Abstract
An intricate network regulates the activities of SIRT1 and PARP1 proteins and continues to be uncovered. Both SIRT1 and PARP1 share a common co-factor nicotinamide adenine dinucleotide (NAD+) and several common substrates, including regulators of DNA damage response and circadian rhythms. We review this complex network using an interactive Molecular Interaction Map (MIM) to explore the interplay between these two proteins. Here we discuss how NAD + competition and post-transcriptional/translational feedback mechanisms create a regulatory network sensitive to environmental cues, such as genotoxic stress and metabolic states, and examine the role of those interactions in DNA repair and ultimately, cell fate decisions.
Keywords: SIRT1, Sirtuins, PARP1, Poly-(ADP) polymerases, Metabolism, Nicotinamide Adenine Dinucleotide (NAD+) competition, Post-translational modifications, Transcriptional regulation, DNA damage repair, Circadian rhythms
Introduction
SIRT1 and PARP1 are enzymes that affect two key post-translational modifications: acetylation and ADP-ribosylation, respectively, for a diverse group of proteins. These enzymes are functionally connected due to their use of a common substrate, nicotinamide adenine dinucleotide (NAD+) [1,2]. Recent studies suggest that these proteins participate in common pathways providing cells with a mechanism for balancing cell survival and death. A well-developed understanding of activity overlap of these proteins may provide insights into the biology of these two proteins as they are actively being pursued as therapeutic targets in a range of conditions, including cancer and metabolic disorders [3-5].
In this review, we look at the role of each of these two proteins using a Molecular Interaction Map (MIM) that visually integrates the experimental findings of the regulatory pathways that surround these proteins, shown in Figure 1. The MIM helps free readers from a linear view of events and gain a better understanding of control loops involved in these pathways [6]. A machine-readable version of the MIM is provided as Additional file 1 viewable using PathVisio-MIM ( http://discover.nci.nih.gov/mim/mim_pathvisio.html) [7]. Additionally, the MIM covers in greater detail the interactions surrounding SIRT1 and PARP1; a complete list of annotations is also provided as Additional file 2.
Figure 1.
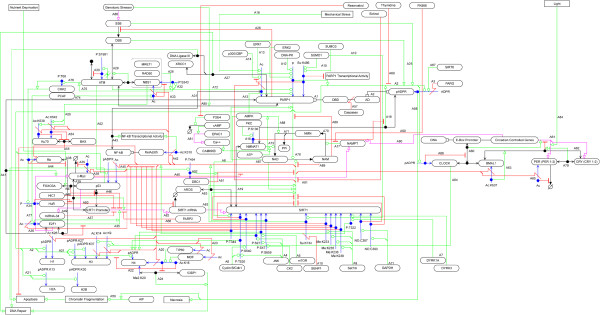
SIRT1 and PARP1 Molecular Interaction Map. Blue: Post-translational modifications, Red: Inhibition and cleavage, Green: Stimulation and catalysis.
Figure 2 shows a modular overview of how the SIRT1 and PARP1 interactions are laid out (an arrow indicates that a molecule or process from the source module has an interaction with a molecule or process in the target module) and Figure 3 provides a legend for reading the MIM notation. Throughout this review readers will see annotation labels in double square brackets and prefixed with a letter that refer to specific interactions in the MIM shown in Figure 1 and Additional file 2. We focus on aspects that alter the activity of these proteins, including: post-translational modifications, co-regulation, NAD + competition and co-regulated targets. Additionally, we discuss outstanding questions that would more accurately describe the relationship between these proteins.
Figure 2.
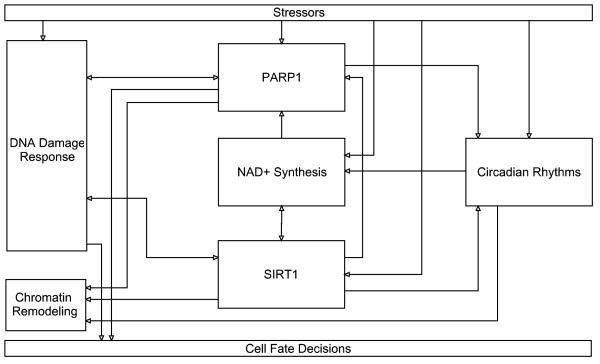
Modular layout diagram of SIRT1 and PARP1 MIM. Arrows connecting modules indicate that a node starting in a module connects to a node in the target module.
Figure 3.
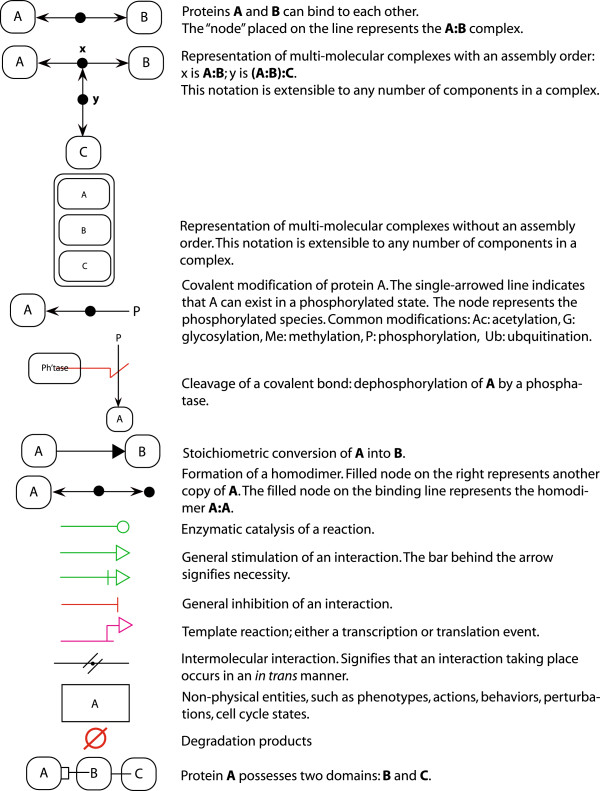
Reference guide for the MIM notation.
