Abstract
The β subunits of voltage-gated calcium channels regulate surface expression and gating of CaV1 and CaV2 α1 subunits and thus contribute to neuronal excitability, neurotransmitter release, and calcium-induced gene regulation. In addition, certain β subunits are targeted into the nucleus, where they interact directly with the epigenetic machinery. Whereas their involvement in this multitude of functions is reflected by a great molecular heterogeneity of β isoforms derived from four genes and abundant alternative splicing, little is known about the roles of individual β variants in specific neuronal functions. In the present study, an alternatively spliced β4 subunit lacking the variable N terminus (β4e) is identified. It is highly expressed in mouse cerebellum and cultured cerebellar granule cells (CGCs) and modulates P/Q-type calcium currents in tsA201 cells and CaV2.1 surface expression in neurons. Compared with the other two known full-length β4 variants (β4a and β4b), β4e is most abundantly expressed in the distal axon, but lacks nuclear-targeting properties. To determine the importance of nuclear targeting of β4 subunits for transcriptional regulation, we performed whole-genome expression profiling of CGCs from lethargic (β4-null) mice individually reconstituted with β4a, β4b, and β4e. Notably, the number of genes regulated by each β4 splice variant correlated with the rank order of their nuclear-targeting properties (β4b > β4a > β4e). Together, these findings support isoform-specific functions of β4 splice variants in neurons, with β4b playing a dual role in channel modulation and gene regulation, whereas the newly detected β4e variant serves exclusively in calcium-channel-dependent functions.
Keywords: Ca2+ channel, Cacnb4, CaV2.1, cerebellar granule cells, hippocampal neurons, lethargic mice
Introduction
Voltage-activated calcium channels control multiple neuronal functions including excitability, synaptic transmission and plasticity, and activity-dependent gene regulation (Catterall and Few, 2008). The cytoplasmic β subunits are essential components of high-voltage-activated calcium channels (CaV1 and CaV2). They regulate surface expression of the channel and thus calcium current density and modulate their gating properties (Buraei and Yang, 2010; Dolphin, 2012). Mammalian genomes encode four β isoforms (β1, β2, β3, and β4), all of which are expressed in the brain. Whereas coexpression in heterologous cells demonstrated promiscuous interactions of all examined α1-β combinations, it is generally accepted that different β subunits endow calcium channels with specific properties and that particular neuronal functions require specific subunit combinations. Accordingly, the lack of specific β isoforms causes distinct neurological phenotypes (Arikkath and Campbell, 2003). For example, mutations of the β4 gene cause ataxia and epilepsy in humans and mice (Burgess et al., 1997; Barclay and Rees, 1999; Hosford et al., 1999; Escayg et al., 2000).
Structurally, calcium channel β subunits resemble membrane-associated guanylate kinase (GK) proteins, with conserved SH3 and GK domains linked by a variable hook region and flanked by variable N- and C-terminal sequences (Hanlon et al., 1999). The GK domain forms the high-affinity binding pocket for the cytoplasmic α interaction domain in the CaV1 and CaV2 α1 subunits (Chen et al., 2004; Opatowsky et al., 2004; Van Petegem et al., 2004). Alternative splicing of the exons encoding the variable regions gives rise to multiple variants of each β isoform, which differ in their endogenous membrane-targeting properties and protein interactions (Buraei and Yang, 2010). These specific properties result in distinct stabilities of channel complexes (Campiglio et al., 2013), different subcellular targeting properties (Xie et al., 2007; Subramanyam et al., 2009; Obermair et al., 2010), and differential functional modulation of calcium currents (Helton and Horne, 2002). Nevertheless, the full complement of β subunits expressed in the brain is still not completely known and our understanding of their specific targeting properties or their specific involvement in particular neuronal functions is still rudimentary.
Moreover, calcium-channel-independent functions of β subunits have been reported. For example, several studies demonstrated targeting of β4 subunits into the nucleus and suggested a direct function in activity-dependent gene regulation (Colecraft et al., 2002; Hibino et al., 2003; Subramanyam et al., 2009; Xu et al., 2011; Tadmouri et al., 2012). A lack of this nonconventional function of the β4 subunit might contribute to the ataxic phenotype in patients and mice with mutations in the β4 gene. However, many aspects of the regulation and function of this new signaling pathway are still controversial.
Here, we report the discovery and characterization of a hitherto unknown β4 splice variant. Our data demonstrate that β4e is the second most abundant β4 variant in cerebellum and that it can interact functionally with CaV2.1 in tsA201 cells and cultured neurons. The three known full-length β4 variants show differential targeting into the distal axon and the nucleus. Its superior capacity to promote CaV2.1 membrane expression and its axonal targeting properties suggest a primary function of the newly found β4e in targeting P/Q-type calcium channels into the nerve terminal. Expression profiling of cerebellar granule cells (CGCs) from lethargic (β4-null) mice individually reconstituted with the β4 splice variants demonstrated that the nuclear β4 subunits specifically regulate neuronal genes, including CaV2.1 and several potassium channels, all of which have been linked previously to ataxia and epilepsy.
Materials and Methods
Primary cultured CGCs.
Cultures from CGCs were grown from postnatal day 7 (P7) BALB/c or lethargic (129/SvJ background) mice of either sex as described previously (Koschak et al., 2007). Neurons were plated on poly-l-lysine-coated coverslips and kept in basal Eagle's medium (Invitrogen) supplemented with 10% FCS, 5 mm KCl, 2 mm glutamine, and 50 g/ml gentamycin. Cytosine βD-arabino furanoside (Ara-C, 10 μm; Sigma-Aldrich) was added 24 h after plating to the neurons to prevent proliferation of non-neuronal cells. Experiments were performed on granule cells differentiating for 7–9 d in vitro.
Primary cultured hippocampal neurons.
Low-density cultures of hippocampal neurons were prepared from 17-d-old embryonic BALB/c or lethargic mice of either sex as described previously (Obermair et al., 2003; Obermair et al., 2004; Kaech and Banker, 2006). Neurons were plated on poly-l-lysine-coated glass coverslips in 60 mm culture dishes at a density of ∼3500 cells/cm2. After plating, cells were allowed to attach for 3–4 h before transferring the coverslips neuron-side down into a 60 mm culture dish with a glial feeder layer. For maintenance, the neurons and glial feeder layer were cultured in serum-free neurobasal medium (Invitrogen) supplemented with Glutamax and B27 supplements (Invitrogen). Ara-C (5 μm) was added 3 d after plating and, once a week, 1/3 of the medium was removed and replaced with fresh maintenance medium.
Plasmids and cloning procedure.
All constructs were cloned into a eukaryotic expression plasmid containing a neuronal chicken β-actin promoter (Obermair et al., 2004). The generation of pβA-β4a-V5, pβA-β4b-V5 and pβA-eGFP has been described previously (Obermair et al., 2004; Subramanyam et al., 2009; Obermair et al., 2010). To construct pβA-β4e-V5, the N terminus of β4e was generated with specific primers and PCR amplified using β4a as a template, thereby introducing an artificial, 5′ HindIII site and a new Kozak sequence, as well as the codons for methionine and alanine in the forward primer (β4eV5-F, 5′-aagcttcctaccatggctgggtcagcagattcctat-3′ and β4eV5-R, 5′-tgtctttatcagctgtgctgg-3′, 850-bp PCR product) and cloned accordingly into pβA-β4a-V5 using HindIII/EcoRV. For generating the viral vectors pHR-βA-β4a, pHR-βA-β4b, and pHR-βA-β4e, the following constructs were used as templates: pβA-β4a-V5, pβA-β4b-V5, and pβA-β4e-V5. The V5 tag was removed by digestion with BglII/SalI. To this end, a stop codon and a SalI site was introduced to the 3′-end of the pβA-βa, pβA-βb, and pβA-βe coding sequence. Genes of interest (pβA-4a, pβA-β4b, or pβA-β4e) were introduced with HindIII/SacI into the pENTRI (Invitrogen) vector and inserted into a custom-built destination vector, pHR-βA-DEST, using LR Clonase II enzyme mixture (GATEWAY; Invitrogen).
Lentiviral transduction and reconstitution of lethargic neurons.
Lentiviruses were produced by transient transfection of confluent 293T cells with the lentiviral expression vectors containing pHR-pβA-β4a, pHR-pβA-β4b, or pHR-pβA-β4e in combination with psPAX2 (packaging plasmid) and the pVSV (envelope plamid) using Metafectene (Biontex Laboratories). The following day, medium was changed to neuronal plating medium and, after 24 and 48 h, supernatants containing the viruses were harvested, sterile filtered (0.20 μm), aliquoted, and stored at −80°C. Cultured hippocampal neurons and cultured CGCs from lethargic mice were transfected with the lentiviral constructs immediately after plating for 4 h. Reconstituted neurons were used for experiments from DIV 1 on.
Transfection of hippocampal neurons.
Expression plasmids were introduced into neurons on day 6 using Lipofectamine 2000-mediated transfection reagent (Invitrogen) as described previously (Obermair et al., 2004). For cotransfection experiments (pβA-β4-V5 and pβA-eGFP), a total amount of 0.75 μg of DNA at a molar ratio of 1:2 was used. Cells were immunostained and analyzed 11–13 d after transfection.
RT-PCR.
RNA was isolated from adult male cerebellum (2 months) and DIV 9 cultured CGCs of BALB/c mice of either sex and RNA concentrations were measured with a NanoDrop 2000 Spectrophotometer. cDNA was prepared as described previously (Schlick et al., 2010). The following primers (MWG Biotec) were used for PCR amplification and identification of β4 splice variants. The reverse primer (β4 R) was identical for all three splice variants: 5′-cactgcgcttggagaatattc-3′; forward primers (F): β4a F, 5′-ctgcatggagttgaagactcg-3′ (yielding a 361 bp product), β4b F, 5′-gcaccacttctaccagcttca-3′ (366 bp), β4e F, 5′-ggtggagtgccagataaagc-3′ (411 bp), and β4a′ F 5′-gactcggaggctgggtca-3′ (346 bp). PCR products were separated in 1% agarose II gels (Amresco) at 50 V. Bands were excised from the gel, DNA was purified using the QIAQuick gel extraction kit, and resulting fragments were sequenced (MWG Biotec).
Quantitative TaqMan RT-PCR.
RNA was isolated from cerebellum, forebrain, and/or hippocampus of 17-d-old embryonic mice of either sex and BALB/c or lethargic male mice (both 2 months old) and from cultured CGCs (BALB/c mice of either sex, DIV 9), as described previously (Schlick et al., 2010). RNA concentrations were measured with a NanoDrop 2000 spectrophotometer. The relative abundance of different CaV subunits and β4 splice variants transcripts was assessed by TaqMan qRT-PCR using a standard curve method as thoroughly described previously (Schlick et al., 2010). The following specific, custom-designed TaqMan gene expression assays were ordered from Life Technologies (R is reverse): β4a F, 5′-tacctgcatggagttgaagact-3′and β4a R, 5′-cgatggcctgcttgtataggaat-3′; β4b F, 5′-cgtcctcgtacggcaagaa-3′ and β4b R, 5′-cctccgggtcgtggtg-3′; and β4e F, 5′-ccgtctctctcagtgccaatg-3′and β4e R, 5′-atggcctgcttgtataggaatctg-3′ and probe sequence are as follows: β4a-FAM 5′-ctgctgacccagcctc-3′; β4b-FAM 5′-tcgggccacctgtgagc-3′; β4x-FAM 5′-ctgacccagccatcaat-3′. The following primers (MWG Biotec, Ebersberg, Germany) were used for standard curve generation using cerebellum cDNA as a template: β4a F, 5′-gcctggtaaatccacaggaa-3′, and β4a R, 5′-caggtttggacttcgctc-3′; β4b F, 5′-gagccgggtagggaagtc-3′, and β4b R, 5′-cctcttccaaggagacatcg-3′; β4e F, 5′-ggtggagtgccagataaagc-3′, and β4e R, 5′-gacttcgctccaggtttg-3′. Standard curves were calculated as described previously (Schlick et al., 2010). Quantitative RT-PCR was performed in triplicate using 20 ng of total RNA equivalents of cDNA and the specific TaqMan gene expression assay in a final volume of 20 μl in TaqMan universal PCR master mix (Applied Biosystems). Data were normalized as described previously (Schlick et al., 2010) and analyzed using the ABI PRISM 7500 sequence detector (Applied Biosystems).
