Abstract
Plp1 gene expression occurs very early in development, well before the onset of myelination, creating a conundrum with regard to the function of myelin proteolipid protein (PLP), one of the major proteins in compact myelin. Using PLP-EGFP mice to investigate Plp1 promoter activity, we found that, at very early time points, PLP-EGFP was expressed in Sox2+ undifferentiated precursors in the spinal cord ventricular zone (VZ), as well as in the progenitors of both neuronal and glial lineages. As development progressed, most PLP-EGFP-expressing cells gave rise to oligodendrocyte progenitor cells (OPCs). The expression of PLP-EGFP in the spinal cord was quite dynamic during development. PLP-EGFP was highly expressed as cells delaminated from the VZ. Expression was downregulated as cells moved laterally through the cord, and then robustly upregulated as OPCs differentiated into mature myelinating oligodendrocytes. The presence of PLP-EGFP expression in OPCs raises the question of its role in this migratory population. We crossed PLP-EGFP reporter mice into a Plp1-null background to investigate the role of PLP in early OPC development. In the absence of PLP, normal numbers of OPCs were generated and their distribution throughout the spinal cord was unaffected. However, the orientation and length of OPC processes during migration was abnormal in Plp1-null mice, suggesting that PLP plays a role either in the structural integrity of OPC processes or in their response to extracellular cues that orient process outgrowth.
Keywords: migration, myelination, oligodendrocyte, OPC, PLP, proteolipid protein
Introduction
Myelin proteolipid protein (PLP) is one of the most abundant components of the myelin sheath in the CNS, comprising ∼50% of the total protein content (Eng et al., 1968). Intriguingly, long before myelin production commences, transcripts encoding PLP, and its shorter isoform DM20, are expressed in a subset of progenitors (LeVine et al., 1990; Timsit et al., 1992; Dickinson et al., 1996). Plp1-expressing cells give rise to oligodendrocytes and astrocytes, and to subtypes of thalamic and medullary neurons (Spassky et al., 1998; Delaunay et al., 2008; Guo et al., 2009; Miller et al., 2009; Michalski et al., 2011). The function of this important myelin structural protein in neuronal and glial progenitors remains unclear, though our previous work (Gudz et al., 2002, 2006) suggests PLP may integrate extracellular signals and regulate migratory behavior of oligodendrocyte progenitor cells (OPCs).
Previous studies investigated Plp1 expression in neuronal and glial progenitors by crossing Plp-Cre mice with floxed reporter strains. This permanently labels Plp1-expressing cells as well as their progeny, which may or may not continue to express Plp1 (Guo et al., 2009; Michalski et al., 2011). By contrast, in PLP-EGFP mice, only cells currently expressing the Plp1 promoter were labeled (Mallon et al., 2002). Thus, we were able to study the dynamics of Plp/Dm20 expression by tracking the migration and fates of embryonic and postnatal cells actively expressing PLP-EGFP. In agreement with earlier studies, both neuronal and glial precursors had robust Plp1 promoter activity at early embryonic stages (indicated by intense EGFP expression). In addition, migratory glial cells continued to display strong Plp1 promoter activity, which was then downregulated in astrocytes. OPCs also downregulated Plp1 promoter activity as they reached the lateral spinal cord, but then upregulated it significantly during postnatal myelination.
There has been debate about the origin of OPCs in the developing CNS, specifically whether early Plp-expressing and platelet-derived growth factor receptor α (PDGFRα)-expressing cells represent two distinct progenitor populations or different stages of a single progenitor (Spassky et al., 1998, 2000; Richardson et al., 2000). Here, we observed that initially all Olig2/PDGFRα-expressing OPCs were PLP-EGFP+. As OPCs moved laterally through the developing spinal cord, PLP-EGFP was downregulated, leading to a mixed population of PLP-EGFP+/PDGFRα−, PLP-EGFP+/PDGFRα+, or PLP-EGFP−/PDGFRα+ cells throughout the cord. Therefore, varying reports of the contributions of each “lineage” to the overall OPC population may have resulted from analyses of this mixed population.
While earlier studies established that the Plp1 promoter and mRNA are expressed in early progenitors (Timsit et al., 1992; Mallon et al., 2002), we report that PLP/DM20 protein is also present in embryonic OPCs. To assess a role for PLP in early neuronal and glial progenitors, we examined their development in Plp1-null mice. During migration, processes of Plp1-null OPCs were disorganized, with many failing to adopt a radial orientation. However, despite abnormal process extension, neither OPC distribution nor the onset of myelination was disrupted in the absence of PLP/DM20.
Materials and Methods
Animals.
Homozygous transgenic Plp-EGFP-3′UTR mice (PLP-EGFP; Mallon et al., 2002) were bred with Plp1-null mice (Klugmann et al., 1997) on a C57BL/6J background (Jackson Labs). The day of insemination, detected by a vaginal plug, was defined as embryonic day (E) 0.5. Embryonic sex (McClive and Sinclair, 2001) and Plp genotypes (Klugmann et al., 1997) were determined by PCR as previously described. Plp1 is on the X chromosome; therefore males carrying the null allele express no PLP/DM20. Plp1-null and wild-type littermate males were used for experiments. All animal procedures were conducted with the approval of the University of Colorado Institutional Animal Care and Use Committee.
Immunostaining.
The following primary antibodies were used: rat anti-PLP/DM20 monoclonal antibody (1:100; clone AA3; Yamamura et al., 1991), rat anti-PDGFRα (1:400; BD PharMingen), rabbit anti-Olig2 (1:10,000; a gift from Dr. Charles Stiles, Harvard University), mouse anti-NeuN (1:400; EMD Millipore), mouse anti-GFAP (1:600), mouse anti-S100β (1:400), rabbit anti-S100β (1:500; Sigma-Aldrich), mouse anti-Nestin (1:120; Abcam), rabbit anti-Sox2 (1:1000; Millipore Bioscience Research Reagents). Pregnant females were killed via cervical dislocation; embryos were removed and immersion fixed in 4% paraformaldehyde in 0.1 m PBS (PFA) overnight at 4°C. Postnatal animals were transcardially perfused with 4% PFA. Spinal cords were removed and postfixed overnight in 4% PFA at 4°C. Tissue was cryoprotected in 30% sucrose for 2–5 nights, depending on age, embedded in O.C.T. embedding medium (Sakura Finetek USA), and sectioned on a cryostat (20 μm; Leica CM1950, Leica-Microsystems). Sections were incubated with 5% normal donkey serum in 0.1 m PBS with 0.3% Triton X-100 for 60 min at room temperature (RT), and then incubated with primary antibodies overnight at 4°C, except for the AA3 antibody, which was incubated for 7 nights at 4°C or overnight at RT on E16.5 tissue. Sections were subsequently incubated with the appropriate fluorescently conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) for 1 h at RT before being stained with Hoechst (1:25,000 in 0.1 m PBS) and coverslipped with Fluoromount-G (SouthernBiotech).
In situ hybridization.
Digoxigenin-labeled cRNA probes (sense and antisense) were prepared using T3-RNA or T7-RNA polymerase. The probe specific for PLP covered the full coding region (Sorg et al., 1987). Fixation and hybridization of fresh frozen cryostat sections was performed as described previously (Fuss et al., 1997), with modifications. Briefly, 20 μm cryostat sections were fixed in 4% PFA in PBS, pH 7.4, and then washed in PBS. Sections were treated with 5 μg/ml proteinase K for 4 min, refixed in 4% PFA for 20 min, washed in PBS, and acetylated for 10 min. After acetylation, sections were prehybridized at 60°C in hybridization buffer (50% formamide, 5× SSC, 50 ng/ml tRNA, 50 μg/ml heparin, 1% SDS). Hybridization of probe (0.13 ng/ml in hybridization buffer) was performed at 60°C overnight. Sections were washed in prewarmed 5× SSC for 30 min at RT, followed by washes in prewarmed 0.2× SSC at 65°C. Bound cRNA was detected using an alkaline phosphatase-coupled antibody to digoxigenin with subsequent color development BM Purple Substrate (Roche Diagnostics).