Review
SIRT1
Sirtuins were originally discovered in yeast where the SIR (Silent Information Regulator) genes are necessary for the repression of silent mating-type loci [1]. The mammalian family of sirtuins consists of 7 proteins, SIRT1-7, which are ubiquitously expressed. Three of the sirtuins, SIRT1, SIRT6, and SIRT7 localize to the nucleus, SIRT2 is mainly localized in the cytoplasm, while the remaining three sirtuins: SIRT3, SIRT4, and SIRT5 are found in the mitochondria [8]. The members of the sirtuin family not only differ in cellular locations, but also in enzymatic function. SIRT1 and SIRT5 are primarily protein deacetylases, SIRT4 and SIRT6 are mono(ADP)-ribosyltransferases, and SIRT2 and SIRT3 exhibit both enzymatic activities; no clear functionality has been attributed to SIRT 7 [9-14]. The conserved catalytic domain of sirtuins is capable of carrying out both deacetylation and ADP-ribosylation activities using NAD+, and it has been suggested that sirtuins may have the potential to carry out either enzymatic activity under the right conditions [15,16]. This review will focus on SIRT1 (EC 3.5.1.-) a member of the sirtuin family that is expressed in many tissues and acts as a NAD + -dependent protein deacetylase [1]. SIRT1 has been implicated in signaling pathways underlying various diseases, including: diabetes, cardiovascular disease, neurodegeneration, cancer, aging, and obesity [17].
PARP1
PARP1 (EC 2.4.2.30) is an NAD + -dependent nuclear ADP-ribosyltransferase with three domains: a DNA binding domain (DBD), an auto-modification domain (AD), and a catalytic domain [[A1]] [18]. The PARP family of proteins are involved in many processes, including: DNA damage response, cell death, cell cycle regulation, and telomere regulation [19]. The main function of PARP1 is the formation of poly(ADP-ribose) (PAR) chains on itself and other proteins [[A2]] [2,20,21]. PARP1 is a transcriptional co-activator where PAR acts as a signal helping to regulate transcription [22]. PAR is quickly cleaved by poly (ADP-ribose) glycohydrolase (PARG) [[A3]] [20]. PARP1 becomes highly activated by DNA strand breaks; electrostatic repulsion between the poly (ADP)-ribose (PAR) chains and DNA eventually leads to its catalytic inactivation [18].
Post-transcriptional regulation of SIRT1 and PARP1
Here we review post-translational modifications that affect the activities of SIRT1 and PARP1.
SIRT1
For a comprehensive review of sirtuin modifications, see Flick and Luscher [23]. Below we describe several of these modifications for SIRT1 and augment this list with additional modifications.
SIRT1 phosphorylation results in both stimulatory and inhibitory effects. Phosphorylation of SIRT1 by JNK occurs at three sites: S27, S47, and T530 in response to oxidative stress that stimulates its deacetylation activity [[A4]] [24]. In contrast, mTOR also phosphorylates SIRT1 in response to oxidative stress, but only at a single site, S47, resulting in the inhibition of SIRT1 [[A5]] suggesting a multi-site phosphorylation regulatory mechanism is in place; such a mechanism may be involved in the regulation of the timing of SIRT1 activity [25,26]. CK2 phosphorylates human SIRT1 at S659 and S661, both in vivo and in vitro, stimulating its deacetylation activity [[A4]] [27,28]. These two phosphorylation sites exist in a region of SIRT1 that are essential for SIRT1 activity both for its catalytic activity and ability to bind to substrates [29]. Cyclin B/Cdk1, a cell cycle-dependent kinase, can phosphorylate SIRT1 at T530 and S540 [[A6]]. Phosphorylation at these two sites decreases the activity of SIRT1 and disrupts progress of the cell cycle [30]. Similar to the case with mTOR and S47, T530 is a site phosphorylated by JNK and may also function as a part of a combinatorial modification program.
Kinases DYRK1A and DYRK3 have been shown to phosphorylate human SIRT1 at T522, stimulating the deacetylation of p53 by SIRT1[[A7]]; phosphorylation at this site increases the rate of product release by SIRT1 [31]. AMPK phosphorylates human SIRT1 at T344 inhibiting its ability to decacetylate p53, a known target of SIRT1 [[A8]] [32]. In addition to phosphorylation, methylation of SIRT1 by Set7/9 at K233, K235, K236, and K238 inhibits the SIRT1-mediated deacetylation of p53 in response to DNA damage [[A9]] [33]. Sumoylation at K734 by SUMO1 increases, whereas desumoylation by SENP1 decreases, the activity of SIRT1 in response to genotoxic stress [[A10]] [34]. In this study, genotoxic stress promoted the association of SIRT1 with SENP1, which may help to inhibit the ability of SIRT1 to promote survival. Additionally, transnitrosylation of SIRT1 by GAPDH at C387 and C390 has been found to inhibit the activity of SIRT1 leading to decreased PGC1α transcriptional activity; PGC1α is an important regulator of metabolism and mitochondrial function [[A11]] [35].
PARP1
The activity of PARP1 can be modulated via post-translational modifications, including phosphorylation, sumoylation, and acetylation. DNA-PK phosphorylates PARP1 though its effect is unknown [[A12]] [36]. Phosphorylation of PARP1 by AMPK has been shown to enhance its activity [[A13]] [37]. This stimulation of PARP1 by AMPK contrasts with the AMPK-mediated inhibition of SIRT1 and suggests one mechanism by which AMPK, a metabolic sensor able to regulate ATP-consuming pathways, may be capable of controlling cell survival given the roles of PARP1 and SIRT1 in response to DNA damage. ERK1/2 has also been shown to phosphorylate PARP1 in neuronal cells and to stimulate the activity of PARP1 in response to DNA damage; inhibition of ERK1/2 results in the inhibition of PARP1-mediated cell death [38]. PARP1 is acetylated by p300/CBP; this acetylation is involved in the activation of NF-κB by PARP1 [[A14]] [39]. PARP1 is sumoylated by SUMO1 and SUMO3 at K486 of PARP1's auto-modification domain. This modification inhibits the ability of p300 to acetylate PARP1 and inhibits the expression of genes that are transcriptionally targeted by PARP1 [[A15]] [40].
