Abstract
Systemic light chain amyloidosis (AL) is one of several protein misfolding diseases and is characterized by extracellular deposition of immunoglobulin light chains in the form of amyloid fibrils [1]. Immunoglobulin (Ig) proteins consist of two light chains (LCs) and two heavy chains (HCs) that ordinarily form a heterotetramer which is secreted by a plasma cell. In AL, however, a monoclonal plasma cell population produces an abundance of a pathogenic LC protein. In this case, not all of the LCs pair with the HCs, and free LCs are secreted into circulation. The LC-HC dimer is very stable, and losing this interaction may result in an unstable LC protein [2]. Additionally, somatic mutations are thought to cause amyloidogenic proteins to be less stable compared to non-amyloidogenic proteins [3-5], leading to protein misfolding and amyloid fibril formation. The amyloid fibrils cause tissue damage and cell death, leading to patient death within 12-18 months if left untreated [6]. Current therapies are harsh and not curative, including chemotherapy and autologous stem cell transplants. Studies of protein pathogenesis and fibril formation mechanisms may lead to better therapies with an improved outlook for patient survival.
Much has been done to determine the molecular factors that make a particular LC protein amyloidogenic and to elucidate the mechanism of amyloid fibril formation. Anthony Fink’s work, particularly with discerning the role of intermediates in the fibril formation pathway, has made a remarkable impact in the field of amyloidosis research. This review provides a general overview of the current state of AL research and also attempts to capture the most recent ideas and knowledge generated from the Fink laboratory.
Keywords: light chain amyloidosis, immunoglobulin light chain, fibril formation, protein misfolding, amyloid
Factors determining amyloidogenicty of light chain proteins
When genes undergo rearrangement in a B cell, either a κ or λ light chain is constructed. A full length LC consists of both a variable domain (VL) and a constant domain (CL). In 85% of AL patients, only the VL is found in amyloid deposits [7]. Intriguingly, although combinatorial diversity allows for thousands of possible sequences, the pathogenic proteins among AL patients most frequently use just a few germline subtypes [8-10], such as the κI O18/O8, κI O12/O2, and κIV B3 families, along with λ1c, λ2a2, λ2b2, λ3r, and λ6a [10-12].
To determine if the germline sequences present a bias toward generating AL proteins that are inherently more amyloidogenic, two studies tested κ and λ germline proteins. Baden et al. compared AL-09, an amyloidogenic protein that has 7 somatic mutations, to a protein derived from its germline, κI O18/O8 [13]. The germline protein was more thermodynamically stable than its amyloidogenic counterpart, and although it was able to form fibrils, its fibril formation kinetics were significantly slower compared to AL-09.
Because the λ6a germline is expressed almost exclusively in AL patients and not in the normal LC repertoire [8-12], del Pozo Yauner et al. hypothesized that this germline would be as unstable as AL proteins. However, experiments with the λ6a germline showed that the protein was more stable than Wil, an amyloidogenic protein from that germline that has 11 somatic mutations [14]. The λ6a germline also had significantly slower fibril formation kinetics than Wil. Although the increase in stability of the λ6a germline protein with respect to its amyloidogenic counterpart is similar to that observed between κI O18/O8 and AL-09, the fibril formation kinetics for λ6a were faster than those for κI O18/O8 (14 hours compared to 216 hours at 37°C). Additionally, λ6a was able to form fibrils in the absence of seeds, while κI O18/O8 required seeds for fibril formation. This may indicate an increase in fibrillogenic propensity for λ6a germline proteins. More studies are necessary to verify that AL-prone germline sequences are more fibrillogenic than normal Ig repertoire germline sequences. These studies may confirm that the mutations are a contributing factor to the loss of stability and increased amyloidogenicity.
The mutational diversity among AL proteins has been well documented, as the somatic hypermutation of immunoglobulins results in a different set of amino acid mutations for the pathogenic protein in each patient. Several studies have compared amino acid sequences of AL proteins, searching for common mutations or mutational regions. In an analysis of 121 κI light chains (37 of which were amyloidogenic), Stevens found four structural features that rendered an LC protein more likely to be amyloidogenic [15]. All of these involved loss or gain of certain residues, including a mutation that introduces a glycosylation site (discussed below), mutations of Arg61 or Ile27b and mutations of Pro residues in β-turns.
A more recent analysis of nearly 100 κ and λ AL light chain sequences catalogued the non-conservative mutations in these proteins and modeled their positions onto known LC structures to correlate structural regions (β-strands or loops) with potentially destabilizing mutations (Poshusta et al., in preparation). This study confirmed that the total number of mutations may be less important as an amyloidogenic determinant than the location of the nonconservative mutations. Additionally, the patients’ free light chain levels, which have been used as an indicator of disease progression [16], were also assessed in a subset of the analyzed protein sequences, revealing a correlation between non-conservative mutations in certain regions and free light chain (FLC) levels. This correlation suggested that patients with initial low FLC levels acquired mutations that rendered these proteins to be more amyloidogenic than patients with higher FLC levels. Analyzing the location of these mutations could further advance understanding of the mechanisms of amyloid formation and lead to a potential indicator for AL disease progression.
Thermodynamic stability influences fibril formation propensity
Mutations have been linked to thermodynamic stability and fibril formation propensity of AL proteins. Amyloidogenic AL proteins are less stable thermodynamically than nonamyloidogenic multiple myeloma (MM) light chain proteins [3-5] . In an analysis linking mutations and stability, Hurle et al. analyzed 36 sequences (18 κ and 18 λ) in search of rare amino acid replacements that occurred in structurally significant regions of the proteins [17]. They then constructed single-point mutants incorporating the rare residues into a nonamyloidogenic Bence Jones LC protein to determine whether the amino acids destabilized the protein significantly enough to induce unfolding. Four of the six mutations were destabilizing, leading to the conclusion that some mutations are involved in amyloidogenicity.
To determine if a single mutation is enough to render a protein amyloidogenic, Davis et al. studied AL protein SMA and MM protein LEN. Only eight residues differ between these two proteins, and each SMA mutation was introduced into LEN to assess the individual effects on fibrillogenesis. Of the mutations tested only P40L, located in a loop region, was able to form thioflavin T (ThT) positive fibrils in unseeded reactions [18]. Although stability data were not reported for these mutants, it is likely that the P40L mutant was less stable than wild-type LEN, as Pro40 (very favorable for loops and turns) is conserved among 98% of all κ and λ germline sequences.
