Abstract
There are currently 1527 known microRNAs (miRNAs) in human, each of which may regulate hundreds or thousands of target genes. miRNA expression levels vary between cell types; for example, miR-302 and miR-290 families are highly enriched in embryonic stem cells, while miR-1 is a muscle specific miRNA. miRNA biosynthesis and function are highly regulated and this regulation may be cell type specific. The processing enzymes and factors that recognize features in sequence and secondary structure of the miRNA play key roles in regulating the production of mature miRNA. Mature miRNA enriched in stem cells control stem cell self-renewal as well as their differentiation. Though specific miRNAs have been shown to control differentiation towards various lineages such as neural or skin cells, some of the most well characterized miRNAs have been found in promoting the formation of cardiac cells. In addition, miRNAs also play a critical role in cardiomyocyte hypertrophy, especially in a pathological context. Such miRNAs are predicted to be therapeutic targets for treating cardiovascular diseases. In this review we will discuss how miRNAs act to maintain the stem cell state and also explore the current knowledge of the mechanisms that regulate miRNAs. Furthermore, we will discuss the emerging roles of miRNAs using cardiomyocyte differentiation and maturation as a paradigm. Emphasis will also be given on some of the less ventured areas such as the role of miRNAs in the physiological maturation of cardiomyocytes. These potentially beneficial miRNAs are likely to improve cardiac function in both in vivo and in vitro settings and thus provide additional strategy to treat heart diseases and more importantly serve as a good model for understanding cardiomyocyte maturation in vitro.
Keywords: MicroRNA, stem cells, cardiomyocyte maturation, mouse, human, hESC, regulation
1. INTRODUCTION
microRNA (miRNA) expression has been found to vary significantly between different cell types and cell states and miRNAs have been found to be important for maintenance of a cell state as well as differentiation. Hence miRNAs are of great interest when studying stem cells or differentiating tissues such as the specification of cardiomyocytes in the formation of heart during embryogenesis and in tissue regeneration in response to cardiac hypertrophy. When investigating miRNA based regulation the key goal has been to identify the targets regulated by the miRNA in question. Recent work on miRNA biogenesis has revealed several regulatory mechanisms from transcription to target silencing that may control specific groups of miRNAs and therefore deserve a closer look. Especially knowing that miRNAs can be causal for a cell state makes investigating the mechanisms behind regulation of miRNA maturation critical. In this review we will discuss what is currently known about the regulatory mechanisms of miRNA biogenesis with an emphasis on the impact of miRNA secondary structure and the features that are of potential interest at the different processing steps. Furthermore, we will review some of the mechanistic roles of miRNAs in stem cell maintenance and elaborate on the recent findings related to miRNAs in cardiomyocyte differentiation and hypertrophy in both in vitro and in vivo settings.
2. REGULATION OF MICRORNA BIOSYNTHESIS
miRNAs are small RNAs that along with an Ago protein form the RNA-induced silencing complex (RISC) and regulate gene expression in the cell by binding to mRNAs. The mechanism is mostly inhibitory although there are cases when miRNA-mRNA target interactions lead to up-regulated translation instead [1]. Primarily the 3′ UTR is targeted but binding of the miRNA to the coding region as well as the 5′UTR of its target has also been reported [2]. In addition each miRNA may target thousands of transcripts, and one mRNA may contain several target sites so the targeting mechanisms show a great deal of complexity. Currently there are 1527 known miRNAs in human and more are constantly being discovered and added to the miRBase database [3]. The mature miRNA on average has a length of 22 nucleotides, although the length varies considerably, which can partly be explained by the structure of the miRNA hairpin which tends to contain several mismatches and accordingly may express some variability in its interactions with processing enzymes.
