Depleting htt did not significantly alter the percentage of red-only autophagosomes compared with control neurons (mock, 62 ± 3.8% vs htt KD, 62 ± 4.4%; Fig. 6E). In addition, htt depletion did not affect late endosome/lysosome motility (Fig. 2H) or density along the axon (mock, 6.4 ± 1.3/100 μm vs htt KD, 6.7 ± 0.9/100 μm; p = 0.84).
Disruption of autophagosome transport by htt depletion leads to accumulation of autophagosomes with undegraded mitochondrial cargo
Previous studies have found that autophagosomes undergo multiple lysosomal fusion events (Eskelinen, 2008; Yu et al., 2010) and that defective autophagosome transport in non-neuronal cells leads to inefficient autophagosome fusion with lysosomes and decreased autophagic cargo degradation (Ravikumar et al., 2005; Jahreiss et al., 2008; Kimura et al., 2008). To determine whether autophagic cargo is inefficiently cleared when autophagosome transport is defective in neurons, we examined DsRed2-mito-labeled mitochondrial degradation in htt-depleted neurons. First, we investigated whether mitochondrial morphology in the axon was disrupted by htt depletion, but found that mitochondria had similar lengths and densities in both control and htt-depleted neurons from either wild-type mice or GFP-LC3 transgenic mice (Fig. 7A,D,E). We also found that the percentage of fragmented mitochondria along the axon, a commonly used indicator of mitochondrial health (Song et al., 2011), was not increased in htt-depleted neurons (Fig. 7F).
To observe mitochondrial cargo fragments cotransporting with autophagosomes, we imaged mitochondria at increased exposure times. We found that at the axon tip, where autophagosomes first form and engulf their cargo, htt depletion had no effect on the flux of mitochondrial engulfment, as measured by autophagosomes positive for mitochondrial fragments leaving the axon tip per micrometer per minute (mock, 0.47 ± 0.19/100 μm/min vs htt KD, 0.35 ± 0.16/100 μm/min; p = 0.63), suggesting that mitochondria are engulfed in autophagosomes at equal rates in both mock and htt-depleted neurons. However, when we examined autophagosomes in the mid-axon, we found that autophagosomes in htt-depleted neurons contained a significantly higher percentage of mitochondrial fragments (21 ± 5.5%) compared with autophagosomes in mock neurons (7 ± 2.7%; p < 0.05; Fig. 7B,C,G), suggesting that mitochondria are not efficiently degraded within autophagosomes in htt-depleted neurons.
To determine whether htt depletion affected the total level of mitochondrial degradation in neurons, we blotted for the mitochondrial marker Hsp60 in lysates from DRG neurons and saw a small net increase in mitochondria protein levels in htt-depleted neurons (1.4 ± 0.3; mean ± SEM, n = 6) normalized to lysates from control neurons. This relatively limited increase is consistent with our observations from live imaging showing that total mitochondrial density along the axon is not altered, although there is an accumulation of undigested mitochondrial fragments in autophagosomes in transit along the axon.
Together, these results suggest that disruption of autophagosome transport induced by htt depletion leads to inefficient degradation of mitochondrial fragments internalized into autophagosomes along the axon. This inefficient degradation does not result from a defect in initial acidification of the autophagosome or from significant alterations in lysosomal localization. Rather, defective autophagosome transport in htt-depleted neurons may lead to decreased encounters and thus decreased fusion with lysosomes along the axon, resulting in inefficient cargo degradation (Kimura et al., 2008).