Another link between thermodynamic stability and fibril formation is found in the recently analyzed κI O18/O8 and λ6a germline proteins. These proteins were significantly more stable than all AL disease proteins that have been studied to date [13-14]. The Tm values (melting temperatures, at which 50% of the proteins are unfolded) for the germline proteins were increased by 15°C and 11.6°C, respectively, over the corresponding AL proteins analyzed in each study. Both κI O18/O8 and λ6a germline proteins had slower fibril formation kinetics than their amyloidogenic counterparts.
Del Pozo Yauner and colleagues incorporated an R25G mutation into the λ6a germline protein (6aJL2-R25G), as this mutation is found in 25% of amyloidogenic λ6 LCs and presumably represents an allotypic variant [14,19-20]. This mutation resulted in a 6°C loss in Tm value for the mutated protein, and 6aJL2-R25G had a much shorter lag time and faster growth rate than the λ6a germline protein. The authors explain that the R25G mutation may affect the structure of complementarity determining region 1 (CDR1), resulting in an altered conformation and increased amyloidogenicity [20].
Further research on the κI O18/O8 germline protein and amyloidogenic AL-09 also connected thermodynamic stability to fibril formation. Baden et al. undertook a systematic restorative mutational analysis of the non-conservative mutations of AL-09, which are all located in the dimer interface [21]. Of the three non-conservative restorative mutations (I34N, Q42K and H87Y), restoring the His87 mutation to the Tyr87 residue found in the germline sequence increased the thermodynamic stability and decreased the fibril formation kinetics to the same levels as κI O18/O8. Significant structural alterations were also observed with this restorative mutant that will be discussed below. Restoring the Asn34 residue had intermediate effects on stability and fibril formation propensity, while reintroducing Lys42 did not appear to alter the thermodynamics to any extent.
In complementary experiments introducing the His87 residue from the amyloidogenic protein into κI O18/O8, this protein was only destabilized half as much as AL-09 (with its 7 mutations). This reciprocal mutant also had intermediate fibril formation kinetics between those measured for κI O18/O8 and AL-09, indicating that this mutation alone may not have been sufficient for the amyloidogenicity observed in AL-09. However, introducing a second mutation into κI O18/O8 (Ile34, in addition to His87) completely destabilized the protein and exhibited the same fast fibril kinetics as amyloidogenic AL-09. Thus, rather than a single mutation that causes amyloidogenesis, it is probable that a combination of destabilizing and compensatory mutations leads to fibrillogenicity among AL proteins.
Structure of AL proteins
Immunoglobulin light chain VL protein structures consist of 9 β-strands (A, B, C, C’, C”, D, E, F, G) arranged into two anti-parallel β-sheets that form a Greek key β-barrel immunoglobulin fold. Interspersed between the β-strands are five loop regions and 3 complementarity determining regions (CDRs). The VLs undergo somatic hypermutation to generate antigenic specificity in an immune response, and CDR regions generally exhibit more mutations than the surrounding framework (FR) regions. Somatic mutations usually do not greatly affect the structure of Ig light chains.
The crystal structures of several Ig VL have been solved, both for amyloidogenic AL and non-amyloidogenic MM proteins [22-28] (Fig. (1)). It appears that the specific somatic mutations accumulated in these structures do not affect the overall topology because all of these structures crystallized as dimers, with only minor variations between each structure.
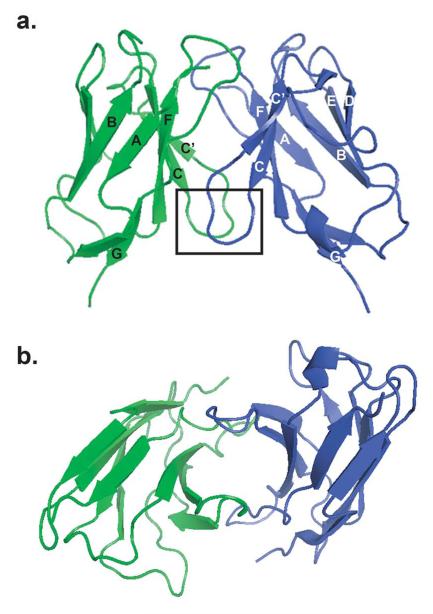
Fig. (1).
Immunoglobulin light chain dimer of a multiple myeloma protein from the κI germline shows the typical structure and dimer interface observed in these proteins (1WTL). a) Two LC monomers (green and blue), each consisting of 9 anti-parallel β-strands (labeled A-G), dimerize. The boxed area indicates the 40-44 loop region discussed in the text that may play a role in amyloidogenicity in AL. b) This view highlights the LC dimer interface in the center. Amino acids from both subunits extend into this interface and interact through hydrogen bonding networks.
Some studies have noted small but potentially significant disruptions in loop regions of AL proteins (Fig. (1)). Schormann et al. analyzed the AL protein BRE, which contains a P40L mutation located in the loop connecting β-strand C with β-strand C’ (40-44 loop) [24]. This region showed the largest structural difference between BRE and non-amyloidogenic protein REI. A recent analysis of two AL proteins, AL-12 and AL-103, also showed small perturbations in loop regions (Randles et al., submitted). AL-12 had small deviations in the 40-44 loop, and both AL-12 and AL-103 had backbone deviations in the CDR3 region (Pro95 loop). Both the 40-44 loop and the Pro95 region are located near the dimer interface and may therefore play a key role in the overall quaternary structure and stability of the protein.
As discussed previously, AL protein AL-09 and its germline counterpart κI O18/O8 differed in their thermodynamic stability and fibril formation capacity [13]. Intriguingly, this study also revealed that the dimer interface of the amyloidogenic protein was rotated almost 90° from the canonical interface observed in other VL proteins. This result led the authors to speculate about the role of structural changes in increasing the amyloidogenic propensity of LC proteins, and, in particular, the role of the dimer interface mutations in the altered AL-09 structure.
In a follow-up study, our laboratory solved the crystal structures of three AL-09 mutants [21]. These experiments revealed that the restorative mutant AL-09 H87Y adopts the canonical VL observed in the κI O18/O8 protein structure. The reciprocal experiments, which introduced mutations from the AL protein into the κI O18/O8 protein, failed to form the altered interface observed in AL-09. However, some backbone disruptions were observed in the 40-44 loop region, which is part of the dimer interface. This is the same region affected in the BRE, AL-12 and AL-103 structures, suggesting that these disruptions could play a common role in triggering amyloidogenesis for light chain proteins.