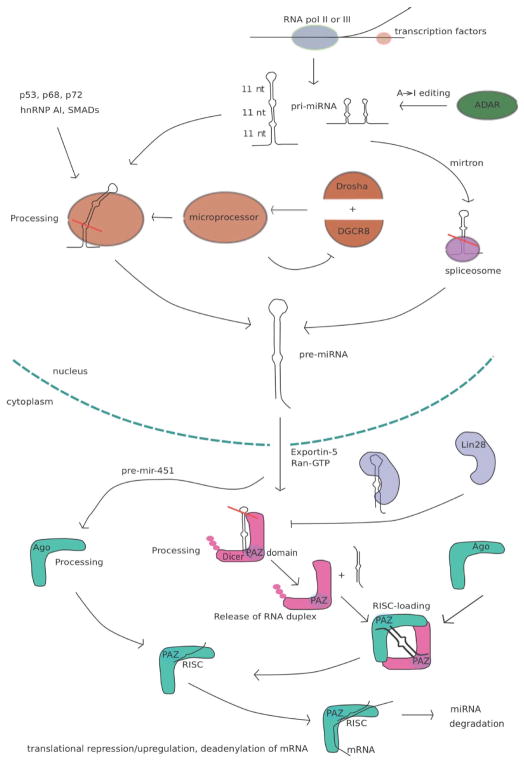
The primary miRNA (pri-miRNA) is transcribed in the nucleus by either RNA polymerase II or III [4, 5], from promoters that share the same regulatory features as those of protein coding genes [6]. Autoregulation of miRNAs through negative feedback loops using transcription factors is a mechanism that helps keep miRNA levels stable and accordingly also its target genes [7]. The pri-miRNA varies in length up to as long as several thousand nucleotides and contains one or several hairpin regions, sometimes in clusters. The hairpins are frequently mismatched, but mostly have the conserved features of 3 double stranded regions, each 11 nucleotides long, a terminal loop, as well as flanking regions below the hairpin (Fig. 1). The size of the terminal loop varies considerably, but in vitro experiments have shown that a a minimum size of 10 nucleotides is necessary for efficient processing by Drosha [8]. Some primary transcripts are subject to editing where single adenosines are converted to inosines, which further may alter the secondary structure [9].
Fig. 1.
Regulation of the microRNA biogenesis pathway.
In the nucleus the pri-miRNAs are recognized and cleaved by the microprocessor, an enzyme complex whose primary components are Drosha and DGCR8. Drosha is the catalytic component, which performs the cleavage, whereas DGCR8 lacks catalytic activity, but appears to do most of the binding to the substrate. The exact structure of the DGCR8-substrate complex is currently unknown, but it has been reported that DGCR8 recognizes the junction between the single stranded and double stranded region at the base of the pri-miRNA hairpin [10]. Structural analysis of DGCR8 also implies that two 11nt double stranded regions are being recognized, and if bound simultaneously on the same pri-miRNA the substrate would have to be bent since the two double-stranded RNA binding domains (dsRBD) of DGCR8 are positioned in an angle relative to each other [11]. The microprocessor cleaves the hairpin approximately 11 bases above the flanks releasing an approximately 22nt long hairpin, called the miRNA precursor (pre-miRNA). The cleavage tends to leave a 2nt overhang although variations do occur. Recent data suggest that Drosha levels may vary by cell type [12] and microprocessor levels can be a limiting factor in miRNA processing as knockdowns of Drosha have resulted in an accumulation of pri-miRNAs whereas Dicer is generally not a rate limiting step which may partly be explained by the levels of pre-miRNA being comparatively low compared to pri-miRNAs [13]. In vitro experiments have also shown that different pri-miRNAs can be processed by Drosha with different efficiencies, where highly conserved miRNAs appeared to be processed more efficiently than the poorly conserved [14]. Further support that different miRNAs are processed with varied efficiency has also been found in vivo, where miRNAs from the same primary transcript turned out to have different expression levels of the mature forms [15]. Besides transcriptional regulation the microprocessor is also self-regulating as it recognizes and cleaves hairpin structures in the DGCR8 mRNA [16]. At the moment it is not know how much variation on Drosha levels is observed in vivo, how this is regulated and whether this is an important, controlling factor for differential processing of microRNAs.
Besides Drosha and DGCR8 the microprocessor consists of other components which help stabilize the complex and facilitate the processing of select miRNAs, e.g. the Smads, the tumor suppressor p53 and the DEAD-box helicases p68 and p72 [17–19]. The heterogeneous ribonucleoprotein AI (hnRNP AI) facilitates the Drosha processing of mir-18a by binding to the loop region of the miRNA hairpin, but interestingly not the other miRNAs that are part of the same cluster [20].