Defective autophagosome transport induced by polyQ-htt leads to inefficient degradation of engulfed mitochondria and accumulation of polyQ-htt aggregates
We next investigated whether cargo degradation was similarly affected by expression of polyQ-htt. We expressed siRNA-resistant wild-type htt (Q23) and polyQ-htt (Q100) in primary neurons depleted of endogenous htt and found that autophagosomes were still able to form at the axon tip (Fig. 8A). Expression of polyQ-htt also did not affect autophagosome density at the axon tip or autophagosome density along the axon in our cultures (Fig. 8A). To determine the effect of autophagosome transport on cargo degradation in polyQ-htt neurons, we again investigated the clearance of DsRed2-mito-labeled mitochondrial fragments by autophagosomes because defects in mitochondrial bioenergetics and dynamics have been observed previously in HD patients and animal models (Costa and Scorrano, 2012). In our neuronal cultures, mitochondria had similar lengths and densities in both WT-htt and polyQ-htt neurons (mitochondrial length: WT-htt, 1.44 ± 0.05 μm vs polyQ-htt, 1.48 ± 0.05 μm; mitochondrial density: WT-htt, 0.18 ± 0.02/μm vs polyQ-htt, 0.20 ± 0.01/μm). The percentage of fragmented mitochondria along the axon was also not altered in polyQ-htt neurons (Fig. 8D).
We also noted that polyQ-htt expression had no effect on the flux of mitochondrial engulfment, as measured by autophagosomes positive for mitochondrial fragments leaving the axon tip per micrometer per minute (WT-htt, 0.41 ± 0.20/100 μm/min vs polyQ-htt, 0.40 ± 0.17/100 μm/min; p = 0.94), indicating that mitochondria are engulfed by autophagosomes at the axon tip at equal rates in neurons expressing either wild-type htt or polyQ-htt. However, when we examined autophagosomes along the axon, we found that expression of polyQ-htt caused a dramatic and significant increase in the percentage of mitochondrial fragments accumulated in autophagosomes traveling along the axon (60 ± 10.7%) compared with WT-htt neurons (13 ± 7.8%; p < 0.05; Fig. 8B,C,E). These results further support the hypothesis that defective autophagosome transport leads to reduced lysosomal fusion events along the axon and, therefore, autophagosomes are unable to maintain an appropriate luminal environment necessary for efficient cargo degradation.
To test this hypothesis, we examined the acidification state of GFP-LC3-labeled autophagosomes in neurons expressing polyQ-htt. For this experiment, we used the pH-sensitive probe LysoTracker Red because we found that this dye labels a broader fraction of autophagosomes in the proximal axon of wild-type neurons (95.1 ± 2.7%) compared with the 62.0 ± 3.8% of autophagosomes in the proximal axon observed to exhibit red-only fluorescence from the tandem mCherry-EGFP-LC3 construct described above. Therefore, we reasoned that loss of LysoTracker Red staining would be a more comprehensive measure of changes in the acidity of this compartment. We examined autophagosomes at three distinct regions along the axon: the axon tip, the mid-axon, and the proximal axon (<100 μm from the cell body). We found that in both WT-htt and polyQ-htt neurons, few autophagosomes were acidified at the axon tip (∼30%), whereas the majority of autophagosomes along the mid-axon were LysoTracker positive (∼90%; Fig. 8G), which is consistent with previous observations in wild-type neurons (Maday et al., 2012). However, when we examined autophagosomes that had reached the proximal axon (closest to the cell body), we found a significant decrease in the percentage of autophagosomes that were LysoTracker positive in polyQ-htt neurons compared with WT-htt neurons (p < 0.01; Fig. 8F,G). These results are consistent with the hypothesis that autophagosomes undergoing disrupted transport in polyQ-htt neurons are unable to maintain an appropriate level of acidification necessary for efficient cargo degradation, possibly due to reduced lysosomal fusion events along the axon.
In HD, both soluble and aggregated pathogenic polyQ-htt are predominantly cleared by autophagy (Ravikumar et al., 2002; Qin et al., 2003). We used live-cell imaging of primary neurons to further examine the role of autophagosomes in clearing polyQ-htt from the distal axon. Both the disease-associated N-terminal fragment of polyQ-htt (GFP-htt-68Q) and full-length polyQ-htt (mCherry-htt-Q100) were observed to cotransport with autophagosomes from the axon tip back to the cell body (Fig. 9A,B). We also saw visible aggregates forming along the cell body and axon and accumulating at the axon tip (Fig. 9C), suggesting that early disruption of autophagosome dynamics may contribute to ineffective autophagic clearance of polyQ-htt in HD neurons, particularly from the distal axon and axon tip.