Immunocytochemistry and image processing.
Neurons were fixed in 4% paraformaldehye/4% sucrose in PBS (pF) at room temperature for 20 min and incubated in 5% normal goat serum in PBS containing 0.2% bovine serum albumin (BSA) and 0.2% Triton X-100 (PBS/BSA/Triton) for 30 min (Obermair et al., 2004). The primary antibodies mouse monoclonal anti-β4 (1:500; NeuroMab, University of California–Davis/National Institutes of Health NeuroMab Facility), mouse monoclonal anti-V5 (1:400; Invitrogen), rabbit polyclonal anti CaV2.1 (1:2000), and anti vGLUT1 (1:20,000; both Synaptic Systems) were applied in PBS/BSA/Triton for 4 h at room temperature (RT), washed in PBS, and then stained with goat anti-rabbit Alexa Fluor 488 and/or goat anti-mouse Alexa Fluor 594 (1:4000; Invitrogen) for 1 h at RT. Where relevant, Hoechst 33342 dye (∼5 μg/ml) was applied to the immunostained neurons for 30 s in PBS/BSA/Triton to label the nuclei. After staining, coverslips were washed and mounted in Vectashield to avoid photo bleaching. Preparations were analyzed on an AxioImager microscope (Carl Zeiss) using a 25× 0.8 numerical aperture (NA), 40× 1.3 NA, and 63× 1.4 NA objectives. Fourteen-bit images were recorded with a cooled CCD camera (SPOT; Diagnostic Instruments) and Metaview image-processing software (Universal Imaging). Figures were arranged in Adobe Photoshop CS6 and, where necessary, linear adjustments were performed to correct black level and contrast.
Nuclear-targeting analysis.
The degree of nuclear targeting in cultured hippocampal neurons from lethargic mice with lentivirus reconstitution of βA-β4a, βA-β4b, or βA-β4e and wild-type (WT) controls was determined by calculating the nucleus/cytoplasm ratio of the anti-β4 fluorescence intensity; the analysis was performed by a semiautomated procedure using a custom-programmed MetaMorph Macro journal. Fourteen-bit image pairs of the anti-β4 immunofluorescence and the corresponding Hoechst stain were acquired using the 63× 1.4 NA objective. The Hoechst stain image was thresholded to trace the nuclei and automatically draw the corresponding regions of interest (nucleus ROI). The corresponding cytoplasm ROI was generated by dilating the nucleus ROI by 30 pixels, yielding a ring of 2.5 μm width. Both ROIs were transferred onto the corresponding anti-β4 fluorescence image and their fluorescence intensities were measured. The intensities of the corresponding nucleus and cytoplasm ROIs were background subtracted and the nucleus/cytoplasm ratio was determined. All experiments were repeated in 3–4 different culture preparations along with WT controls; in total, 30 neurons were analyzed. The experimenter was blinded to the experimental conditions.
Quantification of β4-V5 fluorescent intensity and dendritic and axonal expression.
To analyze the subcellular distribution of the heterologously expressed β4-V5-tagged splice variants, the fluorescence intensity of V5 stain was quantified in cultured hippocampal neurons at DIV 17 as described previously (Obermair et al., 2010). For each condition, 2–10 neurons were analyzed in 3–4 independent culture preparations for each condition.
Quantification of density and fluorescent intensity of CaV2.1 clusters.
To analyze the effects of overexpression of β4 splice variants on the membrane expression of endogenous CaV2.1, fluorescence intensity was measured in cultured neurons at DIV 21. Fourteen-bit grayscale images of the CaV2.1 were acquired. ROIs were drawn on ∼70- to 100-μm-length dendritic segments containing CaV2.1 synapses terminating on dendritic spines. After 2D deconvolution (MetaMorph), images were thresholded to trace the fluorescent clusters using the integrated morphometric analysis option and their average gray values were measured and corrected by background subtraction. For each condition, between 4 and 16 neurons were analyzed in each three independent experiments.
Electrophysiological recordings.
For whole-cell patch-clamp experiments, tsA201 cells were seeded into a 25 cm2 flask and transfected with: 1.5 μg of CaV2.1 (pβA-eGFP-α1A), 1.0 μg of pβA-β4(a, b or e), 1.25 μg of α2δ-1, and 1.25 μg of pUC. Six to 8 h after transfection, the medium was changed; cells were kept at 37°C, 10% CO2, and recorded after 16–20 h. The charge carrier was 2 and 15 mm Ca2+ and 2 and 15 mm Ba2+. Recordings were performed at room temperature as described previously (Koschak et al., 2007; Watschinger et al., 2008) using the following solutions containing the following (in mm): for the internal solution, 135 CsCl, 10 Cs-EGTA, and 1 MgCl2 adjusted to pH 7.4 with CsOH; for the recording solution, 15 BaCl2 or CaCl2, 10 HEPES, 150 choline-Cl, and 1 MgCl2 adjusted to pH 7.4 with CsOH. When 2 mm Ca2+ was used, choline-Cl was increased to 163 mm. To determine the current–voltage relationship, cells were clamped at a holding potential of −98 mV and depolarized for 300 ms to potentials between −78 mV and +72 mV in 10 mV increments. I–V curves were fitted to the equation I = Gmax (V − Vrev)/{1 + exp[(V0.5act-V)/k]}, where Vrev is the extrapolated reversal potential, V is the test potential, I is the peak current amplitude, Gmax is the maximum slope conductance, V0.5,act is the half maximal activation voltage, and k is the slope factor. To guarantee high quality, voltage-clamp currents bigger than 3 nA were excluded from the analysis.
Western blot.
Myotubes of the homozygous dysgenic (mdg/mdg) cell line GLT were cultured and transfected with plasmids pβA-β4a, pβA-β4b, or pβA-β4e as described previously (Powell et al., 1996; Subramanyam et al., 2009). From DIV 7 GLTs and from cerebellum of 2-month-old BALB/c male mice, protein was extracted and homogenized in RIPA buffer containing the following (in mm): 50 Tris-HCl, pH 8, 150 NaCl2, 10 NaF, and 0.5 EDTA, along with 0.10% SDS, 10% glycerol, and 1% igepal with a pestle and mortar. Protein concentrations were determined by Bradford assay (Bio-Rad Laboratories). Ten micrograms of protein from GLTs and 60 μg from cerebellum was loaded per lane onto a 10% Bis-Tris Gel (Novex Invitrogen precast) run at 196 V and 40 mA for 90 min. The blot was performed at 25 V and 100 mA for 3 h at 4°C with a semidry-blot system (Roth). The primary mouse anti-β4 (1:10,000; Neuormab) was applied overnight at at 4°C and HRP-conjugated secondary antibody (Pierce) was incubated for 1 h at room temperature, the development was performed with ECL Supersignal West Pico kit (Pierce) and ImageQuant LAS 4000 was used to visualize the bands.
Affymetrix GeneChip analysis.
The whole-genome gene expression data were obtained at the Expression Profiling Unit of the Medical University Innsbruck using the Affymetrix GeneChip Mouse Genome 430 2.0 Array. Sample preparation was performed according to the manufacturer's protocols. In brief, RNA quantity and purity was determined by optical density measurements (OD 260/280 ratio) and by measuring the RNA integrity using the Agilent Technologies 2100 Bioanalyzer. Then, 500 ng of RNA per sample were processed to generate biotinylated hybridization targets using the Affymetrix GeneChip 3′ IVT Express kit and the Affymetrix GeneChip hybridization, wash, and stain kit. Resulting targets, in total 12.5 μg of fragmented and labeled RNA, were hybridized to the Affymetrix GeneChip Mouse Genome 430 2.0 and stained in an Affymetrix fluidic station 450. Raw fluorescence signal intensities were recorded by an Affymetrix scanner 3000 and image analysis was performed with the Affymetrix GeneChip Command Console software (AGCC). Quality assessment and preprocessing of the microarrays was done in R using the Bioconductor packages affyPLM (Bolstad et al., 2004) and GCRMA (Wu et al., 2004), respectively. Differential gene expression analysis was performed using the limma package (Smyth, 2004). Initial raw data quality controls established that all samples and the corresponding microarrays were of comparably high quality. Nevertheless, principal component and cluster analysis based on the preprocessed expression values indicated strong batch effects between the three cultures that needed to be considered in subsequent bioinformatic analyses. For each probe set, linear models adjusted for experimental batches were fitted to the preprocessed expression values. The extent and significance of differential expression between the individual β4 subunits and the eGFP control were computed based on the individual model fits. The associated p-values were adjusted for multiple hypotheses testing to control the false discovery rate (FDR; Benjamini and Hochberg, 1995). Finally, genes with an M-value >0.7, representing >1.6-fold regulation, and an FDR of smaller than 5% were reported as significantly differentially expressed. The raw and preprocessed microarray data have been submitted to the Gene Expression Omnibus (accession number GSE50822).
Gene ontology.
Gene ontology (GO) analysis was performed in R using software packages from Bioconductor. In particular, the GOstats package (Falcon and Gentleman, 2007) was used to perform conditional hyper geometric testing (Alexa et al., 2006) to enrich for specific GO terms. To obtain the required foreground and background gene sets, probe sets were mapped to their corresponding ENTREZ gene identifiers using the mouse 4302.db annotation package. ENTREZ genes annotated to GO and targeted by at least one probe set with an absolute log2-fold change of >0.7 and a FDR of <10% for at least one of the performed comparisons were used as foreground gene set. The background gene set included all ENTREZ genes annotated to GO and detectable on the microarrays. GO terms with a p-value <0.05 were considered statistically significant.
Statistical analysis.
Results are expressed as means ± SEM except where otherwise indicated. Data were organized and analyzed in Excel, GraphPad, and R statistical software.
Results
Identification of a new splice variant of the calcium channel β4 subunit
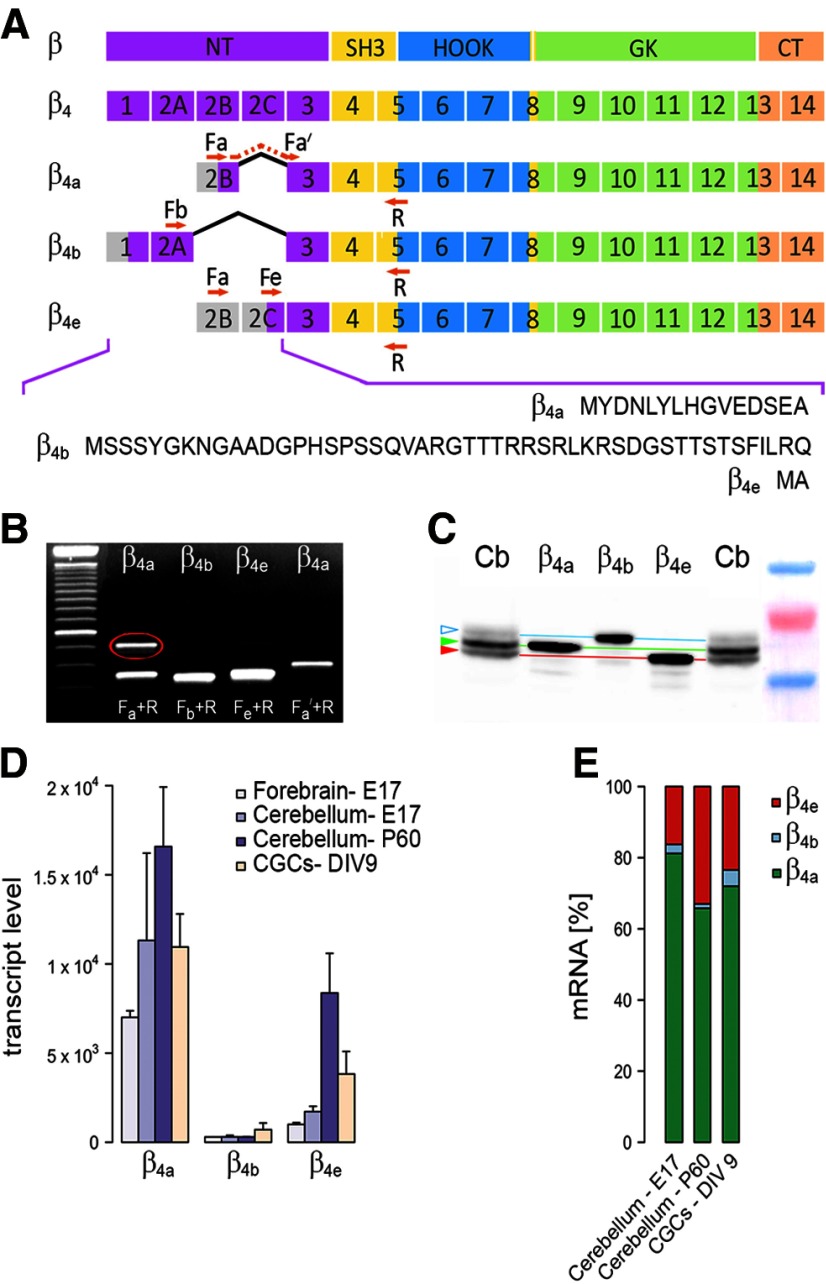
RT-PCR analysis of RNA extracts of cultured mouse CGCs using primer pairs designed to specifically detect β4a and β4b amplified the expected PCR fragments of the known splice variants plus an additional larger fragment with the β4a primer pair Fa/R (Fig. 1A,B). In the Ensemble genome database, the properties of this second PCR fragment matched a hitherto unidentified β4 transcript (ENSMUST00000102761) that, like β4a, starts with exon 2B but then inserts a unique exon 2C before the conserved exon 3 (Fig. 1A). Therefore, we designed new forward primers (Fa′ and Fe) to discriminate between the β4a transcript and the proposed β4 splice variant. Using these specific primer sets, expression of the proposed β4 transcript in CGC extracts was confirmed (Fig. 1B). Sequencing the PCR product of the Fe/R primer pair verified the identity of the new β4 transcript conclusively.
