Abstract
Our current study focuses on the expression of two members of the onecut transcription factor family, One-cut1 (Oc1) and Onecut2 (Oc2), in the developing mouse retina. By immunofluorescence staining, we found that Oc1 and Oc2 had very similar expression patterns throughout retinal development. Both factors started to be expressed in the retina at around embryonic day (E) 11.5. At early stages (E11.5 and E12.5), they were expressed in both the neuroblast layer (NBL) and ganglion cell layer (GCL). As development progressed (from E14.5 to postnatal day [P] 0), expression diminished in the retinal progenitor cells and became more restricted to the GCL. By P5, Oc1 and Oc2 were expressed at very low levels in the GCL. By co-labeling with transcription factors known to be involved in retinal ganglion cell (RGC) development, we found that Oc1 and Oc2 had extensive overlap with Math5 in the NBL, and that they completely overlapped with Pou4f2 and Isl1 in the GCL, but only partially in the NBL. Co-labeling of Oc1 with cell cycle markers confirmed that Oc1 was expressed in both proliferating retinal progenitors and postmitotic retinal cells. In addition, we demonstrated that expression of Oc1 and Oc2 did not require Math5, Isl1, or Pou4f2. Thus, Oc1 and Oc2 may regulate the formation of RGCs in a pathway independent of Math5, Pou4f2, and Isl1. Furthermore, we showed that Oc1 and Oc2 were expressed in both developing and mature horizontal cells (HCs). Therefore the two factors may also function in the genesis and maintenance of HCs. J. Comp. Neurol. 520:952–969, 2012.
INDEXING TERMS: retina, transcription factors, cell differentiation, retinal ganglion cells, horizontal cells, retinal development
The various vertebrate retinal cell types all arise from the naÏve, multipotent retinal progenitor cells (RPCs) in the neural retinal epithelium (Agathocleous and Harris, 2009; Cepko et al., 1996; Livesey and Cepko, 2001). The birth of these retinal cell types follows a distinct temporal order (Young, 1985), and is subject to tight and precise regulation, particularly through gene expression (Agathocleous and Harris, 2009; Mu and Klein, 2004). Genesis of individual retinal cell types is governed by gene regulatory networks (GRNs) composed of pertinent transcription factors and their target genes (Hennig et al., 2008; Mu and Klein, 2004; Swaroop et al., 2010).
We have been interested in the GRN for the retinal ganglion cells (RGCs). RGCs are the first cell type to form in vertebrate retinas, starting at E11.5 in the mouse (Cepko et al., 1996; Young, 1985). RGC formation starts at the center of the optic cup and expands gradually to the periphery (Gan et al., 1999; Hu and Easter, 1999). Development of RGCs from RPCs is a stepwise process. RPCs first become competent for the RGC fate, retract their apical processes, and exit the cell cycle (Hinds and Hinds, 1974). Some of the competent RPCs will then adopt the RGC fate, migrate to the future ganglion cell layer (GCL), and eventually differentiate into fully functional RGCs (Yang et al., 2003). This process is subject to regulation by both extrinsic and intrinsic mechanisms (Brown et al., 1998; Erkman et al., 1996; Kim et al., 2005; Mao et al., 2008; Mu et al., 2008; Wang et al., 2001, 2005; Xiang, 1998; Zhang and Yang, 2001), and a major component of the intrinsic mechanism is gene regulation by specific transcription factors.
Based on phenotypes of gene knockout mice and gene expression profiling, a model for the RGC gene regulatory network was established previously (Mu et al., 2004, 2008, 2005a). In this model, three key transcription factors occupy key node positions: Math5 is upstream and renders the RPCs competent for the RGC fate; Pou4f2 (also known as Brn3b) and Isl1 function after RGCs are born by activating genes essential for differentiation. Pou4f2 and Isl1 regulate two distinct but overlapping sets of genes and define two different gene regulation branches downstream of Math5. Whereas this model provides useful insights into the genetic mechanism governing the formation of RGCs, it also suggests that additional regulators remain to be identified, as many RGC genes are not regulated by the known transcription factors (Mu et al., 2008).
Onecut transcription factors are characterized by a “cut” domain and an atypical homeodomain, and both domains are involved in DNA binding (Iyaguchi et al., 2007; Jacquemin et al., 1999; Lemaigre et al., 1996). This family of transcription factors is conserved in animals and has been found in Caenorhabditis elegans (Cassata et al., 1998), Drosophila (Nguyen et al., 2000), sea urchin (Poustka et al., 2004), zebrafish (Hong et al., 2002; Matthews et al., 2004), frog (Haworth and Latinkic, 2009), and mammals (Jacquemin et al., 1999, 2003b). There are three members in the mouse, Onecut 1(Oc1, also known as Hnf-6), Onecut 2 (Oc2), and Onecut 3 (Oc3) (Francius and Clotman, 2010). These factors, particularly Oc1 and Oc2, have overlapping expression patterns and redundant functions in the development of many tissues, including the liver, pancreas, intestine, and lymphocytes (Bouzin et al., 2003; Clotman et al., 2005, 2002; Dusing et al., 2010; Furuno et al., 2008; Jacquemin et al., 1999, 2003a; Margagliotti et al., 2007; Matthews et al., 2008; Vanhorenbeeck et al., 2007; Zhang et al., 2009). They are also expressed in the central nervous system including the spinal cord, brain, and retina (Francius and Clotman, 2010; Mu et al., 2001), but their functions in these tissues remain undetermined.
In our effort to identify additional transcription factors that are involved in retinal development (particularly RGC differentiation) and occupy key node positions in the RGC GRN, we decided to examine the expression patterns of onecut factors, particularly Oc1 and Oc2, in the developing retina. This was prompted by previous findings that onecut factors are expressed in the retina (Mu et al., 2001), and by our data-mining results of the Genepaint database (Visel et al., 2004), which suggest that Oc1 and Oc2 are expressed in the developing RGCs and their precursors. However, those data were nonsystematic and fragmented. Here we present a detailed analysis of the temporal and spatial expression patterns of Oc1 and Oc2 in the developing mouse retina and their relationships with Math5, Pou4f2, and Isl1. Our results suggest that Oc1 and Oc2 are potential regulators for development of not only RGCs, but also horizontal cells (HCs).
MATERIALS AND METHODS
Animal care
All mice used in this study were from a C57/BL6x129 mixed background. The Math5-, Pou4f2-, and Isl1-null mice have been described before (Gan et al., 1999; Mu et al., 2008; Wang et al., 2001). An HA-tagged Math5 (Math5HA) line was used to visualize the Math5 protein with anti-HA (Fu et al., 2009). In all experiments using mice, the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals was followed, and the procedures were approved by the IACUCs of Roswell Park Cancer Institute and University at Buffalo.
Immunofluorescence
The immunofluorescence procedure followed the protocols of our previous studies (Fu et al., 2006; Mu et al., 2005b). Briefly, embryos (E14.5) or eyes (P0 and older) were fixed for 30 minutes with 4% paraformaldehyde, washed three times with PBST (phosphate-buffered saline [PBS], pH7.4, plus 0.2% Tween 20), and embedded in OCT. The tissues were then sectioned at 16 μm. After 3× 10-minute washes with PBST and blocking with 2% bovine serum albumin (BSA) in PBST for 1 hour, the sections were incubated with primary antibodies for 1 hour, followed by 3× 10-minutes PBST washes. This was then followed by incubation with fluorescent dye-conjugated secondary antibodies and PBST washes. The nuclei were stained with propidium iodide in some cases. The slides were mounted with AquaMount (Lerner Laboratories, Pittsburgh, PA), and images were acquired with a Leica (Deerfield, IL) TCS SP2 confocal microscope. The contrast of images was adjusted in Adobe (San Jose, CA) Photoshop in some cases. The adjustments were made to the same degree for control and mutant sections for the same experiment.
Antibody characterization
Details of the sources of the primary antibodies are given in Table 1. They were characterized either by us or by others as indicated in the references, and are described below:
TABLE 1.
Antibodies Used in This Study
| Antibody | Immunogen | Manufacturer | Host | Clone | Dilution |
|---|---|---|---|---|---|
| Anti-Pou4f2 (anti-brn3)1 | Synthetic peptide corresponding to the C-terminus of human Brn3b 397–410 | Santa Cruz (#sc-6026) | Goat | Polyclonal | 1:200 |
| Anti-HA | YPYDVPDYA (influenza hemagglutinin conjugated to KLH) | Sigma (#H6908) | Rabbit | Polyclonal | 1:200 |
| Anti-HA | YPYDVPDYA (influenza hemagglutinin conjugated to KLH) | GenScript (#A00168) | Goat | Polyclonal | 1:100 |
| Anti-Isl1 (anti-islet1)1 | Recombinant C-terminal (178–349) portion of rat Isl1 | DSHB (#39.4D5) | Mouse | Monoclonal | 1:100 |
| Anti-Oc1 | Bacterial recombinant protein corresponding to amino acids 11–110 of human OC1 (HNF-6) | Santa Cruz (#sc-13050) | Rabbit | Polyclonal | 1:200 |
| Anti-Oc2 | E. coli-derived recombinant human ONECUT2 (Gln185-Thr326) | R&D Systems (#AF6294) | Sheep | Polyclonal | 1:100 |
| Anti-BrdU | DNA labeled with BrdU | Abcam (#Ab6326) | Rat | Monoclonal | 1:200 |
| Anti-PCNA1 | PCNA-protein A fusion protein | Sigma (#P8825) | Mouse | Monoclonal | 1:200 |
| Anti-phospho-histone H3 (Ser10) | Synthetic phosphor-peptide CATKQTARKsTGGKA (KLH-coupled) corresponding to residues surrounding Ser10 of human histone H3 | Cell Signaling Technology (#9706) | Mouse | Monoclonal | 1:200 |
| Anti-calbindin (D28K)1 | Purified bovine kidney calbindin-D28K | Sigma (#C9848) | Mouse | Monoclonal | 1:100 |
| Anti-Lim1/2 | Recombinant rat Lim2 (cross-reacts with Lim1) | DSHB (#4F2) | Mouse | Monoclonal | 1:100 |
Antibodies that are already in the JCN antibody database.