Discussion
Autophagosomes form constitutively at the axon tip in neurons and undergo robust retrograde transport toward the soma (Lee et al., 2011; Maday et al., 2012). Here, we show that a htt/HAP1 complex drives retrograde-directed autophagosome transport by regulating motor protein activity, likely through a scaffolding mechanism. Htt is not required for autophagosome formation or cargo loading in neurons. Instead, defects in htt-mediated autophagosome transport lead to inefficient cargo degradation along the axon. Further, polyQ-htt-expressing neurons demonstrate defective autophagosome transport and inhibited degradative function, potentially contributing to inefficient autophagic polyQ-htt clearance in HD.
How might htt and HAP1 regulate autophagosome transport dynamics? Htt localizes to the outer membrane of autophagosomes (Atwal et al., 2007; Martinez-Vicente et al., 2010). Both htt and HAP1 are present in autophagosome-enriched brain fractions along with microtubule motors and colocalize with LC3-positive autophagosomes along axons of primary neurons. Autophagosome motility is driven by the retrograde dynein-dynactin complex (Jahreiss et al., 2008; Kimura et al., 2008; Lee et al., 2011; Maday et al., 2012) and the anterograde motor kinesin (Maday et al., 2012). We show here that htt is likely not required for motor recruitment to autophagosomes, but rather acts to regulate motor activity. Htt binds directly to dynein (Caviston et al., 2007); here, we found that abolishing the htt/dynein interaction affects the processivity of dynein, resulting in reduced autophagosome run lengths and speeds.
Abolishing the htt/HAP1 interaction also disrupted autophagosome transport, resulting in more stationary autophagosomes. Because HAP1 binds to both dynactin, a required activator for retrograde transport, and kinesin-1, a major motor for anterograde transport (Engelender et al., 1997; Li et al., 1998; McGuire et al., 2006; Twelvetrees et al., 2010), HAP1 may play a key role in coordinating bidirectional motor activity. HAP1 has been shown to regulate kinesin-dependent amyloid precursor protein (APP) transport (McGuire et al., 2006; Yang et al., 2012) and anterograde GABA receptor trafficking (Twelvetrees et al., 2010). Here, we show that depleting HAP1 affected both retrograde and anterograde motility of autophagosomes, resulting in decreased velocities in both directions. In the retrograde direction, the htt/HAP1 complex may favor dynein-mediated motility by enhancing the affinity of the dynein-dynactin interaction through the formation of a quaternary (dynein-dynactin-HAP1-htt) complex. Because depleting HAP1 also prolonged anterograde runs, but at slower speeds, HAP1 may also promote efficient autophagosome transport by limiting the frequency or length of anterograde runs. Because similar changes in autophagosome transport were observed upon depletion of HAP1 and expression of htt-ΔHAP1, htt and HAP1 are likely acting via the same pathway, consistent with previous studies (Gauthier et al., 2004). Together, these results suggest that the htt/HAP1 scaffolding complex promotes efficient retrograde motility by enhancing dynein-dynactin-driven movement and limiting kinesin-driven movement, leading to robust unidirectional autophagosome transport toward the soma (Fig. 10A). The activity of this scaffolding complex may be carefully regulated in vivo by posttranslational modifications by the serine/threonine kinase Akt (Colin et al., 2008) or JNK (cJun N-terminal kinase) activity, which is upregulated in HD (Morfini et al., 2009). Because htt and HAP1 colocalize with a subpopulation of LC3-positive autophagosomes, it is also possible that other scaffolding proteins may regulate the transport of distinct or overlapping subpopulations of autophagosomes; our preliminary data also implicate the JNK-binding scaffolding protein JIP1 in the regulation of autophagosome dynamics (M.M. Fu and E.L.F.H., unpublished observations).