Figure 1.
Expression of calcium-channel β4 splice variants in mouse brain and primary cultured cerebellar granule cells. A, Exon and domain structure of β4 splice variants; the NH2 terminus (purple), the SH3 domain (yellow), the HOOK (blue), the GK domain (green), and the COOH terminus (orange). Alternatively spliced exons 2 are designated 2A, 2B and 2C; positions of specific forward primers (Fa, Fa′, Fb and Fe) and the common reverse primer (R) are indicated. Bottom, Amino acid sequences of the variable N termini of the three β4 splice variants. B, RT-PCR assays using the primers shown in A reveal the two known (β4a and β4b) and a novel (β4e and upper band in lane 1; red circle) splice variant in cultured CGCs. C, Western blot analysis of cerebellar extracts (lanes 1 and 5; Cb) and extracts of dysgenic myotubes transfected with β4a, β4b, or β4e (lanes 2–4) labeled with pan-β4 antibody. The green and red arrowheads indicate the two major β4 bands in cerebellar extracts corresponding to β4a and β4e, respectively; the open arrowhead indicates the position of the faint β4b band (representative blot of n = 3). D, Quantitative (TaqMan) RT-PCR of β4a, β4b and β4e transcripts in embryonic (E17) forebrain and cerebellum, in adult cerebellum (P60), and in CGCs (DIV 9; mean ± SEM, n = 3). E, Relative expression levels of β4a, β4b, and β4e transcripts in cerebellum (E17, P60) and CGCs (DIV 9; n = 3).
Quantitative TaqMan RT-PCR analysis with specific probes for the two known and the newly detected β4 transcripts demonstrated that the new splice variant is amply expressed in extracts of mouse cerebellum and cultured CGCs (Fig. 1D). Because the designations β4a-d were already assigned to other β4 transcripts, we termed the new β4 splice variant β4e. Direct comparison of relative transcript levels in cerebellum (P60) and CGC cultures revealed that, in both preparations, β4a is the most abundant β4 transcript (66% and 72%, respectively), followed by the β4e transcript (33% and 23.5%, respectively; Fig. 1E). Surprisingly, β4b transcript levels were comparatively low, amounting only to 1% and 4.5% of total β4 transcripts in cerebellum and CGCs, respectively (Fig. 1E). qRT-PCR analysis of cerebellar extracts of embryonic day 17 (E17) mice revealed similar expression ratios (β4a 81%; β4b 2.5%; β4e 16.5%) of the three β4 transcripts (Fig. 1D), excluding the possibility that expression levels of the three β4 variants undergo a significant isoform shift upon maturation of the cerebellum.
The β4e transcript contains a translation initiation site near the 3′ end of exon 2C. Therefore, the variable N terminus of the predicted β4e protein is composed of only two residues, methionine and alanine (Fig. 1A). Because the virtual lack of a unique β4e sequence precludes the generation of a splice-variant-specific antibody, we compared the β4 bands labeled in Western blots of cerebellar extracts with those of the three β4 splice variants expressed individually in dysgenic myotubes using a pan-β4 antibody directed against the common C terminus of β4 subunits. The three heterologously expressed β4 splice variants each showed a single band at the expected relative positions (Fig. 1C), indicating that full-length β4a, β4b, and β4e can be discerned readily based on their distinct migration on the gel. In cerebellar extracts, the β4 antibody labeled two strong bands and one faint band. Consistently, the most prominent band corresponded to the position of heterologous β4a, the second most intensive band below corresponded to heterologous β4e, and an upper band corresponding to the position of β4b was barely detectable. Therefore, the relative amounts of the putative β4a, β4e, and β4b subunits in cerebellar extracts resemble the relative amounts of transcript detected with qRT-PCR (Fig. 1D,E). Together, these results demonstrate that neurons express on the mRNA and protein level at least three full-length calcium channel β4 subunits and that, in cerebellum, the newly identified β4e subunit is the second most abundant splice variant.
β4e modulates expression and current properties of CaV2.1
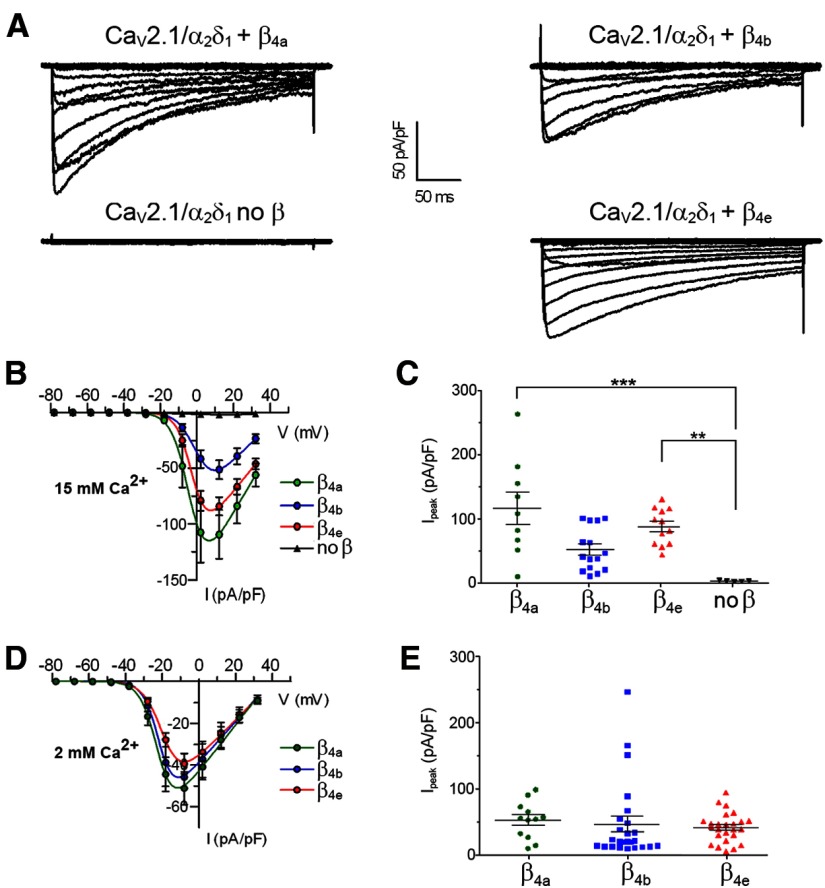
Next, we investigated whether the newly identified β4e variant can interact functionally with calcium channel α1 subunits. When coexpressed with the CaV2.1 α1 subunit in tsA201 cells, β4e modulated the current properties in a way typical for β subunits (Fig. 2A). This indicates that β4e can form a functional complex with CaV2.1 and promotes the surface expression and gating of this calcium channel. Compared with CaV2.1 expressed alone, coexpression of β4e increased the current density on average 40-fold (p < 0.01) and shifted the voltage dependence of activation by >15 mV toward hyperpolarizing potentials (Fig. 2B,C; Table 1). To reveal potential functional differences between the β4 splice variants, calcium and barium currents (at concentrations of 2 and 15 mm) were recorded in tsA201 cells transfected with CaV2.1 plus β4a, β4b, or β4e (Table 1). The current densities showed considerable variability; however, the magnitude of current modulation by β4e was always in the same range as that of the other two β4 splice variants. Most importantly, at physiological extracellular calcium concentrations, both the mean current density and gating properties of all three β4 splice variants were very similar (Fig. 2D,E; Table 1). Therefore, heterologously expressed β4e and CaV2.1 can interact functionally with one another in mammalian cells and the modulatory effects of β4e on P/Q-type calcium currents are not significantly different from those of β4a and β4b.
Figure 2.
Expression of the newly detected β4e splice variants increases the current density of the P/Q type calcium channels. A, Representative whole-cell currents recorded from tsA201 cells transfected with CaV2.1, α2δ-1, and β4a, β4b, β4e or without a β subunit. Cells were depolarized for 300 ms from a holding potential of −98 mV to potentials between −78 mV and +72 mV at 10 mV increments. B–E, CaV2.1 current–voltage relationships and current densities recorded in 15 mm (B, C) or 2 mm calcium (D, E). Note that in B and C, CaV2.1 current densities in cells expressing one of the β4 splice variants were greatly increased compared with those without β subunit (pA/pF at Vmax, no β: 2.2 ± 0.4, β4a: 120.0 ± 26.3, β4b: 52.8 ± 8.9, β4e: 89.8 ± 8.4; means ± SEM). Multiple comparison was performed using a Kruskal–Wallis test with Dunn's post hoc test (***p < 0.001, **p < 0.01). For details of gating properties and number of experiments, see Table 1.
Table 1.
Biophysical parameters and statistical comparison of three β4 splice variants (β4a, β4b, and β4e) in the presence of different charge carriers
| β4a |
β4b |
β4e |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | ±SEM | n | Mean | ±SEM | n | Mean | ±SEM | n | |
| CD (pA/pF) | |||||||||
| 15 mm Ca2+ | 120.0 | 26.3 | 9 | 52.8 | 8.9 | 15 | 89.8 | 8.4 | 12 |
| 15 mm Ba2+ | 101.6 | 25.7 | 4 | 53.7 | 10.0 | 12 | 118.9 | 49.0 | 5 |
| 2 mm Ca2+ | 52.9 | 8.2 | 12 | 47.0 | 12.1 | 25 | 41.8 | 4.6 | 25 |
| 2 mm Ba2+ | 85.7 | 17.9 | 10 | 71.1 | 19.1 | 17 | 59.1 | 10.9 | 18 |
| V50act(mV) | |||||||||
| 15 mm Ca2+ | −1.8 | 1.3 | 9 | 0.2 | 1.1 | 15 | −2.5 | 1.0 | 12 |
| 15 mm Ba2+ | −14.9 | 2.1 | 4 | −13.2 | 1.0 | 12 | −15.6 | 2.0 | 5 |
| 2 mm Ca2+ | −21.2 | 1.1 | 12 | −19.7 | 0.7 | 25 | −19.4 | 1.4 | 25 |
| 2 mm Ba2+ | −30.4* | 1.6 | 10 | −25.1 | 0.8 | 17 | −24.2++ | 1.0 | 18 |
| kact | |||||||||
| 15 mm Ca2+ | 4.3 | 0.3 | 9 | 4.6 | 0.3 | 15 | 3.5 | 0.3 | 12 |
| 15 mm Ba2+ | 4.2 | 0.3 | 4 | 3.7 | 0.4 | 12 | 4.0 | 0.3 | 5 |
| 2 mm Ca2+ | 4.1 | 0.3 | 12 | 4.5 | 0.2 | 25 | 4.1 | 0.2 | 25 |
| 2 mm Ba2+ | 3.7 | 0.3 | 10 | 3.6 | 0.1 | 17 | 3.7 | 0.2 | 18 |
| Vrev (mV) | |||||||||
| 15 mm Ca2+ | 52.2 | 1.1 | 9 | 50.7 | 1.6 | 15 | 55.9**+ | 0.8 | 12 |
| 15 mm Ba2+ | 43.6 | 1.1 | 4 | 43.3 | 1.1 | 12 | 44.8 | 1.6 | 5 |
| 2 mm Ca2+ | 38.4 | 0.4 | 12 | 39.5 | 0.5 | 25 | 39.6 | 0.9 | 25 |
| 2 mm Ba2+ | 29.9 | 1.1 | 10 | 31.5 | 0.7 | 17 | 30.9 | 0.7 | 18 |
CD, current density; V50act, half-maximal voltage of activation; kact, slope of activation curve. Data are presented as means ± SEM. Multiple comparisons was performed using Kruskal–Wallis test with Dunn's post hoc test.