Co-regulation of SIRT1 and PARP1
Cross-modification and transcriptional co-regulation
SIRT1 and PARP1 are transcriptionally and functionally interconnected [41-43]. In SIRT1-deficient mouse cardiomyocytes, Rajamohan et al. in 2009 found increased levels of PARP1 acetylation in response to mechanical stress, suggesting that SIRT1 can deacetylate PARP1 [[A16]] [44]. Whether this interaction occurs during genotoxic stress or other types of stresses remains an open question. No similar modification reaction has been seen on SIRT1 by PARP1 in response to DNA damage. However, SIRT1 is able to negatively regulate the PARP1 promoter, and the SIRT1 promoter has been shown to be under the influence of PARP2 [45,46].
NAD + competition
Another key co-regulatory mechanism between these two proteins is the utilization of nicotinamide adenine dinucleotide (NAD+). It has been suggested by several studies that activation of PARP1 causes a depletion in NAD + levels, which inhibits SIRT1 activity [42-45]. In mammals, NAD + is mainly generated through the salvage pathway; this pathway involves nicotinamide (NAM) as the major precursor in this multi-step process that involves the conversion of NAM into nicotinamide mononucleotide (NMN) and then NMN into NAD+. The rate-limiting protein in the NAM-NMN-NAD + conversion is nicotinamide phosphoribosyltransferase (NAMPT) [[A17]]. PARP1 was shown to have a greater effect on NAD + depletion than SIRT1 in response to the NAMPT inhibitor, FK866 [47]. In a related study, the inhibition of NAMPT by FK866 was shown to produce an effect similar to SIRT1 depletion [48].
Recent evidence suggests that local supplies of NAD + may be important for the enzymatic activities of these two proteins, shown in Figure 4[49-51]. Nicotinamide mononucleotide adenylyltransferase 1 (NMNAT1), which catalyzes the conversion of NMN into NAD + in the synthesis of NAD + via the salvage pathway, can bind to (ADP-ribose) polymers in vitro leading to the stimulation of PARP1 activity; this effect is diminished when NMNAT1 is phosphorylated at S136 by protein kinase C (PKC) [[A18]] [52]. A similar interaction has been shown to occur with SIRT1, whereby SIRT1 binds to NMNAT1 helping to recruit NMNAT1 to specific promoters, which may help stimulate SIRT1 activity [[A19]] [53]. PARP1 activity leads to increased NAM concentrations at DNA damage sites, potentially leading to local inhibition of SIRT1 histone deacetylase activity [54]. Further work is needed to understand the function of these two proteins around chromatin sites occupied beforehand by one of them.
Figure 4.
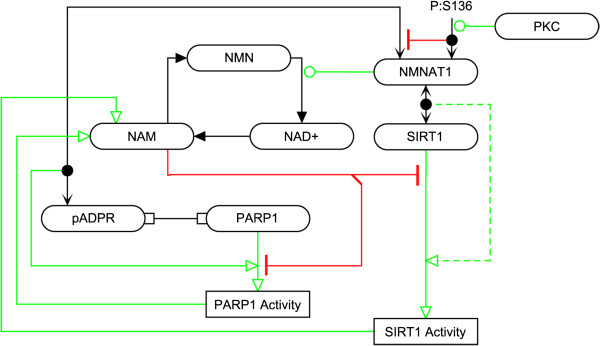
The effect of NMNAT1 binding on SIRT1 and PARP1 activity. Zhang et al. proposed that NMNAT1 may stimulate SIRT1 activity; indicated by the dashed line.
Regulation of common SIRT1 and PARP1 targets
SIRT1/PARP1 response to DNA damage
DNA damage response to both endogenous and exogenous sources is an intricate process that is not fully understood due to the complexity of the potential lesions, the number of proteins involved in both surveillance and repair, the interconnected regulation of proteins involved in the detection and repair of damage, stoppage of the cell cycle, and the potential induction of cell death; reviewed by [55]. SIRT1 and PARP1 play several roles throughout the response to DNA damage from the initial response to final cell fate decisions.
Histones
Both SIRT1 and PARP1 are known to modify histones; deacetylation of histones triggers chromatin compaction and the inhibition of transcription, whereas poly (ADP-ribose) polymers help to relax chromatin. SIRT1 is capable of deacetylating several histone amino acid residues, including H1K26, H3K9, H3K14, and H4K16 [[A20]] [56,57], while PARP1 can modify histones H1, H2AK13, H2BK30, H3K27, H3K37, and H4K16 to possibly regulate transcription [[A21]] [21,58]. It has been suggested that ADP-ribosylation of histone H1 promotes transcription by inhibiting the ability of histone H1 to bind to DNA [59]. Additionally, a competitive interaction has been shown between acetylation and PAR where acetylation of H4K16 inhibits the ADP-ribosylation of histone H4 [[A22]] [21]. Here a potential contradiction in the role of SIRT1 in condensing chromatin arises whereby SIRT1 deacetylation activity could potentially help drive the PARP1 ADP-ribosylation activity on H4K16. Currently, it is known that SIRT1 plays a role in DNA damage repair via histone deacetylation through the deacetylation of the two histone acetyltransferases, TIP60 and MOF, which are able to acetylate histone H4 [[A23]]. Deacetylation of these two proteins promotes their ubiquitin-dependent degradation affecting DNA double-strand break (DSB) repair either through the repression of repair or affecting the choice of repair mechanism (i.e. homologous recombination or non-homologous end joining (NHEJ)) [60-63]. TIP60-dependent acetylation of H4K16 inhibits the binding of 53BP1 to H4K20me2, which promotes non-homologous end joining [[A24]] [62,64]. Further studies are needed to understand how DNA damage might affect modifications on the various histones though it is known that genotoxic stress causes a random redistribution of SIRT1 across the genome with a correlated increase in levels of H1K26 acetylation [65]. However, while PARP1 does localize to DNA strand breaks, it is also not known whether there is any further global redistribution PARP1 or a relationship to the redistribution of SIRT1.