Fibril Formation
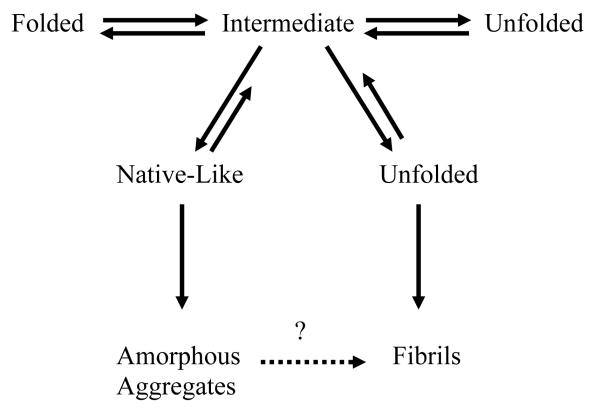
Amyloid fibril formation is initiated by the accumulation of oligomers to form a critical nucleus during the lag phase. After nucleation, the reaction proceeds to the elongation phase which involves the growth of the fibril. In addition to studying the characteristics that make a soluble LC protein more amyloidogenic, a tremendous amount of research has and is currently being done with respect to the factors that affect fibril formation in vitro. These factors include temperature, pH, ionic strength, agitation, protein concentration, and pressure, which all destabilize the protein in order to populate partially folded states that are prone to aggregation [29] (Fig. (2)).
Fig. (2).
Model for protein folding and misfolding with aggregation occurring at the intermediate step. Various factors can destabilize proteins to aid in the formation of either amorphous aggregates or fibrils based on the results from Dr. Fink’s laboratory.
Anthony Fink and his laboratory studied fibril formation of AL protein SMA and MM protein LEN to determine the fibril formation pathway of these two proteins. SMA and LEN are κIV Ig VL proteins [30]. Amyloid formation for these proteins was studied as a function of pH and in the presence of renal solutes, denaturants such as urea and guanidine hydrochloride (GdHCl), and different surfaces.
Experiments at various pH values showed differences between the AL and MM proteins. The rate of fibril formation for SMA was highly accelerated at pH 2 [30]. SMA and LEN formed fibrils with vigorous stirring at pH 2 and the results indicated that SMA had faster kinetics [31]. Amorphous aggregation of SMA was observed in samples from pH 4 to 7, while fibrils were observed in samples at pH ≤3. Thus, SMA formed different partially folded intermediates depending on the pH of the solution. Globular oligomeric deposits were observed with SMA at pH 3 using atomic force microscopy. Annular aggregates were formed at pH 4.5 with low ionic strength whereas at high ionic strength fibrils and amorphous deposits were the predominant species [32]. At pH 7, the fibril formation kinetics of LEN was shorter with lower protein concentrations and increased concentrations of urea (0 to 3 M) [33].
The use of renal solutes shed light onto the destabilizing and compensatory effects that different reagents can have on amyloid formation. Urea, a known protein denaturant, decreased the thermodynamic stability and the fibril formation kinetics of both SMA and LEN while betaine and sorbitol had the opposite effect. A concentration of 1.5 M urea was enough to increase fibril formation of both SMA and LEN [34]. Conversely, the presence of 0.5 M betaine or sorbitol (without urea) partially inhibited SMA fibril formation showing the interplay between stabilizing and denaturing forces that may play a role in physiological environments.
Other denaturant studies indicate that SMA fibril formation kinetics were dependent on the concentration of guanidine hydrochloride (GdHCl) [35]. The reaction at 2 M GdHCl was fastest and the presence of fibrils was confirmed by electron microscopy, whereas amorphous aggregates were formed at lower concentrations of GdHCl. Additionally, GdHCl affected the intermediate structures in fibril formation. Amorphous aggregates formed at 1 M GdHCl present more native-like intermediate structures (followed by circular dichroism spectroscopy), while amyloid fibrils formed at 2 M are generated through an unfolded intermediate.
Another factor that influenced the fibril formation kinetics was protein concentration, in which lower concentrations of LEN had faster kinetics of amyloid formation than higher concentrations [36]. At high protein concentrations, LEN produced off-pathway oligomeric species before fibrils were formed [37]. At low protein concentrations, the off-pathway species were absent [36]. Another way to accelerate the kinetics of fibril formation is to add a “seed” of preformed fibrils to soluble protein solutions to trigger fibril growth [38]. The addition of 5% seeds in a SMA fibril formation reaction decreased the lag time by half when compared to an unseeded reaction [18].
Zhu et al. studied the effect of surfaces on SMA amyloid fibril formation. [39]. They found that on mica surfaces, the rate of fibrillation was faster and the amount of protein required for the reaction was decreased. They also found that the mechanism for fibril growth was different on a mica surface, where protofibrils were observed as compared to the growth in solution where fibrils were present [39]. These surface experiments may be relevant in vivo since AL amyloid deposits are associated with the extracellular matrix in the basement membrane of tissues.
In an effort to understand the role of components of the basement membrane where fibrils deposit, the role of lipids in amyloid formation for AL was recently reported. The results indicated that a higher protein to lipid vesicles ratio slowed SMA amyloid formation kinetics [40]. SMA fibrillation was affected by adding cholesterol to the lipid vesicles; specifically, cholesterol concentrations above 10% had an inhibitory effect. Additionally, calcium ions in the presence of cholesterol and lipid vesicles were shown to decrease SMA fibril formation kinetics depending on the calcium concentration. The same effect was seen with Mg2+ and Zn2+ [40]. This study suggests that amyloid deposition is influenced by the combined effects of cations and membrane surfaces.
Dye binding studies such as thioflavin T fluorescence are commonly used to monitor fibril formation. Differentiating between different species that are formed during fibril formation is not possible with this method, however. Thus, atomic force microscopy imaging was used in order to observe the evolution of different fibrillar species during a fibril formation reaction of SMA with different filament sizes found at different time points during the fibrillation. A model was proposed where two filaments combine to form a protofibril and two protofibrils intertwine to form a type I fibril [41].
