Summary
Poly(ADP-ribose) is a major regulatory macromolecule in the nucleus, where it regulates transcription, chromosome structure and DNA damage repair. Functions in the interphase cytoplasm are less understood. Here we identify a requirement for poly(ADP-ribose) in the assembly of cytoplasmic stress granules, which accumulate RNA-binding proteins that regulate the translation and stability of mRNAs upon stress. We show that poly(ADP-ribose), 6 specific poly(ADP-ribose) polymerases and 2 poly(ADP-ribose) glycohydrolase isoforms are stress granule components. A subset of stress granule proteins, including microRNA-binding Argonaute family members Ago1-4, are modified by poly(ADP-ribose) and such modification increases upon stress – a condition when both microRNA-mediated translational repression and microRNA-directed mRNA cleavage are relieved. Similar relief of repression is also observed upon overexpression of specific poly(ADP-ribose) polymerases or, conversely, upon knockdown of glycohydrolase. We conclude that poly(ADP-ribose) is a key regulator of post-transcriptional gene expression in the cytoplasm.
Introduction
Poly(ADP-ribose), or pADPr, is a macromolecular polymer and post-translational modification best known for its functions in the nucleus (Schreiber et al., 2006). These include DNA damage repair, chromatin remodeling and transcriptional regulation (Krishnakumar and Kraus, 2010). However, increasing evidence suggests that pADPr also functions in the cytoplasm. For example, pADPr is required for the structure and function of the spindle in the mitotic cytoplasm (Chang et al., 2004). Furthermore, the majority of enzymes regulating pADPr synthesis and degradation are localized to the cytoplasm. These include two of the three isoforms of pADPr glycohydrolase (PARG) (Meyer-Ficca et al., 2004), and 5 pADPr polymerase (PARP) family members whose cellular localizations are characterized (Juszczynski et al., 2006; Kickhoefer et al., 1999; Liu et al., 2004; Smith and de Lange, 1999; Yu et al., 2005). Seventeen PARPs exist in humans, all defined by a conserved PARP domain and are classified as pADPr synthesizing, mono-ADP-ribose (mADPr) synthesizing, or enzymatically inactive based primarily on the presence of a triad of histidine, tyrosine and glutamate (HYE) thought to be required for synthesizing the initial mADPr and/or subsequent pADPr subunits (Hottiger et al., 2010; Kleine et al., 2008).
The ability to synthesize mADPr or pADPr is not a prerequisite for PARP function. For example, PARP-13/ZAP (Zinc finger Antiviral Protein) lacks catalytic activity yet inhibit certain viruses by degrading viral RNAs in the cytoplasm (Gao et al., 2002). PARP-13 proteins either lack a PARP domain (PARP-13.2 isoform) or contain a domain lacking key residues of the HYE motif (PARP-13.1 contains YYV). Such lack of catalytic activity does not rule out the possibility that PARP-13 function requires ADPr modification by another PARP. Such trans-modification is common among PARPs from the same subfamily, including modifications between PARP-1 and PARP-2 (Schreiber et al., 2002); and between PARP-5a and PARP-5b (Sbodio et al., 2002). Consistent with PARP-13’s role in regulating RNA in the cytoplasm, pADPr synthesizing and degrading activities were previously identified in cytoplasmic mRNA-protein complexes (mRNPs) (Elkaim et al., 1983; Thomassin et al., 1985). In this study, we discover two functions of pADPr in the cytoplasm – modulating the expression of microRNA (miRNA) targets and assembling mRNP-rich structures called stress granules (SGs).
miRNAs are ~22nt short non-coding RNAs that regulate targets via translational inhibition and mRNA degradation (Fabian et al., 2010). Despite the diverse fates of mRNA targets, all processes are mediated through the miRNA binding protein Argonaute (Ago1-4 in humans). Recent data suggest that the activities of miRNAs can be modulated by multiple mechanisms, including post-translational modifications of Argonaute (Qi et al., 2008; Rudel et al., 2010; Zeng et al., 2008), association with other RNA binding proteins, and re-localization of Argonaute to subcellular locations, such as SGs (reviewed in Leung and Sharp, 2010).
