Abstract
Uveal melanoma (UM) is unique among cancers in displaying reduced endogenous levels of sister chromatid exchange (SCE). Here we demonstrate that FANCD2 expression is reduced in UM and that ectopic expression of FANCD2 increased SCE. Similarly, FANCD2-deficient fibroblasts (PD20) derived from Fanconi anaemia patients displayed reduced spontaneous SCE formation relative to their FANCD2-complemented counterparts, suggesting that this observation is not specific to UM. In addition, spontaneous RAD51 foci were reduced in UM and PD20 cells compared with FANCD2-proficient cells. This is consistent with a model where spontaneous SCEs are the end product of endogenous recombination events and implicates FANCD2 in the promotion of recombination-mediated repair of endogenous DNA damage and in SCE formation during normal DNA replication. In both UM and PD20 cells, low SCE was reversed by inhibiting DNA-PKcs (DNA-dependent protein kinase, catalytic subunit). Finally, we demonstrate that both PD20 and UM are sensitive to acetaldehyde, supporting a role for FANCD2 in repair of lesions induced by such endogenous metabolites. Together, these data suggest FANCD2 may promote spontaneous SCE by influencing which double-strand break repair pathway predominates during normal S-phase progression.
Keywords: FANCD2, homologous recombination, non-homologous end-joining, uveal melanoma, sister chromatid exchange, RAD51
Introduction
Sister chromatid exchange (SCE) occurs spontaneously within cells at a frequency that varies between cell types.1, 2, 3 These spontaneous SCEs are considered a product of normal DNA replication or endogenous DNA damage. In addition, SCEs are induced by a variety of DNA-damaging agents such as mitomycin C (MMC)4 or ionising radiation;5 thus, alterations in DNA repair pathways have been associated with changes in SCE.
The main pathway associated with SCE is homologous recombination (HR); however, depleting cells of HR proteins results in conflicting data. All HR-defective DT40 chicken cells show reduced spontaneous and MMC-induced SCEs;6 however, neither Rad51D mutant CHO nor mouse fibroblast cell lines show reduced spontaneous SCEs relative to their wild-type equivalents.7 In addition, spontaneous SCE is not affected in Rad54 knockout mice but the frequency of MMC-induced SCE is decreased,8 whereas CHO cells deficient in Rad51C show decreased spontaneous and induced SCEs.9 Add to this the fact that human heterozygous carriers of the BRCA2 germline mutation exhibit increased spontaneous SCEs,10 whereas Brca2 knockout mouse embryonic stem cells exhibit reduced spontaneous and MMC-induced SCEs,11 and it becomes obvious that the link between HR and SCE is complex, perhaps with different proteins controlling spontaneous and induced SCEs and interspecies differences.
Another repair pathway associated with alterations in SCE and linked to HR is the Fanconi anaemia (FA) pathway. In FA patients a defect in any of the FA complementation group proteins results in shared clinical and cellular phenotypes, promoting the idea of a common biochemical pathway. One of the characteristics of all FA cells is hypersensitivity to DNA crosslinking agents such as MMC.12 The FA pathway is therefore associated with repair of DNA interstrand crosslinks (ICLs) and FA cells exhibit reduced MMC-induced SCE.13 The association between FA and spontaneous SCE is less clear. However, support for FANCD2 in responding to endogenous damage does exist. During S phase, FANCD2 is monoubiquitinated and spontaneously forms foci in S-phase cells.14 These foci colocalise with BRCA1 and RAD51, suggesting that FANCD2 may be involved in HR in response to endogenous DNA damage that disrupts replication. A known end point of such recombination is SCE;15 thus, it is possible that FANCD2 is also involved in controlling spontaneous SCE formation.
We recently reported that spontaneous SCE in primary cultures of uveal melanoma (UM) and UM-derived cell lines is decreased below normal baseline levels, a phenomenon unique to UM when compared with multiple other cancers.16 Here we demonstrate that complementation of UM cells with FANCD2 increases SCE. In addition, deficiency in FANCD2 also reduced spontaneous SCE in other human cell lines including the FA patient-derived cell line PD20. In both cases, spontaneous RAD51 foci formation was reduced, linking FANCD2 in these instances to the promotion of HR. These data provide insight into the molecular basis of UM, the function of FANCD2 during endogenous replication stress and the mechanism of spontaneous SCE formation.
Results
UM cell lines and short-term primary UM cultures exhibit low levels of FANCD2 protein
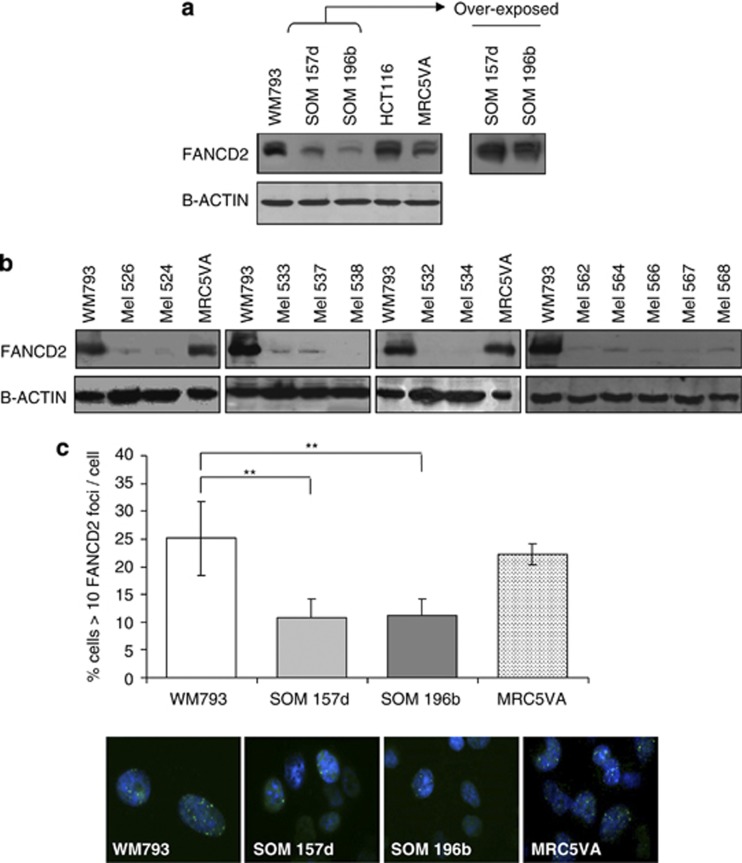
The expression of a panel of proteins involved in DNA repair was determined by western blotting of two established long-term UM cell lines (SOM157d and SOM196b), the cutaneous melanoma cell line WM793 and two other control cell lines routinely used in the lab (HCT116 and MRC5VA). Of the proteins tested (Supplementary Table 1), only FANCD2 expression was reduced in UM compared with all controls (Figure 1a). Consistent with this finding, FANCD2 expression was low in 11 primary UM short-term cultures (STCs) (Figure 1b), and only one primary culture tested showed higher levels (data not shown). In contrast, FANCD2 levels in primary cells from other cancer and normal tissues were not reduced (Supplementary Figure 1), demonstrating that low FANCD2 is not a general finding of primary material. FANCD2 expression is regulated through the cell cycle,14 being monoubiquitinated during the S phase. No differences in proliferation rate or cell cycle progression were found in established UM cell lines SOM196b and SOM157d compared with WM793 or MRC5VA, excluding cell cycle differences or proliferation rates as the direct cause of reduced FANCD2 expression (Supplementary Figure 1). In untreated UM cell lines (SOM157d and SOM196b), both FANCD2S and the monoubiquitinated form FANCD2L were seen, although at lower levels, than control WM793 cells (Figure 1a—more clearly seen in overexposed panel, right). In addition, in SOM196b and SOM157d, the upper FANCD2L band increased in intensity upon treatment with 10 Gy ionising radiation, indicating increased monoubiquitination in response to DNA damage (Supplementary Figure 2). Thus, although FANCD2 expression is reduced in UM, the remaining protein can be monoubiquitinated. In normal replicating cells, monoubiquitination of FANCD2 facilitates the formation of spontaneous FANCD2 foci during S phase of the cell cycle. UM did exhibit spontaneous FANCD2 foci (Figure 1c) but, consistent with lower expression, the number of cells with >10 foci was reduced ∼2.5-fold in each of the UM cell lines compared with WM793 and MRC5VA controls (Student's t-test P<0.01, for WM793 compared with either SOM157d or SOM196b).