++/**p < 0.01; *p < 0.05; */**compared with β4b; +/++compared with β4a.
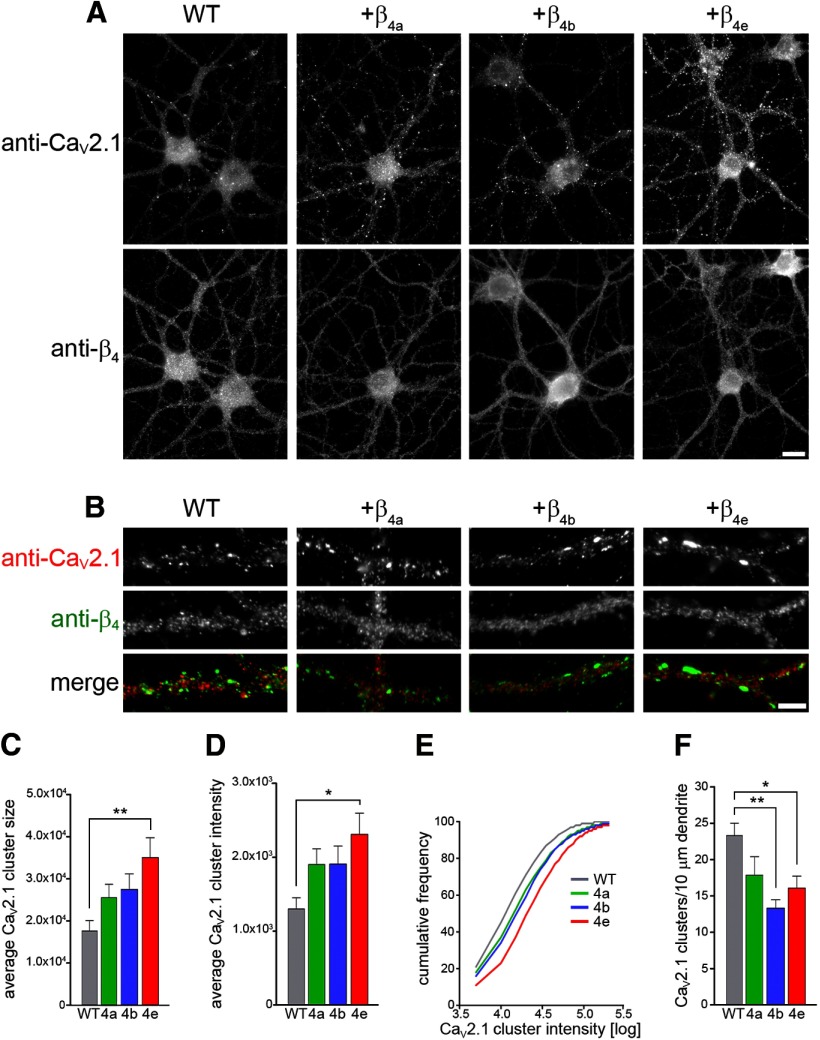
A β-subunit-induced increase in calcium current density is generally interpreted as an increase in expression or the stability of the channel in the membrane. To determine whether β4e can increase surface expression of native CaV2.1 in neurons and to reveal how this property compares to that of β4a and β4b, we overexpressed β4a, β4b, and β4e in cultured hippocampal neurons and analyzed the expression of the endogenous CaV2.1 in synaptic clusters. Figure 3A shows representative immunofluorescence micrographs of differentiated hippocampal neurons (DIV 21) double labeled with antibodies against CaV2.1 and the β4 subunit. The clustered distribution of CaV2.1 corresponds to synapses along axons making contact with the dendrites of the depicted neurons. β4 staining shows a homogenous expression of endogenous plus heterologous β4 subunits in the somata and throughout the neuronal processes. Overexpression of β4a, β4b, or β4e increased the total β4 signal, but did not alter the overall distribution pattern of β4 subunits or the morphology of the transfected neurons. Synaptic clusters of CaV2.1 are visible with and without expression of additional β4 subunits (Fig. 3B). However, synaptic CaV2.1 staining appears more robust in all three overexpressed conditions.
Figure 3.
Overexpression of β4 splice variants increases synaptic expression of CaV2.1 calcium channels in hippocampal neurons. A, Representative micrographs of wild-type hippocampal neurons transfected with the lentiviral constructs βA-β4a, βA-β4b or βA-β4e at plating and immunolabeled with anti-CaV2.1 and anti-β4 at DIV 21. Scale bar, 10 μm. B, Dendritic segments of immunolabled neurons showing the synaptic CaV2.1 clusters. Scale bar, 5 μm. Note that overall expression and distribution of β4 subunits is comparable in wild-type and transfected neurons. C, D, Overexpression of any of the β4 splice variants results in an increase in the average CaV2.1 cluster size (C) and the average cluster intensity (D), which is significant with β4e. **p < 0.01; *p < 0.05. E, Cumulative frequency plots of the CaV2.1 cluster intensity. F, In parallel, the density of clusters along the dendrite decreases. **p < 0.01; *p < 0.05, ANOVA and Tukey post hoc analysis.
This effect was quantified by analyzing the cluster size and fluorescence intensity—two parameters reflecting the number of channels per cluster—and the density of synaptic CaV2.1 along dendrites. Both the average cluster size (Fig. 3C) and the fluorescence intensity (Fig. 3D) were increased upon overexpression of any one of the β4 subunits and this increase was most prominent and statistically significant when β4e was overexpressed in the neurons. The cumulative frequency distribution (Fig. 3E) shows that this increase in CaV2.1-labeling intensity and the difference between the effects of β4a β4b, and β4e occurred homogenously throughout the entire population of CaV2.1 clusters (small to large). However, overexpression of β4a, β4b, or β4e did not increase the density of synaptic CaV2.1 clusters (Fig. 3F), indicating that β4 subunits play no essential role in synapse formation. On the contrary, cluster density was reduced, but this effect might be the result of virtual fusion of closely neighboring clusters upon thresholding as their size increases. Together, these results demonstrate that the newly identified β4e subunit can interact functionally with CaV2.1 in tsA201 cells and in cultured hippocampal neurons. In the neurons, the additional β4 subunits increase the number of CaV2.1 channels in synaptic clusters, but not the number of synaptic clusters themselves. The observation that β4e more potently increased expression of synaptic CaV2.1 than the other examined β4 splice variants suggests that β4e may be the natural partner of this predominantly presynaptic calcium channel in hippocampal neurons.
Three full-length β4 splice variants are distributed differentially in the axons of cultured hippocampal neurons
Because β4a and β4b were shown previously to be targeted differentially in neurons (Vendel et al., 2006), we compared the subcellular distribution of the newly identified β4e with that of β4a and β4b in cultured hippocampal neurons. Overexpression of C-terminally V5-tagged β4 subunits allowed us to analyze specifically the expression of the individual splice variants in neuronal compartments using a V5 antibody. Figure 4A shows that, overall, β4a-V5, β4b-V5, and β4e-V5 were similarly expressed and distributed in the soma and throughout the processes of the neurons. Absolute expression levels differed from cell to cell, but the mean expression levels of the three β4 splice variants in the proximal dendrite were not significantly different from each other (Fig. 4B). Furthermore, intensity analysis showed a uniform decline of all three splice variants in the first 250 μm of the dendrites (Fig. 4C). In addition, overall expression levels in the proximal 250 μm of the axon were similar for all three splice variants, except for an accumulation of certain β4 subunits in the axon hillock (Fig. 4D,E). As described previously (Obermair et al., 2010), β4b-V5 was heavily stained in the axon hillock. Here, we also observed a similar accumulation of β4e-V5 in the axon hillock of many neurons (Fig. 4F,G). However, this phenomenon was not observed in neurons expressing β4a-V5 (Fig. 4F,G).
Figure 4.
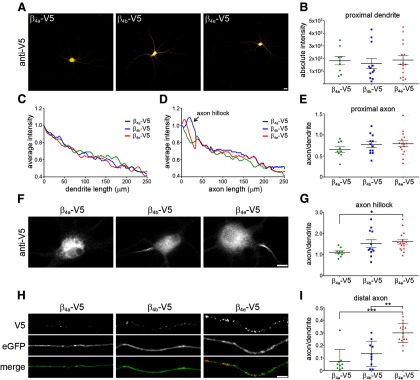
Somatodendritic and axonal distribution pattern of heterologously expressed β4 splice variants in cultures of wild-type hippocampal neurons. Cultured hippocampal neurons were transfected at DIV 6 with β4a-V5, β4b-V5, or β4e-V5, together with eGFP, and labeled with an antibody against the C-terminal V5 epitope (DIV 17). A, Immunostaining of all three V5-tagged β4 splice variants in hippocampal neurons show an overall similar expression pattern. Scale bar, 10 μm. B, C, Quantification of the absolute fluorescence intensity in the first 30 μm of dendrites and the relative decline of fluorescence intensity in 250 μm of dendrites show no difference in expression pattern of the three β4 splice variants. D, E, Quantification of the mean fluorescence intensity in the proximal axon and the first 250 μm of the axon length showed an overall similar distribution pattern; however, as shown in the micrograph (F), a strong accumulation of β4b-V5 and β4e-V5 was observed in the axon hillock (G; *p < 0.05; ANOVA and Tukey post hoc analysis). Scale bar, 5 μm. H, I, In the distal axon (∼1 mm from the soma), all three β4-V5 splice variants displayed a similar clustered staining pattern. However, total axonal expression intensity of β4e was significantly higher than that of β4a and β4b. **p < 0.01; ***p < 0.001, Kruskal–Wallis and ANOVA analysis; 3–4 culture; n = 13–20 neurons). Scale bar, 5 μm.
Importantly, the β4 splice variants also showed a differential expression in the distal axon. Representative example images of distal axon segments indicate higher expression of β4e-V5 compared with β4a-V5 and β4b-V5 (Fig. 4H). Quantification demonstrated that the average fluorescence intensity in the axon 1 mm distant from the soma relative to that of the proximal dendrite (compare Fig. 4B as a reference for overall expression levels of the individual neurons) was >3-fold higher for β4e-V5 compared with β4a-V5 and β4b-V5 (Fig. 4I). This dominant expression of the newly identified β4e variant in the presynaptic compartment of hippocampal neurons is consistent with its higher potency for stimulating CaV2.1 expression in synaptic clusters (Fig. 3).
Three full-length β4 splice variants differ in their activity-dependent nuclear-targeting properties in cultured hippocampal neurons
In addition to differential targeting into presynaptic and postsynaptic neuronal compartments, β4b was shown to be unique among a range of examined β subunits in its ability to localize to the nucleus (Subramanyam et al., 2009). This calcium-channel-independent property of β4b was shown unambiguously in myotubes and hippocampal neurons, but in neurons, the developmental sequence and activity dependence of β4b nuclear targeting is still controversial (Tadmouri et al., 2012). To analyze the nuclear-targeting properties of all three β4 subunit splice variants in hippocampal neurons, we prepared cultures from E17 lethargic (β4-null) mice and reconstituted them individually with untagged β4a, β4b, or β4e using viral transfection. The advantage of using reconstituted lethargic neurons compared with overexpression in wild-type neurons is that the individual β4 splice variants can be analyzed without potential interference of the endogenous β4 subunits and that untagged β4 subunits can be analyzed using the pan-β4 antibody.