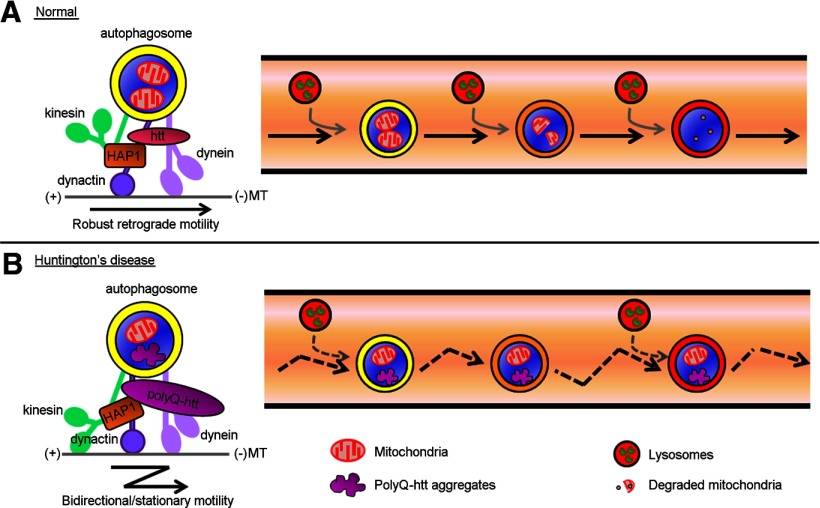
Figure 10.
Model of htt/HAP1's regulation of autophagosome dynamics in neurons. A, Htt and HAP1 regulate the motor activity and processivity of microtubule motors dynein, dynactin, and kinesin on autophagosomes via interactions among htt, HAP1, and neuronal-specific dynein isoforms to drive robust retrograde transport of autophagosomes back to the cell body in neurons along microtubules (MT). Retrograde autophagosome transport is necessary for efficient fusion with lysosomes for degradation of autophagic cargo such as mitochondria. B, In HD, pathogenic polyQ-htt disrupts the htt/HAP1 motor protein complex on autophagosomes via altered polyQ-htt/HAP1 association. This misregulation of motors leads to bidirectional/stationary autophagosome dynamics in HD neurons, thereby disrupting the retrograde transport of autophagosomes necessary for efficient degradation of dysfunctional mitochondria and polyQ-htt.
Interactions of htt with neuronal-specific proteins have been proposed to contribute to selective cell death in HD (Subramaniam et al., 2009). Our observations suggest that both normal and polyQ-htt interact preferentially with the neuronal-specific dynein isoform DIC1A compared with the more ubiquitously expressed dynein isoform DIC2C. Therefore, dynein-driven organelle transport may be selectively impaired in neurons and may contribute to HD neurodegeneration. Previous studies have shown that polyQ-htt expression disrupts the anterograde transport of APP-carrier vesicles and the bidirectional transport of BDNF and mitochondria (Gunawardena et al., 2003; Szebenyi et al., 2003; Gauthier et al., 2004; Lee et al., 2004; Trushina et al., 2004; Chang et al., 2006; Orr et al., 2008; Zala et al., 2008; Her and Goldstein, 2008; Song et al., 2011). Here, we show that polyQ-htt disrupts the predominantly retrograde transport of autophagosomes. Because both wild-type htt and polyQ-htt bind to dynein, autophagosome transport may be disrupted by the increased affinity of polyQ-htt for HAP1 described previously (Li et al., 1995). An alteration in the polyQ-htt/HAP1 motor protein complex may affect either dynein or kinesin motor activity or a combination of both; specifically, a loss of motor coordination may be the basis for the decreased autophagosome motility we have observed in HD neurons (Fig. 10B).
In htt-depleted neurons, we find that autophagosomes still form and still internalize cargo, including ubiquitinated proteins and mitochondria. Autophagosomes also undergo an initial fusion with endosomes, becoming LAMP1 positive upon exit from the axon tip and acidified, as measured by GFP quenching. However, htt depletion caused a significant increase in the fraction of autophagosomes containing undigested mitochondrial fragments. We investigated whether this increase reflected an overall increase in mitophagy, possibly upregulated in response to impaired mitochondrial health. However, we found that mitochondria health, as measured by density, motility, and percent fragmentation, was not affected in htt-depleted neurons. Moreover, the rate of mitochondrial engulfment into autophagosomes at the axon tip was not altered by htt depletion, demonstrating that loss of htt does not increase mitophagic flux. In neurons expressing pathogenic polyQ-htt, we noted an even more pronounced accumulation of undigested mitochondrial fragments within autophagosomes along the axon, again, in the absence of any evidence indicating an increase in mitophagic flux. Together, these observations support the hypothesis that defects in autophagosome transport lead to inefficient cargo degradation (Fig. 10B).