Figure 1.
FANCD2 expression is reduced in UM. (a) Western blot for FANCD2 in the cutaneous melanoma cell line WM793, two UM-derived cell lines SOM157d and SOM196b, the colorectal cancer cell line HCT116 and the transformed fibroblast cell line MRC5VA. (b) Western blot for FANCD2 in primary cells extracted from different UM patients (labelled Mel and then the sample number) and control cells WM793 and MRC5VA. (c) Quantification of endogenous FANCD2 foci formation in WM793, SOM157d, SOM196b and MRC5VA cells. Average and s.d. of at least three repeats is shown. Significance was determined using Student's t-test; **P<0.01. Representative images of immunofluorescent staining for FANCD2 (green) and 4,6-diamidino-2-phenylindole (DAPI; blue) are shown.
Promoter methylation changes allow E2F-1 binding in UM
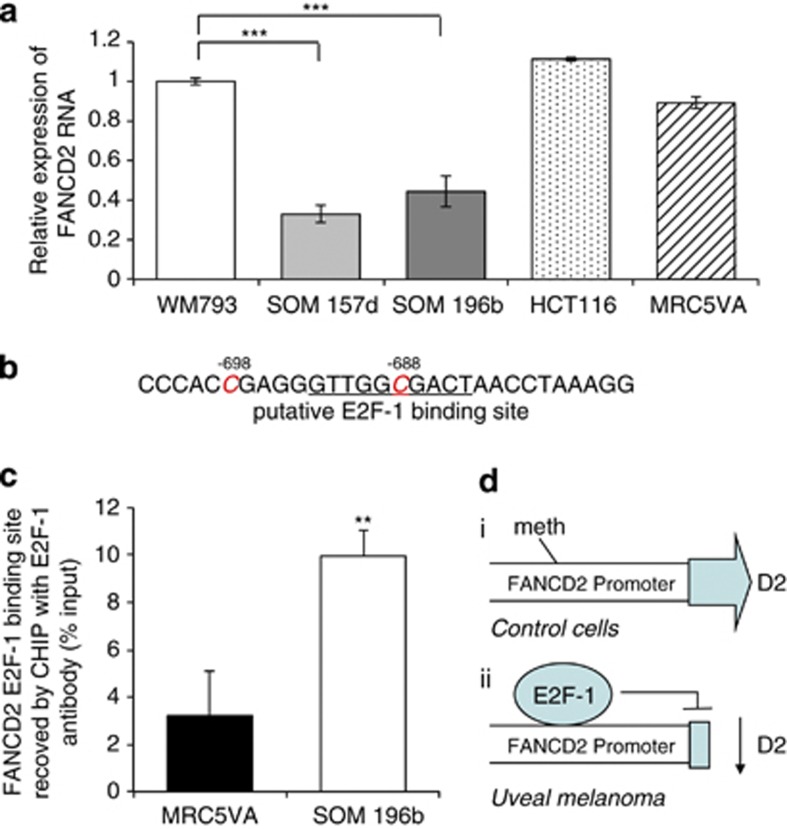
The expression of FANCD2 mRNA was determined by quantitative reverse transcriptase PCR (qRT–PCR) (Figure 2a). FANCD2 mRNA was reduced approximately two-fold in UM cell lines SOM157d and SOM196b compared with the cutaneous melanoma cell line WM793, and MRC5VA and HCT116 controls (Student's t-test P<0.001, for both cell lines compared with each of the controls), suggesting that in UM downregulation of FANCD2 is at the transcriptional level. Control of gene expression in cancer is often altered by changes in promoter methylation. The promoter region 1000 bp upstream of FANCD2 was subject to bisulphite sequencing. Within this region only two changes were detected; in UM cell lines SOM196b and SOM157d cytosine (C) residues 688 and 698 bp upstream of FANCD2 were unmethylated, whereas in each of the control cell lines (WM793, MRC5VA, HCT116, MCF-7, HeLa and U2OS) they were methylated. Consistent with this finding, these residues were unmethylated in all primary UM short-term cultures tested (data not shown). The differentially methylated Cs are close to C698 and within C688, a putative E2F-1 binding site (Figure 2b). Chromatin immunoprecipitation (ChIP) analysis of E2F-1 binding to the site revealed approximately threefold greater E2F-1 binding in the UM cell line SOM196b than in control MRC5VA cells (Figure 2c; Student's t-test P<0.01). Together, these data suggest that in UM demethylation of these specific C residues allows E2F-1 to bind to the FANCD2 promoter. Although E2F-1 is usually associated with gene transactivation,17 it has also been reported as a gene repressor;18, 19, 20, 21, 22, 23 thus, it is possible that increased binding of E2F-1 via altered promoter methylation may explain reduced FANCD2 expression in UM (Figure 2d).
Figure 2.
Promoter methylation and E2F-1 binding is altered in UM. (a) Relative expression of FANCD2 RNA compared with the level in WM793 cells and measured by real-time PCR. Average and s.d. of three repeats is shown. Significance was calculated using Student's t-test; ***P<0.001. (b) FANCD2 promoter region showing C residues −698 and −688 bp upstream of the FANCD2 start site (red) and site of putative E2F binding (underlined). These residues were unmethylated in UM and methylated in control cells. (c) Amount of real-time PCR product spanning the putative E2F binding site immunoprecipitated with E2F-1 antibody relative to input. Average and s.d. of two repeats is shown. Significance was determined using Student's t-test; **P<0.01. (d) Proposed model of dysregulation of expression of FANCD2 in UM; (i) in wild-type cells, the E2F-1 binding site in the promoter is methylated, preventing E2F-1 binding and allowing expression of FANCD2. (ii) In UM, methylation is absent, E2F-1 binds and expression is repressed.