Cell counts and measurements of process lengths and orientations.
Spinal cord sections from wild-type and Plp1-null littermates were matched based on spinal cord segment, and tile-scan images were taken on a Leica SP5 confocal microscope with LAS software (Leica-Microsystems) using either a 25× water-immersion objective (aperture 0.95) or a 40× oil-immersion objective (aperture 0.75–1.25) at RT. Tile-scans were merged using the Mosaic Merge module of the LAS software to reconstruct each spinal cord section. Images were adjusted for contrast and brightness, and cropped in Photoshop (Adobe Systems). Annotations were made in Illustrator (Adobe Systems). For cell counts, images were opened in Fiji (Schindelin et al., 2012) and single-labeled, double-labeled, and triple-labeled cells were counted using the Cell Counter plugin (http://rsbweb.nih.gov/ij/plugins/cell-counter.html, Kurt De Vos). To measure processes, the six longest processes per region per section were measured with the Simple Neurite Tracer plugin (Longair et al., 2011). Process orientation was measured only for bipolar cells from which both full processes could be accurately imaged. Orientation was considered tangential if parallel to midline, radial if the processes would eventually contact both the midline and the pial surface, and all other orientations were classified as “other” (Leber and Sanes, 1995). Measurements were made by a blinded observer.
Western blotting.
Spinal cord extracts were prepared in RIPA buffer containing 1% Triton X-100 with a protease inhibitor mixture (Roche). Proteins were quantified and analyzed by SDS-PAGE, after which proteins were transferred onto PVDF membrane and blocked for 1 h with 5% BSA in TBST. Primary antibodies [myelin basic protein (MBP), SMI94, 1:1000; Covance Research Products; PLP/DM20, clone AA3, 1:100; GAPDH, 14C10, 1:5000; β-tubulin, 1:5000; Cell Signaling Technology] were incubated with the membranes overnight at 4°C. Near-infrared fluorescent secondary antibodies (LI-COR Biosciences) were used at 1:10,000. Proteins were visualized on an Odyssey Infrared Imaging System (LI-COR). MBP levels were normalized to GAPDH or β-tubulin levels for each sample. Quantification was done on 3–5 cords at each time point and the normalized levels of MBP are presented as the mean ± SEM.
Live imaging of OPC migration.
OPCs derived from oligospheres were generated as previously described (Pedraza et al., 2008). Chamber slides (iBidi) were coated with poly-d-lysine (10 μg/ml; Sigma-Aldrich) for 1 h at 37°C, washed, and then coated in fibronectin (10 μg/ml; EMD Millipore) overnight at 37°C. Oligospheres (passage 2–5) were dissociated using NeuroCult (Stem Cell Technologies), plated at a density of 30,000 cells per well in OPC media [DMEM:F12 (Invitrogen Life Technologies), 1× B27 supplement (Invitrogen), PDGF, and FGF (10 ng/ml; Peprotech)], and incubated at 37°C for 24 h. Media was changed just before imaging. Imaging was conducted on an inverted Leica SP5 confocal microscope equipped with a LiveCell imaging chamber (Pathology Devices) for 11 h. Cells were manually tracked using the ImageJ plugin Manual Tracking (http://rsb.info.nih.gov/ij/plugins/track/track.html, Fabrice Cordelires). Tracks of individual cells were analyzed in the Chemotaxis and Migration Tool plugin for ImageJ (iBidi) to determine the average accumulated distance and velocity.
Statistical analysis.
Cell counts were performed on ≥3 spinal cord sections from three wild-type and four Plp1-null embryos across two litters at E14.5, from six wild-type and five Plp1-null embryos across three litters at E16.5, and from five wild-type and four Plp1-null pups across three litters at postnatal day (P) 7. Process measurements were made on three wild-type and three Plp1-null animals at E16.5. Cell migration measurements where made on 291 wild-type cells and 231 Plp1-null cells, across four different experiments. Statistical analyses were performed using one-way ANOVA for group comparisons; a two-tailed, unpaired Student's t test for single comparisons; or a Mann–Whitney U test for population distributions using Prism 6 for Mac OS X (GraphPad Software); p values <0.05 were considered significant.
Results
In the spinal cord, PLP-EGFP-labeled cells in the ventricular zone/subventricular zone (VZ/SVZ) migrated laterally to populate the developing white mater
PLP-EGFP mice were used to track the development of embryonic and postnatal spinal cord oligodendrocytes. In these mice, Plp1 promoter activity drives EGFP expression. At E12.5, robust EGFP expression was present in the VZ/SVZs that surround the central canal of the ventral spinal cord (Fig. 1A). Cells in the ventral spinal cord went on to extend long processes toward the pial surface and migrate laterally (E16.5; Fig. 1B) until they reached the presumptive white matter. By P7, PLP-EGFP expression increased dramatically in white matter regions of myelinated CNS axon tracts (Fig. 1C). Strong transgene expression was also seen in the dorsal root ganglia (Fig. 1A,B), which also express PLP (Wight et al., 1993; Mallon et al., 2002). In situ hybridization of semiadjacent sections demonstrated that Plp/Dm20 mRNA was expressed in the same pattern as PLP-EGFP at E12.5 (D), E16.5 (E), and P7 (F). Although Plp/Dm20 transcripts have been found in the developing spinal cord (Timsit et al., 1992; Dickinson et al., 1996; and others), PLP/DM20 protein has not been observed there. However, incubation of E16.5 sections with PLP/DM20 antibody (AA3) for 7 d at 4°C, or overnight at RT, allowed for detection of PLP/DM20 protein in multiprocessed PLP-EGFP+ cells (Fig. 1G). PLP/DM20 was located in OPC processes and cell bodies, as well as in the distal tips of OPC processes that did not contain the cytosolic EGFP (Fig. 1G, arrow). This suggests that the difficulty in detecting PLP/DM20 protein in bipolar embryonic migratory cells may result from the localization of the protein to the distal tips of processes that cannot be definitively linked to specific cells.
Figure 1.
Dynamic Plp1 expression in the developing spinal cord. A, At E12.5, PLP-EGFP expression (green) was very robust in cells in the VZ. B, At E16.5, fewer cells within the VZ expressed PLP-EGFP, and PLP-EGFP+ cells were dispersed throughout the ventral and dorsal regions of the cord. C, At P7, no cells in the VZ remained PLP-EGFP+ and robust PLP-EGFP expression was seen in the lateral white matter. D–F, PLP/DM20 mRNA expression corresponded to PLP-EGFP expression in the developing spinal cord at E12.5 (D), E16.5 (E), and P7 (F). G, PLP expression in PLP-EGFP+ oligodendrocyte progenitors in the E16.5 spinal cord. Immunostaining for PLP (red) colocalized with EGFP expression in oligodendrocyte progenitor cells in the spinal cord. PLP (red) could be seen in the distal tips of OPCs while PLP-EGFP localized to the cell body as well as processes. Scale bar: A, B, 50 μm; C, 200 μm; G, 10 μm.