Anti-Pou4f2 (Santa Cruz Biotechnology, Santa Cruz, CA): We found that this antibody recognized a single 45-kDa band on Western blot with mouse E14.5 retinal lysates, labeled RGCs on retinal sections in a pattern identical to that previously reported (Gan et al., 1996; Xiang et al., 1995), and gave no signal on Pou4f2-null retinal sections.
Anti-HA (Sigma, St. Louis, MO) and anti-HA (Gen-Script, Piscataway, NJ): These antibodies recognized a single protein of 15 kDa with lysates from Math5HA/HA retinas, but not wild-type ones (our unpublished results), and produced an expression pattern in Math5HA/HA retinal sections similar to that of Math5 mRNA reported previously (Fu et al., 2009).
Anti-Isl1 (DSHB, Iowa City, IA): It recognized a ~38-kDa doublet of protein bands with both E14.5 retinal lysates and lysates from cultured cells transfected with an Isl1-expressing construct (our unpublished results), and recognized RGCs on E14.5 retinal sections in a pattern identical to that previously reported, but yielded no signal on Isl1-null retinal sections (Mu et al., 2008).
Anti-Oc1 (Santa Cruz Biotechnology): It recognized endocrine cells in the pancreas, but the signal disappeared in the Oc1-null mice (Zhang et al., 2009). We also found no signal on the E14.5 retinal sections of Oc-1-null mice. On Western blot, this antibody could detect a single ~53-kDa band with mouse E14.5 and adult brain lysates (our unpublished results). In addition, it yielded a retinal expression pattern similar to that of its mRNA (see Results).
Anti-Oc2 (R&D Systems, Minneapolis, MN): This antibody produced a retinal expression pattern similar to that of its mRNA (see Results). It did not cross-react with Oc1 because its patterns were distinct from those produced by anti-Oc1 (see Fig. 2) and in Oc1-null retinas, although anti-Oc1 detected no signal, the signals from anti-Oc2 did not change (our unpublished data). On Western blot, this antibody recognized a major band of the expected size (~54 kDa) and a minor band of ~100 kDa with E14.5 mouse brain lysates.
Anti-BrdU (Abcam, Cambridge, MA): It recognized S-phase RPCs in a pattern identical to those of previous reports with BrdU pulse-labeled retinas (Dyer et al., 2003; Mu et al., 2005b), but gave no signal with retinas not labeled by BrdU.
Anti-proliferating cell nuclear antigen (PCNA; Sigma): Our results showed that it recognized proliferating progenitor cells, but not differentiated cells, in the retina. The specificity of this antibody has also been characterized by another group (Montgomery et al., 2010). They found that the antibody could pull down PCNA from cell extracts, bind PCNA in competition assays, and yield immunofluorescence patterns identical to those of other commercially available PCNA antibodies.
Anti-p-histone H3 (ser10; Cell Signaling Technology, Danvers, MA): This antibody recognized M-phase RPCs in the expected positions at the apical surface of the retina, but not in other retinal cells (Katoh et al., 2010; Le et al., 2006; Moshiri et al., 2008). On Western blot, it recognized a ~17-kDa protein with E14.5 mouse brain lysates and this band disappeared when the lysates were treated with lambda phage phosphatase (our unpublished results).
Anti-calbindin (D28K) (Sigma): It recognized the horizontal cells in a pattern identical to those previously reported (Poche et al., 2007). On Western blot, this antibody recognized a ~37-kDa doublet with adult mouse brain lysates (our unpublished results).
Anti-Lim1/2 (DSHB): This antibody produced patterns in the retina identical to previous reports of Lim1 expression (Liu et al., 2000b; Poche et al., 2007). Although this antibody can recognize both Lim1 and Lim2, only Lim1 was recognized in the retina because Lim2 is not expressed in the retina as judged by in situ hybridization. On Western blot, this antibody detected a single ~56-kDa band with embryonic brain lysates (our unpublished data).
Figure 2.
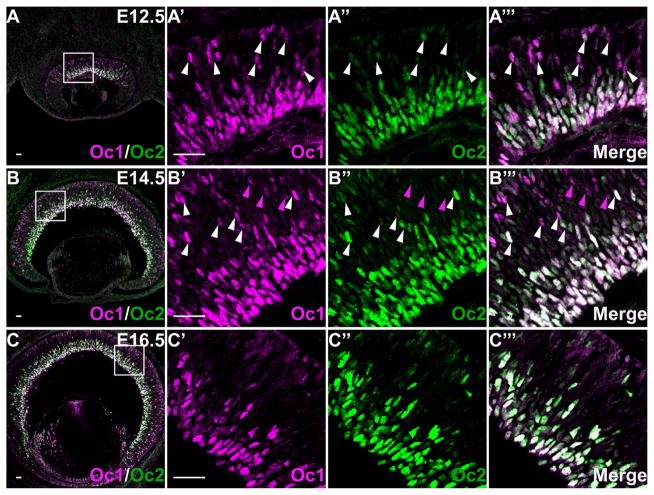
Oc1 and Oc2 have almost identical expression patterns at E12.5, E14.5, and E16.5. A–A‴: At E12.5, Oc1 and Oc2 overlap completely in the GCL and NBL. B–B‴: At E14.5, Oc1 and Oc2 overlap completely in the GCL, but in the NBL, although all Oc2-expressing cells express Oc1, many Oc1-expressing cells do not express Oc2. White arrowheads point to cells expressing both factors in the NBL, and magenta arrowheads indicate cells expressing only Oc1. C–C‴: At E16.5, Oc1 and Oc2 are close to 100% overlapping. Scale bar = 25 μm in A–C, A′ (applies to A′–A‴), B′ (applies to B′–B‴), and C′ (applies to C′–C‴).
Alexa 488- and Alexa 546-conjugated secondary antibodies against IgG of various species (Invitrogen, La Jolla, CA) were used at 1:800 dilutions. They all gave signals in the retina only in the presence of the appropriate primary antibodies, but not when applied to the retinal sections alone.
BrdU labeling
As previously described (Fu et al., 2006; Mu et al., 2005b), 100 μg bromodeoxyuridine (BrdU) per gram body weight was injected into pregnant mice intraperitoneally, and the mice were euthanized 1 hour later. Embryos were embedded in OCT and then sectioned as described above. The sections were treated with 4N HCl for 1.5 hours as previously described and stained with anti-BrdU primary antibody (Abcam, 1:200), together with anti-Oc1, anti-Oc2, or anti-HA. Alexa 488- and Alexa 546-conjugated secondary anti-IgG antibodies against appropriate species were then used to visualize the positive cells. Cell counting was carried out on a computer screen in arbitrary length units in the central region of the retinal sections, and three sections from different animals were counted.
Immunofluorescence of dissociated retinal cells
Immunofluorescence staining of dissociated retinal cells followed a previously published procedure (Dyer et al., 2003). Briefly, retinas of desired stages were dissected, washed with pre-warmed PBS, and treated with trypsin (0.1 mg/ml) for 5 minutes at 37°C. The tissues were then washed with PBS 2× 5 minutes, and triturated with a P1000 pipette tip to break them into single-cell suspensions. Soybean trypsin inhibitor (0.1 mg/ml) was then added to inactivate the trypsin at 37°C for 5 minutes. After further washing with PBS, the cells were resuspended in explants culture medium (45% Ham’s F-12, 45% DMEM, 10% FBS), transferred into chamber slides (Nalgen Nunc, Penfield, NY) coated with poly-D lysine, and incubated for 30 minutes at 37°C. The cells were then fixed with 4% paraformaldehyde for 10 minutes, followed by sequential washing with PBS (5 minutes) and 3% hydrogen peroxide in PBS (5 minutes), and blocked by 2% BSA in PBS containing 0.1% Triton X-100 (1 hour). For BrdU labeling, the cells were also treated with DNase I (2 μg/ml) at 37°C for 45 minutes. The slides were then incubated with primary and secondary antibodies as described above. Brightfield and fluorescent images of the stained slides were taken with a Nikon Eclipse 80i microscope using a SPOT RT3 digital camera (Diagnostic Instruments, Arnold, MD), and total and positive cells in individual view fields were counted in Photoshop on a computer screen. For each experiment, four different view fields from two distinct samples containing at least 500 cells were counted.
In situ hybridization
In situ hybridization followed a protocol described previously by Wallace and Raff with minor modifications (Hufnagel et al., 2010; Wallace and Raff, 1999). Briefly, embryos were fixed in 4% paraformaldehyde overnight at 4°C, embedded in OCT, sectioned at 16 μm, and stored at −80°C until use. Digoxigenin (DIG)-labeled probes were prepared by in vitro transcription and purified as previously described (Mu et al., 2004). The sections were then hybridized with the DIG-labeled probes at 65°C overnight in hybridization solution (1X salt, 50% formamide, 10% dextran sulfate, 1 mg/ml tRNA, 1X Denhardt’s solution). The slides were then washed at 65°C with washing buffer (1X SSC, 50% formamide, 0.1% Tween 20) for 3× 30 minutes and equilibrated with 1X MABT (0.1 M maleic acid, 0.25 M NaCl, 0.1% Tween 20, pH7.5) buffer twice at room temperature, followed by blocking with 20% sheep serum/2% Blocking Reagent (Roche, Indianapolis, IN) in 1X MABT and incubation with alkaline phosphatase (AP)-conjugated anti-DIG (Roche, 1:1,500) at room temperature for overnight. After thorough washing with 1X MABT, the slides were stained with NBT/BCIP until the desired signal intensity was observed. The slides were then mounted and imaged under a Nikon Eclipse 80i microscope using a SPOT RT3 digital camera (Diagnostic Instruments).