We propose that the underlying mechanism linking these two observations is that transport inhibition leads to reduced lysosomal fusion events, which may lead to insufficient accumulation of degradative enzymes within this compartment (Ravikumar et al., 2005; Jahreiss et al., 2008; Kimura et al., 2008) and a failure to maintain the acidified environment required for efficient degradation of engulfed proteins and organelles. In support of this interpretation, defects in autophagosome transport have been associated previously with impaired autophagic clearance and neurodegeneration (Ravikumar et al., 2005; Ikenaka et al., 2013). We found that autophagosomes in htt-depleted neurons still acquire the late endosomal marker LAMP1 and become acidified, suggesting that autophagosome-lysosome fusion is not fully impaired (Jahreiss et al., 2008). Htt depletion also did not affect lysosomal motility or density in neurons compared with observations in non-neuronal cells (Caviston et al., 2011), suggesting that inefficient cargo degradation was not caused by disrupted lysosomal localization. Together, our results indicate that defective autophagosome transport, rather than defects in initial acidification or altered lysosomal localization, lead to reduced lysosomal fusion events and inefficient cargo degradation along the axon. In further support of this interpretation, we found that both WT-htt- and polyQ-htt-expressing neurons had similar levels of LysoTracker-positive autophagosomes in the mid-axon. However, at the proximal axon, polyQ-htt-expressing neurons had lower levels of LysoTracker-positive autophagosomes, suggesting that polyQ-htt both disrupts autophagosome transport and causes a failure to maintain the acidified environment of autophagosomes as they move toward the proximal axon/cell body. The autophagy receptor optineurin (Wild et al., 2011), which binds htt, was found recently to mediate autophagosome maturation via the actin-based motor myosin VI (Tumbarello et al., 2012). Here, we show that htt mediates efficient autophagosome maturation and cargo degradation by regulating microtubule-based motors to promote efficient autophagosome transport and this role is disrupted by expression of expanded polyQ-htt.
In HD models, inefficient clearance of polyQ-htt via autophagy (Ravikumar et al., 2002; Qin et al., 2003) accelerates the formation of toxic polyQ-htt oligomers and ultimately results in cell death (Ravikumar et al., 2004; Sarkar et al., 2007). Because disease-associated fragments and full-length polyQ-htt are cleared from the distal axon by retrograde autophagosome transport in neurons, early defects in transport may lead to inefficient polyQ-htt clearance, which may further clog autophagosomes and inhibit the degradative machinery. In addition, inefficient clearance of mitochondrial fragments may contribute to defective mitochondrial bioenergetics in HD (Costa and Scorrano, 2012). In summary, we have identified htt and HAP1 as regulators of autophagosome dynamics and function in neurons. PolyQ-htt disrupts this complex, causing disrupted autophagosome transport and inefficient clearance of mitochondrial fragments and pathogenic polyQ-htt, potentially contributing to neuronal death in HD patients.
Footnotes
This work was supported by National Institutes of Health (Grant R01 NS060698 to E.L.F.H.) and the Hearst Foundation (2011 Fellowship to Y.C.W.). We thank Swathi Ayloo for autophagosome purification and Mariko Tokito and Karen Wallace for technical assistance.
The authors declare no competing financial interests.