The Drosha cleavage step can also be completely bypassed as is the case with mirtrons, which are processed by the spliceosome instead of the microprocessor [21]. A majority of the miRNAs are located in introns, but only a subset whose pre-miRNA ends are located by the splicing sites are mirtrons [22].
With Drosha selectively processing certain miRNAs with select levels of efficiency, and Drosha levels varying from cell type to cell type regulation of microprocessor levels could be yet another way for the cell to regulate endogenous levels of select miRNAs. Based on the current knowledge of the substrate binding mechanism of the microprocessor the transition state may require the substrate to be significantly bent, in which case the high variability in mismatch compositions of pri-miRNAs significantly could impact the energetic properties of each miRNA during Drosha cleavage since the mismatch composition of an RNA hairpin significantly impacts its flexibility.
After processing by the microprocessor the pre-miRNA is recognized by Exportin5 and its co-factor Ran-GTP and is transported out of the nucleus. In the cytoplasm it is then further processed by a complex consisting of Dicer and TAR RNA binding protein (TRBP). Dicer acts like a ruler and cleaves the miRNA 22nt from the Drosha cleavage site, separating the double stranded portion from the terminal loop leaving a 5′ overhang, typically 2 nt long similarly to the Drosha cleavage step [23]. In vitro experiments have shown that there is some variability to the exact location of the Dicer cleavage site, where miRNAs with many mismatches show less specificity in the location of the cutting site, presumably due to the extra flexibility of the hairpin making the length of the tertiary structure more variable compared to a hairpin with few mismatches [24]. The extra magnitude of miRNAs and target sites that this flexibility may result in is currently unknown. The crystal structure of Dicer has not been determined in human, but comparisons to existing crystal structures of Dicer homologues along with electron microscopy and photocrosslinking experiments have shown that Dicer has the shape of an L where the long arm contains the catalytic center, and the short arm is attached to the TRBP protein which consists of three small domains connected by flexible linkers and appears to stabilize the pre-miRNA as it is being cleaved by Dicer [25]. Following cleavage the resulting RNA duplex is being released and reloaded back onto Dicer/TRBP which along with the argonaute protein Ago2 forms the RISC-loading complex (RLC). The reloaded product is bound in a different position than the substrate, with the less stable end attached to the PAZ domain of Dicer, and the more thermodynamically stable end bound to the PAZ domain of Ago2 (Fig. 1). The strand with its 3′ end bound to the Ago2 PAZ domain is selected to be the guide strand [26]. It was long thought that the miRNA was being immediately loaded into the Ago protein following cleavage, which left the variation in choice of guide strand and passenger strand as somewhat of a mystery. The release of the cleaved miRNA duplex followed by rebinding before RISC loading is an interesting find as it provides an explanation how the guide strand is selected. Specific inhibitors exist for the Dicer cleavage step, such as the protein Lin-28 which binds to miRNAs from the let-7 family, thus not only blocking Dicer, but also in its bound state Lin-28 acts as a wedge on let-7 and disrupt the hairpin structure which may render Dicer unable to recognize the substrate [27].
The RISC complex binds to mRNAs and may cause repression by deadenylation, translational suppression or degradation of the mRNA [28], as well as in some cases translational activation. Ago2 possesses catalytic activity which besides degradation of mRNAs also facilitates the processing of mir-451, which thereby bypasses the Dicer cleavage step [29]. The pre-mir-451 is significantly shorter than most other pre-miRNAs, which may explain why this particular miRNA is singled out from the Dicer cleavage step. It is worth noting that most regulatory mechanisms involving the microprocessor are enhancing the processing of miRNAs, whereas for Dicer the mechanisms tend to be inhibitory, which may share a connection with different miRNA processing efficiencies of the two complexes. As for the role of the miRNA secondary structure in the two processing steps it is worth noting that for Dicer binding and RISC-loading the length of the miRNA and the thermodynamic properties by the Drosha and Dicer cleavage sites seem to be the primary determinants of how the miRNA is being processed. Since the microprocessor may bind the substrate in a bent position the structure and flexibility of the miRNA between the two cutting sites would possibly matter more in Drosha cleavage than in latter steps. In addition, although mismatches are found scattered throughout the hairpin, they are overrepresented at the central region equidistant from the two cleavage sites [30], so there is reason to believe that the secondary structure in that position is of significant importance, and that it would play a role in processing by Drosha rather than Dicer.