In addition to Dr. Fink’s laboratory, other groups have studied fibril formation using different AL and MM proteins. Jto, an MM protein, and Wil, an AL protein, are both light chain proteins from the λ6a germline that differ by 19 amino acids. Fibrils were formed with both Jto and Wil at 37°C, pH 7.5 [3]. Jto fibrils appeared more rigid, were shorter and displayed slower kinetics than fibrils formed by Wil. Similarly, from the κI O18/O8 germline, AL protein BIF and MM protein GAL were compared at 37°C where only BIF formed fibrils [5].
Certain ionic interactions may affect fibrillogenesis and be crucial to maintain the structure and stability of LC proteins. Wall et al. noted an ionic interaction between Asp29 and Arg68 in MM protein Jto, whereas AL protein Wil has neutral amino acids in these positions [42]. To test the importance of this ionic interaction, mutations were made to Jto to introduce the neutral residues (from Wil) at these sites (JtoD29A, JtoR68S). The thermodynamic stabilities of these mutants were the same, and the rate of fibril formation for JtoD29A was the same as that for Jto. However, fibril formation kinetics were much faster for JtoR68S, and an X-ray crystal structure of this mutant revealed several side-chain differences compared to Jto and JtoD29A. These differences changed the electrostatic potential surface and increased the amount of solvent-exposed hydrophobic surface for the protein. These results highlight critical structural features such as ionic interactions that participate in the stability and fibrillogenicity of AL proteins.
Most fibril formation studies are performed with the VL portion of the AL proteins. However, some studies have used full length light chains, which include both the VL and CL. Full length proteins isolated from urine samples from MM, Light Chain Deposition Disease (LCDD) and AL patients were studied to determine the type of aggregate formed by each type of protein. Fibril formation reactions were followed at the Tm for each protein for 72 hours. The results indicated that MM proteins formed spherical species, LCDD formed amorphous aggregates and AL proteins formed fibrils [43].
Hofmeister Series
One factor affecting fibril formation is the addition of salts or ions. The Hofmeister series ranks ions according to their ability to stabilize or destabilize a protein [44-45]. A proof of principle study was done with the κI O18/O8 VL protein AL-12 to determine the role of physiologically relevant anions and cations from the Hofmeister series on protein stability and amyloid fibril formation. The presence of various salts with AL-12 did not affect the secondary structure of the protein [46]. All of the salts studied enhanced AL-12 amyloid formation. Reactions with SO42− and Mg2+ showed the largest enhancement of amyloid formation. In addition, we have recently started a systematic analysis of the effect of different concentrations of NaCl on amyloid formation using two similar amyloidogenic light chains. We have found that while AL-09 readily formed fibrils across a wide range of salt concentrations, the amyloidogenic light chain AL-103 (90% sequence identity to AL-09) showed a roughly inverse dependence of the fibril formation rate on salt concentration (Martin and Ramirez-Alvarado, unpublished observations). These studies with various AL proteins and salts will help determine how sulfate ions enhance amyloid formation and will shed light onto the role of glycosaminoglycan sulfation on fibril formation in vivo.
Glycosaminoglycans (GAGs)
Glycosaminoglycans (GAGs) are a component of the extracellular matrix [47] and have been found extensively in amyloid deposits. They are long, unbranched, negatively charged heterogeneous polysaccharides formed by disaccharides of N-acetylglucosamine or N-acetylgalactosamine and uronic acid. Ohishi et al. found that GAGs are an integral part of AL amyloid fibrils and that the level of GAGs increased 10-fold in tissues from amyloidosis patients, suggesting that GAGs not only play a role interacting with amyloid fibrils but the presence of the fibrils affect GAG levels. [48]. In vitro studies using HPLC chromatography and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) showed an interaction between light chain proteins and various GAGs [49]. In another in vitro study, our laboratory showed that dermatan sulfate accelerated AL-09 amyloid fibril formation, whereas chondroitin sulfate A inhibited fibril formation and yielded a spherical intermediate [50]. Further studies of GAG influence on AL fibrillogenesis via the multiple sulfate moieties or its possible crowding effect on the amyloid fibril reaction may reveal important clues about the mechanism of amyloidogenesis and the role that GAGs play in the in vivo extracellular matrix deposition.
Research is still being done to understand the mechanism of fibril formation for AL proteins. It is important to expand on the currently reported work from Dr. Fink’s lab and others with additional proteins to find commonalities and unique features of fibril formation for the different AL proteins.
Posttranslational modifications and oxidative stress
Posttranslational modifications (PTMs) might also be implicated in amyloidogenicity. Of the amyloidogenic structural risk factors that Stevens identified in κI light chains, N-glycosylation was found in 22 of 121 samples, and 18 of those 22 samples were amyloidogenic [15]. Among these risk factors, none of the light chain germline genes encodes a glycosylation site (N-x-S/T); thus, any putative glycosylation sites are introduced through somatic hypermutation. Of the 18 amyloidogenic glycosylated LCs in this study, most of them (13/18) also had other PTMs (including S-cysteinylation, fragmentation, dimerization and S-sulfonation), so a definitive role for glycosylation is difficult to delineate. Other studies also implicated glycosylation as an important characteristic among amyloidogenic proteins [51-54], and AL proteins were glycosylated more frequently than circulating non-amyloidogenic free LCs [55-56]. Despite this evidence, the precise role of this PTM has yet to be determined.
Another study of nine κI light chains revealed several different PTMs in the full length LC proteins. Each of the proteins studied had at least one type of PTM, and the range of PTMs included N-glycosylation, disulfide-linked dimerization, S-cysteinylation, fragmentation, S-sulfonation, 3-chlorotyrosine formation, and conversion of aspartic acid to pyruvate [57]. The exact relevance of these modifications to AL pathogenesis is unknown, but cysteinylation of other proteins was suggested to induce conformational changes [58-59], which could play a role in misfolding, and chlorotyrosine residues were linked to oxidative damage [60].
Some PTMs found in AL proteins may actually have a protective role against amyloidogenesis. The two most heavily modified proteins in the aforementioned study [57] also included a methionine residue that had been oxidized to methionine sulfoxide. Methionine and cysteine are the most easily oxidized amino acids, and oxidation of a methionine residue could protect other critical residues from damage by reactive oxygen species (ROS) [61]. A study of MM protein LEN showed that the methionine-oxidized form of the protein led to the formation of amorphous aggregates instead of fibrils [62]. Thus, methionine oxidation may offer some protection against amyloidogenic fibril formation for AL proteins. However, because methionine oxidation is a fluctuating process, its antioxidant effect could be overcome by a preponderance of other amyloidogenic factors.