RESULTS
Spatial and temporal expression of Oc1 and Oc2 in the developing retina
By in situ hybridization, we confirmed that Oc1 and Oc2 were expressed not only in RGCs, but also in subset of cells in the neuroblast layer (NBL) at E14.5 (data not shown). In contrast, Oc3 expression was confined to the GCL at a very low level (data not shown). These results suggested that Oc1 and Oc2, but not Oc3, are expressed early in RGC development, and therefore are likely to play more significant roles in the genesis of RGCs. In addition, our inability to obtain a specific anti-Oc3 antibody prevented us from conducting a comprehensive analysis of Oc3. Therefore, we focused our study on Oc1 and Oc2. The anti-Oc1 and Oc-2 antibodies were developed against regions largely nonconserved in these two proteins and were minimally cross-reactive.
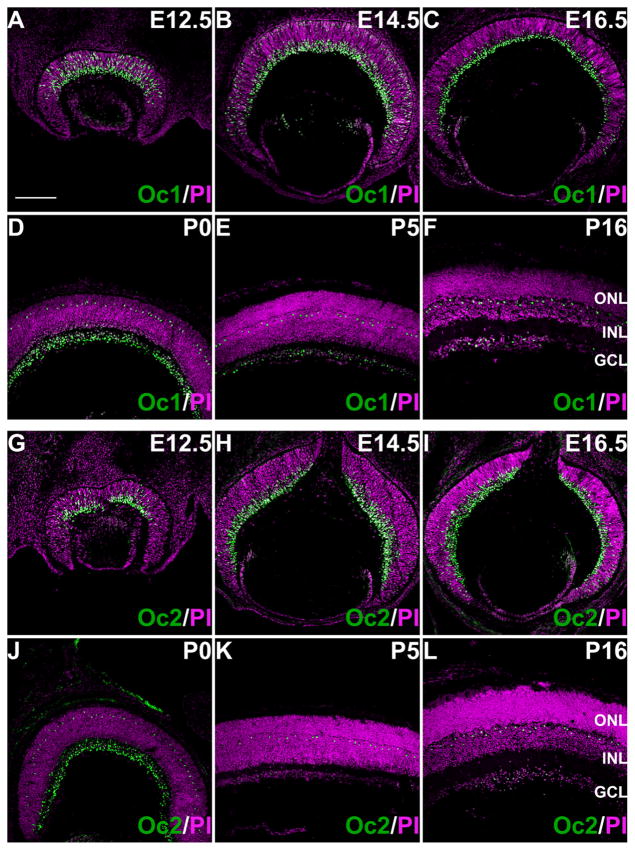
We first performed immunofluorescence staining of Oc1 and Oc2 on retinal sections of different developmental stages. As shown in Figure 1, Oc1 and Oc2 had dynamic, yet very similar expression patterns at all stages of retinal development. Expression of both Oc1 and Oc2 could be detected in the nuclei of mouse retinas at as early as E11.5 (data not shown). At early stages (E11.5 and E12.5) of retinal development, the two transcription factors were both expressed in a subset of, but not all, RPCs in the central retina (Fig. 1A,G), with Oc1 having a higher level than Oc2. Both factors were also expressed strongly in the newly forming GCL (Fig. 1A,G). At E14.5, expression of Oc1 and Oc2 expanded toward the periphery, but became more confined to the GCL (Fig. 1B,H). Nevertheless, both factors, particularly Oc1, could still be detected in the progenitors; this was particularly evident at the peripheral region where neurogenesis lags behind the central region; in the E14.5 RPCs, Oc1 level was still higher than Oc2 and there were more RPCs expressing Oc1 than those expressing Oc2 (Fig. 1B, H). At E16.5, except at the periphery where Oc1 and Oc2 could still be detected in RPCs, their expression became largely confined to the GCL (Fig. 1C,I). Oc1 and Oc2 were still highly expressed in the GCL at P0 (Fig. 1D,J), but decreased markedly afterwards (P5 and P16) (Fig. 1E,F,K,L). In contrast, from P0 through P16, Oc1 and Oc2 were observed in a row of regularly spaced cells located in the outer part of the retina; at P16, these cells were located in the inner nuclear layer (INL) close to the boundary of the INL and outer plexiform layer (OPL; Fig. 1D–F,J–L). The arrangement and location of these cells strongly suggested that they were horizontal cells.
Figure 1.
Oc1 and Oc2 are expressed in the developing and mature mouse retina. A–F: Immunofluorescence staining indicates that Oc1 has dynamic expression patterns during retinal development. At E12.5 (A) and E14.5 (B), Oc1 is expressed in both the ganglion cell layer (GCL) and neuroblast layer (NBL). At E16.5 (C), Oc1 is expressed mostly in the GCL. From P0 to P16, expression of Oc1 in the GCL decreases gradually, but a row of cells with strong Oc1 expression can be observed at the boundary between the INL and OPL. G–L: Oc2 has very similar expression patterns to those of Oc1 throughout retinal development. An obvious difference is that Oc2 has much lower levels in the NBL at E12.5 and E14.5 (G,H). In all panels, green fluorescence is Oc1 or Oc2 and magenta is pseudo color for nuclear counterstaining by propidium iodide. ONL, outer nuclear layer; INL, inner nuclear layer. Scale bar = 150 μm in A (applies to A–L).
In addition, to more accurately examine the proportions of Oc1- and Oc2-expressing cells at different developmental stages, we performed antibody staining on dissociated retinal cells from E12.5 to P16. The percentages of Oc1-positive cells were as follows: E12.5, 19.8 ± 1.0%; E14.5, 14.1 ± 0.4%; E16.5, 6.1 ± 0.1%; P0, 5.1 ± 0.3%; P5, 4.1 ± 0.5%; and P16, 2.5 ± 0.2%. The percentages of Oc2-positive cells were as follows: E12.5, 18.6 ± 1.5%; E14.5, 13.9 ± 1.2%; E16.5, 6.3 ± 0.1%; P0, 4.6 ± 0.5%; P5, 3.8 ± 0.3%; and P16, 2.3 ± 0.3%. These results are consistent with the changes in the expression of Oc1 and Oc2 observed above with retinal sections during development.
The similarity of expression patterns of Oc1 and Oc2 prompted us to further assess the degree of their overlap by co-labeling them at different stages (Fig. 2). At E12.5, they had essentially 100% overlap, but Oc2 was weaker than Oc1 in most cells in the NBL (Fig. 2A–A‴). At E14.5, all cells in the GCL expressed both Oc1 and Oc2 (Fig. 2B–B‴). In the NBL, however, as noted above, there were more cells expressing Oc1 than Oc2, but essentially all cells positive for Oc2 were also positive for Oc1 and these cells accounted for 20.5 ± 1.1% of the Oc1-positive cells (Fig. 2B′–B‴). The dynamic changes of Oc1 and Oc2 in RPCs suggested that their expression coincided with the genesis of RGCs, which starts from the central retinal region at E11.5 and propagates to the periphery to reach a peak at E14.5 (Gan et al., 1999). At later stages including E16.5 (Fig. 2C–C‴), P0, P5, and P16 (data not shown), the overlap of Oc1 and Oc2 remained close to 100% in both the GCL and the presumed horizontal cells, although the expression of both factors in the GCL gradually decreased.