DNA damage signaling pathway
Both SIRT1 and PARP1 are DNA damage responders and the absence of either of these proteins may lead to DNA damage sensitization [66,67]. PARP1 begins to localize to DNA breaks rapidly and becomes activated by binding to DNA breaks. The ADP-ribosylation activity of PARP1 increases 10–500 fold as a result of binding to DNA breaks [[A25]] [2]. Once activated, PARP1 may help repair single strand DNA breaks, preventing their conversion to double-stranded breaks [[A26]] [68]. In addition, PARP1 is involved in DNA repair through its associations with base excision repair (BER) enzymes such as polymerase β, XRCC1 and DNA ligase III by helping these proteins localize to sites of DNA damage [[A27]] [69].
Two early responders of DNA damage linked to SIRT1 and PARP1 regulation are ATM (ataxia telangiectasia, mutated) and CHK2 (checkpoint kinase 2) [70]. The activation of ATM by DNA breaks requires the activation of the MRE11-RAD50-NBS1 (MRN) complex [[A28-A30]]. It has been shown that PARP1 binds to ATM, an interaction that is stimulated by DNA damage, and that the automodification of PARP1 leads to ATM activation [[A31]] [67].
An extended feedback loop has been proposed by Gorospe and de Cabo involving SIRT1 and several key DNA damage repair proteins [70]. In this loop, NBS1 is phosphorylated by ATM in response to genotoxic stress at S343 for the activation of NBS1; to be phosphorylated, it is necessary for NBS1 to be in a hypoacetylated state, which SIRT1 helps to maintain by deacetylating NBS1 [[A32-A33]] [71,72]. CHK2 is activated when T68 is phosphorylated by ATM. CHK2 can then phosphorylate HuR at several sites causing it to dissociate from SIRT1 mRNA, and thereby reduce the half-life of the SIRT1 mRNA [[A34]] [73]. It has been suggested that in repairable DNA damage situations SIRT1 levels are elevated leading to a survival response, but during lethal DNA damage SIRT1 levels can be attenuated by CHK2 through the phosphorylation of HuR that can ultimately result in cell death [70].
SIRT1 is also regulated by c-MYC and E2F1, two proteins involved in cell proliferation, differentiation and apoptosis, through negative feedback loops shown in Figure 5. E2F1, a transcription factor, induces the transcription of SIRT1 [[A35]]. Conversely, E2F1 has been suggested to be a target for SIRT1 deacetylation, which inhibits E2F1 activity [[A36-A37]] [74]. Additionally, the transcriptional activity of E2F1 is inhibited by Retinoblastoma (Rb), which is another substrate of SIRT1 deacetylation; acetylation of Rb has been shown to regulate the binding of Rb to E2F1 [[A38]] [75,76]. Two studies have examined the interactions between SIRT1 and c-MYC producing contradictory results. In one publication, c-MYC over-expression leads to an increase in SIRT1 expression and then deacetylation of c-MYC by SIRT1 leads to the destabilization of c-MYC [[A39]] [77]. In the second publication, neither the induction of SIRT1 expression nor the destabilization of c-MYC was seen following c-MYC activation. Instead, a stabilizing effect on c-MYC due to deacetylation by SIRT1 was found. Also in the second study, Menssen et al. found that c-MYC can induce the transcription of NAMPT and help sequester DBC1, an inhibitor of SIRT1 [[A40]] [78]. Another line of evidence suggesting that SIRT1 may affect NAMPT through a second mechanism involving the circadian clock will be discussed later. There is evidence that PARP1 binds to E2F1 stimulating E2F1-dependent transcription of c-MYC [[A41]] [79]. This presents the possibility that both SIRT1 and PARP1 may be capable of influencing the regulation of NAMPT to affect NAD + levels through c-MYC.
Figure 5.
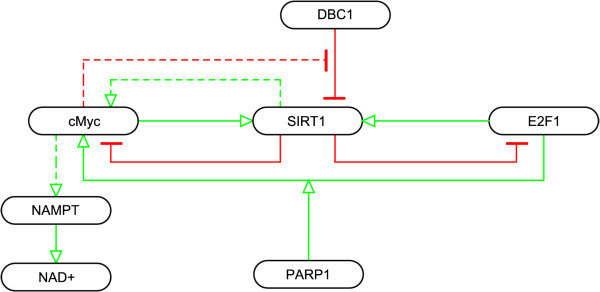
Interactions between SIRT1 and transcription factors c-MYC and E2F1. Solid lines between SIRT1 and c-MYC indicated are interactions from Yuan, Minter-Dykhouse et al. 2009 forming a negative feedback loop, while dashed lines are findings related to SIRT1 and c-MYC from Menssen, Hydbring et al. 2012.
Another co-regulated protein is NF-κB, a regulator of cellular response, including inflammation, to stress. In the case of NF-κB, the effects of SIRT1 and PARP1 are opposing. SIRT1 can deacetylate the RelA/p65 subunit of NF-κB at K310 to inhibit NF-κB transactivation activity [[A42]] [80]. PARP1 is an activator of NF-κB through its direct binding to NF-κB; acetylation of PARP1 by p300/CBP is required for the binding of PARP1 to NF-κB [[A43]] [39].
Given the importance of p53 to apoptotic response, a number of studies have focused on the regulation of p53 by SIRT1. p53 acts as a transcription factor that induces apoptosis and is inhibited by SIRT1 deacetylation [[A44]] [81,82]. SIRT1 has the capability of deacetylating p53 at several sites in mouse embryonic fibroblasts (MEFs) and SIRT1-deficient cells possess hyperacetylated p53; the precise role of p53 acetylation is unclear [[A45]] [83-85]. Several proteins help to modify the interactions of SIRT1 with p53, including p53 (itself), DBC1, AROS, and HIC1, suggesting that it is a cellular imperative to control the inhibition of p53 by SIRT1 under certain conditions. p53 can repress SIRT1 expression during nutrient abundance via p53-binding sites on the SIRT1 promoter. This effect is countered by the transcription factor FOXO3A, which interacts with p53 in an inhibitory fashion during nutrient deprivation [[A46-A47]] [86,87]. Hypermethylated in cancer-1 (HIC1) is a transcriptional repressor of the SIRT1 promoter [[A48]] that helps prevent age-dependent cancers in mice. If HIC1 is inhibited, SIRT1 expression increases, allowing for more efficient inactivation of p53; p53 over-expression leads to the transactivation of HIC1, thus creating a negative feedback loop [88].