Figure 5A shows the qRT-PCR expression profile of high-voltage-activated calcium channel subunits in hippocampus of adult wild-type and lethargic mice. The wild-type expression profile is similar to that previously published by us (Schlick et al., 2010) and expression levels of calcium channel subunits in lethargic mice show little to no differences from WT. This suggests that the lack of the β4 subunits in lethargic mice does not result in compensatory expression of any of the other calcium channel subunits. Apparently, the other three β isoforms, which together make up ∼80% of β transcripts in hippocampus, absorb the lack of β4 subunits. As described previously (Burgess et al., 1997; Lin et al., 1999b), a truncated β4 transcript is still expressed from the mutated β4 gene in lethargic mice. Nevertheless, immunolabeling of WT and lethargic hippocampal neurons with the pan-β4 antibody confirmed the total absence of β4 subunit protein in the lethargic neurons (Fig. 5B). Figure 5C shows the somata of lethargic hippocampal neurons, reconstituted with β4a, β4b, or β4e, fixed, and immunolabeled with the pan-β4 antibody between 1 and 14 days after plating. β4b shows a strong nuclear localization up to day three after plating; thereafter, nuclear targeting of the β4b splice variant declines rapidly. Some neurons expressing β4a also show nuclear targeting during the first days in culture. In contrast, neurons expressing β4e show little to no nuclear targeting at any time of differentiation. To compare quantitatively nuclear targeting of the three β4 splice variants during differentiation of hippocampal neurons, we analyzed the nucleus to cytoplasm ratio of β4 immunolabel and plotted it against the days in culture (Fig. 5D). This time course shows that in the neurons nuclear targeting is limited to the first 4 d in culture and that, during this period, the rank order of the mean nuclear targeting is β4b>β4a>β4e. Because the extent of nuclear targeting varied between individual neurons, we composed a frequency distribution diagram of the nucleus/cytoplasm ratio of β4b at the different time points (Fig. 5E). This graph highlights the prevalence of nuclear targeting in young neurons (DIV 1 and DIV 2), the decline of nuclear targeting at DIV 3, and the lack thereof at 5, 7, and 14 d in culture. Interestingly, little to no β4 nuclear targeting was observed in wild-type neurons. However, this is consistent with the low expression levels of the β4b splice variant in brain (Fig. 1D,E).
Figure 5.
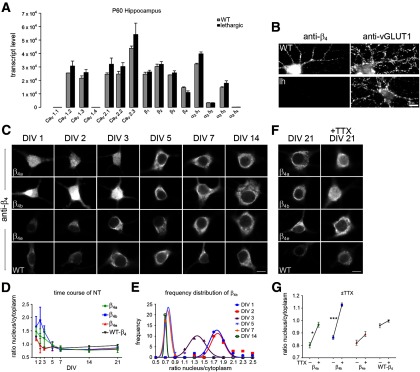
Nuclear-targeting properties of the three β4 splice variants individually expressed in hippocampal neurons of lethargic mice. A, Quantitative RT-PCR revealed similar expression of CaV subunit isoforms in WT and lethargic adult hippocampus (mean ± SEM, n = 3). B, Double immunofluorescence labeling of WT and lethargic hippocampal cultures with anti-β4 and anti-vGLUT1 demonstrated the complete absence of β4 protein in the lethargic neurons. C, Cultured hippocampal neurons from lethargic mice reconstituted with βA-β4a, βA-β4b, or βA-β4e and WT controls immunolabeled with anti-β4. Immature neurons (DIV1, 2, and 3) showed strong nuclear targeting of β4b and, at a lower degree, of β4a; no nuclear targeting of β4e was observed at any developmental stage. D, The nucleus/cytoplasm ratio decreases to <1 within 5 d in culture. E, Frequency distribution analysis of nucleus/cytoplasm ratio of β4b in cultured hippocampal neurons from lethargic mice in different developmental stages showed nuclear targeting only in immature neurons. F, G, Blocking the spontaneous activity in mature neurons by 1 μm TTX for 12 h significantly increased nuclear targeting of β4a and β4b, but not β4e. *p < 0.05; **p < 0.001, unpaired t test. No significant nuclear targeting could be observed in cultures from wild-type hippocampal neurons at any stage of development and with or without TTX. Scale bar, 10 μm.
Because the developmental stage of the decline in β4 nuclear targeting coincided with the onset of spontaneous electric activity of the hippocampal neuron cultures, we analyzed in mature hippocampal neurons the effect of blocking electrical activity on nuclear targeting of all β4 splice variants. Consistent with the developmental time course shown above, representative images of hippocampal neurons differentiated in culture for 21 d show no nuclear targeting of any of the β4 splice variants. However, after 12 h of incubation with the sodium channel blocker TTX (1 μm), β4b and, to a lower extent, β4a accumulated in the neuronal nuclei, whereas β4e did not (Fig. 5F). Quantitative analysis showed that the nucleus/cytoplasm ratio of β4b increased with high significance and that of β4a to a lesser degree but still significantly (Fig. 5G). In contrast, the nucleus/cytoplasm ratio of β4e showed no significant increase in response to TTX treatment. As expected because of the low basal nuclear targeting observed for native β4 subunits in young wild-type cultures, blocking activity with TTX in differentiated (DIV 21) wild-type neurons also did not result in a significant increase of nuclear targeting. Together, these results confirm the nuclear targeting of β4b in young and electrically silent hippocampal neurons. In addition, they demonstrate that, among the three β4 splice variants, β4b shows the highest degree of nuclear targeting, β4a intermediate levels, and the newly identified β4e variant is not targeted into the nucleus at all. This differential subcellular distribution indicates that the three β4 splice variants may differ in their potential to regulate neuronal genes directly.
Differential nuclear targeting of the three β4 splice variants in cultured cerebellar granule cells
The cerebellar cortex expresses the highest levels of calcium channel β4 subunits (Fig. 6A), and the most striking example of β4 nuclear targeting in native neurons has been observed in the cerebellar granular cell layer (Vendel et al., 2006; Subramanyam et al., 2009; Schlick et al., 2010; Ferrandiz-Huertas et al., 2012). To determine the nuclear-targeting properties of the β4 splice variants in the neurons most relevant for β4 function and to establish an appropriate neuronal cell model for subsequent expression profiling, we prepared primary CGC cultures from lethargic mice and reconstituted them individually with β4a, β4b, and β4e using viral transfection immediately after plating.
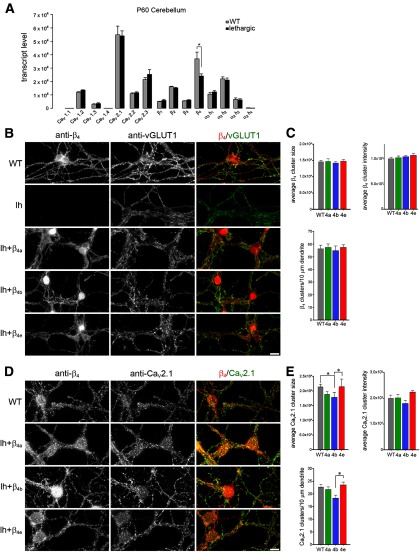
Figure 6.
Expression and distribution of β4 subunits in lethargic cerebellar granule cells reconstituted with β4a, β4b, or β4e. A, Quantitative RT-PCR revealed similar expression of CaV subunit isoforms in WT and lethargic adult cerebellum (mean ± SEM, n = 3). B–E, Cultured cerebellar granule cells from lethargic mice were reconstituted by lentiviral transfection with one of the β4 splice variants, βA-β4a, βA-β4b, or βA-β4e, and immunolabeled with anti-β4, anti-vGLUT1, or anti-β4 and anti-CaV2.1 at DIV 9. B, Wild-type cultures of cerebellar granule cells express β4 subunits in discrete clusters on the processes and around the somata; no expression of β4 subunit was detected in cultures from lethargic mice. C, Quantitative analysis of reconstituted lethargic cultures shows that staining of β4a, β4b, and β4e in the processes is similar to β4 staining in WT controls. However, the three β4 splice variants differ in their localization in nuclei. D, All three β4 splice variants show a similar overall distribution pattern and partial overlap with synaptic CaV2.1 clusters in the soma and along the processes (representative images of 3–4 cultures). E, CaV2.1 cluster size, average cluster intensity, and cluster density along the dendrite were similar to wild-type and to each other, except that size and density of CaV2.1 clusters were reduced upon β4b reconstitution. *p < 0.05, ANOVA and Tukey post hoc analysis. Scale bar, 10 μm.
Quantitative RT-PCR analysis of wild-type mouse cerebellum shows that CaV2.1 and β4 are the predominant α1 and β subunit isoforms, respectively (Fig. 6A; Schlick et al., 2010). In this brain tissue, β4 mRNA amounts to 61% of the total β transcripts. In lethargic cerebellum, β4 transcript levels are significantly reduced to ∼2/3 of control, but no compensatory upregulation of other β-subunit genes can be observed. Cultured CGCs heavily express β4 subunits in the soma and throughout the extensive axonal bundles in a clustered distribution pattern similar to that of presynaptic proteins such as vGLUT1 (Fig. 6B) or RIM1 (data not shown). As expected, CGCs from lethargic mice are entirely devoid of β4 staining. However, this is not accompanied by any apparent changes of the neuronal morphology or the expression and distribution of synaptic proteins. Lentiviral transfection of CGCs with β4a, β4b, or β4e efficiently reconstituted the expression of the β4 splice variants in the great majority of the neurons (Fig. 6B). Quantitative analysis of pan-β4-antibody labeling showed that all three β4 splice variants are expressed at levels comparable to those of total endogenous β4 subunits in wild-type CGCs (Fig. 6C). In addition, the clustered expression patterns in the periphery of the somata and in the axonal bundles were indistinguishable from those of wild-type cultures and no apparent differences between the three β4 splice variants could be discerned (Fig. 6B). In contrast, particularly in β4b-expressing neurons, nuclear staining was conspicuous and occurred more often than in wild-type CGCs or in lethargic CGCs reconstituted with either β4a or β4e. Together, these data indicate that lethargic CGCs can be reconstituted efficiently with the individual β4 splice variants and that β4a, β4b, and β4e are similarly expressed in the membrane but exhibit distinct targeting to the nucleus.
In hippocampal neurons, the newly identified β4e variant displayed increased axonal targeting and a superior capacity to augment surface expression of CaV2.1 (Figs. 4, 5). To analyze the association of the β4 subunits with the predominant presynaptic α1 subunit in CGCs and to reveal potential differences between the three β4 splice variants, we double labeled wild-type and reconstituted CGCs with the pan-β4 antibody and an antibody against CaV2.1 (Fig. 6D). CaV2.1 was localized in discrete clusters throughout the axonal processes of the CGC cultures that were frequently colocalized with presynaptic markers and corresponded to functional release sites as indicated by depolarization-induced FM1–43 dye loading and release (data not shown). The three β4 splice variants were also localized in clusters. Like the clusters of endogenous β4 subunits in wild-type CGCs, these were smaller than the CaV2.1 clusters and more densely distributed throughout the axonal processes. Distance-based colocalization analysis (Bolte and Cordelieres, 2006) confirmed that the β4 distribution in the processes was more extensive than that of CaV2.1 and that the two calcium channel subunits showed only partial colocalization. Importantly, the quantitative analysis did not reveal any significant differences in colocalization of β4a, β4b, or β4e with CaV2.1 in CGCs (data not shown). As expected from the similar expression levels of all β4 splice variants, membrane expression of endogenous CaV2.1 in the reconstituted CGCs was comparable to wild-type levels (Fig. 6E). Interestingly, however, upon reconstitution with β4b CaV2.1, cluster size and density were moderately but significantly reduced.