Co-expression of Oc1 and Oc2 with regulators of RGC development
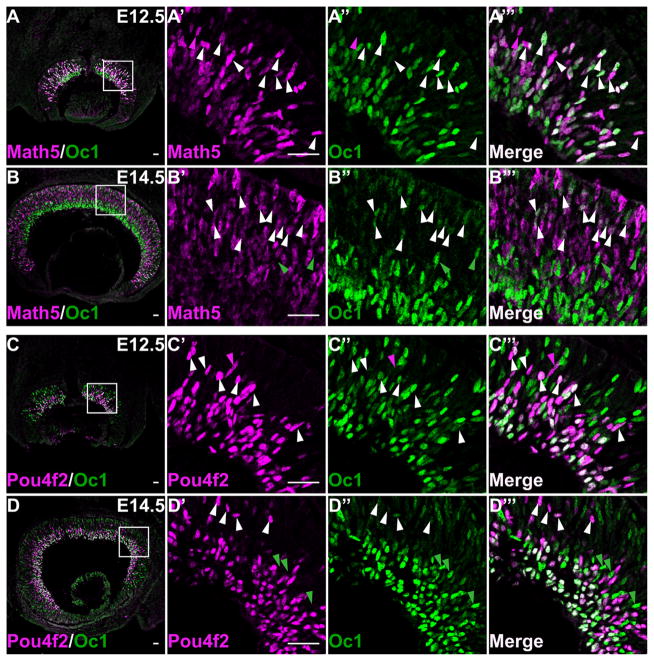
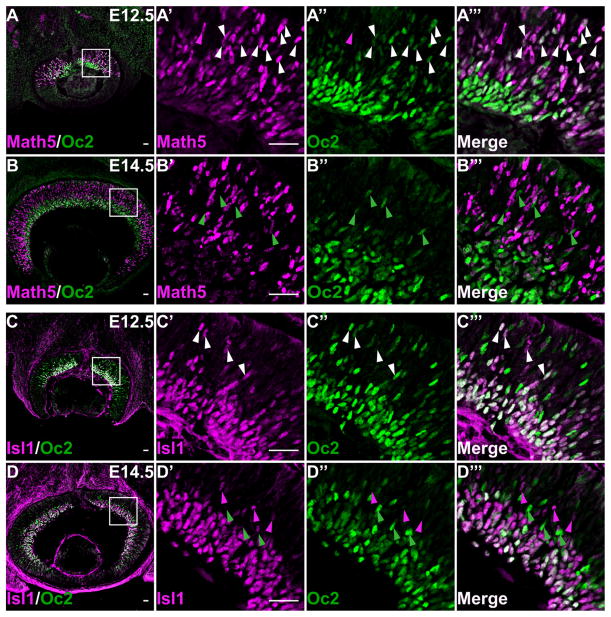
The observation of Oc1 and Oc2 in the GCL and NBL suggested they were expressed in RGCs and their precursors. To confirm this, we performed co-labeling of Oc1 with Math5, Pou4f2, and Isl1, and Oc2 with Math5 and Isl1. We could not perform co-labeling of Oc2 with Pou4f2 because the antibodies were from closely related species (sheep and goat, respectively). Because no high-quality anti-Math5 was available, labeling of Math5 was achieved with anti-HA by using a Math5HA allele we reported previously (Fu et al., 2009). Math5 is expressed in a subset of RPCs competent for RGC fate, but its gene is inactivated soon after the RGC fate is committed; therefore Math5 is detected in the NBL but not in the GCL (Brown et al., 1998) (Figs. 3A,B, 4A,B). At E11.5 and E12.5, both Oc1 and Oc2 almost completely overlapped with Math5 in the NBL of the central retina; all Oc1- and Oc2-positive cells were also Math5 positive, and, despite occasional Math5-positive-only cells, >98% of Math5-positive cells expressed Oc1 and Oc2 (Figs. 3A–A‴, 4A–A‴ and data not shown). In the GCL, Oc1 and Oc2 did not overlap with Math5 because Math5 was turned off (Figs. 3A′–A‴, 4A–A‴). At E14.5, Oc1 and Oc2, but not Math5, continued to be expressed in the GCL (Figs. 3B, 4B). In the NBL, however, the overlap of both Oc1 and Oc2 with Math5 was reduced significantly. Although almost all (90.2 ± 4.3%) Oc1-expressing cells were Math5 positive, a significant proportion (67.9 ± 6.9%) of Math5-expressing cells were Oc1 negative (Fig. 3B′–B‴). As for Oc2, because its level in the NBL became very low at E14.5, a percentage of Oc2-positive cells expressing Math5 could not be reliably obtained, but the vast majority of Math5-positive cells expressed either no or very low levels of Oc2 (Fig. 4B′–B‴). Nevertheless, in the NBL some Oc1-positive/Math5-negative or Oc2-positive/Math5-negative cells could be observed; these cells could be the HC precursors, as described later (Figs. 3B′–B‴, 4B′–B‴). The reduced overlap of Oc1 and Oc2 with Math5 was likely due to the decreased number of Oc1- and Oc2-positive cells in the NBL.
Figure 3.
Oc1 overlaps with Math5 and Pou4f2 in the developing retina. A–A‴, B–B‴: Co-labeling of Oc1 with Math5 at E12.5 and E14.5. At E12.5 (A–A‴), Oc1 and Math5 show almost complete overlap in the NBL with very rare exceptions (magenta arrowhead). At E14.5 (B–B‴), there is still substantial overlap between Math5 and Oc1, but many Math5-positive cells do not express Oc1. Although the great majority of Oc1-positive cells express Math5, a few (green arrowheads) do not. White arrowheads point to examples of cells expressing both factors at both stages. C–C‴, D–D‴: Oc1 and Pou4f2 are co-expressed in both the GCL and NBL at E12.5 and E14.5. In the GCL, Oc1 and Pou4f2 show complete overlap at both stages. In the NBL, most Pou4f2-positive cells express Oc1, but the majority of Oc1-expressing cells do not express Pou4f2. White arrowheads point to cells positive for both factors, magenta arrowheads indicate cells expressing only Pou4f2, and green arrowheads point to possible horizontal cells (HCs) expressing Oc1 but not Pou4f2. Scale bar = 25 μm in A–C, A′ (applies to A′–A‴), B′ (applies to B′–B‴), C′ (applies to C′–C‴), and D′ (applies to D′–D‴).
Figure 4.
Expression of Oc2 overlaps with Math5 and Isl1. A–A‴, B–B‴: Co-labeling of Oc2 with Math5 at E12.5 and E14.5. At E12.5 (A–A‴), similar to Oc1, Oc2 has almost complete overlap with Math5 in the NBL with very rare exceptions (magenta arrowheads). White arrowheads point to examples of cells expressing both Oc2 and Math5. At E14.5 (B–B‴), there is essentially no overlap between Math5 and Oc2. A few Oc2-positive cells (green arrowheads) could be observed in the NBL, which are likely the HC precursor cells. C–C‴, D–D‴: Oc2 and Isl1 are co-expressed in both the GCL and NBL at E12.5, but only in the GCL at E14.5. In the GCL, Oc2 and Isl1 show complete overlap at both stages with only rare exceptions (C′–C‴, D–D‴). In NBL of E12.5 retinas, most Isl1-positive cells express Oc2, but the majority of Oc2-expressing cells do not express Isl1 (C–C‴). At E14.5, there is little overlap between Oc2 and Isl1 in the NBL (D′–D‴). White arrowheads point to cells positive for both factors, magenta arrowheads indicate cells expressing only Isl1, and green arrowhead point to likely HC precursors expressing Oc2 but not Isl1. Scale bar = 25 μm in A–C, A′ (applies to A′–A‴), B′ (applies to B′–B‴), C′ (applies to C′–C‴), and D′ (applies to D′–D‴).
Pou4f2 and Isl1 are two of the earliest RGC markers expressed not only in RGCs that have already migrated to the GCL, but also in newly formed ones still located in the NBL (Mu et al., 2008; Pan et al., 2008). The significance of such expression patterns lies in the fact that the two factors are the earliest regulators in the committed RGC precursors and define two distinct but interacting branches in the RGC gene regulatory network (Mu et al., 2008). Co-staining of Oc1 with Pou4f2 indicated that, at E12.5, all RGCs in the GCL expressed both factors; in contrast, in the NBL, whereas essentially all (98.1 ± 3.2%) Pou4f2-positive cells expressed Oc1, only a subset (24.2 ± 3.8%) of Oc1-positive cells also expressed Pou4f2 (Fig. 3C–C‴). At E14.5, whereas the complete overlap of Oc1 with Pou4f2 in the GCL remained, their overlap decreased in the NBL (Fig. 3D–D‴); about 80.9 ± 4.5% of Pou4f2-positive cells in the NBL expressed some level of Oc1, but only 10.7 ± 2.5% Oc1-positive cells expressed Pou4f2. As expected, we observed similar overlaps of Oc1 with Isl1 in the GCL and NBL at E12.5 and E14.5 (data not shown), because Isl1 and Pou4f2 have essentially identical expression patterns in the retina up to E14.5 (Pan et al., 2008).
Similar to the observations with Oc1, at E12.5, Oc2 and Isl1 completely overlapped in the GCL, but only a proportion (30.9 ± 14.7%) of Oc2-expressing cells in the NBL expressed Isl1 (Fig. 4C–C‴). The co-expression of Oc2 and Isl1 was maintained in the RGCs at E14.5, but there was little overlap of the two factors in the NBL (Fig. 4D–D‴), largely due to the low levels of Oc2 in the NBL. Interestingly, at the outer border of the GCL, cells expressing high levels of Oc1 and Oc2, but negative for Pou4f2 and Isl1, were observed (Figs. 3D′–D‴, 4D′–D‴). These cells were likely HC precursors (see below).
These results suggested that Oc1 and Oc2 were expressed at two stages of RGC formation: in the RGC-competent cells when Math5 was expressed and in the fate-committed RGCs when Math5 was turned off but Pou4f2 and Isl1 were activated.