Micro-RNAs have also been shown to downregulate SIRT1-dependent deacetylation of p53. p53 can stimulate the expression of miRNA-34 [[A49]], which subsequently drives down the expression of SIRT1 lowering SIRT1 availability to inhibit p53. Over 15 micro-RNAs affect the expression of SIRT1 either directly or by decreasing the expression of HuR, which stabilizes SIRT1 mRNA [[A50]] [89,90].
Given the well-studied nature of p53 as a SIRT1 substrate, p53 has been used to characterize SIRT1 inhibitors and activators. In humans, deleted in breast cancer 1 (DBC1) acts as an inhibitor of SIRT1 (an inhibitory effect increased by the phosphorylation of DBC1) and whose effect has been shown to lead to p53 hypoacetylation [[A51-A52]] [29,91-93]. Active Regulator of SIRT1 (AROS) has been shown to bind SIRT1 and help enhance the deacetylation of p53 by SIRT1 [[A53]] [94]. Further studies are needed to understand if the effects on p53 acetylation states are specific to the activities of DBC1 and AROS on SIRT1 or if other substrates of these two proteins are involved.
Much less is known about the interaction between PARP1 and p53. PARP1 helps p53 accumulate in the nucleus by (ADP-ribosyl)ating p53, which prevents p53 nuclear export [95], and there is evidence to suggest that SIRT1 deacetylation activity is capable of blocking p53 nuclear translocation [96].
Cell death
In addition to the role that SIRT1 plays in the inhibition of p53, SIRT1 is capable of deacetylating Ku70 at K539 and K542; the acetylation of Ku70 leads to the dissociation of the Ku70 and Bax helping to trigger apoptosis; Bax is a pro-apoptotic factor that is sequestered by Ku70 [[A54-A55]] [97]. NBS1, which we have discussed here beforehand as being activated via deacetylation by SIRT1, has also been shown to help control the interaction between Ku70 and Bax by stimulating the acetylation of Ku70 [[A56]] [98]. The exact conditions leading to a differential role of SIRT1 on the Ku70 and Bax complex remains to be uncovered.
PARP1 plays a role in cell death pathways (apoptosis or necrosis) in the course of responding to DNA damage. ATP is required for optimal caspase activation, and the depletion of ATP can direct cells between apoptotic and necrotic pathways [99]. During normal apoptosis, PARP1 is cleaved by caspases; the role of these cleaved fragments play is not fully understood [[A57]]. PARP1 cleavage helps prevent energy depletion (NAD + and ATP) in response to severe DNA damage; the extreme loss of NAD + triggers necrosis by reducing cellular ability to synthesize ATP [[A58]] [100]. Cells with severe DNA damage die from necrosis because they are not able to switch away from the necrotic pathway since the kinetics of NAD + depletion are faster than those of PARP1 cleavage [2]. Rapid depletion of NAD + levels by PARP1 reduces SIRT1 activity and inhibits the capability of SIRT1 to deacetylate its targets respond to genotoxic stress [101]. PARP1 has also been implicated in caspase-independent apoptosis, where its activation leads to apoptosis-inducing factor (AIF) release from the mitochondria, which induces nuclear chromatin fragmentation [[A59]] [102].
Circadian rhythms
Recent SIRT1 and PARP1 research has uncovered roles for the two proteins in circadian rhythms creating the possibility for novel interconnections between metabolism, DNA repair, and circadian rhythms (Figure 6). The core circadian machinery involves a transactivating CLOCK/BMAL1 heterodimer, which induces the transcription of a large number of genes, including the cryptochrome (CRY1 and CRY2) and period (PER1, PER2, PER3) genes that form a complex that leads to a negative feedback loop suppressing CLOCK/BMAL1-mediated transcription [[A60-A62]]. Several studies have shown that disruptions in core circadian interactions can lead to alterations in DDR; reviewed in [103].
Figure 6.
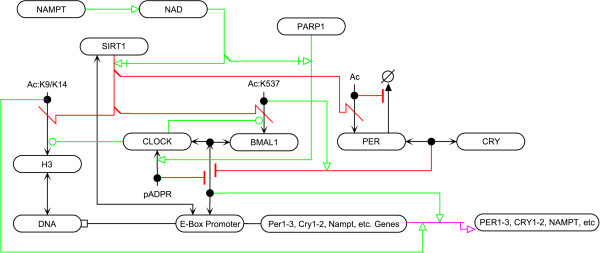
Interactions of SIRT1 and PARP1 with circadian clock components.
SIRT1 deacetylates BMAL1 at K537 destabilizing the interaction between CRY and BMAL1 [[A63]] [104,105]. CLOCK possesses acetyltransferase activity that regulates the transcriptional activity of CLOCK/BMAL1 [106] and is capable of acetylating some of the same locations that SIRT1 deacetylates: H3K9, H3K14, and BMAL1 at K537 [[A64]] [57,105]. SIRT1 has also been shown to deacetylate PER2 destabilizing the protein [[A65]]; it has been hypothesized that acetylation of PER2 at lysine residues prevents their ubiquitination [107]. This presents a dual control mechanism for SIRT1 in the circadian clock where it is capable of balancing transcription through chromatin condensation, but also by disrupting the ability for CRY and PER2 to repress CLOCK/BMAL1 activity. SIRT1 is involved in NAMPT transcriptional regulation, which is under circadian control causing NAD + levels to oscillate as a consequence of NAMPT level oscillation [48,108]. Additionally, it has been shown that PARP1 has rhythmic activity influenced by feeding patterns though further work is necessary to understand the underlying molecular mechanism. PARP1 is capable of ADP-ribosylating CLOCK in a circadian manner disrupting the association between the BMAL1/CLOCK heterodimer and its targets [[A66]] [109]. It remains to be determined whether a regulatory effect exists between SIRT1 or PARP1 and the circadian components during DNA damage.