SGs assemble upon stalled translation initiation (Anderson and Kedersha, 2008), which can be triggered via two pathways: (1) phosphorylation of initiation factor eIF2α, which commonly occurs during cell stresses, such as heat shock or arsenite-mediated oxidative stress, or (2) by addition of translation initiation inhibitors which do not involve the phosphorylation of eIF2α. Here we show that pADPr is important for SG assembly and identify 6 PARPs including PARP-13 that function in these processes. Interestingly, the majority of these PARPs also bind to Ago2, suggesting that they have overlapping activities in miRNA and SG function.
Results
Poly(ADP-ribose) localizes to stress granules in the cytoplasm
Previous work by us and others identified binding interactions between pADPr and cytoplasmic RNA-binding proteins (Chang et al., 2009; Gagne et al., 2008); included among these are SG components. To determine if pADPr is present in SGs, cells were stressed with arsenite then stained with antibodies generated against pADPr (LP96-10) and SG marker eIF3. Four human cell lines were analyzed: retinal primary epithelial cells (RPE-1) and three cancer lines, 293T, U2OS and HeLa (Figure 1A and S1A). In each case, pADPr was enriched in SGs. To confirm, five additional anti-pADPr antibodies were tested (Figure S1B and data not shown). This panel of antibodies includes monoclonal antibody 10H specific for polymers containing ≥ 20 ADP-ribose subunits (Kawamitsu et al., 1984). Each antibody stained the interphase cytoplasm and nucleus in untreated cells and demonstrated a clear co-localization with eIF3-positive puncta in the cytoplasm upon stress, regardless of fixation method (methanol or paraformaldehyde; data not shown). Moreover, such co-localization was also observed in all examined stress conditions that result in SG assembly, including heat shock, glucose deprivation, treatment with the proteasome inhibitor MG132 or with translation initiation inhibitors (pateamine A and hippuristanol) (Figure S1C and data not shown). Together, these data indicate that pADPr is enriched in SGs upon multiple stresses and contain polymers of at least 20 ADP-ribose subunits.
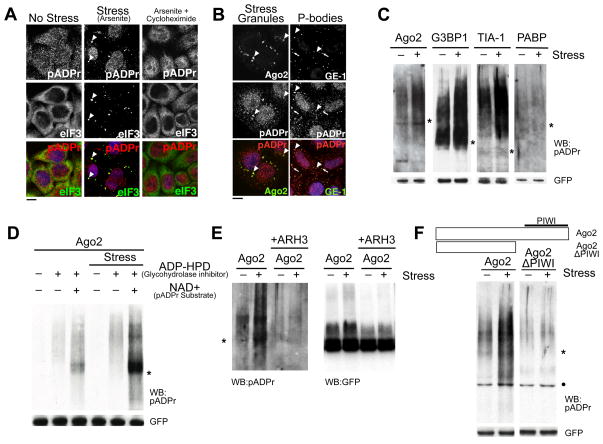
Figure 1. pADPr is enriched in SGs upon multiple types of stresses and modifies specific cytoplasmic RNA-binding proteins dependent on RNA-binding domain.
(A) pADPr staining using LP96-10 antibodies in HeLa cells untreated, or treated with 100 μM arsenite for 60 min, or for 30 min followed by 100 μM arsenite +100 μg/ml cycloheximide for 30 min. Arrowheads, SGs; scale bar, 10 μm. (B) HeLa cells treated with 100 μM arsenite were stained for pADPr, SG component Ago2 (arrowheads) or PB component GE-1 (arrows). DNA was stained with Hoeschst 33342 (blue); scale bar, 10 μm. (C) Immunoprecipitates of 4 GFP-tagged SG-localized RNA-binding proteins from cells treated with or without 20 nM pateamine A were probed for pADPr. (D) Immunoprecipitates of GFP-Ago2 from cells treated with or without 20 nM pateamine A were probed for pADPr. The cell extracts either included or excluded 1 μM ADP-HPD, and with or without 1 mM NAD+ before immunoprecipitation by anti-GFP. (E) pADPr modification of Ago2 from cells treated with or without 20 nM pateamine A was verified by treating the immunoprecipitates with ARH3. The immunoprecipiates were probed for pADPr (left) and GFP (right). (F) Immunoprecipitates of wild-type and PIWI mutant of GFP-Ago2 from cells treated with or without 20 nM pateamine A were probed for pADPr. For panels C-F, cell extracts included 1μM ADP-HPD unless stated otherwise; shown are western blots for pADPr (LP96-10) and GFP levels in each immunoprecipitate. Asterisks indicate the position of the corresponding GFP-tagged RNA-binding protein constructs. Black dots indicate non-specific binding to BSA by LP96-10. See also Figure S1.