Three β4 splice variants display differential nuclear-targeting properties in young and differentiated CGCs
Because the double-labeling experiments indicated that differential nuclear targeting of the three β4 splice variants also occurs in CGCs (Fig. 6B,C), we analyzed this calcium-channel-independent property of specific β4 splice variants in more detail in lethargic CGCs individually reconstituted with β4a, β4b, or β4e (Fig. 7). Wild-type CGCs (at DIV 4 and DIV 9) express β4 subunits in the periphery of the somata (presumably associated with calcium channels in the membrane), but display only weak β4 labeling in the nuclei. Conversely, all three reconstituted cultures display some neurons with strong nuclear β4 staining in addition to the peripheral β4 staining (Fig. 7A). This nuclear localization is most prominent in CGCs reconstituted with β4b, but is also seen to a lesser extent in CGCs reconstituted with β4a. In β4e-expressing CGCs, nuclear staining is rarely observed. Interestingly, in CGCs, β4b nuclear targeting was not restricted to young neurons, but was maintained in well differentiated CGCs at day 9 in culture. Quantifying the frequency at which transfected DIV9 neurons show nuclear localization of β4 label greater than that in the periphery of the somata demonstrated a highly significant difference in β4 nuclear targeting between the β4b variant and β4a or β4e (Fig. 7B). β4b accumulated in the nuclei of 57% of the CGCs, whereas such nuclear localization was observed in only 25% with β4a and 13% with β4e. Electrically silencing the cultures by 12 h application of 1 μm TTX further increased nuclear targeting of β4b 72% (p = 0.04), but did not change nuclear targeting in CGCs reconstituted with β4a or β4e (Fig. 7C). These results demonstrate that nuclear targeting of β4 subunits occurs in CGCs in a splice-variant-specific manner (β4b≫β4a>β4e) and that, in contrast to hippocampal neurons, nuclear targeting of the β4b variant is maintained in differentiated neurons.
Figure 7.
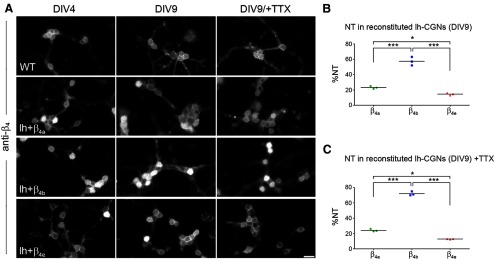
Nuclear-targeting properties of the three β4 splice variants in reconstituted cerebellar granule cells from lethargic mice. Cultured CGCs from lethargic mice were reconstituted by lentiviral transfection with βA-β4a, βA-β4b, or βA-β4e and immunolabeled at DIV 4 and 9 with anti-β4. A, Wild-type and transfected neurons show similar β4 staining in the periphery of the somata. In addition, β4b and, less often, β4a show nuclear localizations at both developmental stages. Scale bar, 10 μm. B, Percentage of reconstituted cultures showing that nuclear targeting of β4b was 2.5 times higher than that of β4a and 4 times higher than that of β4e. ***p < 0.001; *p < 0.05. C, Blocking the spontaneous activity by 1 μm TTX for 12 h at DIV 9 shows a significant increase (p = 0.04) in nuclear targeting of β4b, but no effect on β4a or β4e. ***p < 0.001; *p < 0.05, ANOVA and Tukey post hoc test.
β4 subunits regulate gene expression in CGCs in a splice-variant-specific manner
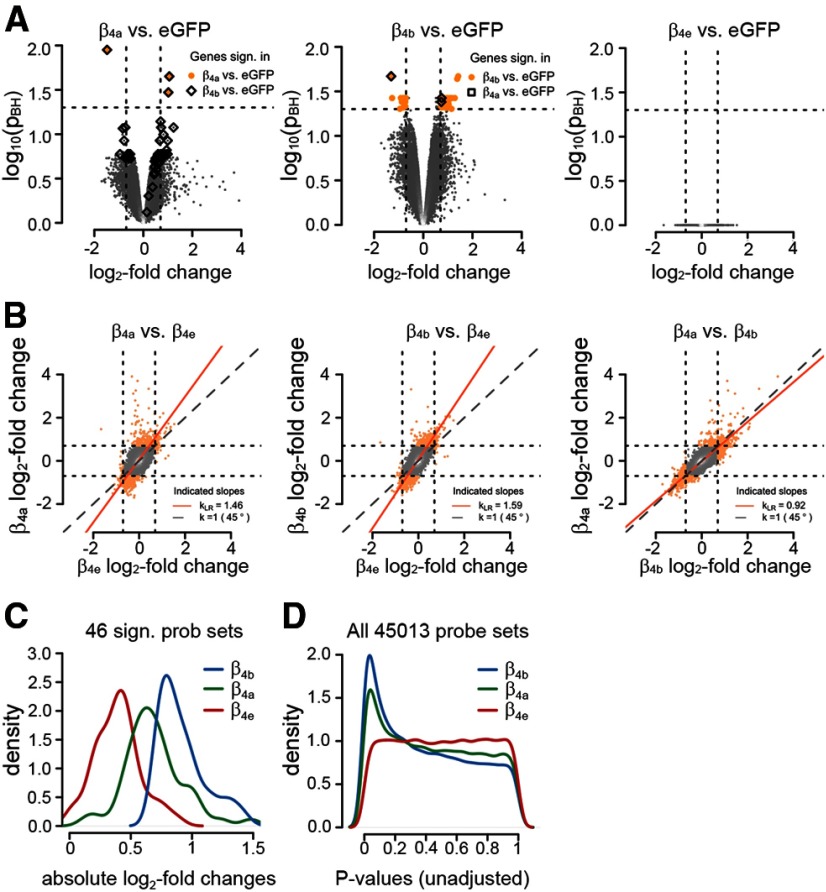
It has been shown previously that β4 subunits interact with nuclear proteins involved in epigenetic control of gene regulation (Hibino et al., 2003; Xu et al., 2011; Tadmouri et al., 2012). Because the capacity to regulate genes by this mechanism is expected to depend on nuclear targeting of the β subunit and because we found that the three full-length β4 splice variants differ in their nuclear-targeting properties, we hypothesized that individual expression of β4a, β4b, or β4e in lethargic CGCs should result in differential regulation of gene expression. Therefore, we performed Affymetrix GeneChip analysis on mRNA extracts from lethargic CGCs reconstituted with β4a, β4b, or β4e and compared the individual expression profiles with that of lethargic CGCs transfected with eGFP as a control (Fig. 8). The microarray experiment was performed in biological triplicates. As hypothesized, expression of individual β4 splice variants in CGCs resulted in the differential upregulation and downregulation of genes (Fig. 8). Volcano plots show substantial gene regulation in β4b-expressing neurons and to a lesser degree in β4a-expressing neurons. Strikingly, no differential regulation of genes was detected in β4e-expressing neurons (Fig. 8A). These differences in gene regulation are also reflected by the numbers of significantly regulated genes. Differential expression analysis of 45,101 probe sets showed that, in CGCs reconstituted with β4a, two genes were significantly upregulated and one gene was significantly downregulated (FDR of 5%, log2-fold change >0.7) compared with eGFP-transfected CGC. In β4b-reconstitued CGCs, 34 genes were significantly upregulated and 12 genes were significantly downregulated. Interestingly, the genes significantly regulated by β4a were also significantly regulated by β4b and most of the genes significantly regulated by β4b were also highly regulated by β4a, although, except for the above three, this did not reach our significance threshold. In contrast, in β4e-reconstituted CGCs no genes were significantly upregulated or downregulated. Therefore, the rank order at which the β4 splice variants regulate genes in the neurons is β4b > β4a > β4e.
Figure 8.
Differential gene expression in CGCs from lethargic mice reconstituted with individual β4 splice variants. RNA was isolated from 9-d-old lethargic CGC cultures reconstituted by lentiviral transfection with βA-β4a, βA-β4b, βA-β4e, or βA-eGFP as a control and analyzed with Affymetrix gene chip mouse genome 430 2.0 arrays (n = 3 separate culture preparations). Dashed lines: horizontal, threshold for significance (pBH < 0.05); vertical, log2-fold change (|M| > 0.7). A, Volcano plots comparing transcript levels in neurons reconstituted with the individual β4 splice variants relative to eGFP controls exhibit the extent and significance of differentially expressed genes. The number of significantly differentially expressed genes (colored data points) is highest with β4b (46), followed by β4a (3), whereas gene expression in β4e-expressing neurons is not significantly different from controls. Significantly regulated genes in β4a are also highly regulated in β4b and vice versa (boxed data points). B, Direct comparison between the splice variants illustrates the differences in overall gene regulation of both β4a and β4b to β4e (least-squares regression [red lines] deviate from 45° axes), whereas differential gene expression in β4a- and β4b-expressing neurons does not differ from each other (scattering along the 45° axes). C, Density plot of the absolute M values shows that differential expression of the 46 significantly regulated genes in β4b (blue) is also higher in β4a (green) than in β4e (red). D, The density blot of raw p-values illustrates that most significant regulation of all genes occurs in β4b, followed by β4a.
Direct comparison of differential expression levels between individual β4 splice variants reveals substantial differences in the extent of gene regulation between β4a or β4b and β4e (Fig. 8B). Conversely, β4a and β4b mostly regulated the same genes in the same direction. This is also illustrated by a density plot of the absolute differential expression levels (absolute M values) of the 46 genes significantly differentially regulated by β4b (Fig. 8C). The same genes are somewhat less, but still highly regulated by β4a. A density plot of the raw p-values for all 45,101 genes reiterates the rank order of gene regulation by the three β4 splice variants (Fig. 8D). The peak of significantly regulated genes is highest in β4b-expressing neurons, lower in β4a-expressing neurons, and nonexistent in β4e-expressing neurons. In total, the bioinformatic analysis of the differentially regulated genes demonstrates that β4 subunits regulate transcription of genes in neurons, that their capacity to do so differs among the three splice variants, and that the rank order of gene regulation (β4b > β4a > β4e) correlates with the rank order of their nuclear-targeting properties.
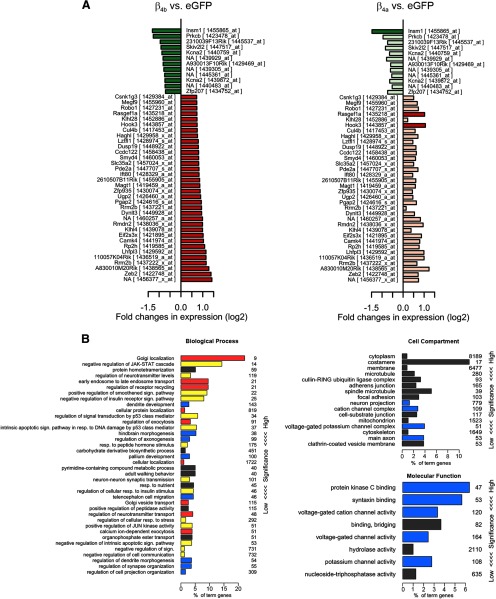
Figure 9A lists the 46 genes significantly upregulated and downregulated in response to expression of β4b and the same set of genes as regulated by β4a. GO analysis (Fig. 9B) demonstrates that β4b (and to a lesser degree β4a) predominantly regulate genes belonging to three groups of biological processes: cellular signaling, membrane/vesicle transport including synaptic transmitter release, and neuronal development. This is consistent with a role of nuclear β4 subunits in regulating genes involved in neuronal activity. Consistent with this function, five of the eight molecular functions revealed by our GO analysis relate to regulation of voltage-gated ion channels and other synaptic proteins. Remarkably, these include CaV2.1, the principal partner of β4 subunits in cerebellar synapses, which is substantially downregulated by β4b and β4a (but not by β4e). This is consistent with the modest reduction of CaV2.1 expression in CGCs reconstituted with β4b (Fig. 6E), and it suggests the existence of a negative feedback loop linking directly the activity of presynaptic calcium channels to their own transcriptional regulation. In active neurons, CaV2.1 and β4 are part of the calcium channel complex that triggers synaptic transmission. In electrically silent neurons, β4 subunits accumulate in the nucleus, where they downregulate the expression of CaV2.1 and other synaptic proteins. Interestingly, β4, CaV2.1 (Fletcher et al., 1996; Burgess et al., 1997; Escayg et al., 2000; Li et al., 2012) and genes of other channels regulated by nuclear β4 subunits (Kcna2, Kcnj12, and Kcnab1) have been linked to ataxia and epilepsy in mice and humans (Young et al., 2009; Xie et al., 2010; Busolin et al., 2011). Therefore, when β4 is mutated, it is possible that the altered regulation of neuronal channels, including CaV2.1, in inactive neurons results in the imbalance of cerebellar network activity and thus in epileptic seizures and motor deficits.
Figure 9.
Genes regulated by β4b and β4a and GO analysis. A, The 46 genes significantly regulated by β4b ranked according to the degree of upregulation (green) and downregulation (red) compared with the regulation of the same genes by β4a (faint colors, p > 0.05 in β4a). B, GO analysis of genes regulated by β4b or β4a by a log2-fold change of >0.7 and a FDR <0.1. The biological processes significantly (p < 0.05) enriched among this set of genes are related to cell signaling (yellow), membrane/vesicle transport including synaptic release (red), and neuronal development (blue). The significantly (p < 0.05) enriched molecular functions are predominantly ion channel and synapse related (blue). Numbers at right indicate the total number of genes of each ontology term.