Interactions with other family members
While the focus of this review has been on the inter-relationships that exist between SIRT1 and PARP1, there is growing evidence that SIRT1 has the capability of interacting with other members of the PARP family of proteins and similarly that PARP1 is capable of interacting with multiple sirtuins. Here we present three cases of these interactions; these interactions range include both direct modifications, as well as transcriptional regulation. First, SIRT6, one of the nuclear sirtuins, plays a role in promoting DNA damage repair by binding and activating PARP1 by mono-ADP-ribosylating PARP1 triggering its auto-ADP-ribosylation activity [[A67]] [110]. Next, the second PARP family member, PARP2, has been shown to inhibit the transcription of SIRT1; the deletion of PARP2 increases overall levels of SIRT1 activity without having to target NAD + levels directly [[A68]] [46]. This finding indicates that inhibitors of PARP proteins may be capable of increasing SIRT1 activity not only via the inhibition of NAD + consumption by PARP family members, but also through the removal of transcriptional inhibition. Lastly, shown recently is that PARP1 increases levels of mitochondrial SIRT3 and that SIRT3 can continue to function under stress conditions because mitochondrial NAD + levels are maintained in the conditions produced by either treatment with methylnitronitrosoguanidine (MNNG), a carcinogen, or N-methyl-D-aspartate (NMDA), a neuronal stressor, even as cytosolic levels of NAD + are depleted by PARP1 [111,112]. This study by Kim et al., did not observe a similar change in SIRT1 protein levels or the expression levels of the other mitochondrial sirtuins, SIRT4 and SIRT5.
Conclusions
Cells respond to DNA damage through coordinated pathways that arrest the cell cycle and repair the damage, and in the presence of severe damage trigger cell death. Both SIRT1 and PARP1 play an intimate role in the regulation of genomic stability, and continued work is necessary for the understanding of the specific contexts in which modulators of SIRT1 and PARP1 activity may be appropriate as therapeutics for cancer and metabolic disorders. The regulation of SIRT1 and PARP1 is controlled by a variety of stimuli, including metabolic, circadian, and genotoxic. Understanding how each of these stimuli affects the regulatory network of these two proteins is vital.
The work of the past several years has trended towards an understanding of the diverse regulatory network surrounding these two proteins, the effects of local levels of their common substrate, NAD+, and the co-regulation of and by the two proteins. There continue to be several unexplored areas. For example, understanding the nature of competitive regulation of acetylation and PAR at a single substrate residue on a single molecule, since both can target lysine residues, remains an open area of research. Another interesting avenue of research remaining to be thoroughly explored is the alteration of the reviewed pathways as an organism ages. Studies have shown age-dependent increases in DNA damage can lead to NAD + depletion [113,114]. In human tissue samples, Massudi et al. found an age-related positive correlation in PARP1 activity in males and negative correlations with SIRT1 activity in males and NAD + levels in both males and females that starts to shed light on the role of these two proteins in the presence of accumulating DNA damage with age [114]. In a related study, Chang et al. showed a reduction in SIRT1 levels over time resulting in alterations in circadian oscillations in mice as they age [115]. Studies such as these suggest that the regulatory network surrounding SIRT1 and PARP1 may undergo large-scale changes over an organism’s lifespan; currently, we do not know the extent of these changes. Additionally, there is a deficit in our knowledge with respect to the influence that DNA damage response may have on circadian regulation and vice versa and the roles that SIRT1 and PARP1 may play. And while this review has focused on evidence of the interactions between the better studied members of their respective protein families, SIRT1 and PARP1, additional work is necessary in our understanding of the roles of the other members of the two protein families and their unique properties and interactions that may play integral roles in the progression of DDR, as well as other processes.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AL wrote the manuscript and drew and annotated the supplemental MIM. MIA and KWK edited the manuscript and MIM. All authors have read and approved the final manuscript.
Supplementary Material
SIRT1/PARP1 MIM GPML (GenMAPP Pathway Markup Language) File. The SIRT1/PARP1 MIM as a machine-readable file that can be read and searched using PathVisio-MIM ( http://discover.nci.nih.gov/mim/mim_pathvisio.html).
SIRT1/PARP1 MIM Annotations Excel File. Table containing annotations for all interactions shown in the SIRT1/PARP1 MIM.
Contributor Information
Augustin Luna, Email: augustin@mail.nih.gov.
Mirit I Aladjem, Email: aladjemm@mail.nih.gov.
Kurt W Kohn, Email: kohnk@dc37a.nci.nih.gov.
Acknowledgements
We would like to acknowledge helpful discussions with Paolo Sassone-Corsi and Myriam Gorospe. This project was supported by the Ford Foundation and the Intramural Research Program of the NIH, Center for Cancer Research, National Cancer Institute.