Complementing with FANCD2 restored SCE levels in UM
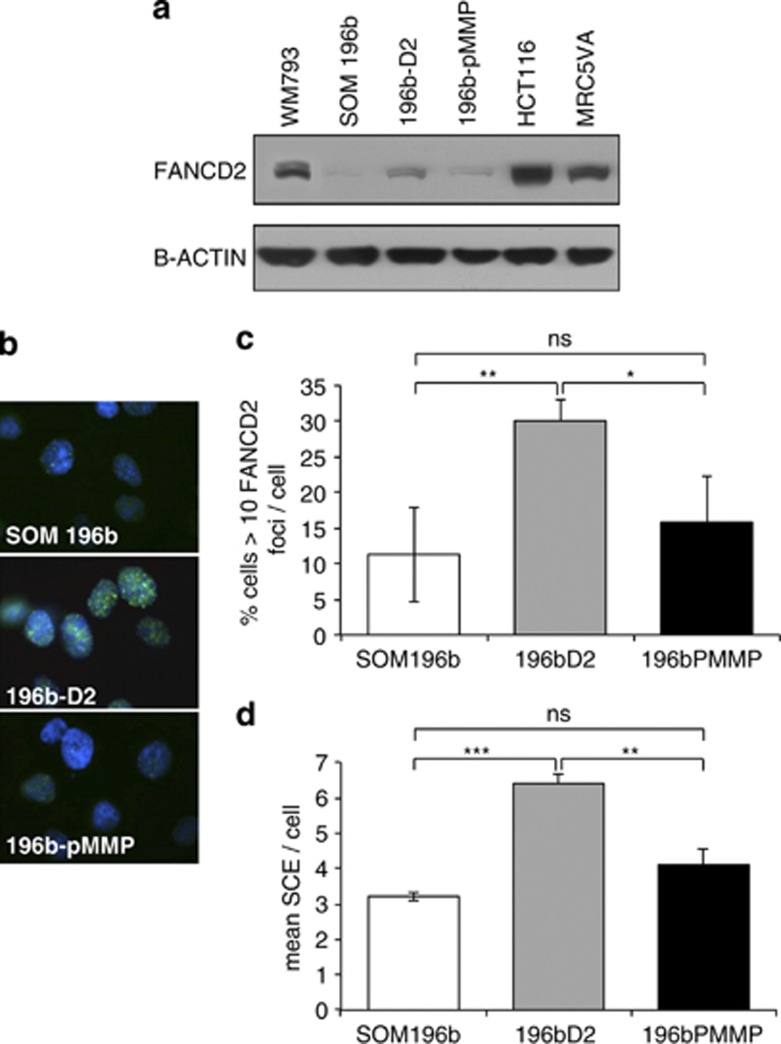
The UM cell line SOM196b was stably transfected with FANCD2 using a retroviral vector containing the FANCD2 gene (196b-D2) or the empty vector control (196b-pMMP). Expression of FANCD2 protein and spontaneous FANCD2 foci formation was increased (Figure 3a–c). Neither expression nor foci formation was significantly altered by the retroviral transfection with the empty vector.
Figure 3.
Complementing UM with FANCD2 overcomes low SCE. (a) Western blot for FANCD2 in the cutaneous melanoma cell line WM793, parental UM-derived cell line SOM196b, SOM196b retrovirally transfected with FANCD2 (196b-D2), SOM196b retrovirally transfected with empty vector (196b-pMMP), the colorectal cancer cell line HCT116 and the transformed fibroblast cell line MRC5VA. (b) Representative images of immunofluorescent staining for FANCD2 (green) and 4,6-diamidino-2-phenylindole (DAPI; blue). (c) Quantification of endogenous FANCD2 foci formation in WM793, SOM196b, 196b-D2 and 196b-pMMP. (d) SCE formation in the parental UM-derived cell line SOM196b, SOM196b retrovirally transfected with FANCD2 (196b-D2), SOM196b retrovirally transfected with empty vector (196b-pMMP). Average and s.d. of three repeats is shown. Significance was calculated using Student's t-test; NS, not significant; *P<0.05, **P<0.01 and ***P<0.001.
Previously, we reported that both primary and established UM cell lines have reduced SCE compared with a panel of normal and cancer cell types,16 suggesting that endogenous DNA damage is handled differently in UM. Here SCE (Figure 3d) increased when SOM196b was complemented with FANCD2 (Student's t-test P<0.001 and P<0.01 for 196bD2 compared with SOM196b and 196b-pMMP, respectively). SCEs in FANCD2-complemented SOM196b were not significantly different to the level seen in cutaneous melanoma and other control cells (data not shown).16 This suggests that FANCD2 expression is influencing previously reported SCE formation in UM.
Spontaneous RAD51 foci formation is reduced in UM, and this is reversed by ectopic expression of FANCD2
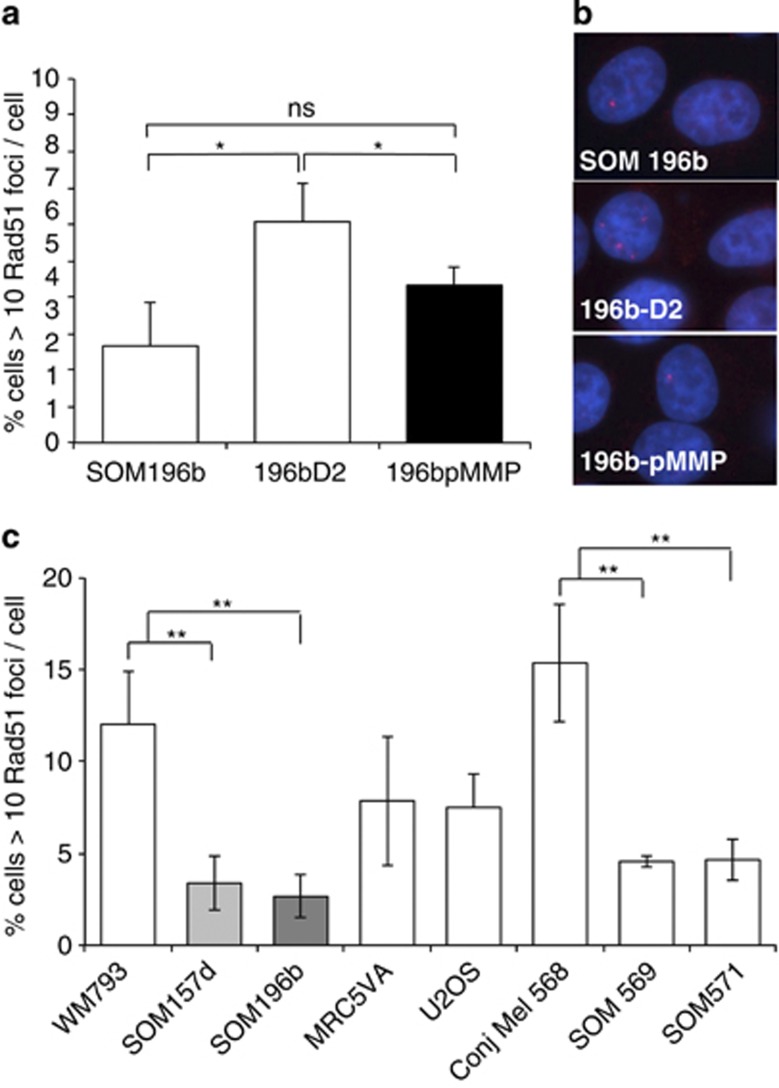
SCE is an end product of HR especially during the repair of collapsed replication forks that can form spontaneously during replication in cells.15 Similarly, FANCD2 is monoubiquitinated and colocalises with RAD51 during S phase,14 suggesting that FANCD2 may facilitate HR in the repair of endogenous damage. Complementing the UM cell line SOM196b with FANCD2 significantly increased the percentage of cells containing >10 RAD51 foci/cell (Figure 4a and b; Student's t-test P<0.05 for 196b-D2 compared with either SOM196b or 196b-pMMP), whereas the addition of the empty vector control did not significantly increase RAD51 foci formation (Figure 4a and b, Student's t-test P=0.27 for 196b-pMMP compared with SOM196b). As RAD51 foci are formed at the onset of HR and a known end product of HR is SCE, RAD51 foci are likely indicative of reduced levels of endogenous HR. These data suggest that the function of FANCD2 in influencing endogenous SCE in UM is to promote RAD51 accumulation at sites of endogenous damage and thus stimulate HR-mediated repair. The levels of RAD51 foci were also reduced in both the primary UM short-term cultures tested (SOM569 and SOM571) compared with control cells (WM793, MRC5VA and U2OS), and a primary short-term conjunctive melanoma (Mel658) (Figure 4c; Students t-test P<0.01 for WM793 compared with either SOM157d or SOM196b and Mel658 compared with either SOM569 or SOM571), demonstrating that this function is preserved in primary tumour material.