The mechanism by which β4b exerts regulation of these genes remains to be shown. Unexpectedly, tyrosine hydroxylase, a gene suggested to be regulated by nuclear β4b in complex with protein phosphatases 2A, the thyroid hormone receptor α, and heterochromatin protein 1 γ (Tadmouri et al., 2012), was not among the β4b-regulated genes in our whole transcriptome analysis of cerebellar neurons. However, tyrosine hydroxylase was also not found among the regulated genes in a subsequent study by the same investigators, in which β4b was expressed in HEK293 cells (Ronjat et al., 2013). Moreover, tyrosine hydroxylase is upregulated in cerebellum of many ataxia mutants, including those of CaV2.1 and α2δ-2 calcium channel subunits (Sawada et al., 1999; Donato et al., 2006; Miki et al., 2008; Li et al., 2012). Together, these observations indicate that upregulation of tyrosine hydroxylase in the lethargic mouse may result from altered calcium channel functions rather than from the nuclear function of β4b.
Discussion
Here, we discovered the expression of a third, hitherto unidentified N-terminal splice variant of the calcium channel β4 subunit isoform in mouse cerebellum. We also demonstrate that the newly identified β4e subunit interacts functionally with CaV2.1 in heterologous cells and in neurons, that the three β4 splice variants are differentially targeted into axons and neuronal nuclei, that nuclear targeting is a prerequisite for differential regulation of genes by β4 splice variants in cerebellar granule cells, and that nuclear β4b specifically regulates CaV2.1 and several potassium channel genes, all of which have been linked to ataxia and epilepsy.
So far, published evidence suggested the existence of two N-terminal β4 splice variants (β4a and β4b), each with additional variations in the C terminus (giving rise to β4c and β4d, respectively; Buraei and Yang, 2010). Our present study provides evidence for the expression of another N-terminal β4 splice variant resulting from usage of a third alternative exon 2 (2C). Like β4a, the β4e transcript skips exons 1 and 2a and starts with exon 2b. However, different from β4a, the β4e transcript includes an additional exon 2c, which contains another translation initiation site near its 3′ end. Therefore, the variable N-terminal domain of the predicted β4e protein is only two residues (met, ala) long and β4e (473 aa) is shorter than β4a (468 aa) and β4b (519 aa). This was confirmed by our Western blot analysis of recombinant β4 variants, and a corresponding native protein band was detected in cerebellar extracts. Both quantitative RT-PCR and Western blot analysis indicate that β4e is the second most abundant β4 variant expressed in mouse cerebellum. Its gel migration just below β4a and detection with an antibody directed against a C-terminal epitope further indicate that the native β4e variant expressed in mouse cerebellum contains all C-terminal exons, thus representing the third full-length β4 splice variant expressed in mammalian brain.
The newly identified β4e subunit interacts functionally with neuronal calcium channels. When coexpressed with CaV2.1 in tsA201 cells, β4e normalizes P/Q-type calcium currents. Like β4a and β4b, β4e increased the current density by 40-fold and shifted the voltage dependence of activation toward hyperpolarized potentials. This is consistent with previous reports demonstrating that the N terminus of the β4 subunit is not essential for modulating channel gating (Vendel et al., 2006). However, we did not consistently observe splice variant-specific differences in modulation of current properties (Helton and Horne, 2002). In cultured hippocampal neurons, overexpression of β4e increased the size and fluorescence intensity of presynaptic CaV2.1 clusters, demonstrating that β4e can interact functionally with CaV2.1 channels in their native neuronal environment. Previously, we demonstrated differential effects of a range of β-subunit isoforms on membrane expression of CaV1.2 channels in hippocampal neurons (Obermair et al., 2010). Interestingly, β4b (the only β4 isoform examined at that time) had little effect on CaV1.2 membrane expression compared with β1 and β2 isoforms, suggesting that β4 subunits are poor partners for l-type channel in the somatodendritic compartment. In contrast, here, we find robust effects of β4e on expression of CaV2.1 in axons, which is suggestive of a predominant presynaptic function of this β4 splice variant. Vendel et al. (2006) found previously that β4a is highly expressed in the molecular layer of the cerebellar cortex and that its N terminus specifically interacts with synaptotagmin 1, both indicating a presynaptic function of β4a in parallel fiber–Purkinje cell synapses. Here, we show that β4e is even more highly expressed in the distal axons of hippocampal neurons and is the most potent β4 variant in enhancing CaV2.1 surface expression. Therefore, it is tempting to speculate that β4e and β4a are both expressed in the presynaptic compartment but serve complementary functions in channel targeting and calcium-dependent neurotransmitter secretion, respectively.
Conversely, the newly identified β4e subunit exhibited poor nuclear-targeting properties in neurons. The finding that the β4e splice variant, which essentially lacks the variable N terminus, shows the lowest degree of nuclear targeting is consistent with the importance of N-terminal sequences in determining the accumulation of β4 subunits in the nucleus (Subramanyam et al., 2009). In hippocampal neurons and cultured CGCs, the β4b splice variant showed the highest degree of nuclear targeting and β4a intermediary levels. The observation that β4a can also be targeted into the nucleus—although at a substantially lower level as β4b—is consistent with our own previous findings (Subramanyam et al., 2009) and with independent studies showing nuclear targeting of β4c, a truncated β4 splice variant sharing the N terminus with β4a (Hibino et al., 2003; Xu et al., 2011). Together, these findings demonstrate that the specific N-terminal sequences determine the distinct nuclear-targeting properties of the three full-length β4 splice variants, but do not exclude alternative mechanisms by which β subunits can be imported into the nucleus. As described previously (Subramanyam et al., 2009), β4 nuclear targeting was limited to young and electrically silent hippocampal cultures, indicating that, in spontaneously active hippocampal neurons, nuclear export mechanisms prevail over nuclear import. Interestingly, Tadmouri et al. (2012) recently reported exactly the opposite and explain the discrepancy between their and our previous findings (Subramanyam et al., 2009) with a possible interference of the V5 tag with the nuclear-targeting mechanisms. Our present experiments exclude this possibility by demonstrating nuclear targeting of untagged β4 splice variants in young and electrically silenced hippocampal neurons from lethargic mice. In CGCs, β4 nuclear targeting persisted throughout differentiation in vitro, suggesting that regulation of β4 nuclear targeting differs between neuronal cell types. Moreover, this explains our previous findings of β4 nuclear targeting in granule cells in adult mouse and rat cerebellum (Subramanyam et al., 2009). Surprisingly, however, our quantitative RT-PCR analysis revealed very low levels of β4b expression relative to β4a and β4e. Therefore, the previously observed nuclear targeting of native β4 in cerebellum may arise, at least in part, from nuclear targeting of the β4a splice variant, which exhibits only ∼1/3 as much nuclear targeting as β4b but is 65% more highly expressed in adult cerebellar cortex.
Splice-variant-specific differences in nuclear-targeting properties inevitably will result in different functions of the β4 splice variants in neurons. Because of their interactions with proteins of the epigenetic machinery, a role of β4 subunits in transcriptional regulation has been proposed (Hibino et al., 2003; Xu et al., 2011; Tadmouri et al., 2012). Indeed, heterologous expression of wild-type and C-terminally truncated β4b in HEK cells resulted in differential regulation of genes, possibly as a consequence of their distinct nuclear-targeting properties (Ronjat et al., 2013). In the present study, we tested this hypothesis in cerebellar neurons from lethargic mice individually reconstituted with the three β4 splice variants. We chose cultured CGCs because these neurons expressed the highest levels of β4 subunits and displayed β4 nuclear targeting in the native brain tissue (Subramanyam et al., 2009). Our immunofluorescence analysis verified that, in the reconstituted neurons, all three β4 variants were expressed at levels comparable to total β4 in wild-type neurons and that they were normally distributed throughout the neurons. Therefore, these cultures differed primarily with regard to the nuclear targeting of the three β4 splice variants. Consequently, lethargic CGCs reconstituted with β4a, β4b, or β4e represent an optimal cell system with which to study potential differences in gene regulation by the three β4 splice variants in differentiated neurons.
Because all three β4 splice variants interacted similarly with CaV2.1 calcium channels and because gene regulation was only observed with β4 splice variants that also showed nuclear targeting, our GeneChip expression analysis provides compelling evidence demonstrating that calcium channel β subunits regulate gene expression in neurons independently of their calcium-channel-related functions. Reconstitution of CGCs with individual β4 splice variants resulted in the upregulation and downregulation of neuronal genes and the number of regulated genes correlated directly with the nuclear-targeting properties of the splice variants. Earlier analysis of microarray data from wild-type and lethargic mouse cerebellum revealed numerous differentially expressed genes (Tadmouri et al., 2012). Whereas this indicated a role of β4 subunits in transcriptional regulation, altered gene regulation in the null-mutant mice might result from altered calcium channel functions or from secondary effects related to the lethargic phenotype (Hosford et al., 1999; Lin et al., 1999a; Khan and Jinnah, 2002). Interestingly, in our study, gene expression of lethargic CGCs reconstituted with β4e was not different from lethargic controls, even though β4e expression and localization were similar to that in wild-type neurons and β4e-dependent augmentation of CaV2.1 membrane expression indicated normal calcium-channel-related functions of this β4 splice variant. Apparently, the reconstitution of β4 calcium channel functions alone in β4e-transfected CGCs did not result in altered gene expression. In contrast, expression of the β4 splice variants that showed nuclear targeting (β4b and β4a) resulted in differential regulation of genes and nuclear targeting and gene regulation correlated quantitatively with one another. Thus, it is very likely that the differences in gene regulation result from the different nuclear-targeting properties of the β4 splice variants. The fact that β4b and β4a mostly regulated the same set of genes further indicates that, in the nucleus, both β4 splice variants activate the same regulatory mechanism. Therefore, the differences in the variable N terminus of the β4 splice variants determine the differences in nuclear targeting but not the mechanism of gene regulation itself. This is in agreement with findings showing that the interactions of β4 variants with proteins of the epigenetic machinery, such as heterochromatin protein-1γ (Hibino et al., 2003; Xu et al., 2011) or the regulatory subunit of protein phosphatases-2A (Tadmouri et al., 2012), are sensitive to truncations of the C terminus but not of the N terminus.
In summary, this study supports the notion that members of the heterogeneous family of calcium channel β subunits serve differential functions in the brain. These involve both calcium channel functions and calcium-channel-independent functions such as the regulation of genes through interactions with nuclear proteins. Among the three β4 splice variants, the newly discovered β4e subunit primarily serves calcium-channel-dependent functions in the presynaptic compartment, whereas the β4b variant and, to a lesser degree, β4a are also targeted into the nuclei of developing and electrically silent neurons, where they regulate the expression of genes directly, including the principal channel partner of β4 in cerebellar synapses, CaV2.1, and other ion channels that have been linked to ataxia and epilepsy.
Footnotes
This work was supported by the Austrian Science Fund (Grants P23479, P24079, W1101, F4406, and F4408). We thank Anita Kofler and Barbara Gschirr for excellent technical support with the gene chip analysis and Benedikt Nimmervoll for help with the FM dye experiments.
The authors declare no competing financial interests.