References
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Plant Physiol Plant Mol Biol. 2004;4:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;4:249–268. doi: 10.1042/0264-6021:3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;4:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba JM, de Cabo R, Alcain FJ. A patent review of sirtuin activators: an update. Expert Opin Ther Pat. 2012;4:355–367. doi: 10.1517/13543776.2012.669374. [DOI] [PubMed] [Google Scholar]
- Canto C, Sauve AA, Bai P. Crosstalk between poly(ADP-ribose) polymerase and sirtuin enzymes. Mol Aspects Med. 2013;4(6):1168–1201. doi: 10.1016/j.mam.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A, Karac EI, Sunshine M, Chang L, Nussinov R, Aladjem MI, Kohn KW. A formal MIM specification and tools for the common exchange of MIM diagrams: an XML-based format, an API, and a validation method. BMC Bioinformatics. 2011;4:167. doi: 10.1186/1471-2105-12-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A, Sunshine ML, van Iersel MP, Aladjem MI, Kohn KW. PathVisio-MIM: PathVisio plugin for creating and editing Molecular Interaction Maps (MIMs) Bioinformatics. 2011;4(15):2165–2166. doi: 10.1093/bioinformatics/btr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Bio Cell. 2005;4(10):4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD + -dependent tubulin deacetylase. Mol Cell. 2003;4(2):437–444. doi: 10.1016/S1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;4(2):149–159. doi: 10.1016/S0092-8674(01)00527-X. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G. et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;4(5):941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;4(22):21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;4(14):13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;4(6):703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;4(37):5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;4(7):735–745. doi: 10.1016/S0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Rahman S, Islam R. Mammalian Sirt1: insights on its biological functions. Cell Commun Signal. 2011;4:11. doi: 10.1186/1478-811X-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem. 1993;4:22575–22580. [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;4(8):882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biology R. 2006;4:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;4(19):6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;4(5):417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick F, Lüscher B. Regulation of sirtuin function by posttranslational modifications. Frontiers Pharm. 2012;4:29. doi: 10.3389/fphar.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PloS ONE. 2009;4(12):e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar C, Brummer A, Alberghina L, Hofer T. Timing control in regulatory networks by multisite protein modifications. Trends Cell Biol. 2010;4(11):634–641. doi: 10.1016/j.tcb.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Back JH, Rezvani HR, Zhu Y, Guyonnet-Duperat V, Athar M, Ratner D, Kim AL. Cancer cell survival following DNA damage-mediated premature senescence is regulated by mammalian target of rapamycin (mTOR)-dependent Inhibition of sirtuin 1. J Biol Chem. 2011;4(21):19100–19108. doi: 10.1074/jbc.M111.240598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PLoS ONE. 2009;4(8):e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschoernig B, Mahlknecht U. Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochem Biophys Res Commun. 2009;4(3):372–377. doi: 10.1016/j.bbrc.2009.02.085. [DOI] [PubMed] [Google Scholar]
- Kang H, Suh JY, Jung YS, Jung JW, Kim MK, Chung JH. Peptide switch is essential for Sirt1 deacetylase activity. Mol Cell. 2011;4(2):203–213. doi: 10.1016/j.molcel.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, Scrable H. Phosphorylation regulates SIRT1 function. PloS ONE. 2008;4:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;4(17):13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Wong LL, Tse EY, Liu HF, Leong VY, Lee JM, Hardie DG, Ng IO, Ching YP. AMPK promotes p53 acetylation via phosphorylation and inactivation of SIRT1 in liver cancer cells. Cancer Res. 2012;4(17):4394–4404. doi: 10.1158/0008-5472.CAN-12-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang D, Zhao Y, Tu B, Zheng Z, Wang L, Wang H, Gu W, Roeder RG, Zhu WG. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1) Proc Natl Acad Sci USA. 2011;4(5):1925–1930. doi: 10.1073/pnas.1019619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;4:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, Law L, Hester LD, Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;4(11):1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J Biol Chem. 1998;4(23):14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- Walker JW, Jijon HB, Madsen KL. AMP-activated protein kinase is a positive regulator of poly(ADP-ribose) polymerase. Biochem Biophys Res Commun. 2006;4(1):336–341. doi: 10.1016/j.bbrc.2006.01.145. [DOI] [PubMed] [Google Scholar]
- Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson R. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc Natl Acad Sci USA. 2006;4:7136–7141. doi: 10.1073/pnas.0508606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;4:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- Messner S, Schuermann D, Altmeyer M, Kassner I, Schmidt D, Schär P, Müller S, Hottiger MO. Sumoylation of poly(ADP-ribose) polymerase 1 inhibits its acetylation and restrains transcriptional coactivator function. FASEB. 2009;4:3978–3989. doi: 10.1096/fj.09-137695. [DOI] [PubMed] [Google Scholar]
- Zhang J. Are poly(ADP-ribosyl)ation by PARP-1 and deacetylation by Sir2 linked? Bioessays. 2003;4:808–814. doi: 10.1002/bies.10317. [DOI] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD + depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;4(52):43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- Kolthur Seetharam U, Dantzer F, Mcburney MW, Murcia GD, Sassone-corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;4(8):873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol. 2009;4:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH. et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;4(4):461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H. et al. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011;4(4):450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittelli M, Formentini L, Faraco G, Lapucci A, Rapizzi E, Cialdai F, Romano G, Moneti G, Moroni F, Chiarugi A. Inhibition of nicotinamide phosphoribosyltransferase: cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J Biol Chem. 2010;4:34106–34114. doi: 10.1074/jbc.M110.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone Corsi P. Circadian control of the NAD + salvage pathway by CLOCK-SIRT1. Science. 2009;4:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;4:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Yao J, Dumond ME, Krishnakumar R, Ruhl DD, Ryu KW, Gamble MJ, Kraus WL. Regulation of poly (ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD + synthase. J Biol Chem. 2012;4(15):12405–12416. doi: 10.1074/jbc.M111.304469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H-S, Sang W-W, Wang Y-O, Liu W. Nicotinamide phosphoribosyltransferase/sirtuin 1 pathway is involved in human immunodeficiency virus type 1 Tat-mediated long terminal repeat transactivation. J Cell Biochem. 2010;4:1464–1470. doi: 10.1002/jcb.22704. [DOI] [PubMed] [Google Scholar]