Figure 4.
Spontaneous RAD51 foci formation is reduced in UM. (a) Quantification of RAD51 foci in the parental UM-derived cell line SOM196b, SOM196b retrovirally transfected with FANCD2 (196b-D2) and SOM196b retrovirally transfected with empty vector (196b-pMMP). (b) Representative images from (a). (c) Quantification of RAD51 foci in the cutaneous melanoma cell line WM793, two UM-derived cell lines SOM157d and SOM196b, the transformed fibroblast cell line MRC5VA, the osteocarcinoma cell line U2OS, primary cells from a conjunctive melanoma (conj Mel 568) and primary cells from two UMs (SOM569 and SOM571). Average and s.d. of three repeats is shown. Significance was calculated using Student's t-test; NS, not significant; *P<0.05 and **P<0.01. SCE is expressed as the number of SCE observed/diploid genome. Where polyploidy existed, numbers were adjusted accordingly.
Spontaneous levels of SCE and RAD51 foci are reduced in FANCD2-deficient FA patient cells
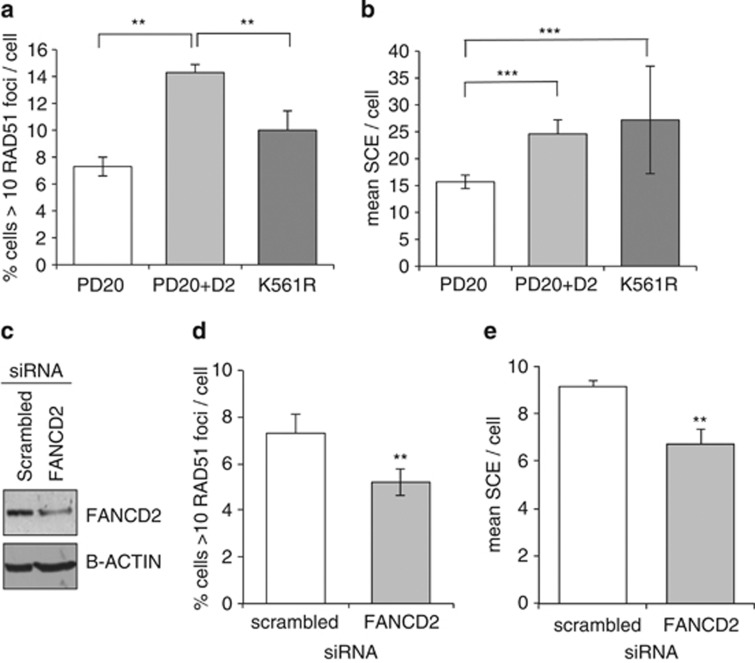
FANCD2 deficiency is classically studied in the genetic disorder FA. In the FA patient-derived cell line PD20 (deficient in FANCD2),24 there was a lower percentage of cells containing >10 RAD51 foci/cell (Figure 5a; Student's t-test P<0.01) and lower rates of spontaneous SCE (Figure 5b; Student's t-test P<0.01) than in the FANCD2-complemented PD20-D2 cell line. These data suggest that FANCD2 is also involved in controlling spontaneous HR and SCE in FA patients during the normal S phase.
Figure 5.
Endogenous RAD51 foci and SCE levels are decreased in FANCD2-deficient PD20 and FANCD2-depleted MRC5VA cells. (a) Quantification of RAD51 and (b) SCE formation in the FA-derived cell line PD20 and wild-type FANCD2 (PD20-D2) and FANCD2 monoubiquitination mutant (PD20-K561R) complemented PD20 cells. (c) Western blot for FANCD2 in MRC5VA cells treated for 48 h with scrambled or FANCD2 siRNA. (d) Quantification of RAD51 foci and (e) SCE formation in FANCD2-depleted MRC5VA cells. Average and s.d. of three repeats is shown. Significance was calculated using Student's t-test; **P<0.01 and ***P<0.001. SCE is expressed as the number of SCE observed/diploid genome. Where polyploidy existed, numbers were adjusted accordingly.
Interestingly, when PD20 cells, which complemented with the monoubiquitination mutant K561R,25 were compared with the data above, they displayed intermediate levels of RAD51 foci formation, and individual cells had unexpectedly varied SCE rates (range 6–53 SCE/cell) when compared with PD20 (range 6–32) and PD20-D2 (range 11–41). The average SCE/cell in PD20-K561R was 27.2 (corrected for ploidy), similar to the average of 24.62 seen in PD20-D2. The K561 mutant however (unlike PD20 and PD20-D2) displayed high levels of spontaneous hyperploidy and aberrant chromosomes, including rings and interchanges (Supplementary Figure 3). Thus, although FANCD2 is required to promote RAD51 to sites of endogenous damage and promotes normal S-phase-associated SCE, if FANCD2 which cannot be monoubiquitinated is present, then high levels of spontaneous genomic instability exist. Thus, there may be an additional specific function for monoubiquitinated FANCD2 at endogenous DNA lesions, and additional factors that affect SCE formation, although perhaps a note of caution should be taken in interpreting data from cells with such high levels of genetic instability.
Spontaneous levels of SCE and RAD51 foci are reduced in MRC5VA cells depleted of FANCD2
MRC5VA cells were depleted of FANCD2 using small interfering RNA (siRNA; Figure 5c). Depletion was not complete, but RAD51 and SCE were still reduced (Figures 5d and e; Student's t-test P<0.01 in both cases), again suggesting that FANCD2 can indeed regulate endogenous SCE in cells.