References
- Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualizes in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005;1:23–36. doi: 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Ross JL, Antony SM, Tokito M, Holzbaur EL. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci U S A. 2007;104:10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP, Zajac AL, Tokito M, Holzbaur EL. Huntingtin coordinates the dynein-mediated dynamic positioning of endosomes and lysosomes. Mol Biol Cell. 2011;22:478–492. doi: 10.1091/mbc.E10-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DT, Rintoul GL, Pandipati S, Reynolds IJ. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol Dis. 2006;22:388–400. doi: 10.1016/j.nbd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Colin E, Zala D, Liot G, Rangone H, Borrell-Pagès M, Li XJ, Saudou F, Humbert S. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa V, Scorrano L. Shaping the role of mitochondria in the pathogenesis of Huntington's disease. EMBO J. 2012;31:1853–1864. doi: 10.1038/emboj.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, Holzbaur EL, Ross CA. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL. New insights into the mechanisms of macroautophagy in mammalian cells. Int Rev Cell Mol Biol. 2008;266:207–247. doi: 10.1016/S1937-6448(07)66005-5. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pagès M, Dompierre JP, Rangone H, Cordelières FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, Saudou F. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B, Sintasath L, Bonini NM, Goldstein LS. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/S0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME. Huntington's disease: seeing the pathogenic process through a genetic lens. Trends Biochem Sci. 2006;31:533–540. doi: 10.1016/j.tibs.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Her LS, Goldstein LS. Enhanced sensitivity of striatal neurons to axonal transport defects induced by mutant huntingtin. J Neurosci. 2008;28:13662–13672. doi: 10.1523/JNEUROSCI.4144-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenaka K, Kawai K, Katsuno M, Huang Z, Jiang YM, Iguchi Y, Kobayashi K, Kimata T, Waza M, Tanaka F, Mori I, Sobue G. dnc-1/dynactin 1 knockdown disrupts transport of autophagosomes and induces motor neuron degeneration. PLoS One. 2013;8:e54511. doi: 10.1371/journal.pone.0054511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33:109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kuta A, Deng W, Morsi El-Kadi A, Banks GT, Hafezparast M, Pfister KK, Fisher EM. Mouse cytoplasmic dynein intermediate chains: identification of new isoforms, alternative splicing and tissue distribution of transcripts. PLoS One. 2010;5:e11682. doi: 10.1371/journal.pone.0011682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sato Y, Nixon RA. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J Neurosci. 2011;31:7817–7830. doi: 10.1523/JNEUROSCI.6412-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Yoshihara M, Littleton JT. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington's disease. Proc Natl Acad Sci U S A. 2004;101:3224–3229. doi: 10.1073/pnas.0400243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Gutekunst CA, Hersch SM, Li XJ. Interaction of huntingtin-associated protein with dynactin P150Glued. J Neurosci. 1998;18:1261–1269. doi: 10.1523/JNEUROSCI.18-04-01261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Li H, Torre ER, Li XJ. Expression of huntingtin-associated protein-1 in neuronal cells implicates a role in neuritic growth. Mol Cell Neurosci. 2000;16:168–183. doi: 10.1006/mcne.2000.0858. [DOI] [PubMed] [Google Scholar]
- Li XJ, Li SH, Sharp AH, Nucifora FC, Jr, Schilling G, Lanahan A, Worley P, Snyder SH, Ross CA. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire JR, Rong J, Li SH, Li XJ. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J Biol Chem. 2006;281:3552–3559. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfini GA, You YM, Pollema SL, Kaminska A, Liu K, Yoshioka K, Björkblom B, Coffey ET, Bagnato C, Han D, Huang CF, Banker G, Pigino G, Brady ST. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci. 2009;12:864–871. doi: 10.1038/nn.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan J, Köchl R, Watson R, Collinson LM, Jefferies HB, Tooze SA. In vitro reconstitution of fusion between immature autophagosomes and endosomes. Autophagy. 2009;5:676–689. doi: 10.4161/auto.5.5.8378. [DOI] [PubMed] [Google Scholar]