PLP-EGFP-labeled precursors of multiple lineages are present in the VZ/SVZ of the E12.5 spinal cord
To identify the Plp1-expressing cells in the E12.5 VZ/SVZ, we studied the expression of markers for neuronal and glial precursors in PLP-EGFP embryos (Fig. 2). Many PLP-EGFP+ cells immediately adjacent to the central canal were immunopositive for Sox2 (Fig. 2A), indicating that they were yet undifferentiated multipotent precursors (Avilion et al., 2003; Ellis et al., 2004; Masui et al., 2007). A small group of Pax6-expressing cells, which give rise to ventral interneurons and motor neurons (Ericson et al., 1997), partially overlapped with the PLP-EGFP domain. In particular, some Pax6+/PLP-EGFP+ cells (Fig. 2B, arrows) were at the dorsal-most limit of the PLP-EGFP domain of the VZ/SVZ. Because a subset of OPCs are known to express Nkx2.2 (Qi et al., 2001; Fu et al., 2002), we also examined Nkx2.2 expression. Although occasional Nkx2.2+/PLP-EGFP+ cells were seen in the VZ (Fig. 2C, arrow), Nkx2.2+ cells were predominantly located outside of the PLP-EGFP-expressing domain. Olig2 expression was studied because OPCs arise from Olig2-expressing cells of the motor neuron progenitor (pMN) domain (Lu et al., 2002; Park et al., 2002). At E12.5, Olig2+ cells were concentrated in the VZ/SVZ of the pMN, and all were positive for PLP-EGFP (Fig. 2D, arrows; Table 1). Another marker of OPCs, PDGFRα, was expressed by a subset of cells delaminating out of the VZ, which were also positive for PLP-EGFP (Fig. 2E, arrows). As reported previously (Delaunay et al., 2008), Nestin+/PLP-EGFP+ radial glia were also seen, with long processes that extended conical end feet to the pial surface (see Fig. 7). These cells are known to differentiate into astrocytes, with a small subset differentiating into oligodendrocytes (McMahon and McDermott, 2001; Fogarty et al., 2005; McDermott et al., 2005).
Figure 2.
PLP-EGFP is expressed in multiple progenitor populations in the VZ of the E12.5 spinal cord. A, Many PLP-EGFP-expressing cells (green) were unspecified Sox2-positive (red) progenitors (arrows). B, At the dorsal limit of PLP-EGFP expression in the VZ, a small number of cells (arrows) were Pax6-positive (red). C, Most Nkx2.2 cells (red) were negative for PLP-EGFP at this stage, although within the VZ, some double-labeled cells were present (arrow). D, All Olig2-expressing cells (red) in the VZ of the pMN domain were positive for PLP-EGFP (arrows). E, In the same section as D, all PDGFRα-positive cells (pseudocolored red) seen delaminating from the VZ also labeled with PLP-EGFP. Dashed boxes indicate regions shown at higher magnification at right.
Table 1.
PLP-EGFP expression in neurons and glia during development of spinal corda
| Percentage of cells expressing PLP-EGFP | E12.5 | E14.5 | E16.5 | P7 |
|---|---|---|---|---|
| Olig2+/PDGFRα+ OPCs (% PLP-EGFP) | 100% | 93.5 ± 0.2% | 73.7 ± 10.9% | 45.0 ± 12.8% |
| NeuN+ neurons (% PLP-EGFP) | n.d. | 3.5 ± 0.5% | 0.9 ± 0.3% | 1.2 ± 0.5% |
| GFAP+ astrocytes (% PLP-EGFP) | n.d. | n.d. | 39.5 ± 2.5% | 24.5 ± 1.8% |
aData indicate the number of OPCs, neurons, or astrocytes that expressed PLP-EGFP. Values represent the average ± SEM. n.d., Not determined.
Figure 7.
Radial glia and OPCs were generated normally in E12.5 Plp1-null spinal cords. A, B, PLP-EGFP+ (green) and Nestin+ (red) radial glia at E12.5 (A) wild-type spinal cord and (B) Plp1-null cord. C, PDGFRα+ OPCs in the VZ of E12.5 wild-type spinal cord. D, Generation of PDGFRα+ OPCs was not disrupted in Plp1-null spinal cords. Scale bar: B, D, 50 μm.
PLP-EGFP was expressed early and then downregulated by migratory OPCs before being upregulated during myelination
At E14.5, PLP-EGFP+ cells were still present in the ventral VZ surrounding the central canal. However, many PLP-EGFP+ cells had migrated out of the SVZ into the dorsal and ventral intermediate zones (IZs) and they began to populate the lateral presumptive white matter (pWM; Fig. 3A). In addition to the PLP-EGFP+ cells in the ventral cord, PLP-EGFP+ cells were in the VZ/SVZ of the dorsal cord at E14.5 as well (Fig. 3A). At this stage, these PLP-EGFP+ cells in the dorsal VZ did not label with Olig2 or PDGFRα, but likely represent the OPC progenitors that arise later in the dorsal spinal cord (Fogarty et al., 2005; Vallstedt et al., 2005; Cai et al., 2007). Around the central canal, many PLP-EGFP+ cells continued to express Sox2 (Fig. 3B), indicating that they remained undifferentiated precursors. All of the Olig2+/PDGFRα+ cells in the VZ were PLP-EGFP+ and nearly all Olig2+/PDGFRα+ cells outside of the VZ/SVZ continued to express PLP-EGFP (Fig. 3C,D, white arrows; Table 1). Some PLP-EGFP+ cells in the IZ were negative for both Olig2 and PDGFRα (Fig. 3D,E). These cells may represent early progenitors outside of the oligodendrocyte lineage, e.g., neurons and astrocytes, a small percentage of which also express the Plp1 promoter (see Figs. 5, 6). Many PLP-EGFP+ cells at this stage were proliferative, as determined by Ki67 immunostaining (data not shown).
Figure 3.
Olig2+, PDGFRα+ oligodendrocyte progenitor cells expressed PLP-EGFP throughout the E14.5 spinal cord. A, PLP-EGFP expression (green) in the cervical spinal cord at E14.5. White boxes indicate regions shown in C–E. B, PLP-EGFP+ progenitors within the VZ continued to express Sox2 (red). C, In the dorsal spinal cord, OPCs immunolabeled with Olig2 (red) and PDGFRα (blue) also expressed PLP-EGFP. D, E, In the ventral cord, PLP-EGFP was expressed by Olig2+ cells in the ventral VZ (D) as well as by OPCs throughout the lateral cord (E). White arrows indicate examples of triple-labeled cells. Asterisks indicate blood vessels. Scale bars: A, 25 μm; B, 100 μm; C–E, 50 μm.
Figure 5.
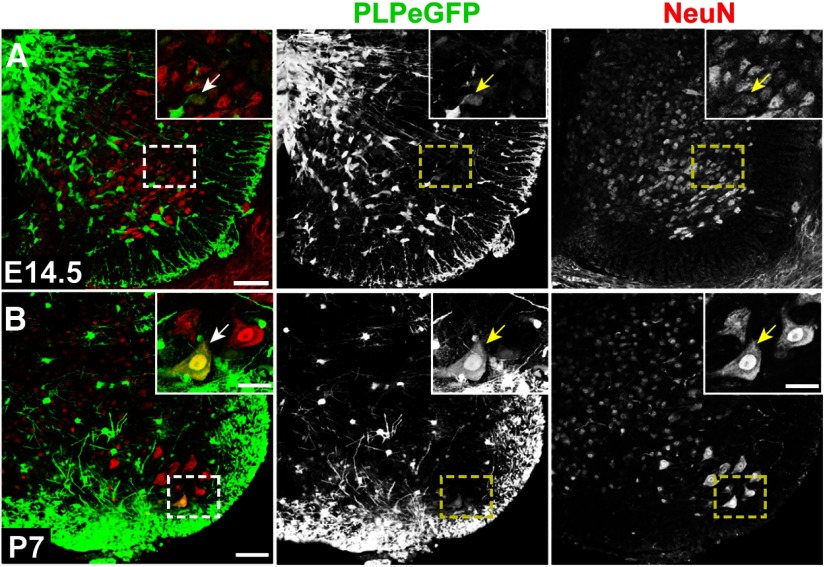
PLP-EGFP expression in neurons in the embryonic and postnatal spinal cord. A, PLP-EGFP expression (green) in NeuN+ (red) neurons (arrow, inset) in the ventral horn of the spinal cord at E14.5. B, At P7, PLP-EGFP was very rarely seen in NeuN+ (red) neurons (arrow, inset). Dashed boxes indicate regions shown in inset. Scale bars: A, B, 50 μm; inset, 10 μm.
Figure 6.