UM- and FANCD2-deficient FA patient cells are sensitive to acetaldehyde
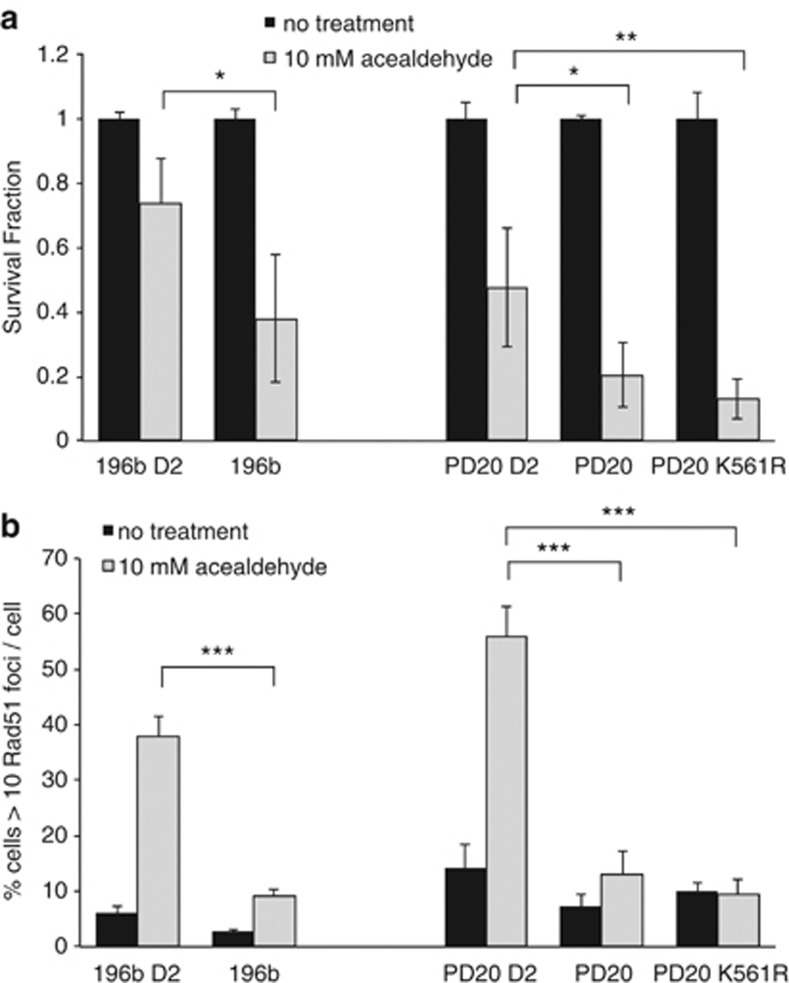
The FA pathway is usually associated with the repair of DNA ICL damage. Studying ICL repair in UM is complicated by the fact they are deficient in cytP450 and thus resistant to MMC.26 However, here sensitivity to the FA activating DNA crosslinking/damaging agent acetaldehyde27, 28, 29 was tested. Both UM and PD20 cell lines were sensitive to acetaldehyde compared with FANCD2-complemented counterparts (Figure 6a—Student's t-test P<0.05 in both cases). PD20-K561R cells were also sensitive to acetaldehyde (Student's t-test P<0.01) as compared with wild-type FANCD2-complemented cells. Consistent with this acetaldehyde-induced RAD51 foci formation was reduced in PD20-deficient UM and FA cells, as well as in FANCD2-K561R-complemented cells (Figure 6b—Student's t-test P<0.001 in all cases).
Figure 6.
UM- and FANCD2-deficient FA patient cells are sensitive to acetaldehyde. (a) Fraction of cells surviving compared with equivalent untreated control 4 days after exposure to 10 mM acetaldehyde and (b) quantification of RAD51 foci formation, in the parental UM-derived cell line SOM196b, SOM196b retrovirally transfected with FANCD2 (196b-D2), the FA-derived cell line PD20 and wild-type FANCD2 (PD20-D2) and FANCD2 monoubiquitination mutant (PD20-K561R) complemented PD20 cells. *P<0.05, **P<0.01 and ***P<0.001.
Discussion
Here, we found that the low spontaneous SCE phenotype seen in UM can be reversed by restoring expression of FANCD2, suggesting that FANCD2 is involved in the regulation of spontaneous SCE. In support of this, we demonstrated that knocking down expression of FANCD2 by siRNA reduces SCE formation in MRC5VA cells. Furthermore, we saw that the PD20 cell line derived from a FA patient lacking FANCD2 also had reduced spontaneous SCE when compared with the FANCD2-complemented counterpart (PD20-D2 cell line). There are no previous reports of spontaneous SCE in PD20 or any other FANCD2-deficient patient material, and there have been no reports of spontaneous SCE levels following siRNA-mediated FANCD2 depletion in human cells. However, the background levels of SCE reported in early work on FA patients when the complementation groups were not known suggests that spontaneous SCE in FA compared with normal controls is unchanged or reduced.13, 30, 31, 32, 33, 34, 35, 36 It would therefore be of great interest to examine background SCE in FA patient samples of known complementation groups to further elucidate the role of the FA pathway in spontaneous SCE. In addition, we show that monoubiquitination mutants of FANCD2 have reduced RAD51 foci but similar SCE formation to FANCD2-proficient cells; furthermore, K561R mutation appears to have a larger effect on genomic instability than reduced expression of wild-type FANCD2. The role of monoubiquitination of FANCD2 in the absence of exogenous DNA damaging agents is not understood and warrants further investigation.
Studies in other mammalian cell lines support our findings in human cells, and CHO cells defective in several different FA proteins are reported to have reduced or unchanged levels of SCE.9, 37 However, in DT40 chicken cells, mutation of FA genes including FANCD2 result in increased spontaneous SCE,38, 39 whereas similar to mammalian cells DT40 cells defective in core HR proteins do show reduced SCE.6 Our data indicate that the CHO model is more closely matched to human disease. DT40 cells are thought to be immortalised in a state of somatic hypermutation,40 and thus they represent a highly specialized cell type, where certain functions of recombination may not be conserved.
Using FA patient-derived FANCD2-deficient cells or wild-type human cells depleted of FANCD2 using siRNA, FANCD2 has been previously shown to influence double-strand break-induced HR in a reporter plasmid assay,41 but spontaneous levels of HR were not reported. During replication, unrepaired endogenous DNA damage in the form of persistent single-strand breaks is converted into double-strand breaks collapsing replication forks. We have previously shown that the majority of spontaneous HR results in SCE, and that this arises from such collapsed forks.15 Our data linking FANCD2, RAD51 foci formation and SCE indicate a role for FANCD2 in promoting HR at spontaneously collapsed replication forks. In support of this, FANCD2 is highly regulated through the cell cycle, is required for normal cell cycle progression and colocalises with BRCA1 and RAD51 to form distinct nuclear foci specifically during the S phase.14 In addition, FANCD2-depleted Xenopus extracts are unable to restart collapsed replication forks, whereas replication restart following stalled forks is unaffected.42 Here we demonstrate for the first time that FANCD2-deficient human disease-derived cells are sensitive to acetaldehyde, which has recently been implicated as the endogenous substrate responsible for FA pathway activation27, 28, 29 and is a known DNA–protein and DNA–DNA crosslinking agent.43 Our data indicate that as in other model systems human FANCD2 is required for repair of acetaldehyde-induced DNA damage; thus, in addition to collapsed forks, FANCD2 may promote spontaneous SCE in response to endogenous lesions induced by such metabolites.
How FANCD2 regulates HR is unclear, and one mechanism suggested is that following ICL damage, FANCD2 is required for the recruitment of HR factors to sites of repair; here, we show that spontaneous RAD51 foci formation is reduced in FANCD2-deficient cells, suggesting that FANCD2 may be required for the assembly of HR factors during spontaneous recombination. It has also been suggested that the function of FANCD2 following ICL damage is to suppress non-homologous end-joining (NHEJ),44, 45 as evidenced by the fact that inhibiting NHEJ rescues the MMC-sensitive FA phenotype. Inhibition of DNA-PKcs (DNA-dependent protein kinase, catalytic subunit; a component of the NHEJ pathway) increased SCE in the UM cell line SOM196b and in the FA cell line PD20 (Supplementary Figure 4), suggesting that spontaneous SCE can also be increased by inhibiting NHEJ. Thus, the function of FANCD2 to influence NHEJ after ICL may also be conserved during normal replication when endogenous damage collapses replication forks. If NHEJ becomes the predominant repair pathway in the absence of FANCD2, this could contribute to genomic instability in UM and FA.