On the level of miRNA-mRNA target interactions SNPs have been shown to provide some diversity among miRNAs as they do in the rest of the genome [31]. Due to the limited size of the miRNAs a single SNP may result in completely different targeting networks, or even complete deactivation of the miRNA. The miRNA target interactions may also be subject to inhibition by mRNA-binding factors that sterically interfer with miRNA-mRNA binding [32]. The targeting is also affected by competition as has been shown through transfection of small RNAs, which caused upregulation of genes targeted by endogenous miRNAs [33].
The decay and lifespan of miRNAs remain somewhat of a mystery in animals as the nucleases that control miRNA degradation remain to be identified. The average miRNA half- life has been estimated to 119 hours [34], but there is plenty of variation, which implies that the mechanisms behind miRNA degradation affect miRNAs selectively, rather than collectively. The selective degradation of miRNA groups is further supported by experiments where a few miRNAs have rapidly depleted upon sudden environmental changes, such as the loss of cell adhesion [35] and upon the activation of neuronal cells [36].
3. MICRORNA FUNCTION
miRNAs are required for stem cell state and self-renewal as well as their differentiating progeny. In this review we briefly summarize the current knowledge of miRNA function in embryonic stem cells and will further concentrate on one of the ESC differentiation progeny, cardiomyocyte differentiation.
3.1. miRNAs in the Stem Cell State
Stem cells have the capacity to self-renew and regenerate throughout the lifetime of an organism. While adult stem cells can be multipotent, the embryonic stem cells (ESC) are pluripotent cells that can regenerate all germ layers in an organism. It is therefore critical to understand the regulation of self-renewal and the control of embryonic stem cell state and differentiation. During recent years it has become clear that miRNAs play a key role in these processes, in normal and disease development [37–41]. This first became clear when stem cell differentiation was shown to be impaired in Dicer deficient cells displaying the overall importance of the miRNA pathway in ESC [42–44]. Further miRNA profiling and sequencing analysis revealed that ESCs display a specific group of miRNAs [45–49]. Interestingly, the key stem cell transcription factors, Oct4, Sox2 and Nanog bind to the promoters of these miRNA genes [50]. The functional experiments have revealed that the overall reduction of miRNAs in ESC attenuates cell proliferation and that cell growth can be restored to a large extent through a rescue involving overexpression of key miRNAs [48, 51, 52]. Among others, miR-92b, miR-290, miR-195 and miR-372 have shown to be key regulators of the process [48, 51, 52]. Furthermore, the miRNA profile changes significantly at a very early stage during stem cell differentiation, such as by decreasing levels of the miR-302, -17, and -515 families, and the miR-371–373 cluster [49].
miRNAs have also been shown to control the differentiation of ESCs. Interestingly, let-7, a key differentiation miRNA that is silenced in ESCs by Lin-28, has a capacity to downregulate stem cell factors [53]. Let-7 regulation by lin28 is an example of miRNA’s posttranscriptional/biosynthetic control that critically defines which cells can express the mature form of this key cell fate regulator.
The importance of miRNAs in stem cell maintenance is further supported by experiments showing the ability of the miR-302–367 cluster to cause a reversal to a stem cell like state when overexpressed in somatic cells [54, 55]. These so called induced pluripotent stem cells (iPSC) have an increased expression of the miR-371–373 and miR-17–92 cluster compared to the cells they are derived from [56]. The miR-290 cluster, which has an overall high expression in mouse embryonic stem cells, helps maintain the stem cell state by targeting the NF-κB subunit p65 (RelA) which is known to promote differentiation, giving further insight to the pathways of miRNA regulation of the stem cell state [57].
The powerful potential of the miRNA regulatory networks needs to be taken into consideration while studying differentiating cells and the overall strong conservation of miRNAs and their regulatory networks make them a suitable mechanism to study in model organisms while investigating the development of tissues [58–61]. When investigating the role of miRNAs in cell states the focus has often been on what target genes are regulated by miRNAs, but as more is being discovered about the regulatory mechanisms of the miRNA biogenesis modifiers of certain groups of miRNAs will be of interest regarding their role in triggering differentiation as well as cell state maintenance.