PLP-EGFP expression in astrocytes in the embryonic and postnatal spinal cord. A–C, At E16.5 (A), PLP-EGFP (green) expression was seen in GFAP+ (red) astrocyte progenitors (arrowheads) in the cord, including in bipolar (B) and process-bearing (C) GFAP+ astrocytes. D, Note robust expression of PLP (blue) in oligodendrocytes (D, double-headed arrows) but not in astrocytes (B, C, arrowheads). E, At P7, very low levels of PLP-EGFP were seen in a fraction of GFAP+ (red) astrocytes (F, G, arrowheads), although PLP-EGFP levels were much higher in oligodendrocytes (F, double-headed arrows) that also expressed PLP (blue). E–G, Some astrocytes (E) expressed low levels of PLP-EGFP in their cell bodies (F, G, arrowheads) although many astrocytes were not GFP+, and no astrocytes expressed PLP (blue). Dashed boxes in A and E depict regions of shown in B–D and F–G. Scale bars: A, E, 100 μm; (in G) B–D, F, G, 10 μm.
At E16.5, cells continued to migrate laterally away from the VZ/SVZ (Fig. 4A). Although many Olig2+/PDGFRα+ OPCs throughout the dorsal (Fig. 4B) and ventral spinal cord (Fig. 4D) continued to express PLP-EGFP (Fig. 4B–D, single-headed arrows), there were also Olig2+/PDGFRα+ OPCs that no longer expressed PLP-EGFP (Fig. 4B–D, double-headed arrows), particularly in the lateral pWM. Very few Olig2+ cells remained in the VZ (Fig. 4C). While we were not able to follow the fate of individual PLP-EGFP+ cells over time, our analysis suggests that as OPCs arise from the VZ, they first express PLP-EGFP before expressing Olig2 and PDGFRα. Olig2 expression was maintained and PDGFRα expression increased as PLP-EGFP-expressing OPCs moved away from the SVZ. OPCs farther away from the VZ decreased PLP-EGFP expression, until finally Olig2+/PDGFRα+ OPCs no longer expressed PLP-EGFP.
Figure 4.
Oligodendrocyte progenitor cells downregulated PLP-EGFP expression as they migrated laterally at E16.5 and in the P7 cord. A, PLP-EGFP expression (green) in the cervical spinal cord at E16.5. B, In the dorsal cord, some Olig2+/PDGFRα+ OPCs no longer express PLP-EGFP (double-headed arrows) while others continued to express PLP-EGFP (single-headed arrows). C, In the VZ, fewer cells expressed PLP-EGFP at E16.5. Within the IZ, many Olig2+/PDGFRα+ OPCs expressed PLP-EGFP (single-headed arrows), while more laterally located OPCs were PLP-EGFP-negative (double-headed arrows). D, In the pWM, no PLP-EGFP expression was seen in the most lateral Olig2+/PDGFRα+ OPCs (double-headed arrows). E, Many PDGFRα+ (blue) and Olig2+ (red) OPCs in the gray matter were PLP-EGFP-negative or expressed very low levels of PLP-EGFP (arrows), while myelinating oligodendrocytes had robust PLP-EGFP expression and nuclear Olig2 expression (red, inset). F, PLP-EGFP colocalized with MBP expression (red), indicating these cells are mature myelinating oligodendrocytes. Scale bars: A, 100 μm; B–D, 50 μm; E, inset, 10 μm; F, 50 μm.
By P7, OPCs in white matter regions had matured into myelinating oligodendrocytes that expressed high amounts of PLP-EGFP (Fig. 4E,F) and MBP (Fig. 4F). At this stage, myelinating oligodendrocytes were present throughout the lateral white matter as well as the gray matter of the spinal cord, and these Olig2+ oligodendrocytes expressed intense PLP-EGFP (Fig. 4E, inset). In the gray matter, Olig2+/PDGFRα+ OPCs were present and many were PLP-EGFP-negative (Fig. 4E). Some Olig2+/PDGFRα+ OPCs did express PLP-EGFP in their cell bodies, but at very low levels compared with myelinating (MBP+) oligodendrocytes (Fig. 4F, Table 1). Low-level expression of PLP-EGFP in postnatal OPCs could be residual from earlier expression or the beginning of upregulation of Plp activity in preparation for myelination. Collectively, our analysis of PLP-EGFP expression in the developing spinal cord suggests that the Plp1 gene is expressed by early progenitors and migrating OPCs. OPCs then downregulate Plp1 until they mature into myelinating oligodendrocytes, at which time they dramatically upregulate Plp1 as they myelinate axons.
In the developing spinal cord, PLP-EGFP was expressed by neurons and astrocytes
PLP-EGFP was also seen in progenitor cell populations other than developing oligodendrocytes (Table 1). In agreement with Delaunay et al., (2008), PLP-EGFP expression was detected in a small subset of NeuN+ neurons (Fig. 5A) in the ventral horn at E14.5. PLP-EGFP intensity was much lower in neurons (Fig. 5A, inset, arrow) than in OPCs (Figs. 3, 4), suggesting that neurons were downregulating Plp1. Although rare, PLP-EGFP+ neurons (Fig. 5B, inset, arrow) were still seen occasionally in the spinal cord at P7, but PLP was not detected in these cells (data not shown).
PLP-EGFP expression was also observed in immature GFAP+ astrocytes at E16.5 (Fig. 6A–D). At E16.5, PLP-EGFP was seen in both bipolar migratory astrocytes near the lateral white matter (Fig. 6B, arrowheads) and in process-bearing astrocytes in the gray matter (Fig. 6C, arrowheads). PLP, as detected by the AA3 antibody, was not detected in astrocytes, but was confined to multiprocess-bearing OPCs in the ventral white matter (Fig. 6D, double-headed arrows) and dorsal columns. By P7, many GFAP+ astrocytes no longer expressed PLP-EGFP+ (Fig. 6E–G). In GFAP+ astrocytes that did continue to express PLP-EGFP (Fig. 6F,G, arrowheads), the intensity of EGFP was much lower than in neighboring oligodendrocytes (Fig. 6F, double-headed arrows), and PLP-EGFP was confined to the soma, suggesting that astrocytes were downregulating PLP promoter activity as they matured. We quantified the number of GFAP+ astrocytes expressing PLP-EGFP and found a surprising number of astrocytes labeled with PLP-EGFP: ∼40% at E16.5 and 25% at P7 (Table 1). We also used S100β+, which labels immature astrocytes and subtypes of mature astrocytes (Wang and Bordey, 2008), to identify cell bodies of GFAP+ astrocytes (data not shown). Because cells of the oligodendrocyte lineage can also express S100β (Deloulme et al., 2004), only S100β+ cells that were negative for Olig2 were counted as astrocytes. We found that although S100β only labeled ∼13% of the astrocytes at E16.5, nearly all S100β+/GFAP+ astrocytes (96.4 ± 3.7%) expressed PLP-EGFP. At P7 only 35.7 ± 7.5% of the S100β+ astrocytes had detectable PLP-EGFP in their cell bodies. Intriguingly, it appears that the subset of astrocytes that express S100β in the embryo express the greatest amount of PLP-EGFP, which is consistent with the concept that PLP-EGFP is expressed in immature cells, such as immature astrocytes that express S100β+ (Wang and Bordey, 2008). Importantly, PLP was not detected in astrocytes at either E16.5 (Fig. 6A–D) or P7 (Fig. 6E,F), suggesting that residual EGFP label was from earlier plp/dm20 promoter activity. These results indicated that although the plp/dm20 promoter is active in neuron and astrocyte progenitors, it diminishes with development; PLP is not produced in these cell types but rather is restricted to cells of the oligodendroglial lineage.