Co-expression of Oc1 and Oc2 with horizontal cell markers
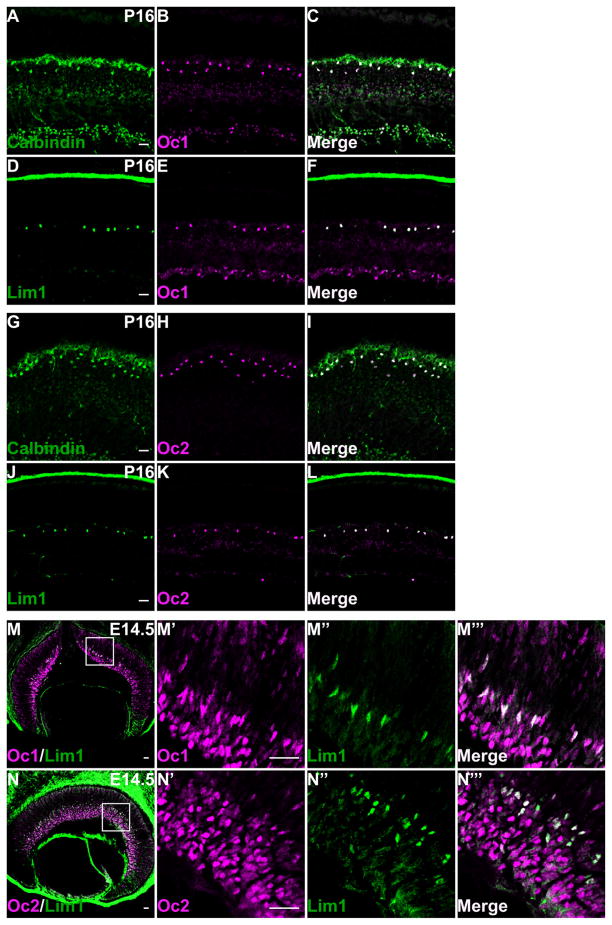
We next wanted to confirm that the row of cells expressing Oc1 and Oc2 in the outer retina after P0 were indeed HCs as indicated by their spacing and positions. To that end, we performed co-staining of Oc1 and Oc2 with two HC markers, Lim1 and calbindin (D28K) at P16. Lim1 is a lim-homeodomain transcription factor and is expressed exclusively in developing and mature HCs (Liu et al., 2000b; Poche et al., 2007) (Fig. 5D,J). Calbindin is a calcium-binding protein expressed in all HCs and a subset of amacrine cells (Dyer et al., 2003; Poche et al., 2007) (Fig. 5A,G). Oc1 showed complete overlap with both Lim1 and calbindin in the HCs (Fig. 5A–F). Essentially all HCs, as indicated by their expression of Lim1 and calbindin, expressed Oc1, and conversely all Oc1 expressing cells in the outer INL were positive for Lim1 and calbindin. By co-staining these two HC markers with Oc2, a similar degree of overlap was observed (Fig. 5G–L). These data confirmed that the Oc1/2-positive cells observed in the outer retinal region after P0 were indeed HCs, and established Oc1 and Oc2 as novel HC markers. Like RGCs, horizontal cells form during the first wave of retinal neurogenesis, which peaks at around E14.5 (Cepko et al., 1996; Young, 1985). We therefore further examined whether Oc1 and Oc2 were expressed in the precursors of HC at this stage.
Figure 5.
Oc1 and Oc2 are expressed in developing and mature HCs. A–F: Co-labeling of Oc1 with Lim1 and calbindin (28kD), two HC markers, show that all HCs express Oc1 in the mature (P16) retina. G–L: Oc2 is expressed in all HCs at P16, as demonstrated by the complete overlap of Oc2 with Lim1 and Calbindin. M–M‴: At E14.5, all HC precursors (Lim1-positive cells) express Oc1 at the boundary between the GCL and NBL. N–N″″: All Lim1-expressing HC precursors express Oc2. Scale bar = 25 μm in A (applies to A–C), D (applies to D–F), G (applies to G–I), J (applies to J–L), M, N, M′ (applies to M′–M‴), and N′ (applies to N′–N‴).
This was achieved by co-labeling with Lim1, because Lim1 is expressed at the onset of HC genesis (Poche et al., 2007) (Fig. 5M,N). As previously reported, we observed expression of Lim1 in a small group of cells neighboring the GCL on the apical side (Fig. 5M″,N″), which were the HC precursor cells. More importantly, all these cells were positive for both Oc1 and Oc2, but negative for the RGC marker Pou4f2 (Fig. 5M′–M‴, 5N′–N‴, and data not shown). These results indicated that Oc1 and Oc2 were activated at the early stages of HC development and maintained through adulthood, implying potential roles in HC specification, differentiation, and maintenance.
Oc1, Oc2 and Math5 are expressed in proliferating RPCs
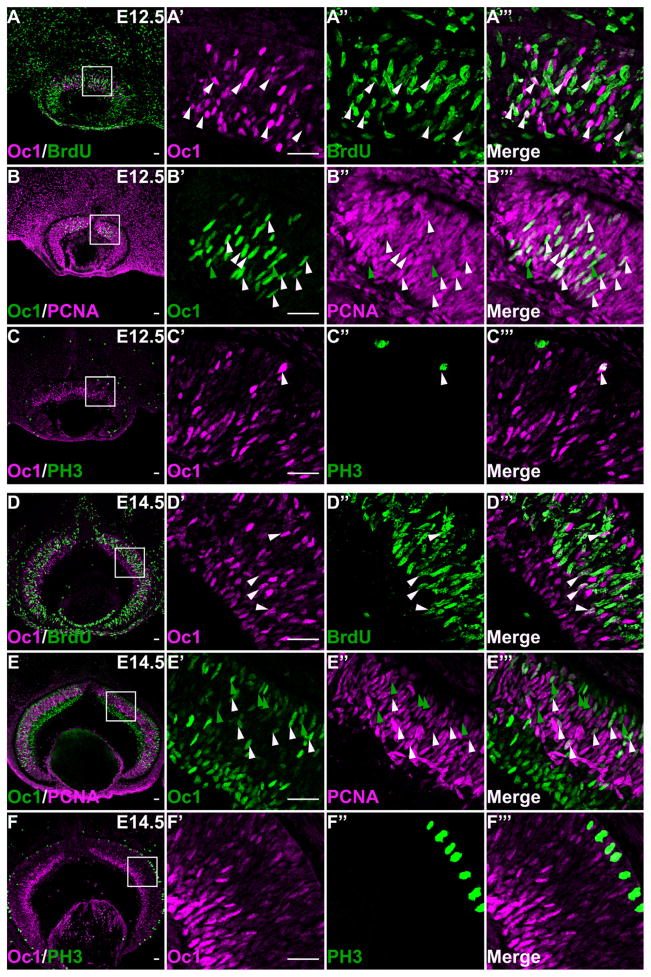
Because Oc1 and Oc2 were detected in both differentiated cells (RGCs and HCs) and RPCs, it seemed likely that they were expressed not only in postmitotic cells but also in proliferating progenitors. To confirm this, we examined the relationship of Oc1 and Oc2 expression with the cell cycle by co-labeling with markers for proliferating RPCs. We used BrdU pulse-labeling to mark S-phase cells that were located at the center of the NBL, anti-PCNA to label all proliferating cells in the NBL, and anti-phosphohistone H3 (PH3) to label M-phase cells located at the apical surface of the neural retina (Fig. 6). At E12.5, all Oc1-expressing cells in the newly forming GCL were BrdU negative, as expected, but 44.4 ± 9.1% of Oc1-expressing cells were BrdU positive in the NBL (Fig. 6A–A‴). For PCNA, a vast majority (95.8 ± 1.6%) of Oc1-expressing cells in the NBL were also PCNA positive at E12.5, but some Oc1-positive/PCNA-negative cells were observed (Fig. 6B–B‴), which were likely those exiting the cell cycle for differentiation. At this stage, a subset of M-phase (PH3-positive) cells also expressed Oc1 (Fig. 6C–C‴). All these data suggested that at the early stages of retinal development, a substantial proportion of Oc1-expressing cells were proliferating RPCs.
Figure 6.
Oc1 is expressed in both proliferating and postmitotic cells in the retina. A–A‴: At E12.5, a subset of Oc-1 expressing cells in the NBL co-labels with BrdU, but those in the newly forming GCL do not. B–B‴: At E12.5, most Oc1-expressing cells in the NBL are PCNA positive. C–C‴: Some M-phase (PH3-positive) cells express Oc1 at E12.5. D–D‴: At E14.5, many Oc1-expressing cells in the NBL still co-label with BrdU, but the percentage decreases significantly. E–E‴: Both PCNA-positive and -negative cells can be observed in the E14.5 NBL. F–F‴: Essentially no PH3-positive cells express Oc1 at E14.5. In all panels, white arrowheads indicate examples of double-labeled cells, and green arrowheads point to cells expressing only Oc1, but not the other marker. Scale bar = 25 μm in A–F, A′ (applies to A′–A‴), B′ (applies to B′–B‴), C′ (applies to C′–C‴), D′ (applies to D′–D‴), E′ (applies to E′–E‴), and F′ (applies to F′–F‴).
In contrast, at E14.5, many more Oc1-expressing cells became postmitotic, as demonstrated by the reduced overlaps of Oc1 with the cell cycle markers (Fig. 6D–D‴, E–E‴); only 19.8 ± 4.0% of Oc1-positive cells were BrdU positive, 44.2 ± 2.5% were PCNA positive, and very few, if any, PH3-positive cells expressed Oc1. Additionally, we performed BrdU/Oc1 co-labeling with dissociated retinal cell,s and, consistent with the above observations, we found that 35.5 ± 4.0% of Oc1-positive cells were BrdU positive at E12.5. This number dropped to 4.8 ± 1.0% at E14.5, and to close to 0% at E16.5. Note that these numbers are lower than those obtained with retinal sections because Oc1-positive cells included both RPCs and RGCs. These data suggested that as development advanced, expression of Oc1 shifted from mostly in proliferating RPCs to more in postmitotic cells.
We also performed co-labeling of these three proliferation markers with Oc2 on retinal sections and observed similarly that some Oc2-expressing cells were proliferating cells at E12.5, but very few at E14.5 (data not shown). Staining of dissociated retinal cells for Oc2 revealed similar results; 34.1 ± 4.7% of Oc2-positive cells were BrdU positive at E12.5, but close to 0% was found at both E14.5 and E16.5. This was expected considering the degree of overlaps of Oc1 and Oc2 expression at these stages (Fig. 2).