Although we have demonstrated a role of FANCD2 in influencing SCE, it is unlikely to be the overriding factor in all cancers. Indeed, we have found a conjunctival melanoma (Mel568) that had low FANCD2 expression (not shown) but had the highest level of RAD51 foci formation (Figure 4c) and had normal SCE (not shown). These data demonstrate the complex nature of primary tumours compared with the study of transformed cells. Thus, although FANCD2 can influence SCE, its regulation may vary in different tumours.
Epigenetic events that alter gene expression are important in the tumourigenesis of many sporadic cancers. Downregulation of FANCD2 expression was shown at the level of transcription and we demonstrate a loss of methylation within a predicted E2F-1 binding site in the FANCD2 promoter region. Methylation of several promoters has been shown to prevent E2F-1 binding;46 consistent with this, we saw increased binding of E2F-1 to the unmethylated FANCD2 promoter region in UM. Although E2F-1 is usually considered an activator of gene expression,17 it can also suppress transcription.18, 19, 21, 23 Thus, we conclude that the most likely reason for reduced expression of FANCD2 in UM is demethylation of the promoter leading to increased E2F-1 binding and inhibition of transcription. There are increasing numbers of reports describing changes in the FA/BRCA pathway in sporadic cancer;47, 48, 49 however, this is the first time that an aberrant methylation of the FANCD2 promoter has been reported in human cancer, and it will be of interest to see if other sporadic cancers contain the same epigenetic modification.
Although changes in methylation have not been seen previously, lack of FANCD2 has been linked to tumourigenesis. FANCD2 knockout mice develop breast tumours,50 FANCD2 expression is absent in 10–20% of sporadic and BRCA1-related breast cancers51 and low expression of FANCD2 in breast cancer is associated with poor patient outcome.52 In addition, polymorphisms of FANCD2 have been associated with increased sporadic breast cancer.53 Together, these data suggest that lack of, or reduced expression of, FANCD2 can contribute to tumour development or progression. It is therefore likely that reduced FANCD2 expression in UM contributes to tumourigenesis, perhaps by diverting repair of endogenous DNA damage away from the error-free process of HR into NHEJ.
UMs are divided into subgroups based on characteristic genetic abnormalities in chromosomes 1, 3, 6 and 8. These genetic changes also influence clinical outcome.54, 55, 56, 57 Previously we reported that low SCE was not specific to any clinical subtype or genetic pathway.16 Thus, it is likely to represent an early change in the development of the tumour. Similarly, here the primary UMs included were a mixture of tumours representing a range of genetic backgrounds (Supplementary Table 2). Low expression of FANCD2 was common to most UMs (11/12 primary STCs), regardless of the chromosome 3 copy number status. As monsomy 3 is a relevant prognostic change in UM, and FANCD2 resides at 3p25, the low expression of FANCD2 in all UMs is the first time the same genetic defect has been seen in UM, regardless of previously identified characteristic chromosomal abnormality or of clinical subtype.
Deficiency in FA proteins is classically associated with an inability to repair interstrand crosslinks and a decrease in crosslink-induced SCE. However, the wide spectrum of tissues affected in FA patients suggests that it is unlikely that all disease symptoms are because of exposure to exogenous crosslinking agents. Our data that FA-derived PD20 cells have low SCE, suggest that an inability to repair collapsed endogenous DNA damage is likely to contribute to the pathogenesis of FA as well as UM.58
Materials and methods
Cell lines and primary cultures
Established Sheffield Ocular Melanoma (SOM) cell lines, SOM157d and SOM196b, were used alongside control cell lines. All cell lines were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin sulphate (100 μg/ml) at 37 °C under 5% CO2. Short-term cultures established from primary UMs were grown as described previously.16 Informed patient consent was obtained from UM patients and protocols followed the principles of the Declaration of Helsinki (SSREC94/247). Established UM cell lines were confirmed as authentic by fluorescence in situ hybridisation before and during the study, looking for characteristic abnormalities in chromosomes 1, 3, 6 and 8.54, 55, 56 PD20 and PD20-D2 cells were a gift from Alan D'Andrea (Dana-Farber Cancer Institute, Boston, MA, USA). PD20 K651R cells were a gift from Toshiyasu Taniguchi (Fred Hutchinson Cancer Research Center, Seattle, WA, USA). Other cells were obtained from ATCC (Manassas, VA, USA).
Complementation of UM with FANCD2 using retroviral vectors
Lipofectamine 2000 (Life Technologies Ltd, Paisley, UK) was used to transfect the 293GPG packaging cell line with pMMP-empty and pMMP-FANCD2 retroviral vectors. The growth medium was changed daily, and at 96, 120, 144 and 168 h post transfection, the pseudotype virus was collected. Then, 2 ml of virus was added to SOM196b cells in the presence of 24 μg/ml polybrene and left for 24 h. Media were then changed for fresh media without virus and cells were left for 24 h to recover. Virus was then added again and the procedure repeated over a period of 7 days. Then, 3 μg/ml puromycin was added to cells to select for successfully complemented cells.
siRNA
siGenome smart pool siRNA against FANCD2 was purchased from Thermo Scientific (Lafayette, CA, USA). Scrambled siRNA was purchased from Dharmacon (Lafayette, CA, USA). MRC5VA cells were transfected with 100 nM scrambled or FANCD2 siRNA, using Oligofectamine Reagent (Life Technologies Ltd) according to the manufacturer's instructions. Cells were then cultured in normal growth medium for 24 h before adding 0.24 nM 5-bromodeoxyuridine (Sigma-Aldrich Company Ltd, Gillingham, UK), and 48 h later, cells were harvested for SCE analysis.
Western blotting
SDS–PAGE and western blotting were performed as described previously.26 Antibodies used were rabbit anti-FANCD2 (1:10 000, Novus Biologicals, Ltd, Cambridge, UK) and rabbit anti-β-actin (1:2000, Sigma-Aldrich Company Ltd).
Immunofluorescence
Cells were fixed in 3% paraformaldehyde in phosphate-buffered saline containing 0.1% Triton X-100 and washed four times in phosphate-buffered saline containing 0.15% bovine serum albumin and 0.1% Triton X-100 for 15 min before incubation with either rabbit polyclonal anti-RAD51 antibody (1:500, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or mouse anti-FANCD2 (1:1000, Santa Cruz Biotechnology) in 3% goat serum for 16 h at 4 °C. The coverslips were subsequently washed (as above) followed by 1 h of incubation at room temperature with Cy-3-conjugated goat anti-rabbit or mouse IgG antibody (Life Technologies Ltd) as required at a concentration of 1:500. DNA was stained with 1 μg/ml 4,6-diamidino-2-phenylindole in mounting solution (Life Technologies Ltd). Images were obtained with a Zeiss LSM 510 inverted confocal microscope (Zeiss, Cambridge, UK) using planapochromat 63X/NA 1.4 oil immersion objective and excitation wavelengths 488, 546 and 630 nm. The frequencies of cells containing foci were determined in at least three separate experiments and at least 50 nuclei were counted in each experimental repeat. Each experiment was repeated at least three times.
Real-time PCR
Primers for FANCD2 were designed to amplify the longer FANCD2 isoform from transcript sequence NM-033084. The β-actin primers were also used to amplify β-actin complementary DNA from each sample as a reference gene. Primers were:
FANCD2: forward 5′-CATGGCTGTTCGAGACTTCA-3′ and
reverse 5′-GACACAAGGCTGCTTCATCA-3′.