Oxidation effects are particularly relevant to the study of AL proteins because oxidative stress has been linked both to amyloid fibril deposits and to the mechanism of cell death [63-64]. In a study by Ando et al., AL amyloid deposits stained positively for 4-hydroxy-2-nonenal (HNE), a lipid peroxidation product that indicates oxidative injury [65]. This result could not differentiate whether oxidative stress was involved in amyloid formation or if the fibrils triggered an oxidative stress reaction after deposition. However, a more recent study indicated that oxidative stress caused by soluble amyloidogenic AL proteins plays a role in cell death. Brenner et al. examined the effects of cardiac AL proteins on cardiomyocytes and found that introducing the amyloidogenic proteins caused an increase in intracellular reactive oxygen species and upregulation of a redox-sensitive protein (heme oxygenase-1) [66]. In addition, the contractility and relaxation of the cardiomyocytes was impaired, directly linking these soluble light chain proteins to cardiomyopathy in AL patients.
Cell Toxicity Studies
Recent studies have been conducted using cardiac cells (primary rat fibroblasts and cardiomyocytes) to determine the mechanism of toxicity of light chains. Primary cardiac fibroblasts were shown to internalize amyloidogenic LCs and alter the production and sulfation pattern of proteoglycans, inducing an injury-like cellular response. Internalization did not appear to be mediated by the presence of GAGs on the cell membrane [67]. Monis et al. have demonstrated that cardiac fibroblasts internalize light chains through a fluid phase endocytic mechanism (pinocytosis). They also found that protein size and concentration as well as temperature affect the internalization of the light chains [68].
The effect of soluble light chain proteins has also been studied in renal mesangial cells. Keeling et al. found that AL and LCDD proteins cause different phenotypic transformations in mesangial cells. LCDD light chains were catabolized in the early endosomes compared to AL LCs which were transported to the lysosomes [69]. The transformation of the mesangial cells was also different. Cells treated with LCDD LCs underwent a myofibroblastic phenotypic transformation whereas AL light chains induced a macrophage-like phenotype. Clathrin played a key role in the interactions of light chains and human mesangial cells and in the endocytosis of the AL LCs [70]. These interactions may provide target areas to prevent pathological outcomes.
Model System
Currently, there is no animal model for AL. Arendt and co-workers recently established the first amyloidogenic human cell line system, ALMC-1 and ALMC-2 [71]. Arendt et al. used plasma cells from an AL patient isolated both pre- (ALMC-1) and post- (ALMC-2) stem cell transplant. They were successful in establishing cell culture conditions for the growth and propagation of the cell lines. These cell lines secrete full length λ6a LC protein. While there was some genetic variation between ALMC-1 and ALMC-2, the protein sequences from both cell lines are 100% identical. The protein secreted from these cell lines was fully folded with a β-sheet structure, was as stable as other full length proteins [43], and had the ability to form amyloid fibrils in vitro. These cell lines are a valuable tool because this is the only human derived system that secretes a significant amount of protein for biophysical studies. We expect that future studies using these cell lines will advance our understanding of the cell biology microenvironment and its possible role in the misfolding of light chain proteins.
Amyloid Inhibitors
While much research is being conducted to discern the mechanism of fibril formation and cell toxicity, other efforts have focused on the inhibition of fibrillation to prevent amyloid deposition. These inhibition studies have been conducted using molecular chaperones, peptides corresponding to a portion of the VL, monoclonal antibodies and small molecules.
In the normal endoplasmic reticulum (ER) folding environment of plasma cells, molecular chaperones such as BiP prevent aggregation of light chains [72]. BiP binds to early folding intermediates of LCs. Mutations that affect the folding of the VL displayed increased association with BiP (reviewed in [18]). These results prompted Davis and co-workers to study the effect of BiP on amyloid formation in vitro. Their results showed that BiP slowed the fibril formation reaction in vitro and inhibited protein aggregation in the cell [18] . Peptides from SMA corresponding to the regions binding to BiP were shown to act as effective inhibitors of fibril formation. In a similar study, Dul et al. showed that overexpression of Hsp70 improved solubility and decreased aggregation of SMA. SMA peptides corresponding to the Hsp70 binding site also inhibited fibril assembly in vitro and prevented aggregation of SMA in COS cells [73]. These studies showed that peptides corresponding to the binding sites of BiP and Hsp70, which target amino acids 65-70 of the light chain protein, can act as inhibitors of SMA fibrillation which gives clues to regions that may be involved in the fibril formation process.
In other inhibition studies, a murine monoclonal antibody (mAB 11-1F4) that binds to light chain fibrils and not soluble proteins was generated and characterized [74-75]. Immunohistochemical analysis revealed mAB 11-1F4 recognized light chain fibrils despite their VL subgroup. The specificity of this antibody for AL fibrils (κI, κII, κIV, λ1,λ3, λ6, λ8) was shown by Europium-Linked Immunosorbant Assay (EuLISA) where an EC50 value (concentration of antibody at half maximum binding) for binding was ~130 ± 39 nM [75]. The interaction of mAB 11-1F4 with native and fibrillar light chain LEN components were also checked by EuLISA and the antibody had similar avidity with both components. However, the fibrils had a ~2 fold reduction in signal [75]. Peptide mapping was used to determine the cryptic epitope which is located in the first 18 amino acids of the variable light chain domain and a prolyl residue at position 8 is necessary. A competition EuLISA was set up with mAB 11-1F4 and recombinant Wil fibrils were inhibited by a 50-fold molar excess of soluble LEN (1-22) peptide [75].
Small molecules have also been tested in search of fibril formation inhibitors. Congo red is a histological dye that binds to amyloid fibrils and presents a green birefringence under polarized light [76]. AL-09 fibril formation was inhibited by Congo red at a 1:1 molar ratio [50]. In contrast, Congo red did not inhibit fibril formation of SMA suggesting some specificity in the role of Congo red as an inhibitor [77]. More research is needed to find effective fibril inhibitors for a variety of AL proteins both in vitro and using cell culture systems.