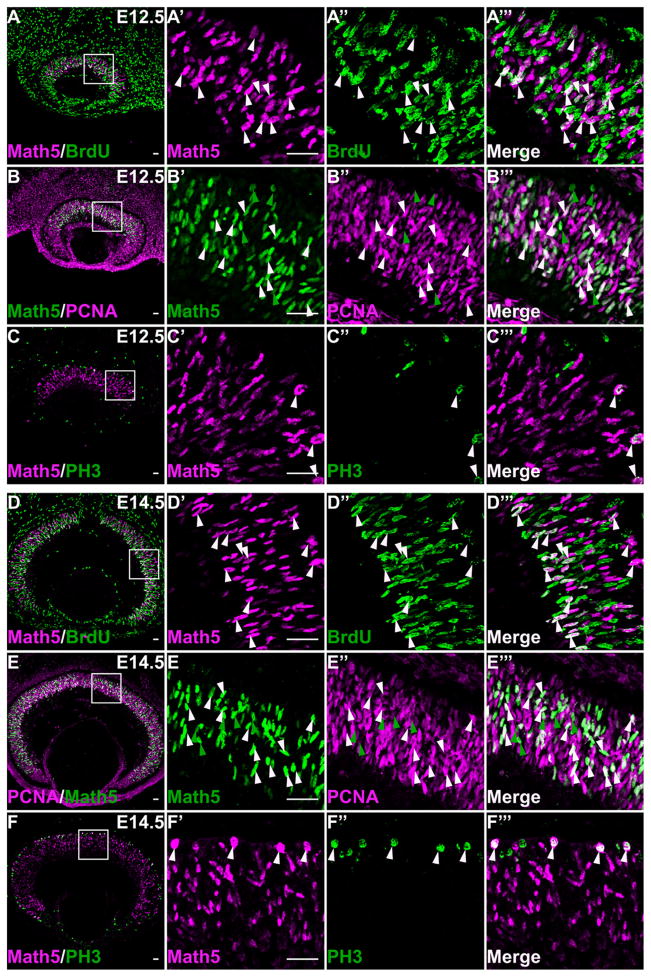
Our findings that Oc1and Oc2 had significant overlap with Math5, and were expressed in a substantial subset of proliferating RPCs, prompted us to further examine whether Math5 was also expressed in proliferating RPCs. By using a Math5lacZ knock-in allele, Yang et al. (2003) first reported previously that Math5-expressing cells were postmitotic. However, recent findings suggest that this may not be the case (Kiyama et al., 2011). To further clarify this issue, which is important for understanding the cellular events leading to RGC formation, we performed co-labeling experiments of Math5 with the above-mentioned cell cycle markers. At E12.5, 46.9 ± 8.0% of Math5-expressing cells were BrdU positive, and 91.8 ± 2.1% were PCNA positive (Fig. 7A,B). In addition, about one-fourth of PH3-positive cells also expressed Math5 (Fig. 7C). These observations confirmed that Math5-expressing cells were indeed largely proliferating RPCs. At E14.5, unlike Oc1 and Oc2, the majority of Math5-expressing cells remained proliferating. At this stage, 37.4 ± 1.7% of Math5-positive cells were BrdU positive, 93.7 ± 1.8% were PCNA positive, and many mitotic cells (PH3 positive) remained Math5 positive (Fig. 7D–F). These data clearly demonstrated that Math5 was expressed in proliferating RPCs.
Figure 7.
Math5 is expressed in proliferating RPCs. A–A‴: At E12.5, many Math5-expressing cells are BrdU positive. B–B‴: Most Math5-positive cells co-label with PCNA at E12.5. C–C‴: Many PH3-positive cells express Math5. D–D‴: At E14.5, still a sizable proportion of Math5-expressing cells remain BrdU positive. E–E‴: Both PCNA-positive and -negative Math5-expressing cells are seen in the NBL of E14.5 retina. F–F‴: Math5 expression can still be detected in many PH3-positive M-phase cells at E14.5. Some double-labeled cells are indicated by white arrowheads, and green arrowheads indicate examples of cells positive with Math5 only. Scale bar = 25 μm in A–F, A′ (applies to A′–A‴), B′ (applies to B′–B‴), C′ (applies to C′–C‴), D′ (applies to D′–D‴), E′ (applies to E′–E‴), and F′ (applies to F′–F‴).
Oc1 and Oc2 are independent of known regulators of RGC development
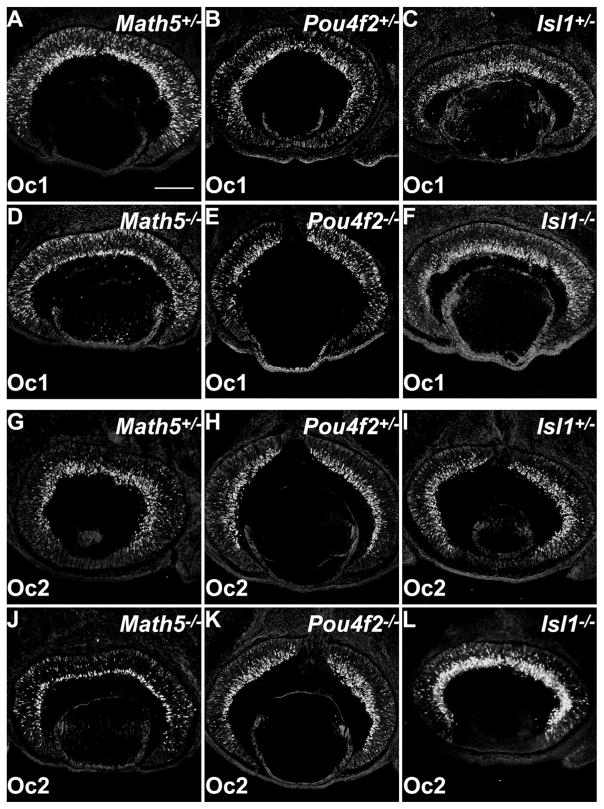
The expression patterns of Oc1 and Oc2 and their co-expression with Math5, Pou4f2, and Isl1 suggested that the two factors participate in regulating RGC formation. Thus it was important to know the genetic relationships of Oc1 and Oc2 with Math5, Pou4f2, and Isl1. For that purpose, we analyzed the expression of Oc1 and Oc2 in retinas mutant for Math5, Pou4f2, and Isl1 at E12.5 and E14.5. The results from E12.5 and E14.5 were essentially the same; we therefore only present the data from E14.5. In Math5-null retinas, compared with the heterozygous ones, there were essentially no changes in the levels and patterns of Oc1 and Oc2 expression in the NBL (Fig. 8A,D,G,J). In the inner most regions of Math5-null retinas where RGCs normally reside, only a narrow band of strongly Oc1- and Oc2-expressing cells could be detected (Fig. 8D,J). This was expected because very few RGCs develop in Math5-null retinas. The narrow band of Oc1/ Oc2-expressing cells were likely developing horizontal cells, which are normally located at the boundary between the GCL and the NBL (Fig. 5M,N); absence of the GCL might have resulted in their re-location to the innermost layer in the mutant retina.
Figure 8.
Expression of Oc1 and Oc2 is largely not regulated by Math5, Pou4f2, and Isl1. A–F: Math5-, Pou4f2-, and Isl1-null retinas (D–F) and their respective heterozygous controls (A–C) were stained with anti-Oc1. Except for Math5-null, in which the GCL is missing, the expression patterns and levels of Oc1 do not change in the mutant retinas compared with the controls. G–L: Math5-, Pou4f2-, and Isl1-null retinas (J–L) and their respective heterozygous controls (G–I) were stained with anti-Oc2. In Math5-null (J), although the GCL is missing, Oc2 expression in the NBL does not change. In Pou4f2-null, the pattern and level of Oc2 expression are the same as the control (H,K). In Isl1-null, the pattern of Oc2 expression did not change, but its level is much higher than the control (I,L). Scale bar = 150 μm in A (applies to A–L.
In Pou4f2-null retinas, the expression patterns of Oc1 and Oc2 were the same as those in heterozygous retinas, and no obvious changes in their expression levels were detected (Fig. 8B,E,H,K). In Isl1-null retinas, both Oc1 and Oc2 were observed in patterns identical to those in the heterozygous retinas (Fig. 8C,F,I,L). There was no change in Oc1 level in Isl1-null retinas (Fig. 8F), but a markedly higher level of Oc2 was seen in the mutant in both the NBL and the GCL (Fig. 8L), suggesting that Oc2, but not Oc1, is subject to repression by Isl1. These results indicated that, except that Oc2 seems to be repressed by Isl1, Oc1 and Oc2 were largely independent of the three known regulators of RGC development.
DISCUSSION
Oc1 and Oc2 are expressed in two stages of RGC development
In search of novel regulators in the GRN for mouse RGC development, we systemically analyzed the expression patterns of Oc1 and Oc2 through retinal development. In addition, we compared the expression of Oc1 and Oc2 with three transcription factors, Math5, Pou4f2, and Isl1, which are known to regulate two different stages of RGC development (Mu et al., 2008 2005a). Math5 is expressed in a subset of RPCs and functions in rendering these progenitor cells competent for the RGC fate, but not all Math5-expressing cells adopt the RGC fate (Brown et al., 1998, 2001; Wang et al., 2001; Yang et al., 2003). In comparison, Pou4f2 and Isl1 are the earliest RGC markers and initiation of their expression is concurrent with the specification of the RGC fate. Their expression continues in RGCs throughout adulthood (Mu et al., 2008; Pan et al., 2008; Xiang et al., 1995). These two factors therefore function not only in the differentiation and maintenance of RGCs (Erkman et al., 2000; Gan et al., 1996, 1999; Mu et al., 2004), but may also be required for their initial fate specification (Liu et al., 2000a; Qiu et al., 2008). Math5 has a transient overlap with Pou4f2 and Isl1 in newly forming RGCs, defining the transition from competent RPCs to specified RGCs (Fu et al., 2009).