β-actin: forward 5′-ACACCCCAGCCATGTACGTAGCC-3′ and
reverse 5′-AAGAGCCTCAGGGCAACGGAACC-3′.
Samples were mixed with SYBR Green PCR master mix (Life Technologies Ltd) and 10 μM primers. Real-time PCR was then carried out (1 × 50 °C for 2 min, 1 × 95 °C for 10 min, 40 × 95 °C for 15 s followed by 60 °C for 1 min) using the Applied Biosystems (Carlsbad, CA, USA) 7900HT fast real time PCR system. Images and data were documented using the SDS Enterprise Database software (Applied Biosystems by Life Technologies).
Bisulphite sequencing
Genomic DNA was extracted from MRC5VA and SOM196b cells using GenElute Mammalian Genomic DNA Minipep kit (Sigma-Aldrich Company Ltd). It was then subject to sodium bisulphite modification using the CpGenome DNA modification kit (Merck Millipore, Billerica, MA, USA). Three sets of primers were designed to amplify a 1000-bp region of the FANCD2 promoter from the modified DNA:
Set 1: forward F1 5′-TTTTATTTTAGGAAGGGAAATG-3′
reverse R1 5′-AGGATTATTTAGAGGTAGATGTTGGA-3′
Set 2: forward F2 5′-TGATTTTTATTTGTTTATGAGGGAG-3′
reverse R2 5′-AGGATTATTTAGAGGTAGATGTTGGA-3′
Set 3: forward F3 5′-TGGGTAGGATTATTTAGAGGTAGATG-3′
reverse R3 5′-TTTTTTGTGGTTTAATTTTTAAGTT-3′.
PCR conditions were as follows: 1 × 95 °C for 4 min, 5 × (95 °C for 30 s, 54 °C for 90 s and 72 °C for 2 min), 25 × (95 °C 30 s, 54 °C for 90 s and 72 °C for 90 s) and 1 × 72 °C for 4 min. The same primers were used for sequencing at a concentration of 1 μM using an ABI 3730 capillary sequencer. Data were analysed using Sequencher 4.1.
Chromatin immunoprecipitation
ChIP was carried out using 2.5 μg of E2F-1 antibody (Santa Cruz Biotechnology) and the EZ ChIP Assay kit (Merck Millipore) according to the manufacturers' instructions. DNA was analysed by real-time PCR using FANCD2 primers in order to amplify a 377-bp region of the FANCD2 promoter.
forward 5′-CACCCTAGGAAGGGAAATGATA-3′
reverse 5′-TCTATGAGGGAGGTACTGTTA-3′.
Analysis was performed using the ΔΔCT method where samples were normalised to the INPUT and an endogenous control (glyceraldehyde-3-phosphate dehydrogenase).
Analysis of cultures for SCE
To visualise SCEs, cultures were incubated with 0.24 nM 5-bromodeoxyuridine (Sigma-Aldrich Company Ltd) for two cell cycles, and SCE analysis was performed as described previously.16 Exchanges observed in each cell were defined by an exchange of dark-stained (Hoechst-bound TT-rich) segments with light, bleached (5-bromodeoxyuridine incorporated) segments. Staining variation where chromatids clearly twisted around one another was excluded from the exchange rate count. For each cell line, at least 30 cells were analysed on each of at least 3 separate occasions. The mean, median and range of SCEs observed per diploid cell were calculated for each sample. Where polyploidy was seen, SCEs were adjusted to compensate.
Cell proliferation assays
1 × 105 cells were plated into T25 flasks 4 h before treatment with 10 mM acetaldehyde. Cells were then left for 4 days before trypsinisation, Trypan blue staining and counting of live cells. Survival fraction was calculated as a fraction of cells surviving compared with the equivalent untreated control. On each occasion, eight counts of each condition were taken and an average calculated. The experiment was repeated three times and average and s.d. of the survival fractions calculated.
Acknowledgments
We thank Professors Toshiyasu Taniguchi and Alan D'Andrea for providing cells, the FANCD2-expressing retroviral vectors and for their technical advice. They also thank Professors Tomas Lindahl and Mark Meuth and Dr Spencer Collis for critical reading of the manuscript and discussion. We are grateful for funding from an RCUK fellowship to HB and MRC studentship to PG and for general funding from Weston Park Hospital Cancer Appeal (CA95), The Sarcoma Trust, Yorkshire Eye Research and Yorkshire Cancer Research (S305PA).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Miranda M, Ligas C, Amicarelli F, D'Alessandro E, Brisdelli F, Zarivi O, et al. Sister chromatid exchange (SCE) rates in human melanoma cells as an index of mutagenesis. Mutagenesis. 1997;12:233–236. doi: 10.1093/mutage/12.4.233. [DOI] [PubMed] [Google Scholar]
- Goyanes V, Fernandez JL, Pereira S, Campos A, Rodriguez E, Gonzalez M, et al. Frequency of sister chromatid exchanges in humans at 14–16 week foetal stage and at birth. Cytobios. 1994;77:67–72. [PubMed] [Google Scholar]
- Kato H, Stich HF. Sister chromatid exchanges in ageing and repair-deficient human fibroblasts. Nature. 1976;260:447–448. doi: 10.1038/260447a0. [DOI] [PubMed] [Google Scholar]
- Nagasawa H, Fornace D, Little JB. Induction of sister-chromatid exchanges by DNA-damaging agents and 12-O-tetradecanoyl-phorbol-13-acetate (TPA) in synchronous Chinese hamster ovary (CHO) cells. Mutat Res. 1983;107:315–327. doi: 10.1016/0027-5107(83)90173-2. [DOI] [PubMed] [Google Scholar]
- Wojcik A, Bruckmann E, Obe G. Insights into the mechanisms of sister chromatid exchange formation. Cytogenet Genome Res. 2004;104:304–309. doi: 10.1159/000077507. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- Dronkert ML, Beverloo HB, Johnson RD, Hoeijmakers JH, Jasin M, Kanaar R. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol Cell Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godthelp BC, Wiegant WW, van Duijn-Goedhart A, Scharer OD, van Buul PP, Kanaar R, et al. Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res. 2002;30:2172–2182. doi: 10.1093/nar/30.10.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Zitzmann S, Westermann F, Arnold K, Brouwers S, Schwab M, et al. Increased rates of spontaneous sister chromatid exchange in lymphocytes of BRCA2+/− carriers of familial breast cancer clusters. Cancer Lett. 2004;210:85–94. doi: 10.1016/j.canlet.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee CA, Gavish H, Shannon WR, Buchwald M. Cloning of cDNAs for Fanconi's anaemia by functional complementation. Nature. 1992;356:763–767. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- Latt SA, Stetten G, Juergens LA, Buchanan GR, Gerald PS. Induction by alkylating agents of sister chromatid exchanges and chromatid breaks in Fanconi's anemia. Proc Natl Acad Sci USA. 1975;72:4066–4070. doi: 10.1073/pnas.72.10.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh L, Gravells P, Canovas D, Ul-Hassan A, Rennie IG, Bryant H, et al. Atypically low spontaneous sister chromatid exchange formation in uveal melanoma. Genes Chromosomes Cancer. 2011;50:34–42. doi: 10.1002/gcc.20829. [DOI] [PubMed] [Google Scholar]
- Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Chen Y, Hu W, Wu M. E2F1 inhibits MDM2 expression in a p53-dependent manner. Cell Signal. 2011;23:193–200. doi: 10.1016/j.cellsig.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Croxton R, Ma Y, Cress WD. Differences in DNA binding properties between E2F1 and E2F4 specify repression of the Mcl-1 promoter. Oncogene. 2002;21:1563–1570. doi: 10.1038/sj.onc.1205232. [DOI] [PubMed] [Google Scholar]