Conclusions
Recent AL research has focused on factors that enhance or inhibit fibril formation, the contributions of the germline and somatic mutations to amyloidogenicity and structural studies examining the effects of mutations in AL proteins. Sequence comparisons continue to yield intriguing correlations between the identity, location or modification of certain residues and amyloidogenicity. Efforts to delineate a mechanism of amyloid fibril formation in AL have focused largely on a small cohort of proteins, and expanding these studies to include more AL proteins may confirm these results and enhance our understanding of the disease pathogenesis.
Although much progress has been made in the field of AL research, several challenges remain. Developing an animal model for AL would permit countless experimental opportunities. Although such a model remains elusive, the ALMC cell lines have the potential to advance our understanding of disease mechanisms. Further studies examining toxicity in a variety of cells could yield valuable information about the mechanism of cell death and the nature of the toxic species in AL. Determining the toxic species will inform the design of therapeutics that can target this species for prevention or elimination.
Cell lines and the development of an animal model could also provide the platform to test novel amyloid fibril inhibitors. Recent research has examined the role of molecular chaperones and small molecules in inhibition and future studies could include high throughput screening for small molecule inhibitors or targeted testing of other cofactors that interact with AL proteins. Combined with studies investigating the toxic species in AL, inhibition studies may be the key to new treatment options for AL patients.
Abbreviations
- AL
light chain amyloidosis
- Ig
immunoglobulin
- LC
light chain
- HC
heavy chain
- VL
light chain variable domain
- CL
light chain constant domain
- MM
multiple myeloma
- Tm
melting temperature
- ThT
thioflavin T
- CDR
complementarity determining region
- FR
framework region
- GdHCl
guanidine hydrochloride
- LCDD
light chain deposition disease
- GAG
glycosaminoglycan
- PTM
posttranslational modification
References
- [1].Buxbaum JN. Trends Biochem Sci. 2003;28:585–92. doi: 10.1016/j.tibs.2003.09.009. [DOI] [PubMed] [Google Scholar]
- [2].Stevens FJ, Westholm FA, Solomon A, Schiffer M. Proc Natl Acad Sci U S A. 1980;77:1144–8. doi: 10.1073/pnas.77.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wall J, Schell M, Murphy C, Hrncic R, Stevens FJ, Solomon A. Biochemistry. 1999;38:14101–8. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- [4].Stevens PW, Raffen R, Hanson DK, Deng YL, Berrios-Hammond M, Westholm FA, Murphy C, Eulitz M, Wetzel R, Solomon A, et al. Protein Sci. 1995;4:421–32. doi: 10.1002/pro.5560040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kim Y, Wall JS, Meyer J, Murphy C, Randolph TW, Manning MC, Solomon A, Carpenter JF. J Biol Chem. 2000;275:1570–4. doi: 10.1074/jbc.275.3.1570. [DOI] [PubMed] [Google Scholar]
- [6].Pepys MB. Philos Trans R Soc Lond B Biol Sci. 2001;356:203–10. doi: 10.1098/rstb.2000.0766. discussion 210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Olsen KE, Sletten K, Westermark P. Biochem Biophys Res Commun. 1998;245:713–6. doi: 10.1006/bbrc.1998.8515. [DOI] [PubMed] [Google Scholar]
- [8].Comenzo RL, Zhang Y, Martinez C, Osman K, Herrera GA. Blood. 2001;98:714–20. doi: 10.1182/blood.v98.3.714. [DOI] [PubMed] [Google Scholar]
- [9].Perfetti V, Casarini S, Palladini G, Vignarelli MC, Klersy C, Diegoli M, Ascari E, Merlini G. Blood. 2002;100:948–53. doi: 10.1182/blood-2002-01-0114. [DOI] [PubMed] [Google Scholar]
- [10].Abraham RS, Geyer SM, Price-Troska TL, Allmer C, Kyle RA, Gertz MA, Fonseca R. Blood. 2003;101:3801–8. doi: 10.1182/blood-2002-09-2707. [DOI] [PubMed] [Google Scholar]
- [11].Solomon A, Frangione B, Franklin EC. J Clin Invest. 1982;70:453–60. doi: 10.1172/JCI110635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ozaki S, Abe M, Wolfenbarger D, Weiss DT, Solomon A. Clin Immunol Immunopathol. 1994;71:183–9. doi: 10.1006/clin.1994.1070. [DOI] [PubMed] [Google Scholar]
- [13].Baden EM, Owen BA, Peterson FC, Volkman BF, Ramirez-Alvarado M, Thompson JR. J Biol Chem. 2008;283:15853–60. doi: 10.1074/jbc.M705347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].del Pozo Yauner L, Ortiz E, Sanchez R, Sanchez-Lopez R, Guereca L, Murphy CL, Allen A, Wall JS, Fernandez-Velasco DA, Solomon A, Becerril B. Proteins. 2008;72:684–92. doi: 10.1002/prot.21934. [DOI] [PubMed] [Google Scholar]
- [15].Stevens FJ. Amyloid. 2000;7:200–11. doi: 10.3109/13506120009146835. [DOI] [PubMed] [Google Scholar]