Compared with these three known regulators, Oc1 and Oc2 demonstrate distinct and novel expression patterns. At the early stages (e.g., E12.5), they are mostly expressed in the Math5-expressing RPCs, suggesting that they play a role in establishing the competence for RGC fate in RPCs and that they may do so by collaborating with Math5. However, unlike Math5, whose expression is turned off in RGCs, expression of Oc1 and Oc2 continues in specified RGCs where Pou4f2 and Isl1 are expressed. Moreover, as development progresses, most Math5-expressing cells stop expressing Oc1 and Oc2. Oc1 and Oc2 become more restricted to the GCL and are co-expressed mostly with Pou4f2 and Isl1. Their continued expression in RGCs suggests that Oc1 and Oc2 also function in the specification and differentiation of RGCs. Therefore, Oc1 and Oc2 may function in two stages of RGC development, first in establishing the competence in RPCs, and then in RGC fate-specification and RGC differentiation. Because the GCL also contains displaced amacrine cells, it remains possible that Oc1 and Oc2 also function in these cells. Notably, Oc1 and Oc2 are essentially turned off in adult RGCs, indicating that they don’t play significant roles in the maintenance of RGCs in adulthood.
Relationship of Oc1 and Oc2 with known regulators in the RGC GRN
In the model of RGC GRN we previously established, Math5, Pou4f2, and Isl1 occupy key node positions at two levels (Mu et al., 2005a, 2008). The positions they occupy reflect their roles in the different stages of RGC development and their relationships to each other. Math5 is upstream and Pou4f2 and Isl1 are downstream. Pou4f2 and Isl1 interact with each other to co-regulate some downstream genes. The fact that Oc1 and Oc2 co-express with these three transcription factors at the two levels of RGC GRN indicates that they function at both stages of RGC development.
Our findings that Oc1 and Oc2 are largely not regulated by Math5, Pou4f2, or Isl1 imply that the two factors represent additional nodes at the two levels and define a novel and distinct pathway in the RGC GRN. At the Math5 level, Oc1 and Oc2 may regulate the properties of RPCs for the RGC fate, but further studies are needed to confirm this. At the Pou4f2 and Isl1 level, we previously demonstrated that many RGC genes are not regulated by either Pou4f2 or Isl1. Those genes must be regulated by additional key node transcription factors that define pathways parallel to those represented by Pou4f2 and Isl1. It is likely that Oc1 and Oc2 are such factors regulating RGC genes not regulated by Pou4f2 and/or Isl1; similar to Pou4f2 and Isl1, they may do so either directly or by regulating lower-tier transcription factor genes (Mao et al., 2008; Mu et al., 2008). Oc1 and Oc2 are likely to play redundant roles and define a shared pathway in RGC development, because the two factors have highly similar expression patterns in the retina, are highly homologous, particularly in their DNA-binding domain, and bind to identical cis elements on target genes (Iyaguchi et al., 2007; Jacquemin et al., 1999; Lemaigre et al., 1996). Obviously further investigation is needed to understand the roles of Oc1 and Oc2 in RGC development and to confirm that they indeed define a novel gene regulatory pathway in the RGC GRN.
Oc1/2, Math5, and cell cycle exit during RGC development
Consistent with a recent report (Kiyama et al., 2011), we found that Math5, along with Oc1 and Oc2, is expressed in proliferating RPCs. This finding revised the view that Math5 is expressed exclusively in postmitotic retinal cells (Yang et al., 2003). This diversion from the earlier results may be because in the previous studies the LacZ protein from a knockin Math5 allele, but not the native Math5 protein, was analyzed, and because LacZ has different cellular dynamics from Math5. Expression of Math5 is necessary, but not sufficient for RGC generation, because although mutation of Math5 leads to failure of RGC development, Math5-expressing cells adopt essentially all retinal cell fates (Yang et al., 2003). Therefore, Math5 defines a subpopulation of proliferating RPCs competent for the RGC fate. Oc1 and Oc2 are co-expressed with Math5 in these cells during early retinal development; they therefore may participate in the establishment of this competence state. Questions that arise from these findings include what roles Math5 and Oc1/2 play in the exit of cell cycle during RGC development and how cell cycle exit and RGC fate specification are coordinated. Analysis of Math5-null retinas showed that p27 expression and cell cycle progression were affected, but only temporarily (Le et al., 2006). In zebrafish, Ath5-expressing cells were shown by live imaging to divide only one more time (Poggi et al., 2005), and Ath5 was suggested to be activated in the G2 phase of the last cell cycle. However, our results clearly show that Math5 is already expressed in S phase. Thus it is not clear how many times Math5-expressing cells will continue to divide in the mouse. Previous studies clearly indicate a role for Math5 in cell cycle exit, but the underlying mechanism is unknown. Furthermore, it is not known how cell cycle exit is coupled with fate determination of RGCs, because Math5-expressing cells can adopt any retinal fate. Resolving these issues will require creative experimental design and novel genetic models.
Roles of Oc1 and Oc2 in HC development
Generation of HCs occurs during the first wave of retinal neurogenesis at around E14.5 in the mouse. Horizontal precursor cells first appear at the boundary between the newly forming GCL and the neuroblast layer (Poche et al., 2007). As development progresses, these precursors migrate toward the apical side, differentiate into fully mature HCs, and eventually reside in the outer side of the INL of the mature retina, with their processes synapsing with photoreceptors in the OPL (Poche et al., 2007). Although generation of HCs follows roughly the same timeline as RGCs, it is regulated by a distinct gene regulatory network in which several key transcription factors, including FoxN4, Prox1, Ptf1a, and Lim1, have been identified. FoxN4, Prox1, and Ptf1a are all required for the HC fate, with FoxN4 being upstream and Prox1 and Ptf1a downstream (Dyer et al., 2003; Fujitani et al., 2006; Li et al., 2004; Nakhai et al., 2007). Lim1, in contrast, is required for the migration of HCs to the correct position (Poche et al., 2007). Our observation that Oc1 and Oc2 are expressed in both developing and mature HCs suggests they play a role in HC development and maintenance. They may be involved in either the initial birth of HCs or the various aspects of differentiation, migration, and survival. Oc1 and Oc2 may carry out these functions by working together with the other transcription factors mentioned above. Further studies should be directed toward understanding their roles and their relationship with the other transcription factors in HC development.
Acknowledgments
Grant sponsors: The Glaucoma Foundation (to X.M.), The Whitehall Foundation (to X.M.), Grant sponsor: National Eye Institute; Grant number: EY020545 (to X.M.); Grant sponsor: Research to Prevent Blindness (unrestricted grant to the Department of Ophthalmology of University at Buffalo).
We thank Nadean Brown for providing a detailed in situ hybridization protocol, and Travis X. Liu and Tad Kaczyn-ski for technical support. We are also grateful for stimulating discussions with members of the Developmental Genomics Group at the Center of Excellence and the Department of Ophthalmology at University of Buffalo.