OPC development in Plp1-null mice
Due to the high levels of Plp1 promoter activity seen in migratory neuronal, oligodendroglial, and astroglial progenitors, it was tempting to speculate that PLP/DM20 may serve a role in progenitor cell migration. To examine the role of PLP/DM20 in the migration and development of oligodendrocytes, the PLP-EGFP reporter (Mallon et al., 2002) was crossed into mice carrying the Plp1-null allele (Klugmann et al., 1997). OPC generation, process extension, and migration were examined in vivo at E12.5, E14.5, E16.5, and P7 in both Plp1-null PLP-EGFP and wild-type PLP-EGFP male littermates. At E12.5, PLP-EGFP-expressing cells arose comparably in the ventral VZ of both wild-type and Plp1-null mice, and PLP-EGFP+ processes emanated radially from these VZ/SVZ cells (Fig. 7). As in wild-type mice (Fig. 7A), Nestin+ radial glia in Plp1-null mice extended processes from the VZ, and radial glia end feet appeared to contact the pial surface normally (Fig. 7B). In both wild-type (Fig. 7C) and Plp1-null (Fig. 7D) spinal cords, PDGFRα-expressing cells arose from the ventral VZ in the same spatiotemporal manner.
We next looked at the distribution of OPCs in the dorsal and ventral spinal cord in wild-type and Plp1-null mice at E14.5 and at E16.5. No differences in the total number of Olig2+/PDGFRα+ OPCs or their distribution throughout the dorsal and ventral cord were seen at either E14.5 (data not shown) or E16.5 (Fig. 8). Intriguingly, in wild-type mice, PLP-EGFP+ cells emanated long processes from the SVZ that were radially oriented (Fig. 8A), while PLP-EGFP+ processes in Plp1-null mice appeared shorter and much more disorganized (Fig. 8B). To analyze the organization of process extension, the orientation of PLP-EGFP+ processes was determined and classified as tangential, radial, or other (i.e., random; see Materials and Methods). Plp1-null mice had a significant increase in the number of cell processes with other orientations compared with wild-type mice (p = 0.013, t test, two-tailed; Fig. 8D). Measurements of the PLP-EGFP+ process lengths were made in both the ventral and dorsal regions of the spinal cord (Fig. 8E). Interestingly, PLP-EGFP+ processes in the ventral region of the E16.5 cord were much longer than PLP-EGFP+ processes in the dorsal region of the cord. Within the ventral cord, the PLP-EGFP+ processes were significantly shorter in Plp1-null mice, compared with wild type (Fig. 8E). No significant difference was seen in the lengths of dorsal PLP-EGFP+ processes between Plp1-null and wild-type mice. Despite the differences in process length, the total number of Olig2+/PDGFRα+ OPCs in different regions of the ventral and dorsal cord, as well as their overall distribution, was similar in both wild-type and Plp1-null mice at E16.5 (Fig. 8C).
Figure 8.

In Plp1-null spinal cords, OPCs were distributed normally, although cells had disorganized processes in vivo and migrated more quickly in vitro. A, In wild-type spinal cords at E16.5, PLP-EGFP (green) cells extended radially orientated processes toward the pial surface. B, In Plp1-null cords at E16.5, processes of PLP-EGFP cells were shorter and more randomly aligned. Many of these cells also expressed PDGFRα (blue) and Olig2 (red) as indicated by white arrows. Red arrows indicate Olig2+/PDGFRα+ cells that were negative for PLP-EGFP. Yellow arrows indicate PLP-EGFP+ cells that were negative for Olig2 and PDGFRα. Asterisks indicate blood vessels. C, The total number of Olig2+/PDGFRα+ OPCs and their distribution throughout the cord was the same in wild-type and Plp1-null cords. D, In Plp1-null spinal cords, there was a significant increase (**p < 0.0001, t test, 2-tailed) in the number of randomly aligned PLP-EGFP+ processes compared with wild type. E, PLP-EGFP+ processes were significantly shorter in the ventral cord of Plp1-null animals compared with wild type (*p = 0.013, t test, 2-tailed), but did not differ in the dorsal cord. Graphs represent group means ± SEM. F, The population of Plp1-null OPCs migrated significantly faster in live imaging analysis of cell migration in vitro compared with wild-type OPCs (‡p < 0.01, Mann–Whitney U test). G, Plp1-null OPCs also migrated over greater distances compared with wild-type OPCs (‡p < 0.01, Mann–Whitney U test). Frequency histograms represent the total number of cells tracked (291 wild type and 231 Plp1 null) across four experiments. Scale bar, 50 μm.
We have previously shown that PLP forms complexes with membrane receptors, including αv integrin, to influence OPC migration on fibronectin (Gudz et al., 2002, 2006). Fibronectin is expressed along radial glial processes (Sheppard et al., 1995; Stettler and Galileo, 2004), and thus could be a substrate along which neurons and glial cells migrate in vivo. We analyzed the migration of OPCs generated from both wild-type and Plp1-null neurospheres (Fig. 8F,G) using live imaging analysis of OPC migration on fibronectin-coated chamber slides. Cells were tracked and the velocity and distance of migration were quantified with the Chemotaxis and Migration Plugin (iBidi). We generated frequency histograms to determine the distributions of cell velocities (Fig. 8F) and accumulated distances (Fig. 8G) within the populations of wild-type and Plp1-null cells. For velocity, the medians of wild-type and Plp1-null cells were 0.179 and 0.202 μm/min respectively, and the median distances traveled were 107.8 μm (wild type) and 125.8 μm (Plp1-null). We ran a Mann–Whitney's U test to evaluate the difference in the migration velocities and accumulated distances between wild-type and Plp1 cells. We found that Plp1-null cells were significantly faster (p = 0.0041) and migrated farther (p = 0.0006) than wild-type cells on fibronectin. Studies to understand the mechanisms underlying these differences are ongoing.
Despite the absence of the major myelin protein, Plp1-null mice are still able to myelinate axons and produce compact myelin around axons (Klugmann et al., 1997; Griffiths et al., 1998; Jurevics et al., 2003). To assess whether the absence of PLP impacts the onset of myelination in Plp1-null mice, spinal cords from early postnatal mice were examined by immunohistochemistry and Western blot for PLP and MBP (Fig. 9). By P7, robust PLP expression throughout the cord overlapped with PLP-EGFP expression (Fig. 9A). EGFP filled the entire cell body and many of the larger processes of oligodendrocytes, while PLP was in processes, but mostly excluded from oligodendrocyte cell bodies. As expected, no PLP immunoreactivity was seen in Plp1-null cords by immunostaining (Fig. 9B) or Western blot (Fig. 9E), but MBP expression overlapped PLP-EGFP expression in both wild-type (Fig. 9C) and Plp1-null cords (Fig. 9D). The total numbers of oligodendrocytes and OPCs throughout the cord were unchanged in Plp1-null mice at P7 compared with wild type (data not shown). Given the expression of PLP-EGFP outside of the oligodendroglial lineage, we counted total numbers of astrocytes and neurons in wild-type and Plp1-null cords at P7, and found no change in Plp1-null animals (data not shown). We also examined glial activation of GFAP+ astrocytes and Iba1+ microglia (data not shown), and these were similarly unchanged in Plp1-null mice compared with wild type. Quantification of MBP levels by Western blots of Plp1-null and wild-type spinal cord lysates revealed no change in MBP expression at P4 (data not shown) or P7 (Fig. 9E).
Figure 9.

Normal levels of MBP in P7 spinal cords of Plp1-null mice. A, Wild-type PLP-EGFP+ oligodendrocytes expressed PLP (blue) in their processes throughout the gray and white matter. B, No PLP was detected in Plp1-null cords. C, D, MBP was present in PLP-EGFP+ oligodendrocytes in both wild-type (C) and Plp1-null (D) cords. E, Western blot analysis of P7 wild-type and Plp1-null spinal cord lysates for PLP/DM20, MBP, and β-tubulin. Quantification of MBP levels normalized to β-tubulin was similar between wild-type and Plp1-null mice at P7. n.s., Not significantly different.