- Berger F, Lau C, Ziegler M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc Natl Acad Sci USA. 2007;4:3765–3770. doi: 10.1073/pnas.0609211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, Sauve AA, Kraus WL. Enzymes in the NAD + salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem. 2009;4:20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszewski M, Szumiel I. Sirtuins (histone deacetylases III) in the cellular response to DNA damage–facts and hypotheses. DNA Repair (Amst) 2005;4:1306–1313. doi: 10.1016/j.dnarep.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;4(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;4:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;4:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;4(1):8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;4(5864):819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen J. SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J Biol Chem. 2010;4(15):11458–11464. doi: 10.1074/jbc.M109.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Ling H, Yuan Z, Fang B, Bloom G, Fukasawa K, Koomen J, Chen J, Lane WS, Seto E. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol Cell Biol. 2012;4(14):2823–2836. doi: 10.1128/MCB.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;4(3):317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ. et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;4(14):3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao KY, Mizzen CA. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J Mol Cell Biol. 2013;4(3):157–165. doi: 10.1093/jmcb/mjs066. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park S, Hartlerode A, Stegmuller J, Hafner A, Loerch P. et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;4:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita N, Takami Y, Kimura M, Tachiiri S, Ishiai M, Nakayama T, Takata M. Role of NAD-dependent deacetylases SIRT1 and SIRT2 in radiation and cisplatin-induced cell death in vertebrate cells. Genes Cells. 2005;4(4):321–332. doi: 10.1111/j.1365-2443.2005.00836.x. [DOI] [PubMed] [Google Scholar]
- Aguilar Quesada R, Munoz Gamez JA, Martin Oliva D, Peralta A, Valenzuela MT, Matinez Romero R, Quiles Perez R, Menissier De Murcia J, De Murcia G, Ruiz De Almodovar M. et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol Biol. 2007;4:29. doi: 10.1186/1471-2199-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse BC, Dianova II, Parsons JL, Dianov GL. Poly(ADP-ribose) polymerase-1 modulates DNA repair capacity and prevents formation of DNA double strand breaks. DNA Repair. 2008;4:932–940. doi: 10.1016/j.dnarep.2008.03.017. [DOI] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M. et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;4(14):7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M, de Cabo R. AsSIRTing the DNA damage response. Trends Cell Biol. 2008;4:77–83. doi: 10.1016/j.tcb.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Seto E. A functional link between SIRT1 deacetylase and NBS1 in DNA damage response. Cell Cycle. 2007;4:2869–2871. doi: 10.4161/cc.6.23.5026. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;4(1):149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Pullmann R Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA. et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;4(4):543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD. et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;4:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Wong S, Weber JD. Deacetylation of the retinoblastoma tumour suppressor protein by SIRT1. Biochem J. 2007;4(3):451–460. doi: 10.1042/BJ20070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham D, Munro S, Soloway J, O'Connor DP, La Thangue NB. DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep. 2006;4(2):192–198. doi: 10.1038/sj.embor.7400591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;4:203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Luscher B, Larsson LG, Hermeking H. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci USA. 2012;4(4):E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Samara R, Espinoza LA, Hassa PO, Hottiger MO, Smulson ME. PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during re-entry of quiescent cells into S phase. Oncogene. 2003;4(52):8460–8471. doi: 10.1038/sj.onc.1206897. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye R, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;4:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;4(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X-J, Finkel T, Shen D-W, Yin J-J, Aszalos A, Gottesman MM. SIRT1 contributes in part to cisplatin resistance in cancer cells by altering mitochondrial metabolism. Mol Cancer Res. 2008;4:1499–1506. doi: 10.1158/1541-7786.MCR-07-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;4:123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;4:137–148. doi: 10.1016/S0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;4(1):28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L, Zhou H, Xia Y, Wang H, Gao G, Chen B, Liu Q, Shao C, Gong Y. Serum withdrawal up-regulates human SIRT1 gene expression in a p53-dependent manner. J Cell Mol Med. 2009;4(10):4176–4184. doi: 10.1111/j.1582-4934.2008.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;4(5704):2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;4(3):437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;4:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M. MicroRNA Regulation of SIRT1. Frontiers Physiol. 2012;4:68. doi: 10.3389/fphys.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse J-P, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;4:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Luo K, Liu T, Lou Z. Regulation of SIRT1 activity by genotoxic stress. Genes Dev. 2012;4:791. doi: 10.1101/gad.188482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-E, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;4:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Kim E, Kho J, Kang M, Um S. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;4:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol. 2007;4:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;4(3):241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;4:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Iijima K, Muranaka C, Kobayashi J, Sakamoto S, Komatsu K, Matsuura S, Kubota N, Tauchi H. NBS1 regulates a novel apoptotic pathway through Bax activation. DNA Repair. 2008;4:1705–1716. doi: 10.1016/j.dnarep.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Leist M, Single B, Castoldi AF, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J Exp Med. 1997;4(8):1481–1486. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DW, Dawson TM, Dawson VL. Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol Res. 2005;4:5–14. doi: 10.1016/j.phrs.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Furukawa A, Tada-Oikawa S, Kawanishi S, Oikawa S. H2O2 accelerates cellular senescence by accumulation of acetylated p53 via decrease in the function of SIRT1 by NAD + depletion. Cell Physiol Biochem. 2007;4(1–4):45–54. doi: 10.1159/000104152. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4(167):ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana S, Mahmood S. Circadian rhythm and its role in malignancy. J Circadian Rhythms. 2010;4:3. doi: 10.1186/1740-3391-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;4:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD + -dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;4:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;4:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;4:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong H-K, Chong JL, Buhr ED, Lee C. et al. Circadian clock feedback cycle through NAMPT-mediated NAD + biosynthesis. Science. 2009;4:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;4:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;4:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lu HF, Alano CC. Neuronal Sirt3 protects against excitotoxic injury in mouse cortical neuron culture. PLoS ONE. 2011;4(3):e14731. doi: 10.1371/journal.pone.0014731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alano CC, Tran A, Tao R, Ying W, Karliner JS, Swanson RA. Differences among cell types in NAD(+) compartmentalization: a comparison of neurons, astrocytes, and cardiac myocytes. J Neurosci Res. 2007;4(15):3378–3385. doi: 10.1002/jnr.21479. [DOI] [PubMed] [Google Scholar]
- Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Poljak A, Grant R. Age related changes in NAD + metabolism oxidative stress and Sirt1 activity in wistar rats. PloS ONE. 2011;4(4):e19194. doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massudi H, Grant R, Braidy N, Guest J, Farnsworth B, Guillemin GJ. Age-associated changes in oxidative stress and NAD(+) metabolism in human tissue. PloS ONE. 2012;4(7):e42357. doi: 10.1371/journal.pone.0042357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;4(7):1448–1460. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SIRT1/PARP1 MIM GPML (GenMAPP Pathway Markup Language) File. The SIRT1/PARP1 MIM as a machine-readable file that can be read and searched using PathVisio-MIM ( http://discover.nci.nih.gov/mim/mim_pathvisio.html).
SIRT1/PARP1 MIM Annotations Excel File. Table containing annotations for all interactions shown in the SIRT1/PARP1 MIM.