- Croxton R, Ma Y, Song L, Haura EB, Cress WD. Direct repression of the Mcl-1 promoter by E2F1. Oncogene. 2002;21:1359–1369. doi: 10.1038/sj.onc.1205157. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Packham G, Nip J, Fee BE, Hiebert SW, Zambetti GP, et al. Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene. 2001;20:6983–6993. doi: 10.1038/sj.onc.1204892. [DOI] [PubMed] [Google Scholar]
- Phillips AC, Ernst MK, Bates S, Rice NR, Vousden KH. E2F-1 potentiates cell death by blocking antiapoptotic signaling pathways. Mol Cell. 1999;4:771–781. doi: 10.1016/s1097-2765(00)80387-1. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Matsumura I, Ezoe S, Satoh Y, Sakamaki T, Albanese C, et al. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappaB activity that facilitates MnSOD-mediated ROS elimination. Mol Cell. 2002;9:1017–1029. doi: 10.1016/s1097-2765(02)00522-1. [DOI] [PubMed] [Google Scholar]
- Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Gravells P, Hoh L, Canovas D, Rennie IG, Sisley K, Bryant HE. Resistance of uveal melanoma to the interstrand cross-linking agent mitomycin C is associated with reduced expression of CYP450R. Br J Cancer. 2011;104:1098–1105. doi: 10.1038/bjc.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- Abraham J, Balbo S, Crabb D, Brooks PJ. Alcohol metabolism in human cells causes DNA damage and activates the Fanconi anemia-breast cancer susceptibility (FA-BRCA) DNA damage response network. Alcohol Clin Exp Res. 2011;35:2113–2120. doi: 10.1111/j.1530-0277.2011.01563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Auerbach AD, Wolman SR. Susceptibility of Fanconi's anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–496. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- Gebhart E, Kysela D, Matthee H, Nikol M. Cytogenetic analyses utilizing various clastogens in two sibs with Fanconi anemia, their relatives, and control individuals. Hum Genet. 1985;69:309–315. doi: 10.1007/BF00291647. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Schmid W. The rate of sister chromatid exchanges parallel to spontaneous chromosome breakage in Fanconi's anemia and to trenimon-induced aberrations in human lymphocytes and fibroblasts. Humangenetik. 1975;29:201–206. doi: 10.1007/BF00297624. [DOI] [PubMed] [Google Scholar]
- Howell RT. Sister chromatid exchange evaluation as an aid to the diagnosis and exclusion of Fanconi's anaemia by induced chromosome damage analysis. J Med Genet. 1991;28:468–471. doi: 10.1136/jmg.28.7.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotna B, Goetz P, Surkova NI. Effects of alkylating agents on lymphocytes from controls and from patients with Fanconi's anemia. Studies of sister chromatid exchanges, chromosome aberrations, and kinetics of cell division. Hum Genet. 1979;49:41–50. doi: 10.1007/BF00277685. [DOI] [PubMed] [Google Scholar]
- Porfirio B, Dallapiccola B, Mokini V, Alimena G, Gandini E. Failure of diepoxybutane to enhance sister chromatid exchange levels in Fanconi's anemia patients and heterozygotes. Hum Genet. 1983;63:117–120. doi: 10.1007/BF00291529. [DOI] [PubMed] [Google Scholar]
- Sperling K, Wegner RD, Riehm H, Obe G. Frequency and distribution of sister-chromatid exchanges in a case of Fanconi's anemia. Humangenetik. 1975;27:227–230. doi: 10.1007/BF00278349. [DOI] [PubMed] [Google Scholar]
- Wilson JB, Johnson MA, Stuckert AP, Trueman KL, May S, Bryant PE, et al. The Chinese hamster FANCG/XRCC9 mutant NM3 fails to express the monoubiquitinated form of the FANCD2 protein, is hypersensitive to a range of DNA damaging agents and exhibits a normal level of spontaneous sister chromatid exchange. Carcinogenesis. 2001;22:1939–1946. doi: 10.1093/carcin/22.12.1939. [DOI] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, Namikoshi K, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol Cell Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstedde JM, Reynaud CA, Humphries EH, Olson W, Ewert DL, Weill JC. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Stone S, Hoatlin ME, Gautier J. Fanconi anemia proteins stabilize replication forks. DNA Repair (Amst) 2008;7:1973–1981. doi: 10.1016/j.dnarep.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenti Garcia C, Mechilli M, Proietti De Santis L, Schinoppi A, Kobos K, Palitti F. Relationship between DNA lesions, DNA repair and chromosomal damage induced by acetaldehyde. Mutat Res. 2009;662:3–9. doi: 10.1016/j.mrfmmm.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci USA. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyakhovich A, Surralles J. Disruption of the Fanconi anemia/BRCA pathway in sporadic cancer. Cancer Lett. 2006;232:99–106. doi: 10.1016/j.canlet.2005.07.038. [DOI] [PubMed] [Google Scholar]
- Neveling K, Kalb R, Florl AR, Herterich S, Friedl R, Hoehn H, et al. Disruption of the FA/BRCA pathway in bladder cancer. Cytogenet Genome Res. 2007;118:166–176. doi: 10.1159/000108297. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Groep P, Hoelzel M, Buerger H, Joenje H, de Winter JP, van Diest PJ. Loss of expression of FANCD2 protein in sporadic and hereditary breast cancer. Breast Cancer Res Treat. 2008;107:41–47. doi: 10.1007/s10549-007-9534-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudland PS, Platt-Higgins AM, Davies LM, de Silva Rudland S, Wilson JB, Aladwani A, et al. Significance of the Fanconi anemia FANCD2 protein in sporadic and metastatic human breast cancer. Am J Pathol. 2010;176:2935–2947. doi: 10.2353/ajpath.2010.090779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso E, Milne RL, Fernandez LP, Zamora P, Arias JI, Benitez J, et al. FANCD2 associated with sporadic breast cancer risk. Carcinogenesis. 2006;27:1930–1937. doi: 10.1093/carcin/bgl062. [DOI] [PubMed] [Google Scholar]
- Sisley K, Rennie IG, Parsons MA, Jacques R, Hammond DW, Bell SM, et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer. 1997;19:22–28. doi: 10.1002/(sici)1098-2264(199705)19:1<22::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Patel KA, Edmondson ND, Talbot F, Parsons MA, Rennie IG, Sisley K. Prediction of prognosis in patients with uveal melanoma using fluorescence in situ hybridisation. Br J Ophthalmol. 2001;85:1440–1444. doi: 10.1136/bjo.85.12.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damato B, Dopierala J, Klaasen A, van Dijk M, Sibbring J, Coupland SE. Multiplex ligation-dependent probe amplification of uveal melanoma: correlation with metastatic death. Invest Ophthalmol Vis Sci. 2009;50:3048–3055. doi: 10.1167/iovs.08-3165. [DOI] [PubMed] [Google Scholar]
- Harbour JW. Molecular prognostic testing and individualized patient care in uveal melanoma. Am J Ophthalmol. 2009;148:823–9 e1. doi: 10.1016/j.ajo.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q, Andreassen PR. Fanconi anemia proteins and endogenous stresses. Mutat Res. 2009;668:42–53. doi: 10.1016/j.mrfmmm.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.