SGs can be disassembled by cycloheximide which shifts the equilibrium of mRNA pools from stalled initiation complexes at SGs to polyribosomes (Anderson and Kedersha, 2008). Upon addition of cycloheximide to arsenite-treated cells, pADPr no longer localized as punctate structures (Figure 1A, rightmost panel), suggesting that the pADPr originally at SGs relocated with mRNA and/or their binding proteins. Thus, pADPr co-localization with other mRNA binding proteins was further analyzed (Anderson and Kedersha, 2008). pADPr co-localized with miRNA-binding protein Ago2, RNA decay factor G3BP1, translational suppressor TIA-1 and poly(A) binding protein PABP (Figure 1B and S1D). In contrast, no significant pADPr co-localization was observed with GE-1, a marker for P-bodies, another cytoplasmic structure enriched in RNA-binding proteins (Figure 1B, S1E and legends for statistics). Thus, pADPr localization to SGs is not the result of non-specific binding of pADPr with mRNA binding proteins.
pADPr modification of cytoplasmic RNA-binding proteins requires the presence of an RNA-binding domain
While pADPr modifications occur primarily on PARP proteins, pADPr also modifies other targets (Schreiber et al., 2006). Since Ago2, G3BP1, TIA-1 and PABP co-localized with pADPr at SGs, we examined their pADPr modification status. Proteins were expressed as GFP-fusions, immunoprecipitated from extracts of either unstressed or stressed cells in the presence of PARG-specific inhibitor ADP-HPD, then immunoblotted for pADPr. If the protein is modified, a slower migrating material stained positively with anti-pADPr is expected and would appear as a “smear” due to heterogeneity in the length of polymer attached (Figure 1C). Such pADPr-positive material was observed for Ago2, G3BP1, and TIA-1, but not PABP. Upon stress, Ago2, G3BP1 and TIA-1, but not PABP, exhibited significantly increased pADPr modification (Figure 1C). Similarly, other Argonaute family members, Ago1, Ago3 and Ago4 are modified by pADPr during non-stress conditions suggesting that all Argonaute proteins are actively regulated by pADPr in unstressed cells and such modification increases upon stress (Figure S1F).
The presence of pADPr modification was further verified for Ago2 in four different ways. First, endogenous Ago2 was immunoprecipitated from unstressed and stressed cells and probed for pADPr (Figure S1G). Similar to GFP-Ago2, endogenous Ago2 exhibited moderate amounts of pADPr modification during non-stress conditions and the modification increased upon stress. Second, the pADPr staining associated with GFP-Ago2 under non-stress and stress conditions decreased when PARG-specific inhibitor ADP-HPD was omitted, and increased upon addition of NAD+, a substrate for ADPr synthesis, suggesting that pADPr modification of Ago2 is actively regulated even in in vitro conditions (Figure 1D). Third, treatment of pADPr glycohydrolase ARH3, but not RNase A, eliminated the anti-pADPr signal from the immunoprecipitates (Figure 1E and data not shown). These slow-migrating, pADPr-positive materials were also GFP-positive, confirming that the observed signal was derived from GFP-Ago2 (Figure 1E). Finally, the ADP-ribosylating activities in the GFP-Ago2 immunoprecipitates were further examined by incubating with increasing concentrations of NAD+ containing trace amounts of radioactive NAD+. The reactions were then resolved via SDS-PAGE and visualized by autoradiography (Figure S1H). A heterogeneous mobility of radioactive signal was observed immediately above where GFP-Ago2 migrated (denoted by asterisk). The intensity of this signal was directly dependent on the concentration of NAD+ added. These data suggest that PARPs associated with Ago2 in the immunoprecipitates can synthesize pADPr. Importantly, addition of the general PARP inhibitor 3-aminobenzamide (3-AB) to the reactions completely eliminated any incorporation of radioactivity in the Ago2 precipitates, suggesting that the signal observed is due to ADP-ribosylating activities (Figures S1H).