LITERATURE CITED
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Bouzin C, Clotman F, Renauld JC, Lemaigre FP, Rousseau GG. The onecut transcription factor hepatocyte nuclear factor-6 controls B lymphopoiesis in fetal liver. J Immunol. 2003;171:1297–1303. doi: 10.4049/jimmunol.171.3.1297. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassata G, Kagoshima H, Pretot RF, Aspock G, Niklaus G, Burglin TR. Rapid expression screening of Caenorhabditis elegans homeobox open reading frames using a two-step polymerase chain reaction promoter-gfp reporter construction technique. Gene. 1998;212:127–135. doi: 10.1016/s0378-1119(98)00137-1. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jacquemin P, Roskams T, Rousseau GG, Lemaigre FP. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development. 2002;129:1819–1828. doi: 10.1242/dev.129.8.1819. [DOI] [PubMed] [Google Scholar]
- Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849–1854. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusing MR, Maier EA, Aronow BJ, Wiginton DA. Onecut-2 knockout mice fail to thrive during early postnatal period and have altered patterns of gene expression in small intestine. Physiol Genomics. 2010;42:115–125. doi: 10.1152/physiolgenomics.00017.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Erkman L, McEvilly RJ, Luo L, Ryan AK, Hooshmand F, O’Connell SM, Keithley EM, Rapaport DH, Ryan AF, Rosenfeld MG. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- Erkman L, Yates PA, McLaughlin T, McEvilly RJ, Whisenhunt T, O’Connell SM, Krones AI, Kirby MA, Rapaport DH, Bermingham JR, O’Leary DD, Rosenfeld MG. A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron. 2000;28:779–792. doi: 10.1016/s0896-6273(00)00153-7. [DOI] [PubMed] [Google Scholar]
- Francius C, Clotman F. Dynamic expression of the One-cut transcription factors HNF-6, OC-2 and OC-3 during spinal motor neuron development. Neuroscience. 2010;165:116–129. doi: 10.1016/j.neuroscience.2009.09.076. [DOI] [PubMed] [Google Scholar]
- Fu X, Sun H, Klein WH, Mu X. Beta-catenin is essential for lamination but not neurogenesis in mouse retinal development. Dev Biol. 2006;299:424–437. doi: 10.1016/j.ydbio.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Kiyama T, Li R, Russell M, Klein WH, Mu X. Epitope-tagging Math5 and Pou4f2: new tools to study retinal ganglion cell development in the mouse. Dev Dyn. 2009;238:2309–2317. doi: 10.1002/dvdy.21974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, Fujikado T, Magnuson MA, Xiang M, Wright CV. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Furuno K, Ikeda K, Hamano S, Fukuyama K, Sonoda M, Hara T, Sasazuki T, Yamamoto K. Onecut transcription factor OC2 is a direct target of T-bet in type-1 T-helper cells. Genes Immun. 2008;9:302–308. doi: 10.1038/gene.2008.18. [DOI] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Wang SW, Huang Z, Klein WH. POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev Biol. 1999;210:469–480. doi: 10.1006/dbio.1999.9280. [DOI] [PubMed] [Google Scholar]
- Haworth KE, Latinkic B. Expression of Xenopus tropicalis HNF6/Onecut-1. Int J Dev Biol. 2009;53:159–162. doi: 10.1387/ijdb.072472ke. [DOI] [PubMed] [Google Scholar]
- Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds JW, Hinds PL. Early ganglion cell differentiation in the mouse retina: an electron microscopic analysis utilizing serial sections. Dev Biol. 1974;37:381–416. doi: 10.1016/0012-1606(74)90156-0. [DOI] [PubMed] [Google Scholar]
- Hong SK, Kim CH, Yoo KW, Kim HS, Kudoh T, Dawid IB, Huh TL. Isolation and expression of a novel neuron-specific onecut homeobox gene in zebrafish. Mech Dev. 2002;112:199–202. doi: 10.1016/s0925-4773(01)00647-5. [DOI] [PubMed] [Google Scholar]
- Hu M, Easter SS. Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Dev Biol. 1999;207:309–321. doi: 10.1006/dbio.1998.9031. [DOI] [PubMed] [Google Scholar]
- Hufnagel RB, Le TT, Riesenberg AL, Brown NL. Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev Biol. 2010;340:490–503. doi: 10.1016/j.ydbio.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyaguchi D, Yao M, Watanabe N, Nishihira J, Tanaka I. DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6. Structure. 2007;15:75–83. doi: 10.1016/j.str.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Lannoy VJ, Rousseau GG, Lemaigre FP. OC-2, a novel mammalian member of the ONECUT class of homeodomain transcription factors whose function in liver partially overlaps with that of hepatocyte nuclear factor-6. J Biol Chem. 1999;274:2665–2671. doi: 10.1074/jbc.274.5.2665. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Lemaigre FP, Rousseau GG. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev Biol. 2003a;258:105–116. doi: 10.1016/s0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Pierreux CE, Fierens S, van Eyll JM, Lemaigre FP, Rousseau GG. Cloning and embryonic expression pattern of the mouse Onecut transcription factor OC-2. Gene Expr Patterns. 2003b;3:639–644. doi: 10.1016/s1567-133x(03)00110-8. [DOI] [PubMed] [Google Scholar]
- Katoh K, Omori Y, Onishi A, Sato S, Kondo M, Furukawa T. Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J Neurosci. 2010;30:6515–6526. doi: 10.1523/JNEUROSCI.0771-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–1930. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- Kiyama T, Mao CA, Cho JH, Fu X, Pan P, Mu X, Klein WH. Overlapping spatiotemporal patterns of regulatory gene expression are required for neuronal progenitors to specify retinal ganglion cell fate. Vision Res. 2011;51:251–259. doi: 10.1016/j.visres.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 2006;295:764–778. doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Lemaigre FP, Durviaux SM, Truong O, Lannoy VJ, Hsuan JJ, Rousseau GG. Hepatocyte nuclear factor 6, a transcription factor that contains a novel type of homeodomain and a single cut domain. Proc Natl Acad Sci U S A. 1996;93:9460–9464. doi: 10.1073/pnas.93.18.9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Liu W, Khare SL, Liang X, Peters MA, Liu X, Cepko CL, Xiang M. All Brn3 genes can promote retinal ganglion cell differentiation in the chick. Development. 2000a;127:3237–3247. doi: 10.1242/dev.127.15.3237. [DOI] [PubMed] [Google Scholar]
- Liu W, Wang JH, Xiang M. Specific expression of the LIM/homeodomain protein Lim-1 in horizontal cells during retinogenesis. Dev Dyn. 2000b;217:320–325. doi: 10.1002/(SICI)1097-0177(200003)217:3<320::AID-DVDY10>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Mao CA, Kiyama T, Pan P, Furuta Y, Hadjantonakis AK, Klein WH. Eomesodermin, a target gene of Pou4f2, is required for retinal ganglion cell and optic nerve development in the mouse. Development. 2008;135:271–280. doi: 10.1242/dev.009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margagliotti S, Clotman F, Pierreux CE, Beaudry JB, Jacquemin P, Rousseau GG, Lemaigre FP. The Onecut transcription factors HNF-6/OC-1 and OC-2 regulate early liver expansion by controlling hepatoblast migration. Dev Biol. 2007;311:579–589. doi: 10.1016/j.ydbio.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Matthews RP, Lorent K, Russo P, Pack M. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–259. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Matthews RP, Lorent K, Pack M. Transcription factor onecut3 regulates intrahepatic biliary development in zebrafish. Dev Dyn. 2008;237:124–131. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- Montgomery JE, Parsons MJ, Hyde DR. A novel model of retinal ablation demonstrates that the extent of rod cell death regulates the origin of the regenerated zebrafish rod photoreceptors. J Comp Neurol. 2010;518:800–814. doi: 10.1002/cne.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri A, Gonzalez E, Tagawa K, Maeda H, Wang M, Frishman LJ, Wang SW. Near complete loss of retinal ganglion cells in the math5/brn3b double knockout elicits severe reductions of other cell types during retinal development. Dev Biol. 2008;316:214–227. doi: 10.1016/j.ydbio.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Semin Cell Dev Biol. 2004;15:115–123. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Mu X, Zhao S, Pershad R, Hsieh TF, Scarpa A, Wang SW, White RA, Beremand PD, Thomas TL, Gan L, Klein WH. Gene expression in the developing mouse retina by EST sequencing and microarray analysis. Nucleic Acids Res. 2001;29:4983–4993. doi: 10.1093/nar/29.24.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Beremand PD, Zhao S, Pershad R, Sun H, Scarpa A, Liang S, Thomas TL, Klein WH. Discrete gene sets depend on POU domain transcription factor Brn3b/Brn-3.2/POU4f2 for their expression in the mouse embryonic retina. Development. 2004;131:1197–1210. doi: 10.1242/dev.01010. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Beremand PD, Thomas TL, Klein WH. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev Biol. 2005a;280:467–481. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Liang S, Maeda H, Frishman LJ, Klein WH. Ganglion cells are required for normal progenitor-cell proliferation but not cell-fate determination or patterning in the developing mouse retina. Curr Biol. 2005b;15:525–530. doi: 10.1016/j.cub.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci U S A. 2008;105:6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GI, Schmid RM. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134:1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- Nguyen DN, Rohrbaugh M, Lai Z. The Drosophila homolog of Onecut homeodomain proteins is a neural-specific transcriptional activator with a potential role in regulating neural differentiation. Mech Dev. 2000;97:57–72. doi: 10.1016/s0925-4773(00)00431-7. [DOI] [PubMed] [Google Scholar]
- Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135:1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poche RA, Kwan KM, Raven MA, Furuta Y, Reese BE, Behringer RR. Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J Neurosci. 2007;27:14099–14107. doi: 10.1523/JNEUROSCI.4046-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggi L, Vitorino M, Masai I, Harris WA. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J Cell Biol. 2005;171:991–999. doi: 10.1083/jcb.200509098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka AJ, Kuhn A, Radosavljevic V, Wellenreuther R, Lehrach H, Panopoulou G. On the origin of the chordate central nervous system: expression of onecut in the sea urchin embryo. Evol Dev. 2004;6:227–236. doi: 10.1111/j.1525-142X.2004.04028.x. [DOI] [PubMed] [Google Scholar]
- Qiu F, Jiang H, Xiang M. A comprehensive negative regulatory program controlled by Brn3b to ensure ganglion cell specification from multipotential retinal precursors. J Neurosci. 2008;28:3392–3403. doi: 10.1523/JNEUROSCI.0043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhorenbeeck V, Jenny M, Cornut JF, Gradwohl G, Lemaigre FP, Rousseau GG, Jacquemin P. Role of the Onecut transcription factors in pancreas morphogenesis and in pancreatic and enteric endocrine differentiation. Dev Biol. 2007;305:685–694. doi: 10.1016/j.ydbio.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace VA, Raff MC. A role for Sonic hedgehog in axon-to-astrocyte signalling in the rodent optic nerve. Development. 1999;126:2901–2909. doi: 10.1242/dev.126.13.2901. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dakubo GD, Thurig S, Mazerolle CJ, Wallace VA. Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development. 2005;132:5103–5113. doi: 10.1242/dev.02096. [DOI] [PubMed] [Google Scholar]
- Xiang M. Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev Biol. 1998;197:155–169. doi: 10.1006/dbio.1998.8868. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somato-sensory neurons. J Neurosci. 1995;15:4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ables ET, Pope CF, Washington MK, Hipkens S, Means AL, Path G, Seufert J, Costa RH, Leiter AB, Magnuson MA, Gannon M. Multiple, temporal-specific roles for HNF6 in pancreatic endocrine and ductal differentiation. Mech Dev. 2009;126:958–973. doi: 10.1016/j.mod.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Yang XJ. Regulation of retinal ganglion cell production by Sonic hedgehog. Development. 2001;128:943–957. doi: 10.1242/dev.128.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]