- [16].Dispenzieri A, Lacy MQ, Katzmann JA, Rajkumar SV, Abraham RS, Hayman SR, Kumar SK, Clark R, Kyle RA, Litzow MR, Inwards DJ, Ansell SM, Micallef IM, Porrata LF, Elliott MA, Johnston PB, Greipp PR, Witzig TE, Zeldenrust SR, Russell SJ, Gastineau D, Gertz MA. Blood. 2006;107:3378–83. doi: 10.1182/blood-2005-07-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hurle MR, Helms LR, Li L, Chan W, Wetzel R. Proc Natl Acad Sci U S A. 1994;91:5446–50. doi: 10.1073/pnas.91.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Davis PD, Raffen R, Dul LJ, Vogen MS, Williamson KE, Stevens JF, Argon Y. Immunity. 2000;13:433–42. doi: 10.1016/s1074-7613(00)00043-1. [DOI] [PubMed] [Google Scholar]
- [19].Ch’ang LY, Yen CP, Besl L, Schell M, Solomon A. Mol Immunol. 1994;31:531–6. doi: 10.1016/0161-5890(94)90040-x. [DOI] [PubMed] [Google Scholar]
- [20].del Pozo Yauner L, Ortiz E, Becerril B. Proteins. 2006;62:122–9. doi: 10.1002/prot.20779. [DOI] [PubMed] [Google Scholar]
- [21].Baden EM, Randles EG, Aboagye AK, Thompson JR, Ramirez-Alvarado M. J Biol Chem. 2008 doi: 10.1074/jbc.M804822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang DB, Chang CH, Ainsworth C, Brunger AT, Eulitz M, Solomon A, Stevens FJ, Schiffer M. Biochemistry. 1994;33:14848–57. doi: 10.1021/bi00253a024. [DOI] [PubMed] [Google Scholar]
- [23].Epp O, Lattman EE, Schiffer M, Huber R, Palm W. Biochemistry. 1975;14:4943–52. doi: 10.1021/bi00693a025. [DOI] [PubMed] [Google Scholar]
- [24].Schormann N, Murrell JR, Liepnieks JJ, Benson MD. Proc Natl Acad Sci U S A. 1995;92:9490–4. doi: 10.1073/pnas.92.21.9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pokkuluri PR, Solomon A, Weiss DT, Stevens FJ, Schiffer M. Amyloid. 1999;6:165–71. doi: 10.3109/13506129909007322. [DOI] [PubMed] [Google Scholar]
- [26].Alim MA, Yamaki S, Hossain MS, Takeda K, Kozima M, Izumi T, Takashi I, Shinoda T. Clin Exp Immunol. 1999;118:344–8. doi: 10.1046/j.1365-2249.1999.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bourne PC, Ramsland PA, Shan L, Fan ZC, DeWitt CR, Shultz BB, Terzyan SS, Moomaw CR, Slaughter CA, Guddat LW, Edmundson AB. Acta Crystallogr D Biol Crystallogr. 2002;58:815–23. doi: 10.1107/s0907444902004183. [DOI] [PubMed] [Google Scholar]
- [28].Roussel A, Spinelli S, Deret S, Navaza J, Aucouturier P, Cambillau C. Eur J Biochem. 1999;260:192–9. doi: 10.1046/j.1432-1327.1999.00136.x. [DOI] [PubMed] [Google Scholar]
- [29].Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson CM. Proc Natl Acad Sci U S A. 1999;96:3590–4. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Khurana R, Gillespie JR, Talapatra A, Minert LJ, Ionescu-Zanetti C, Millett I, Fink AL. Biochemistry. 2001;40:3525–35. doi: 10.1021/bi001782b. [DOI] [PubMed] [Google Scholar]
- [31].Khurana R, Souillac PO, Coats AC, Minert L, Ionescu-Zanetti C, Carter SA, Solomon A, Fink AL. Amyloid. 2003;10:97–109. doi: 10.3109/13506120309041731. [DOI] [PubMed] [Google Scholar]
- [32].Zhu M, Han S, Zhou F, Carter SA, Fink AL. J Biol Chem. 2004;279:24452–9. doi: 10.1074/jbc.M400004200. [DOI] [PubMed] [Google Scholar]
- [33].Souillac PO, Uversky VN, Millett IS, Khurana R, Doniach S, Fink AL. J Biol Chem. 2002;277:12657–65. doi: 10.1074/jbc.M109230200. [DOI] [PubMed] [Google Scholar]
- [34].Kim YS, Cape SP, Chi E, Raffen R, Wilkins-Stevens P, Stevens FJ, Manning MC, Randolph TW, Solomon A, Carpenter JF. J Biol Chem. 2001;276:1626–33. doi: 10.1074/jbc.M007766200. [DOI] [PubMed] [Google Scholar]
- [35].Qin Z, Hu D, Zhu M, Fink AL. Biochemistry. 2007;46:3521–31. doi: 10.1021/bi061716v. [DOI] [PubMed] [Google Scholar]
- [36].Souillac PO, Uversky VN, Millett IS, Khurana R, Doniach S, Fink AL. J Biol Chem. 2002;277:12666–79. doi: 10.1074/jbc.M109229200. [DOI] [PubMed] [Google Scholar]
- [37].Souillac PO, Uversky VN, Fink AL. Biochemistry. 2003;42:8094–104. doi: 10.1021/bi034652m. [DOI] [PubMed] [Google Scholar]
- [38].Harper JD, Lansbury PT., Jr. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- [39].Zhu M, Souillac PO, Ionescu-Zanetti C, Carter SA, Fink AL. J Biol Chem. 2002;277:50914–22. doi: 10.1074/jbc.M207225200. [DOI] [PubMed] [Google Scholar]
- [40].Meng X, Fink AL, Uversky VN. J Mol Biol. 2008;381:989–99. doi: 10.1016/j.jmb.2008.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ionescu-Zanetti C, Khurana R, Gillespie JR, Petrick JS, Trabachino LC, Minert LJ, Carter SA, Fink AL. Proc Natl Acad Sci U S A. 1999;96:13175–9. doi: 10.1073/pnas.96.23.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wall JS, Gupta V, Wilkerson M, Schell M, Loris R, Adams P, Solomon A, Stevens F, Dealwis C. J Mol Recognit. 2004;17:323–31. doi: 10.1002/jmr.681. [DOI] [PubMed] [Google Scholar]
- [43].Sikkink LA, Ramirez-Alvarado M. Amyloid. 2008;15:29–39. doi: 10.1080/13506120701815324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cacace MG, Landau EM, Ramsden JJ. Q Rev Biophys. 1997;30:241–77. doi: 10.1017/s0033583597003363. [DOI] [PubMed] [Google Scholar]
- [45].Zhang Y, Furyk S, Bergbreiter DE, Cremer PS. J Am Chem Soc. 2005;127:14505–10. doi: 10.1021/ja0546424. [DOI] [PubMed] [Google Scholar]
- [46].Sikkink LA, Ramirez-Alvarado M. Biophys Chem. 2008;135:25–31. doi: 10.1016/j.bpc.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bosman FT, Stamenkovic I. J Pathol. 2003;200:423–8. doi: 10.1002/path.1437. [DOI] [PubMed] [Google Scholar]