Discussion
These studies demonstrated that Plp1 promoter activity in the spinal cord was biphasic, first occurring very early in the embryonic neuroepithelium and then again postnatally in myelinating oligodendrocytes. PLP-EGFP was expressed in undifferentiated precursors of the spinal cord as well as by neuronal, astroglial, and oligodendroglial progenitors. Within the oligodendroglial lineage, PLP-EGFP expression was very dynamic. Initially, in the ventral VZ, all Olig2+/PDGFRα+ OPCs expressed PLP-EGFP. Olig2+/PDGFRα+ cells lost PLP-EGFP expression as they migrated laterally toward the pWM. PLP-EGFP remained downregulated in postnatal OPCs, while myelinating oligodendrocytes strongly upregulated PLP-EGFP. Biphasic activity of the Plp promoter was also reported in Plp-cre;Z/EG mice in which the lag between Cre-recombinase expression and recombination leading to GFP expression could be used to identify earlier-born and later-born cells (Delaunay et al., 2008). Our data suggest that oligodendrocytes in the spinal cord arise from a single cell lineage, with differential expression of PLP/DM20 and PDGFRα at distinct stages. The biphasic expression of PLP-EGFP could explain some of the past controversy regarding the origin of OPCs (Spassky et al., 1998; 2000; Richardson et al., 2000). Previous reports found Pdgfrα transcripts in cells that were Plp1-negative and proposed that these cells were of two separate OPC lineages. Given the low levels of Plp1 mRNA and protein at embryonic stages (Ivanova et al., 2003; this study), it may be difficult to detect them directly. Using the Plp1 promoter and 3′UTR to drive high levels of EGFP, we were able to clearly identify Plp1 promoter activity in many embryonic cells. Our results suggest that because of the biphasic expression of PLP-EGFP, the embryonic PDGFRα+ population that was positive for PLP-EGFP was less differentiated than the PDGFRα+ OPCs that were PLP-EGFP-negative. It is possible that the PLP-EGFP-negative OPCs might never have expressed PLP-EGFP, and migrated out earlier than the PLP-EGFP population. However, this is unlikely since at the earliest stages examined all Olig2+/PDGFRα+ cells in the VZ/SVZ were PLP-EGFP+. Given that PLP and GFP may have different half lives within the same cell (Delaunay et al., 2008, their Discussion), it is possible that some EGFP+ cells may no longer be producing PLP. In fact we were unable to detect PLP in PLP-EGFP-labeled neurons or astrocytes. However we have shown that at both E16.5 and P7, PLP is present in the processes of PLP-EGFP-expressing OPCs. At later stages, PLP, but not DM20, continues to be expressed by a subset of NG2+ cells in the brain (Mallon et al., 2002; Ye et al., 2003), which presumably give rise to myelinating oligodendrocytes.
In the absence of PLP expression, there was mild disruption of process extension in ventral PLP-EGFP-expressing cells; processes were shorter and often disorganized, with fewer processes aligned radially. Despite abnormal process development, no differences in OPC numbers, distribution, or onset of myelination were seen in Plp1-null spinal cords. Within the spinal cord, OPCs arise from either the ventral or dorsal regions. The generation of ventrally derived OPCs is dependent on Sonic hedgehog (Shh), while dorsal OPCs arise independently of Shh signaling, possibly dependent on fibroblast growth factor or bone morphogenic factor instead (for review, see Richardson et al., 2006). Ventrally and dorsally derived oligodendrocytes preferentially myelinate different tracts in both the brain and spinal cord (Tripathi et al., 2011); furthermore, dorsally derived oligodendrocytes are more likely to repopulate lesions and remyelinate axons after focal demyelination (Zhu et al., 2011). Although we could not distinctly fate-map dorsally versus ventrally derived OPCs in our PLP-EGFP mice, we did see PLP-EGFP expression in both populations. The ventrally derived PLP-EGFP+ cells displayed shorter processes in Plp1-null mice, while the dorsally derived PLP-EGFP+ cells did not, suggesting that these two populations respond to different environmental cues to regulate process extension.
At E12.5, many PLP-EGFP+ cells in the VZ were outside of the pMN domain, from which Olig2+ cells arise. Cells from a variety of lineages expressed PLP-EGFP, including neurons (Pax6), oligodendrocytes and motor neurons (Olig2+), and unspecified precursors (Sox2). As in other studies (Delaunay et al., 2008; Guo et al., 2009; Michalski et al., 2011), we found that the Plp1 promoter was active in neurons and astrocytes. Our findings that ∼25% of the GFAP+ astrocytes at P7 are labeled with PLP-EGFP are very consistent with the findings of other groups that used the PLP promoter to label cells in the CNS (Guo et al., 2009; Michalski et al., 2011; PLP-CreERT2; PLP-CreERT2; and PLP-Cre). They reported that 33% of astrocytes in the P4 spinal cord (Michalski et al., 2011), 12% of astrocytes in the P15 spinal cord (Guo et al., 2009), and 11% in the P28 spinal cord (Michalski et al., 2011) had PLP promoter activity. The downregulation of PLP-EGFP and the absence of PLP in neurons and astrocytes indicate that many of the early PLP-EGFP-expressing cells in the VZ downregulate PLP-EGFP and adopt nonoligodendroglial cell fates. This expression pattern raises the question of whether PLP/DM20 plays a role in specifying oligodendroglial versus astroglial or neuronal cell identity, in addition to its role in myelination. To determine whether PLP/DM20 regulated precursor fate specification, total numbers of neurons, astrocytes, and oligodendrocytes, as well as the percentage that expressed PLP-EGFP, were assessed in both wild-type and Plp1-null spinal cords. We did not detect any differences in the numbers of astrocytes, neurons, oligodendrocytes, or their distributions throughout the cord, indicating that PLP/DM20 was not essential for cell fate specification, survival, or migration. In vitro studies have reported that reduced levels of PLP can reduce apoptosis and increase oligodendrocyte cell survival, and cultures from Plp1-null mice produce twice as many oligodendrocytes as cultures from wild-type mice (Skoff et al., 2004). We saw no changes in OPC numbers in our studies, nor have other studies of Plp1-null mice reported such increases, but it may be that in vivo there is not enough trophic support for excess oligodendrocytes, keeping numbers constant.
The dynamic expression of the Plp1 promoter, high in early progenitors, low at intermediate stages, and then high again postnatally, suggests two roles for PLP: an early role in migratory progenitors, followed by a later role in mature oligodendrocytes during myelination. However, the question of PLP's role in nonmyelinating cell types persists. PLP/DM20 proteins resemble the tetraspanins (Boucheix and Rubinstein, 2001; Rubinstein, 2011) and our previous in vitro studies indicate PLP may function similarly by providing a nucleating scaffold that enables the interaction of other proteins, including integrins, thereby integrating extracellular signaling and intracellular pathway activation (Gudz et al., 2002, 2006). PLP-EGFP+ cell processes in the ventral spinal cord of Plp1-null mice had shorter and often disorganized processes relative to wild-type controls, supporting a role for PLP signaling in migration and/or process extension. In live imaging studies of OPC migration on fibronectin, a ligand for αv integrins, Plp1-null OPCs migrated faster and farther than wild-type OPCs. PLP can form a complex with αv integrin and GluR2, an AMPA receptor subunit, and the association of PLP with αv integrin increases upon stimulation with glutamate or glutamate agonist (Gudz et al., 2002, 2006). Although in the current study we did not stimulate cells in our live imaging migration assays, there may be, in the absence of PLP, reduced clustering of integrins in OPCs on fibronectin, causing decreased binding and increased migration speed. While our in vivo results indicate PLP/DM20 is nonessential for OPC migration and developmental myelination, it could be important in the context of adult remyelination after injury. After focal demyelination in the adult mouse corpus callosum, OPCs migrating from the SVZ have been shown to form functional glutamatergic synapses with demyelinated axons (Etxeberria et al., 2010). Whether Plp1-null OPCs would be able to make such connections and effectively remyelinate demyelinated axons remains unknown.