- Myers KR, Lo KW, Lye RJ, Kogoy JM, Soura V, Hafezparast M, Pfister KK. Intermediate chain subunit as a probe for cytoplasmic dynein function: biochemical analyses and live cell imaging in PC12 cells. J Neurosci Res. 2007;85:2640–2647. doi: 10.1002/jnr.21213. [DOI] [PubMed] [Google Scholar]
- Orr AL, Li S, Wang CE, Li H, Wang J, Rong J, Xu X, Mastroberardino PG, Greenamyre JT, Li XJ. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjørkøy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo R, Molina-Calavita M, Poizat G, Keryer G, Humbert S, Saudou F. pARIS-htt: an optimised expression platform to study huntingtin reveals functional domains required for vesicular trafficking. Mol Brain. 2010;3:17. doi: 10.1186/1756-6606-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlson E, Jeong GB, Ross JL, Dixit R, Wallace KE, Kalb RG, Holzbaur EL. A switch in retrograde signaling from survival to stress in rapid-onset neurodegeneration. J Neurosci. 2009;29:9903–9917. doi: 10.1523/JNEUROSCI.0813-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Salata MW, Dillman JF, 3rd, Torre E, Lye RJ. Identification and developmental regulation of a neuron-specific subunit of cytoplasmic dynein. Mol Biol Cell. 1996a;7:331–343. doi: 10.1091/mbc.7.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Salata MW, Dillman JF, 3rd, Vaughan KT, Vallee RB, Torre E, Lye RJ. Differential expression and phosphorylation of the 74-kDa intermediate chains of cytoplasmic dynein in cultured neurons and glia. J Biol Chem. 1996b;271:1687–1694. doi: 10.1074/jbc.271.3.1687. [DOI] [PubMed] [Google Scholar]
- Power D, Srinivasan S, Gunawardena S. In-vivo evidence for the disruption of Rab11 vesicle transport by loss of huntingtin. Neuroreport. 2012;23:970–977. doi: 10.1097/WNR.0b013e328359d990. [DOI] [PubMed] [Google Scholar]
- Qin ZH, Wang Y, Kegel KB, Kazantsev A, Apostol BL, Thompson LM, Yoder J, Aronin N, DiFiglia M. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum Mol Genet. 2003;12:3231–3244. doi: 10.1093/hmg/ddg346. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O'Kane CJ, Brown SD, Rubinsztein DC. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- Rong J, McGuire JR, Fang ZH, Sheng G, Shin JY, Li SH, Li XJ. Regulation of intracellular trafficking of huntingtin-associated protein-1 is critical for TrkA protein levels and neurite outgrowth. J Neurosci. 2006;26:6019–6030. doi: 10.1523/JNEUROSCI.1251-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O'Kane CJ, Schreiber SL, Rubinsztein DC. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, Masliah E, Ellisman M, Rouiller I, Schwarzenbacher R, Bossy B, Perkins G, Bossy-Wetzel E. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strømhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335:217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G, Morfini GA, Babcock A, Gould M, Selkoe K, Stenoien DL, Young M, Faber PW, MacDonald ME, McPhaul MJ, Brady ST. Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron. 2003;40:41–52. doi: 10.1016/S0896-6273(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- Trushina E, Dyer RB, Badger JD, 2nd, Ure D, Eide L, Tran DD, Vrieze BT, Legendre-Guillemin V, McPherson PS, Mandavilli BS, Van Houten B, Zeitlin S, McNiven M, Aebersold R, Hayden M, Parisi JE, Seeberg E, Dragatsis I, Doyle K, Bender A, Chacko C, McMurray CT. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol. 2012;14:1024–1035. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL, MacAskill AF, Rostaing P, Lumb MJ, Humbert S, Triller A, Saudou F, Yan Z, Kittler JT. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron. 2010;65:53–65. doi: 10.1016/j.neuron.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dötsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yang GZ, Yang M, Lim Y, Lu JJ, Wang TH, Qi JG, Zhong JH, Zhou XF. Huntingtin associated protein 1 regulates trafficking of the amyloid precursor protein and modulates amyloid beta levels in neurons. J Neurochem. 2012;122:1010–1022. doi: 10.1111/j.1471-4159.2012.07845.x. [DOI] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zala D, Colin E, Rangone H, Liot G, Humbert S, Saudou F. Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Hum Mol Genet. 2008;17:3837–3846. doi: 10.1093/hmg/ddn281. [DOI] [PubMed] [Google Scholar]
- Zhang J, Twelvetrees AE, Lazarus JE, Blasier KR, Yao X, Inamdar NA, Holzbaur EL, Pfister KK, Xiang X. Establishing a novel knock-in mouse line for studying neuronal cytoplasmic dynein under normal and pathologic conditions. Cytoskeleton. 2013;70:215–227. doi: 10.1002/cm.21102. [DOI] [PMC free article] [PubMed] [Google Scholar]