3.2. miRNAs in Cardiac Development
Heart formation requires complex regulatory interactions among various cell types from several lineages including cardiomyocytes, endothelial and vascular cells and fibroblasts. Such coordinated interaction is achieved by the tight control of gene-expression among these cell types. Enrichment of miRNAs in specific cardiac cell types and further genetic ablation and anti-sense mediated knockdown studies have clearly indicated that miRNAs have important regulatory functions in controlling such complex cardiac gene expression. Few examples of such miRNAs and their functions in cardiac development are reviewed here.
The two striated muscle specific miRNAs that are widely conserved and robustly expressed in adult heart are miR-1 and miR-133a. These are derived from a common bicistronic precursor [62, 63]. The mature forms of mir-1/mir-133a are encoded by two conserved loci, mir-1-1/mir-133a-2 and mir-1-2/133a-1, located on chromosome 2 and 18 respectively [64]. Two MADS-box transcription factors, myocyte enhancer factor -2 (MEF2) and serum response factor (SRF) directly regulate the cardiac transcription of miR-1/miR-133a pairs [63]. While SRF can activate the transcription of mir1/mir-133 transcript by binding to a serum response element located on the enhancer region of miR-1/miR-133 [63], MEF2 promotes the expression of these miRNAs via an intragenic muscle specific enhancer [65]. Studies in mice have shown that miR-1 begins to be expressed in E8.5 mouse and continues to increase throughout its development. Overexpression of miR-1 diminishes cardiac growth by targeting a basic helix-loop-helix protein, Hand2, which is involved in ventricular myocyte expansion [63]. Mice lacking miR-1 die at E10.5 due to ventricular hypoplasia and decreased trabeculation of the left ventricle [63]. Chen et al. (2006) demonstrated that in cultured myoblasts, miR-1 promotes myoblast differentiation by translationally repressing histone deacetylase 4 (HDAC4) which in turn is a repressor MEF2 dependent muscle specific genes. In contrast to miR-1, miR-133 stimulates myoblast proliferation mostly by targeting SRF, which is important in muscle proliferation as well as activation of mir-1/mir-133 transcript thus creating a negative feedback loop [62]. Studies in mouse and human ESCs have revealed that miR-1/miR-133 pair promotes the specification of the mesodermal cell fate while suppressing differentiation into the ectodermal and neuroectodermal lineages [66]. Although they work in concert to suppress the non-muscle lineages, miR-1 and miR-133 have antagonistic role on further differentiation into muscle lineages - whereas miR-1 promotes differentiation of ESCs into cardiac fate most likely by inhibiting the notch ligand Delta-like (Dll-1), miR-133 inhibits their differentiation into cardiac muscle [66]. Since miR-1 and miR-133 are processed from the same bicistronic precursor, their opposite actions in each differentiation state begs for further analysis of differential processing.
The only cardiac patterning miRNA known till date is miR-138. Studies in zebra fish heart have clearly shown that miR-138 is specifically expressed in the ventricular chamber [67]. Fish lacking mir-138 function demonstrate expansion of atriaventricular canal (AVC) gene expression and failure of ventricular cardiomyocytes to fully mature thus implying the role of miR-138 in cardiac patterning. miR-138 in part functions by repressing the retinoic acid pathway. This results in repression of the AVC restricted cell adhesion molecule called cspg2 encoding for versican [67]. Thus miR-138 maintains cardiac patterning in zebra fish by controlling gene expression in discrete domains of the heart. It’s yet to be seen if this function of miR-138 stands true in the mammalian four chambered heart.
Analyses of miRNAs expressed in undifferentiated ESCs and differentiating cardiomyocytes were recently published in two independent large scale profiling studies, one using mouse ES cells and the other with human ES cells [68, 69]. A selection of miRNAs that are enriched during the differentiation of ESCs to cardiomyocytes in both the systems are shown in Table 1. Interestingly, a considerable number of miRNAs upregulated in differentiating human cardiomyocytes are also enriched in differentiating mouse cardiomyocytes (unpublished observation, KTK and HRB; Table 1). This suggests that the miRNA signature expression pattern is overall conserved in the differentiating cardiomyocytes of the two model systems at least in a cultured setting. This list reveals miRs with known roles in cardiomyocyte development such as miR-1, 133, etc. Interestingly, a number of miRs that have not been explored in the cardiomyocyte differentiation program including 125a-5p, 652 and 103 also are revealed from these two studies suggesting that the function of several new miRNAs during cardiogenesis still remain to be elucidated.