Other in vitro studies also suggest a role for PLP in OPC process extension. For example, PLP antisense treatment of OPCs results in abnormal process development and reduced membrane formation (Yang and Skoff, 1997), while overexpression of PLP in Oli-Neu cells results in increased process extension and branching (Werner et al., 2013). Despite the process abnormalities of Plp1-null OPCs, we did not see any dramatic differences in the distribution of OPCs throughout the developing spinal cords of Plp1-null mice. Redundant mechanisms in OPC development may exist such that OPC migration and myelination were minimally disrupted. Indeed, in the absence of PLP, expression of a PLP-homolog, glycoprotein M6B, is increased and able to partially compensate for the loss of PLP during myelination (Werner et al., 2013).
Although its role in OPC migration and myelination appears dispensable, PLP clearly has an important role in supporting normal axonal function. Plp1-null animals generate normal numbers of oligodendrocytes and produce compact myelin that contacts and wraps axons, but over time develop axonal swellings, problems with retrograde axonal transport, and eventual axonal degeneration (Boison and Stoffel, 1994; Rosenbluth et al., 1996; Klugmann et al., 1997; Griffiths et al., 1998; Yool et al., 2001). PLP is necessary for the transport of certain proteins, such as the NAD+-dependent deacetylase, sirtuin 2, and cholesterol into CNS myelin, and PLP-deficient myelin has reduced cholesterol content (Krämer-Albers et al., 2006; Werner et al., 2007, 2013). PLP is in exosomes released by oligodendrocytes, which can be taken up by neurons and other glia (Krämer-Albers et al., 2007; Fitzner et al., 2011; Frühbeis et al., 2013). The axonal degeneration seen in Plp1-null mice likely results from a loss of trophic or metabolic support, either from altered myelin structural integrity, perturbed exosome trafficking, or an as yet undefined form of oligodendrocyte-axonal communication. Future studies on interactions between oligodendrocytes and neurons in Plp1-null animals may reveal additional undiscovered roles for PLP.
Footnotes
This work was supported by National Institutes of Health Grant NS25304 (W.B.M.), a National Multiple Sclerosis Society postdoctoral fellowship (D.E.H.), and an undergraduate fellowship (R25GM097633; E.M.V.). We would like to acknowledge Jared Ahrendsen for help with the PLP in situ analysis.
The authors declare no competing financial interests.
References
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison D, Stoffel W. Disruption of the compacted myelin sheath of axons of the central nervous system in proteolipid protein-deficient mice. Proc Natl Acad Sci U S A. 1994;91:11709–11713. doi: 10.1073/pnas.91.24.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134:1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- Delaunay D, Heydon K, Cumano A, Schwab MH, Thomas JL, Suter U, Nave KA, Zalc B, Spassky N. Early neuronal and glial fate restriction of embryonic neural stem cells. J Neurosci. 2008;28:2551–2562. doi: 10.1523/JNEUROSCI.5497-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloulme JC, Raponi E, Gentil BJ, Bertacchi N, Marks A, Labourdette G, Baudier J. Nuclear expression of S100B in oligodendrocyte progenitor cells correlates with differentiation toward the oligodendroglial lineage and modulates oligodendrocytes maturation. Mol Cell Neurosci. 2004;27:453–465. doi: 10.1016/j.mcn.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Dickinson PJ, Fanarraga ML, Griffiths IR, Barrie JM, Kyriakides E, Montague P. Oligodendrocyte progenitors in the embryonic spinal cord express DM-20. Neuropathol Appl Neurobiol. 1996;22:188–198. doi: 10.1111/j.1365-2990.1996.tb00894.x. [DOI] [PubMed] [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Eng LF, Chao FC, Gerstl B, Pratt D, Tavaststjerna MG. The maturation of human white matter myelin. Fractionation of the myelin membrane proteins. Biochemistry. 1968;7:4455–4465. doi: 10.1021/bi00852a042. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/S0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Etxeberria A, Mangin JM, Aguirre A, Gallo V. Adult-born SVZ progenitors receive transient synapses during remyelination in corpus callosum. Nat Neurosci. 2010;13:287–289. doi: 10.1038/nn.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, Simons M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Science. 2011;124:447–458. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- Fogarty M, Richardson WD, Kessaris N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development. 2005;132:1951–1959. doi: 10.1242/dev.01777. [DOI] [PubMed] [Google Scholar]
- Frühbeis C, Fröhlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Möbius W, Goebbels S, Nave KA, Schneider A, Simons M, Klugmann M, Trotter J, Krämer-Albers EM. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Qi Y, Tan M, Cai J, Takebayashi H, Nakafuku M, Richardson W, Qiu M. Dual origin of spinal oligodendrocyte progenitors and evidence for the cooperative role of Olig2 and Nkx2.2 in the control of oligodendrocyte differentiation. Development. 2002;129:681–693. doi: 10.1242/dev.129.3.681. [DOI] [PubMed] [Google Scholar]
- Fuss B, Baba H, Phan T, Tuohy VK, Macklin WB. Phosphodiesterase I, a novel adhesion molecule and/or cytokine involved in oligodendrocyte function. J Neurosci. 1997;17:9095–9103. doi: 10.1523/JNEUROSCI.17-23-09095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths I, Klugmann M, Anderson T, Yool D, Thomson C, Schwab MH, Schneider A, Zimmermann F, McCulloch M, Nadon N, Nave KA. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science. 1998;280:1610–1613. doi: 10.1126/science.280.5369.1610. [DOI] [PubMed] [Google Scholar]
- Gudz TI, Schneider TE, Haas TA, Macklin WB. Myelin proteolipid protein forms a complex with integrins and may participate in integrin receptor signaling in oligodendrocytes. J Neurosci. 2002;22:7398–7407. doi: 10.1523/JNEUROSCI.22-17-07398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudz TI, Komuro H, Macklin WB. Glutamate stimulates oligodendrocyte progenitor migration mediated via an αv integrin/myelin proteolipid protein complex. J Neurosci. 2006;26:2458–2466. doi: 10.1523/JNEUROSCI.4054-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Ma J, McCauley E, Bannerman P, Pleasure D. Early postnatal proteolipid promoter-expressing progenitors produce multilineage cells in vivo. J Neurosci. 2009;29:7256–7270. doi: 10.1523/JNEUROSCI.5653-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Nakahira E, Kagawa T, Oba A, Wada T, Takebayashi H, Spassky N, Levine J, Zalc B, Ikenaka K. Evidence for a second wave of oligodendrogenesis in the postnatal cerebral cortex of the mouse. J Neurosci Res. 2003;73:581–592. doi: 10.1002/jnr.10717. [DOI] [PubMed] [Google Scholar]
- Jurevics H, Hostettler J, Sammond DW, Nave KA, Toews AD, Morell P. Normal metabolism but different physical properties of myelin from mice deficient in proteolipid protein. J Neurosci Res. 2003;71:826–834. doi: 10.1002/jnr.10544. [DOI] [PubMed] [Google Scholar]
- Klugmann M, Schwab MH, Pühlhofer A, Schneider A, Zimmermann F, Griffiths IR, Nave KA. Assembly of CNS myelin in the absence of proteolipid protein. Neuron. 1997;18:59–70. doi: 10.1016/S0896-6273(01)80046-5. [DOI] [PubMed] [Google Scholar]
- Krämer-Albers EM, Gehrig-Burger K, Thiele C, Trotter J, Nave KA. Perturbed interactions of mutant proteolipid protein/DM20 with cholesterol and lipid rafts in oligodendroglia: implications for dysmyelination in spastic paraplegia. J Neurosci. 2006;26:11743–11752. doi: 10.1523/JNEUROSCI.3581-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer-Albers EM, Bretz N, Tenzer S, Winterstein C, Möbius W, Berger H, Nave KA, Schild H, Trotter J. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: Trophic support for axons? Proteomics Clin Appl. 2007;1:1446–1461. doi: 10.1002/prca.200700522. [DOI] [PubMed] [Google Scholar]