References
- Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Arikkath J, Campbell KP. Auxiliary subunits: essential components of the voltage-gated calcium channel complex. Curr Opin Neurobiol. 2003;13:298–307. doi: 10.1016/S0959-4388(03)00066-7. [DOI] [PubMed] [Google Scholar]
- Barclay J, Rees M. Mouse models of spike-wave epilepsy. Epilepsia. 1999;40:17–22. doi: 10.1111/j.1528-1157.1999.tb00894.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Bolstad BM, Collin F, Simpson KM, Irizarry RA, Speed TP. Experimental design and low-level analysis of microarray data. Int Rev Neurobiol. 2004;60:25–58. doi: 10.1016/S0074-7742(04)60002-X. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Buraei Z, Yang J. The ss subunit of voltage-gated Ca2+ channels. Physiol Rev. 2010;90:1461–1506. doi: 10.1152/physrev.00057.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DL, Jones JM, Meisler MH, Noebels JL. Mutation of the Ca2+ channel beta subunit gene Cchb4 is associated with ataxia and seizures in the lethargic (lh) mouse. Cell. 1997;88:385–392. doi: 10.1016/S0092-8674(00)81877-2. [DOI] [PubMed] [Google Scholar]
- Busolin G, Malacrida S, Bisulli F, Striano P, Di Bonaventura C, Egeo G, Pasini E, Cianci V, Ferlazzo E, Bianchi A, Coppola G, Elia M, Mecarelli O, Gobbi G, Casellato S, Marchini M, Binelli S, Freri E, Granata T, Posar A, Parmeggiani A, Vigliano P, Boniver C, Aguglia U, Striano S, Tinuper P, Giallonardo AT, Michelucci R, Nobile C. Association of intronic variants of the KCNAB1 gene with lateral temporal epilepsy. Epilepsy Res. 2011;94:110–116. doi: 10.1016/j.eplepsyres.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Campiglio M, Di Biase V, Tuluc P, Flucher BE. Stable incorporation versus dynamic exchange of beta subunits in a native Ca2+ channel complex. J Cell Sci. 2013;126:2092–2101. doi: 10.1242/jcs.jcs124537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Few AP. Calcium channel regulation and presynaptic plasticity. Neuron. 2008;59:882–901. doi: 10.1016/j.neuron.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Chen YH, Li MH, Zhang Y, He LL, Yamada Y, Fitzmaurice A, Shen Y, Zhang H, Tong L, Yang J. Structural basis of the alpha1-beta subunit interaction of voltage-gated Ca2+ channels. Nature. 2004;429:675–680. doi: 10.1038/nature02641. [DOI] [PubMed] [Google Scholar]
- Colecraft HM, Alseikhan B, Takahashi SX, Chaudhuri D, Mittman S, Yegnasubramanian V, Alvania RS, Johns DC, Marbán E, Yue DT. Novel functional properties of Ca(2+) channel beta subunits revealed by their expression in adult rat heart cells. J Physiol. 2002;541:435–452. doi: 10.1113/jphysiol.2002.018515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolphin AC. Calcium channel auxiliary alpha2delta and beta subunits: trafficking and one step beyond. Nat Rev Neurosci. 2012;13:542–555. doi: 10.1038/nrn3311. [DOI] [PubMed] [Google Scholar]
- Donato R, Page KM, Koch D, Nieto-Rostro M, Foucault I, Davies A, Wilkinson T, Rees M, Edwards FA, Dolphin AC. The ducky(2J) mutation in Cacna2d2 results in reduced spontaneous Purkinje cell activity and altered gene expression. J Neurosci. 2006;26:12576–12586. doi: 10.1523/JNEUROSCI.3080-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, De Waard M, Lee DD, Bichet D, Wolf P, Mayer T, Johnston J, Baloh R, Sander T, Meisler MH. Coding and noncoding variation of the human calcium-channel beta4-subunit gene CACNB4 in patients with idiopathic generalized epilepsy and episodic ataxia. Am J Hum Genet. 2000;66:1531–1539. doi: 10.1086/302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Ferrandiz-Huertas C, Gil-Mínguez M, Luján R. Regional expression and subcellular localization of the voltage-gated calcium channel beta subunits in the developing mouse brain. J Neurochem. 2012;122:1095–1107. doi: 10.1111/j.1471-4159.2012.07853.x. [DOI] [PubMed] [Google Scholar]
- Fletcher CF, Lutz CM, O'Sullivan TN, Shaughnessy JD, Jr, Hawkes R, Frankel WN, Copeland NG, Jenkins NA. Absence epilepsy in tottering mutant mice is associated with calcium channel defects. Cell. 1996;87:607–617. doi: 10.1016/S0092-8674(00)81381-1. [DOI] [PubMed] [Google Scholar]
- Hanlon MR, Berrow NS, Dolphin AC, Wallace BA. Modelling of a voltage-dependent Ca2+ channel beta subunit as a basis for understanding its functional properties. FEBS Lett. 1999;445:366–370. doi: 10.1016/S0014-5793(99)00156-8. [DOI] [PubMed] [Google Scholar]
- Helton TD, Horne WA. Alternative splicing of the beta 4 subunit has alpha1 subunit subtype-specific effects on Ca2+ channel gating. J Neurosci. 2002;22:1573–1582. doi: 10.1523/JNEUROSCI.22-05-01573.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino H, Pironkova R, Onwumere O, Rousset M, Charnet P, Hudspeth AJ, Lesage F. Direct interaction with a nuclear protein and regulation of gene silencing by a variant of the Ca2+-channel beta 4 subunit. Proc Natl Acad Sci U S A. 2003;100:307–312. doi: 10.1073/pnas.0136791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosford DA, Lin FH, Wang Y, Caddick SJ, Rees M, Parkinson NJ, Barclay J, Cox RD, Gardiner RM, Hosford DA, Denton P, Wang Y, Seldin MF, Chen B. Studies of the lethargic (lh/lh) mouse model of absence seizures: regulatory mechanisms and identification of the lh gene. Adv Neurol. 1999;79:239–252. [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Khan Z, Jinnah HA. Paroxysmal dyskinesias in the lethargic mouse mutant. J Neurosci. 2002;22:8193–8200. doi: 10.1523/JNEUROSCI.22-18-08193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschak A, Obermair GJ, Pivotto F, Sinnegger-Brauns MJ, Striessnig J, Pietrobon D. Molecular nature of anomalous l-type calcium channels in mouse cerebellar granule cells. J Neurosci. 2007;27:3855–3863. doi: 10.1523/JNEUROSCI.4028-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhou Y, Tian X, Kim TY, Ito N, Watanabe K, Tsuji A, Niimi K, Aoyama Y, Arai T, Takahashi E. New ataxic tottering-6j mouse allele containing a Cacna1a gene mutation. PLoS One. 2012;7:e44230. doi: 10.1371/journal.pone.0044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Wang Y, Hosford DA. Age-related relationship between mRNA expression of GABA(B) receptors and calcium channel beta4 subunits in cacnb4lh mice. Brain Res Mol Brain Res. 1999a;71:131–135. doi: 10.1016/S0169-328X(99)00161-8. [DOI] [PubMed] [Google Scholar]
- Lin F, Barun S, Lutz CM, Wang Y, Hosford DA. Decreased (45)Ca(2)(+) uptake in P/Q-type calcium channels in homozygous lethargic (Cacnb4lh) mice is associated with increased beta3 and decreased beta4 calcium channel subunit mRNA expression. Brain Res Mol Brain Res. 1999b;71:1–10. doi: 10.1016/S0169-328X(99)00141-2. [DOI] [PubMed] [Google Scholar]
- Miki T, Zwingman TA, Wakamori M, Lutz CM, Cook SA, Hosford DA, Herrup K, Fletcher CF, Mori Y, Frankel WN, Letts VA. Two novel alleles of tottering with distinct Ca(v)2.1 calcium channel neuropathologies. Neuroscience. 2008;155:31–44. doi: 10.1016/j.neuroscience.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermair GJ, Kaufmann WA, Knaus HG, Flucher BE. The small conductance Ca2+-activated K+ channel SK3 is localized in nerve terminals of excitatory synapses of cultured mouse hippocampal neurons. Eur J Neurosci. 2003;17:721–731. doi: 10.1046/j.1460-9568.2003.02488.x. [DOI] [PubMed] [Google Scholar]
- Obermair GJ, Szabo Z, Bourinet E, Flucher BE. Differential targeting of the l-type Ca2+ channel alpha 1C (CaV1.2) to synaptic and extrasynaptic compartments in hippocampal neurons. Eur J Neurosci. 2004;19:2109–2122. doi: 10.1111/j.0953-816X.2004.03272.x. [DOI] [PubMed] [Google Scholar]
- Obermair GJ, Schlick B, Di Biase V, Subramanyam P, Gebhart M, Baumgartner S, Flucher BE. Reciprocal interactions regulate targeting of calcium channel beta subunits and membrane expression of alpha1 subunits in cultured hippocampal neurons. J Biol Chem. 2010;285:5776–5791. doi: 10.1074/jbc.M109.044271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opatowsky Y, Chen CC, Campbell KP, Hirsch JA. Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain. Neuron. 2004;42:387–399. doi: 10.1016/S0896-6273(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Powell JA, Petherbridge L, Flucher BE. Formation of triads without the dihydropyridine receptor alpha subunits in cell lines from dysgenic skeletal muscle. J Cell Biol. 1996;134:375–387. doi: 10.1083/jcb.134.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronjat M, Kiyonaka S, Barbado M, De Waard M, Mori Y. Nuclear life of the voltage-gated Cacnb4 subunit and its role in gene transcription regulation. Channels (Austin) 2013;7:119–125. doi: 10.4161/chan.23895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada K, Komatsu S, Haga H, Sun XZ, Hisano S, Fukui Y. Abnormal expression of tyrosine hydroxylase immunoreactivity in cerebellar cortex of ataxic mutant mice. Brain Res. 1999;829:107–112. doi: 10.1016/S0006-8993(99)01347-5. [DOI] [PubMed] [Google Scholar]
- Schlick B, Flucher BE, Obermair GJ. Voltage-activated calcium channel expression profiles in mouse brain and cultured hippocampal neurons. Neuroscience. 2010;167:786–798. doi: 10.1016/j.neuroscience.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Subramanyam P, Obermair GJ, Baumgartner S, Gebhart M, Striessnig J, Kaufmann WA, Geley S, Flucher BE. Activity and calcium regulate nuclear targeting of the calcium channel beta4b subunit in nerve and muscle cells. Channels (Austin) 2009;3:343–355. doi: 10.4161/chan.3.5.9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmouri A, Kiyonaka S, Barbado M, Rousset M, Fablet K, Sawamura S, Bahembera E, Pernet-Gallay K, Arnoult C, Miki T, Sadoul K, Gory-Faure S, Lambrecht C, Lesage F, Akiyama S, Khochbin S, Baulande S, Janssens V, Andrieux A, Dolmetsch R, Ronjat M, Mori Y, De Waard M. Cacnb4 directly couples electrical activity to gene expression, a process defective in juvenile epilepsy. EMBO J. 2012;31:3730–3744. doi: 10.1038/emboj.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Petegem F, Clark KA, Chatelain FC, Minor DL., Jr Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain. Nature. 2004;429:671–675. doi: 10.1038/nature02588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendel AC, Terry MD, Striegel AR, Iverson NM, Leuranguer V, Rithner CD, Lyons BA, Pickard GE, Tobet SA, Horne WA. Alternative splicing of the voltage-gated Ca2+ channel beta4 subunit creates a uniquely folded N-terminal protein binding domain with cell-specific expression in the cerebellar cortex. J Neurosci. 2006;26:2635–2644. doi: 10.1523/JNEUROSCI.0067-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watschinger K, Horak SB, Schulze K, Obermair GJ, Wild C, Koschak A, Sinnegger-Brauns MJ, Tampe R, Striessnig J. Functional properties and modulation of extracellular epitope-tagged Ca(V)2.1 voltage-gated calcium channels. Channels (Austin) 2008;2:461–473. doi: 10.4161/chan.2.6.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. doi: 10.1198/016214504000000683. [DOI] [Google Scholar]
- Xie G, Harrison J, Clapcote SJ, Huang Y, Zhang JY, Wang LY, Roder JC. A new Kv1.2 channelopathy underlying cerebellar ataxia. J Biol Chem. 2010;285:32160–32173. doi: 10.1074/jbc.M110.153676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Li X, Han J, Vogt DL, Wittemann S, Mark MD, Herlitze S. Facilitation versus depression in cultured hippocampal neurons determined by targeting of Ca2+ channel Cavbeta4 versus Cavbeta2 subunits to synaptic terminals. J Cell Biol. 2007;178:489–502. doi: 10.1083/jcb.200702072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lee YJ, Holm JB, Terry MD, Oswald RE, Horne WA. The Ca2+ channel beta4c subunit interacts with heterochromatin protein 1 via a PXVXL binding motif. J Biol Chem. 2011;286:9677–9687. doi: 10.1074/jbc.M110.187864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CC, Stegen M, Bernard R, Müller M, Bischofberger J, Veh RW, Haas CA, Wolfart J. Upregulation of inward rectifier K+ (Kir2) channels in dentate gyrus granule cells in temporal lobe epilepsy. J Physiol. 2009;587:4213–4233. doi: 10.1113/jphysiol.2009.170746. [DOI] [PMC free article] [PubMed] [Google Scholar]