Table 1.
Summary of Regulated miRs in Differentiating Cardiomyocytes from Mouse and Human ES Cells
| Species | Upregulated miRs | Downregulated miRs |
|---|---|---|
| mouse | 1, 21, 23b, 26a, 28a, 30a, 30b, 30c, 30d, 34a, 99b, 103, 125a-5p, 133a, 133b, 143, 145, 210, 298, 652 | 20b, 92a, 106a, 290-3p, 290-5p, 292-5p, 293, 467a, 501-3p |
| human | 1, 21, 26a, 26b, 30a, 30b, 30c, 99b, 125a-5p, 126, 129-3p, 133a, 133b, 145, 148a, 181b, 652 | 17, 20a, 20b, 106a, 106b, 124, 182, 183, 183*, 200c, 205, 302c, 302c* |
Information in this table was extracted from Wilson et al. (2010) and Gan et al. (2011).
3.3. miRNAs in Cardiac Hypertrophy
The adult heart undergoes hypertrophic growth and cardiac remodeling to compensate for sustaining cardiac output and impairing cardiac function. Cardiac hypertrophy has two forms: a) physiological, where the heart enlarges in healthy individuals following rigorous exercise and is not associated with any cardiac damage and b) pathological, where the size of the heart initially increases to compensate for the damage to cardiac tissue, but subsequently declines in its function.
Pathological hypertrophy is the phenotype endpoint that has been mostly studied in relation to miRNAs to date. In animal models of pathological hypertrophy, whole arrays of miRNAs have indicated that separate miRNAs are upregulated, downregulated or remain unchanged [70–75]. Among those that are frequently reported to be upregulated with hypertrophy include miR-208, miR-21, miR-125, miR-129 and miR-195, whereas miR-1, miR-133, miR-29, miR-30 and miR-150 have often found to be downregulated. Invitro experiments using either overexpression or knockdown of miRNAs in cultured cardiomyocytes indicate that a subset of these miRNAs is indeed actively involved in cardiomyocyte hypertrophy. Reports from Olson lab have elegantly demonstrated that mir-208 family, encoded by an intron of the α-MHC (mir-208a) and β- MHC (mir-208b) is required for cardiac hypertrophy [76, 77]. Transgenic overexpression of miR-208a in the heart results in pronounced repression of miR-208a regulatory targets- thyroid hormone associated protein 1 (THRAP1) and myostatin, 2 regulators of muscle growth and hypertrophy and thus is sufficient to induce hypertrophic growth in mice heart [76, 77]. Most notably, miR-208a null mice fail to upregulate β- MHC in response to stress and inhibition of T3 signaling in the adult heart thus suggesting that miR-208b is a downstream regulator of miR-208a. A molecular hallmark of hypertrophy is the reactivation of a set of fetal cardiac genes including β- MHC in the adult heart [76]. Interestingly, a recent study has demonstrated that therapeutic inhibition of miR-208a during hypertension-induced heart failure in rats by subcutaneous delivery of antimir-208a prevents the pathological myosin switching while improving cardiac function, overall health and survival [78]. Thus, miR-208a appears to be a promising candidate to be used as a potent therapeutic target for the modulation of cardiac function and remodeling during heart disease progression. Similarly, miR-195 was sufficient to drive pathological cardiac growth when overexpressed in transgenic mice [75]. It will be interesting to test whether miR-195 target, Wee1, a negative regulator of G2-M state of cell cycle might play a role in this context as it did in hESCs [48]. By contrast to miR-195 and miR-208, in vitro over expression of miR-150 and miR-181b, which are downregulated in cardiac hypertrophy, resulted in reduced cardiomyocyte size [75]. In the same line, overexpression of miR-133 by infecting both neonatal and adult mouse myocytes reduced the hypertrophic response to agonist stimulation [79]. Conversely, in vivo knockdown of miR-133 by antisense RNA oligonucleotide appeared sufficient to induce significant hypertrophic growth of the heart compared with saline treated animals [79] suggesting an active role of miR-133 in inhibiting cardiac hypertrophy.