Since pADPr modification was observed on mRNA-binding proteins Ago2, G3BP1 and TIA-1, we next tested whether their modification requires mRNA binding. To test this, fragments of each protein were expressed at levels comparable to wild-type, immunoprecipitated from unstressed and stressed cells and immunoblotted for pADPr. In all cases, pADPr modification required the presence of an mRNA-binding domain, such as Ago2’s PIWI domain (Figure 1F) or RRM in G3BP1 and TIA-1 (Figure S1I and J) in both non-stress and stress conditions. These data suggest that either the pADPr modification site is within or proximal to the RNA-binding domains of these proteins, or that the modification begins with the activation of PARP(s) that is/are associated via mRNAs.
Specific PARPs and PARG isoforms associate with cytoplasmic mRNP complexes
We reasoned that PARPs responsible for such modification are likely associated with these cytoplasmic RNA-binding proteins during non-stress conditions and these associations may be retained in SGs upon stress. Thus, SGs were used as a surrogate to identify such PARPs. We screened a library of GFP fusions (Vyas et al., in preparation) to 17 of the 18 human PARPs (PARP-13.2 isoform included, but not PARP-14 because the full length cDNA was unavailable) for co-localization with known SG markers (Figure 2A, S2A and data not shown). Five out of 17 PARPs localized to SGs – PARP-5a, -12, -13.1, -13.2 and -15 (Figure S2B). Specific SG localization of these PARPs was confirmed using live-cell imaging. In each case, GFP-tagged PARPs strongly co-localized with RFP-G3BP1 at the assembling SGs (Movie S1). To further rule out over-expression artifacts, we repeated the co-localization screen in HeLa (Figure 2B) and RPE1 cells (Figure S2C) using a library of affinity purified and characterized antibodies against each PARP (Vyas et al, in preparation). Antibody staining confirmed the GFP-PARP screen results and also identified PARP-14 as a SG protein (confirmed via two distinct antibodies) (Figure 2B and S2C and data not shown). Consistent with the GFP localization screen, the remaining 12 PARPs did not localize to SGs endogenously, confirming these 6 PARPs as SG proteins (hereafter referred as SG-PARPs; Figure S2B).
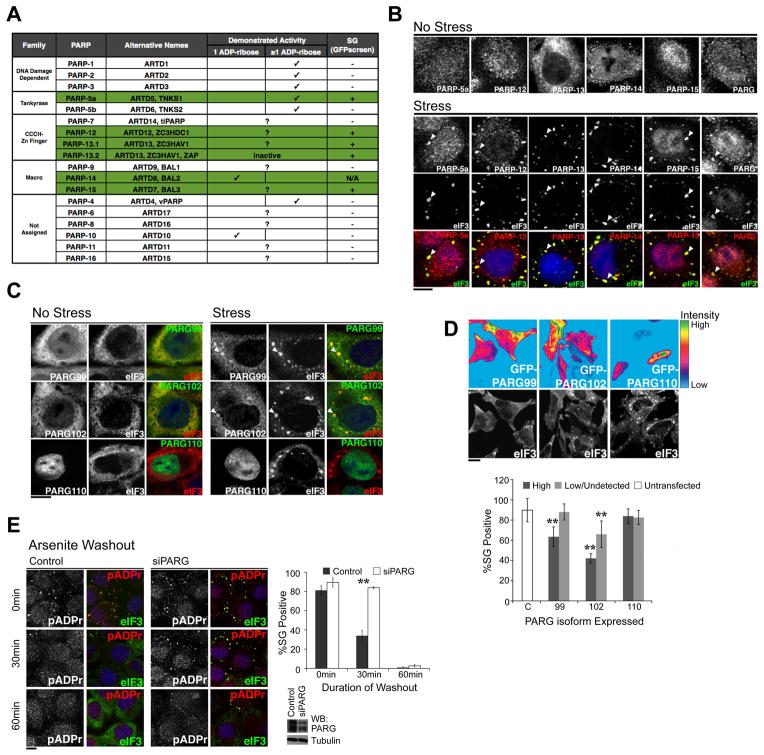
Figure 2. Specific PARPs and PARG isoforms localize in the cytoplasmic SGs and the level of PARG modulates the kinetics of SG assembly and disassembly.