- [48].Ohishi H, Skinner M, Sato-Araki N, Okuyama T, Gejyo F, Kimura A, Cohen AS, Schmid K. Clin Chem. 1990;36:88–91. [PubMed] [Google Scholar]
- [49].Jiang X, Myatt E, Lykos P, Stevens FJ. Biochemistry. 1997;36:13187–94. doi: 10.1021/bi970408h. [DOI] [PubMed] [Google Scholar]
- [50].McLaughlin RW, De Stigter JK, Sikkink LA, Baden EM, Ramirez-Alvarado M. Protein Sci. 2006;15:1710–22. doi: 10.1110/ps.051997606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Engvig JP, Olsen KE, Gislefoss RE, Sletten K, Wahlstrom O, Westermark P. Scand J Immunol. 1998;48:92–8. doi: 10.1046/j.1365-3083.1998.00352.x. [DOI] [PubMed] [Google Scholar]
- [52].Dwulet FE, O’Connor TP, Benson MD. Mol Immunol. 1986;23:73–8. doi: 10.1016/0161-5890(86)90173-2. [DOI] [PubMed] [Google Scholar]
- [53].Foss GS, Nilsen R, Cornwell GC, 3rd, Husby G, Sletten K. Scand J Immunol. 1998;47:348–54. doi: 10.1046/j.1365-3083.1998.00311.x. [DOI] [PubMed] [Google Scholar]
- [54].Omtvedt LA, Bailey D, Renouf DV, Davies MJ, Paramonov NA, Haavik S, Husby G, Sletten K, Hounsell EF. Amyloid. 2000;7:227–44. doi: 10.3109/13506120009146437. [DOI] [PubMed] [Google Scholar]
- [55].Omtvedt LA, Husby G, Cornwell GG, 3rd, Kyle RA, Sletten K. Scand J Immunol. 1997;45:551–6. doi: 10.1046/j.1365-3083.1997.d01-431.x. [DOI] [PubMed] [Google Scholar]
- [56].Holm E, Sletten K, Husby G. Biochem J. 1986;239:545–51. doi: 10.1042/bj2390545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Connors LH, Jiang Y, Budnik M, Theberge R, Prokaeva T, Bodi KL, Seldin DC, Costello CE, Skinner M. Biochemistry. 2007;46:14259–71. doi: 10.1021/bi7013773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen W, Yewdell JW, Levine RL, Bennink JR. J Exp Med. 1999;189:1757–64. doi: 10.1084/jem.189.11.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Watarai H, Nozawa R, Tokunaga A, Yuyama N, Tomas M, Hinohara A, Ishizaka K, Ishii Y. Proc Natl Acad Sci U S A. 2000;97:13251–6. doi: 10.1073/pnas.230445397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mohiuddin I, Chai H, Lin PH, Lumsden AB, Yao Q, Chen C. J Surg Res. 2006;133:143–9. doi: 10.1016/j.jss.2005.10.008. [DOI] [PubMed] [Google Scholar]
- [61].Levine RL, Moskovitz J, Stadtman ER. IUBMB Life. 2000;50:301–7. doi: 10.1080/713803735. [DOI] [PubMed] [Google Scholar]
- [62].Hu D, Qin Z, Xue B, Fink AL, Uversky VN. Biochemistry. 2008;47:8665–77. doi: 10.1021/bi800806d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Merlini G, Westermark P. J Intern Med. 2004;255:159–78. doi: 10.1046/j.1365-2796.2003.01262.x. [DOI] [PubMed] [Google Scholar]
- [64].Schubert D, Behl C, Lesley R, Brack A, Dargusch R, Sagara Y, Kimura H. Proc Natl Acad Sci U S A. 1995;92:1989–93. doi: 10.1073/pnas.92.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ando Y, Nyhlin N, Suhr O, Holmgren G, Uchida K, el Sahly M, Yamashita T, Terasaki H, Nakamura M, Uchino M, Ando M. Biochem Biophys Res Commun. 1997;232:497–502. doi: 10.1006/bbrc.1996.5997. [DOI] [PubMed] [Google Scholar]
- [66].Brenner DA, Jain M, Pimentel DR, Wang B, Connors LH, Skinner M, Apstein CS, Liao R. Circ Res. 2004;94:1008–10. doi: 10.1161/01.RES.0000126569.75419.74. [DOI] [PubMed] [Google Scholar]
- [67].Trinkaus-Randall V, Walsh MT, Steeves S, Monis G, Connors LH, Skinner M. Am J Pathol. 2005;166:197–208. doi: 10.1016/S0002-9440(10)62244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Monis GF, Schultz C, Ren R, Eberhard J, Costello C, Connors L, Skinner M, Trinkaus-Randall V. Am J Pathol. 2006;169:1939–52. doi: 10.2353/ajpath.2006.060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Keeling J, Teng J, Herrera GA. Lab Invest. 2004;84:1322–38. doi: 10.1038/labinvest.3700161. [DOI] [PubMed] [Google Scholar]
- [70].Teng J, Russell WJ, Gu X, Cardelli J, Jones ML, Herrera GA. Lab Invest. 2004;84:440–51. doi: 10.1038/labinvest.3700069. [DOI] [PubMed] [Google Scholar]
- [71].Arendt BK, Ramirez-Alvarado M, Sikkink LA, Keats JJ, Ahmann GJ, Dispenzieri A, Fonseca R, Ketterling RP, Knudson RA, Mulvihill EM, Tschumper RC, Wu X, Zeldenrust SR, Jelinek DF. Blood. 2008;112:1931–41. doi: 10.1182/blood-2008-03-143040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Stevens FJ, Argon Y. Semin Cell Dev Biol. 1999;10:443–54. doi: 10.1006/scdb.1999.0315. [DOI] [PubMed] [Google Scholar]
- [73].Dul JL, Davis DP, Williamson EK, Stevens FJ, Argon Y. J Cell Biol. 2001;152:705–16. doi: 10.1083/jcb.152.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Solomon A, Weiss DT, Wall JS. Cancer Biother Radiopharm. 2003;18:853–60. doi: 10.1089/108497803322702824. [DOI] [PubMed] [Google Scholar]
- [75].O’Nuallain B, Allen A, Kennel SJ, Weiss DT, Solomon A, Wall JS. Biochemistry. 2007;46:1240–7. doi: 10.1021/bi0616605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sipe JD, Cohen AS. J Struct Biol. 2000;130:88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- [77].Kim YS, Randolph TW, Manning MC, Stevens FJ, Carpenter JF. J Biol Chem. 2003;278:10842–50. doi: 10.1074/jbc.M212540200. [DOI] [PubMed] [Google Scholar]