In contrast to pathological hypertrophy, the role of miRNAs in physiological hypertrophy has been a less explored area. So far, only one study has demonstrated that rats subjected to exercise training and transgenic mice with selective cardiac overexpression of a constitutively active mutant of the Akt kinase had reduced levels of the muscle specific miRNAs, mir-1 and mir-133 [79]. In line with this finding, miR-1 and miR-133 were found to be downregulated in the plantaris muscle of mice in response to functional overload [80]. Since the miR-1/miR-133 pair shows similar behavior during pathological hypertrophy, it appears to participate in a general hypertrophic program. Although both these hypertrophic models may be considered compensatory in that the heart biochemically and physiologically adjusts to cellular alterations that occur according to the severity of the overload, yet the factors determining the two kinds and their progression towards the phenotypes are very different [81]. Studies related to the physiological maturation of hESC- derived cardiomyocytes in the in vitro settings are beginning to gain more attention especially due to their importance in transplantation studies to treat heart diseases and for their utility for drug toxicity screens. In understanding the role of miRNAs in the model of pathological hypertrophy leading to cardiovascular diseases, we have seen that the in vivo studies have often complimented for the in vitro studies. Thus in this context, understanding the mechanistic role of miRNAs and their targets during physiological cardiac hypertrophy and in vitro cardiomyocyte maturation will be highly significant.
CONCLUSIONS
Despite current insights into the secondary structure, biogenesis and the regulation of miRNAs, our understanding of how miRNAs function in various developmental and disease pathways is far from complete and numerous conceptual and experimental questions still remain. To date, only a handful of the hundreds of miRNAs expressed in cardiovascular system have been functionally analyzed. The ability of these miRNAs to fine-tune gene expression programs portends their importance in many facets of cardiac biology. It’s a virtual certainty that many unexplored roles of miRNAs in control of normal and abnormal cardiac function are awaiting discovery. Recent advances in understanding the role of miRNAs in pathological cardiac hypertrophy has indeed opened up a pandora of opportunities for using such miRNAs as drug therapeutics during cardiac failure and as potential agonists for physiological maturation in the in vitro setting. Taken together, understanding the miRNA biology and its regulation undoubtedly offers promising and untapped opportunities for elucidating developmental pathways and treating diseases.
Acknowledgments
We thank members of the Ruohola-Baker lab for helpful discussions. This work was partly supported by Tietze Award to KK, Schultz Fellowship to HS and NIH R01GM083867-03S1 to KK and R01GM083867, R01GM097372 and 1P01GM081619 to HRB.
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
References
- 1.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 2.Lee I, Ajay SS, Yook JI, et al. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19(7):1175–83. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4(4):e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103(7):2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24(1):138–48. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blow MJ, Grocock RJ, van Dongen S, et al. RNA editing of human microRNAs. Genome Biol. 2006;7(4):R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305(5689):1434–7. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 12.Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24(17):1951–60. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 14.Feng Y, Zhang X, Song Q, Li T, Zeng Y. Drosha processing controls the specificity and efficiency of global microRNA expression. Biochim Biophys Acta. 2011;1809(11–12):700–7. doi: 10.1016/j.bbagrm.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuchida A, Ohno S, Wu W, et al. miR-92 is a key oncogenic component of the miR-17–92 cluster in colon cancer. Cancer Sci. 2011;102(12):2264–71. doi: 10.1111/j.1349-7006.2011.02081.x. [DOI] [PubMed] [Google Scholar]
- 16.Han J, Pedersen JS, Kwon SC, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136(1):75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda T, Yamagata K, Fujiyama S, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9(5):604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 20.Michlewski G, Guil S, Semple CA, Caceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32(3):383–93. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130(1):89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macrae IJ, Zhou K, Li F, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311(5758):195–8. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 24.Starega-Roslan J, Krol J, Koscianska E, et al. Structural basis of microRNA length variety. Nucleic Acids Res. 2010;39(1):257–68. doi: 10.1093/nar/gkq727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang HW, Noland C, Siridechadilok B, et al. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009;16(11):1148–53. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell. 2011;43(1):110–21. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147(5):1080–91. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 29.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465(7298):584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 31.Gong J, Tong Y, Zhang HM, et al. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat. 2011;33(1):254–63. doi: 10.1002/humu.21641. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125(6):1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 33.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27(6):549–55. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gantier MP, McCoy CE, Rusinova I, et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011;39(13):5692–703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YK, Yeo J, Ha M, Kim B, Kim VN. Cell adhesion-dependent control of microRNA decay. Mol Cell. 2011;43(6):1005–14. doi: 10.1016/j.molcel.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 36.Krol J, Busskamp V, Markiewicz I, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141(4):618–31. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 37.Fang K, Qian F, Chen YQ. MicroRNAs as regulators in normal hematopoietic and leukemia stem cells: current concepts and clinical implications. Curr Mol Med. 2012;12(5):536–46. doi: 10.2174/156652412800620002. [DOI] [PubMed] [Google Scholar]