(A) Summary of SG localization screen of PARP family. Green shading indicates SG-PARPs as determined by GFP-PARP fusions or PARP specific antibodies. (B) HeLa cells were treated with or without 100 μM arsenite for 60 min and stained using antibodies against SG-PARPs and PARG. (C) HeLa cells expressing GFP-tagged PARG isoforms were treated with or without 250 μM arsenite for 30 min. (D) Overexpression of cytoplasmic PARG isoforms inhibits SG assembly. Experiment performed as in panel C, but heat map shows level of GFP-PARG isoforms (99, 102, 110) compared with untransfected control (C). Accompanying graph shows quantitation of image data; ≥200 cells for each condition from at least six independent fields. Cells with GFP intensity above background are classified as ‘High’ while cells with intensity indistinguishable from background levels as ‘Low/Undetected’. Paired t-test p < 0.01 (**), derived from comparison to untransfected control; error bars indicates SD. (E) pADPr hydrolysis is required for SG disassembly. Shown are representative images taken 0, 30, and 60 min after washout of 30 min 100 μM arsenite treatment in control and PARG knockdown HeLa cells. Quantitation: ≥100 cells for each condition, n = 3. Paired t-test p < 0.01 (**) and error bars indicate SD. Accompanying blot shows level of PARG knockdown with tubulin as loading control. For panels B-E, pADPr were stained by LP96-10 antibodies, SGs (arrowheads) by anti-eIF3 and DNA by Hoechst 33342 (blue); scale bars = 10 μm. See also Figure S2 and Movie S1.
We next examined the localization of pADPr hydrolyzing enzyme PARG. Immunostaining suggests that a fraction of endogenous PARG localizes to the cytoplasm (Figure 2B). Similar to the SG-PARPs, PARG was enriched in SGs upon stress (Figure 2B). Three major PARG isoforms exist: PARG99, PARG102 and PARG110 (Figure S2D). To determine which localize to SGs, GFP fusions of each were screened. Both PARG99 and PARG102 strongly co-localized with SGs upon stress and were dispersed in the cytoplasm during non-stress conditions. PARG110 was nuclear and unaffected by stress (Figure 2C). While nuclear PARG110 is known to function in DNA damage repair (Schreiber et al., 2006), PARG99 and PARG102 isoforms are functionally uncharacterized; thus their identification at SGs uncovers potential function for these cytoplasmic isoforms.
The presence of 6 PARPs and 2 PARG isoforms in SGs suggests that the pADPr concentration may be dynamically regulated there. Overexpression of each SG-PARP resulted in de novo assembly of SGs without altering eIF2α phosphorylation levels (Figure S2E). This suggests that these enzymes and/or their product, pADPr, play a structural role in SG function rather than causing a general impairment of translation via eIF2α regulation. Overexpression of cytoplasmic PARG99 or PARG102 inhibited SG assembly while overexpression of nuclear PARG110 had no effect (Figure 2D). Moreover, 3 distinct siRNAs targeting the first exon of PARG102 coding region individually delayed SG disassembly (Figure 2E and S2F). No such effect was observed upon knock down of a nuclear pADPr glycohydrolase ARH3 (Figure S2G). Interestingly, all tested proteins that can nucleate SGs upon overexpression, including Ago2, TIA-1 and G3BP1, but not non-nucleator PABP (Anderson and Kedersha, 2008), serve as acceptors of pADPr modification (Figure 1C), suggesting a strong correlation between polymer synthesis and SG assembly. Consistent with this, no pADPr modification was identified on mutants of TIA-1 and G3BP1 that are dominant negative in SG formation (Kedersha et al., 1999; Tourriere et al., 2003; Figure S1I–J). Taken together, these data suggest that both assembly and maintenance of SG structure depends on regulating pADPr concentration locally in the cytoplasm.