- Leber SM, Sanes JR. Migratory paths of neurons and glia in the embryonic chick spinal cord. J Neurosci. 1995;15:1236–1248. doi: 10.1523/JNEUROSCI.15-02-01236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVine SM, Wong D, Macklin WB. Developmental expression of proteolipid protein and DM20 mRNAs and proteins in the rat brain. Dev Neurosci. 1990;12:235–250. doi: 10.1159/000111853. [DOI] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD. Simple neurite tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/S0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Mallon BS, Shick HE, Kidd GJ, Macklin WB. Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J Neurosci. 2002;22:876–885. doi: 10.1523/JNEUROSCI.22-03-00876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- McClive PJ, Sinclair AH. Rapid DNA extraction and PCR-sexing of mouse embryos. Mol Reprod Dev. 2001;60:225–226. doi: 10.1002/mrd.1081. [DOI] [PubMed] [Google Scholar]
- McDermott KW, Barry DS, McMahon SS. Role of radial glia in cytogenesis, patterning and boundary formation in the developing spinal cord. J Anat. 2005;207:241–250. doi: 10.1111/j.1469-7580.2005.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SS, McDermott KW. Proliferation and migration of glial precursor cells in the developing rat spinal cord. J Neurocytol. 2001;30:821–828. doi: 10.1023/A:1019693421778. [DOI] [PubMed] [Google Scholar]
- Michalski JP, Anderson C, Beauvais A, De Repentigny Y, Kothary R. The proteolipid protein promoter drives expression outside of the oligodendrocyte lineage during embryonic and early postnatal development. PLoS ONE. 2011;6:e19772. doi: 10.1371/journal.pone.0019772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Kangas CD, Macklin WB. Neuronal expression of the proteolipid protein gene in the medulla of the mouse. J Neurosci Res. 2009;87:2842–2853. doi: 10.1002/jnr.22121. [DOI] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. Olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Pedraza CE, Monk R, Lei J, Hao Q, Macklin WB. Production, characterization, and efficient transfection of highly pure oligodendrocyte precursor cultures from mouse embryonic neural progenitors. Glia. 2008;56:1339–1352. doi: 10.1002/glia.20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Smith HK, Sun T, Pringle NP, Hall A, Woodruff R. Oligodendrocyte lineage and the motor neuron connection. Glia. 2000;29:136–142. doi: 10.1002/(SICI)1098-1136(20000115)29:2<136::AID-GLIA6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth J, Stoffel W, Schiff R. Myelin structure in proteolipid protein (PLP)-null mouse spinal cord. J Comp Neurol. 1996;371:336–344. doi: 10.1002/(SICI)1096-9861(19960722)371:2<336::AID-CNE12>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Rubinstein E. The complexity of tetraspanins. Biochem Soc Trans. 2011;39:501–505. doi: 10.1042/BST0390501. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard AM, Brunstrom JE, Thornton TN, Gerfen RW, Broekelmann TJ, McDonald JA, Pearlman AL. Neuronal production of fibronectin in the cerebral cortex during migration and layer formation is unique to specific cortical domains. Dev Biol. 1995;172:504–518. doi: 10.1006/dbio.1995.8034. [DOI] [PubMed] [Google Scholar]
- Skoff RP, Saluja I, Bessert D, Yang X. Analyses of proteolipid protein mutants show levels of proteolipid protein regulate oligodendrocyte number and cell death in vitro and in vivo. Neurochem Res. 2004;29:2095–2103. doi: 10.1007/s11064-004-6882-0. [DOI] [PubMed] [Google Scholar]
- Sorg BA, Smith MM, Campagnoni AT. Developmental expression of the myelin proteolipid protein and basic protein mRNAs in normal and dysmyelinating mutant mice. J Neurochem. 1987;49:1146–1154. doi: 10.1111/j.1471-4159.1987.tb10005.x. [DOI] [PubMed] [Google Scholar]
- Spassky N, Goujet-Zalc C, Parmantier E, Olivier C, Martinez S, Ivanova A, Ikenaka K, Macklin W, Cerruti I, Zalc B, Thomas JL. Multiple restricted origin of oligodendrocytes. J Neurosci. 1998;18:8331–8343. doi: 10.1523/JNEUROSCI.18-20-08331.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Olivier C, Perez-Villegas E, Goujet-Zalc C, Martinez S, Thomas JI, Zalc B. Single or multiple oligodendroglial lineages: a controversy. Glia. 2000;29:143–148. doi: 10.1002/(SICI)1098-1136(20000115)29:2<143::AID-GLIA7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Stettler EM, Galileo DS. Radial glia produce and align the ligand fibronectin during neuronal migration in the developing chick brain. J Comp Neurol. 2004;468:441–451. doi: 10.1002/cne.10987. [DOI] [PubMed] [Google Scholar]
- Timsit SG, Bally-Cuif L, Colman DR, Zalc B. DM-20 mRNA is expressed during the embryonic development of the nervous system of the mouse. J Neurochem. 1992;58:1172–1175. doi: 10.1111/j.1471-4159.1992.tb09378.x. [DOI] [PubMed] [Google Scholar]
- Tripathi RB, Clarke LE, Burzomato V, Kessaris N, Anderson PN, Attwell D, Richardson WD. Dorsally and ventrally derived oligodendrocytes have similar electrical properties but myelinate preferred tracts. J Neurosci. 2011;31:6809–6819. doi: 10.1523/JNEUROSCI.6474-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallstedt A, Klos JM, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45:55–67. doi: 10.1016/j.neuron.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Wang DD, Bordey A. The astrocyte odyssey. Prog Neurobiol. 2008;86:342–367. doi: 10.1016/j.pneurobio.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, Orfaniotou F, Dhaunchak A, Brinkmann BG, Möbius W, Guarente L, Casaccia-Bonnefil P, Jahn O, Nave KA. Proteolipid protein is required for transport of sirtuin 2 into CNS Myelin. J Neurosci. 2007;27:7717–7730. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner HB, Krämer-Albers EM, Strenzke N, Saher G, Tenzer S, Ohno-Iwashita Y, De Monasterio-Schrader P, Möbius W, Moser T, Griffiths IR, Nave KA. A critical role for the cholesterol-associated proteolipids PLP and M6B in myelination of the central nervous system. Glia. 2013;61:567–586. doi: 10.1002/glia.22456. [DOI] [PubMed] [Google Scholar]
- Wight PA, Duchala CS, Readhead C, Macklin WB. A myelin proteolipid protein-LacZ fusion protein is developmentally regulated and targeted to the myelin membrane in transgenic mice. J Cell Biol. 1993;123:443–454. doi: 10.1083/jcb.123.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura T, Konola JT, Wekerle H, Lees MB. Monoclonal antibodies against myelin proteolipid protein: identification and characterization of two major determinants. J Neurochem. 1991;57:1671–1680. doi: 10.1111/j.1471-4159.1991.tb06367.x. [DOI] [PubMed] [Google Scholar]
- Yang X, Skoff RP. Proteolipid protein regulates the survival and differentiation of oligodendrocytes. J Neurosci. 1997;17:2056–2070. doi: 10.1523/JNEUROSCI.17-06-02056.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Bagnell R, D'Ercole AJ. Mouse NG2+ oligodendrocyte precursors express mRNA for proteolipid protein but not its DM-20 variant: a study of laser microdissection-captured NG2+ cells. J Neurosci. 2003;23:4401–4405. doi: 10.1523/JNEUROSCI.23-11-04401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yool DA, Klugmann M, McLaughlin M, Vouyiouklis DA, Dimou L, Barrie JA, McCulloch MC, Nave KA, Griffiths IR. Myelin proteolipid proteins promote the interaction of oligodendrocytes and axons. J Neurosci Res. 2001;63:151–164. doi: 10.1002/1097-4547(20010115)63:2<151::AID-JNR1007>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Whittemore SR, Devries WH, Zhao X, Kuypers NJ, Qiu M. Dorsally derived oligodendrocytes in the spinal cord contribute to axonal myelination during development and remyelination following focal demyelination. Glia. 2011;59:1612–1621. doi: 10.1002/glia.21203. [DOI] [PubMed] [Google Scholar]