- 38.Mathieu J, Zhang Z, Zhou W, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71(13):4640–52. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGregor RA, Choi MS. microRNAs in the regulation of adipogenesis and obesity. Curr Mol Med. 2011;11(4):304–16. doi: 10.2174/156652411795677990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stadler BM, Ruohola-Baker H. Small RNAs: keeping stem cells in line. Cell. 2008;132(4):563–6. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong J. Emerging roles of microRNA-22 in human disease and normal physiology. Curr Mol Med. 2012;12(3):247–58. doi: 10.2174/156652412799218886. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 43.Kanellopoulou C, Muljo SA, Kung AL, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102(34):12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bar M, Wyman SK, Fritz BR, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26(10):2496–505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5(2):351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 47.Morin RD, O’Connor MD, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–21. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi J, Yu JY, Shcherbata HR, et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8(22):3729–41. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stadler B, Ivanovska I, Mehta K, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19(7):935–50. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–33. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sengupta S, Nie J, Wagner RJ, Yang C, Stewart R, Thomson JA. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells. 2009;27(7):1524–8. doi: 10.1002/stem.84. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40(12):1478–83. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463(7281):621–6. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anokye-Danso F, Trivedi CM, Juhr D, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao B, Bao X, Liu L, et al. MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem. 2011;286(19):17359–64. doi: 10.1074/jbc.C111.235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18(5):749–58. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luningschror P, Stocker B, Kaltschmidt B, Kaltschmidt C. miR-290 cluster modulates pluripotency by repressing canonical NF-kappaB signaling. Stem Cells. 2012;30(4):655–64. doi: 10.1002/stem.1033. [DOI] [PubMed] [Google Scholar]
- 58.Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331(1):57–66. doi: 10.1007/s00441-007-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435(7044):974–8. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 60.Reynolds S, Ruohola-Baker H. StemBook [Internet] Cambridge (MA): Harvard Stem Cell Institute; 2008. The role of microRNAs in germline differentiation. [PubMed] [Google Scholar]
- 61.Yu JY, Reynolds SH, Hatfield SD, et al. Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development. 2009;136(9):1497–507. doi: 10.1242/dev.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 64.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci USA. 2005;102(52):18986–91. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu N, Williams AH, Kim Y, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA. 2007;104(52):20844–9. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ivey KN, Muth A, Arnold J, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2(3):219–29. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci USA. 2008;105(46):17830–5. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gan L, Schwengberg S, Denecke B. MicroRNA profiling during cardiomyocyte-specific differentiation of murine embryonic stem cells based on two different miRNA array platforms. PLoS One. 2011;6(10):e25809. doi: 10.1371/journal.pone.0025809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson KD, Hu S, Venkatasubrahmanyam S, et al. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499. Circ Cardiovasc Genet. 2010;3(5):426–35. doi: 10.1161/CIRCGENETICS.109.934281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen JF, Murchison EP, Tang R, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2008;105(6):2111–6. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng Y, Ji R, Yue J, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170(6):1831–40. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ikeda S, Kong SW, Lu J, et al. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31(3):367–73. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 73.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100(3):416–24. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 74.Tatsuguchi M, Seok HY, Callis TE, et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42(6):1137–41. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103(48):18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Callis TE, Pandya K, Seok HY, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119(9):2772–86. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–9. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 78.Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–47. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613–8. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 80.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102(1):306–13. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 81.Wikman-Coffelt J, Parmley WW, Mason DT. The cardiac hypertrophy process. Analyses of factors determining pathological vs physiological development. Circ Res. 1979;45(6):697–707. doi: 10.1161/01.res.45.6.697. [DOI] [PubMed] [Google Scholar]