Stress or specific PARP overexpression alleviates miRNA-mediated repression
To identify a possible function for pADPr modification of RNA-binding proteins on their activities, we focused on Argonautes due to their critical function in regulating >60% of all mRNAs (Bartel, 2009). Given that Argonaute localization to SGs is dependent on miRNAs (Leung et al., 2006) and pADPr modification of Ago2 increases upon stress, we examined miRNA activity during stress. miRNA activity was monitored using a luciferase reporter that contains 6 bulged sites for a siRNA, siCXCR4, in its 3′UTR (Doench et al., 2003). The luciferase protein contains a destabilization signal that reduces its half-life to 20 minutes so that any change in expression is rapidly monitored. Under non-stress conditions, the luciferase construct was repressed 15-fold by targeting siCXCR4 relative to a control siRNA. Consistent with Bhattacharyya et al., 2006, we observed a relief of miRNA-mediated silencing under stress conditions. Under stress conditions, the relative fold repression is reduced to ~5-fold (Figure 3A). Notably, following these stresses, the translation rate was globally reduced; however, the expression of the luciferase mRNA targeted by siCXCR4 decreased to a lesser extent than the non-targeted reporter.
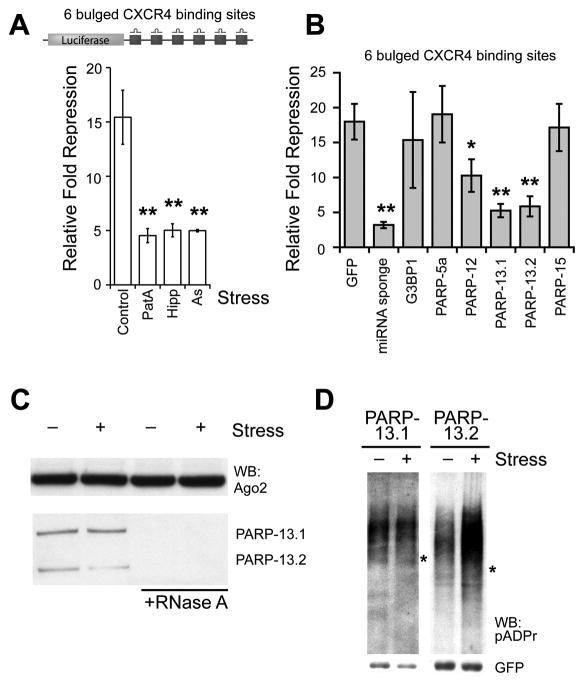
Figure 3. Stress or PARP-13 overexpression alleviates miRNA-mediated repression.
(A) miRNA activity assay in untreated 293T cells or cells treated with 30 nM pateamine A (PatA), 1 μM hippuristanol (Hipp) or 250 μM arsenite (As) for 2 hr, where relative fold repression was measured as the activity of luciferase (upper panel) in the presence of the targeting siRNA normalized to a control siRNA; n = 3. (B) miRNA activity assay upon overexpression of SG-PARPs. Relative fold repression was measured as in panel A; n = 4. For panels A and B, error bars indicate SD; paired t-test p < 0.05 (*) and < 0.01 (**). (C) Antibodies against endogenous Ago2 was used for immunoprecipitation from cytoplasmic extract of HeLa cells treated with or without 250 μM arsenite for 30 min. On the right, the extract was pre-treated with 200 μg/ml RNase A for 20 min at 25°C. (D) Immunoprecipitates of GFP-tagged PARP-13.1 or -13.2 from cells treated with or without 20 nM pateamine A for 30 min were probed for pADPr, where cell extracts included 1 μM ADP-HPD. Asterisks indicate the position of the corresponding GFP-tagged PARPs. See also Figure S3.
As SGs can be assembled via SG-PARP overexpression in the absence of exogenous stress, we next examined whether miRNA activity is correlated with SG induction or specific SG-PARP overexpression (Figure 3B). Interestingly, overexpression of PARP-13.1 or PARP-13.2 each resulted in a ~3 fold decrease in repression of miRNA activity and overexpression of PARP-12 a modest ~1.8 fold decrease whereas overexpression of other SG-PARPs or another SG nucleator G3BP1 had no effect. As controls, overexpression of GFP had no effect while the repression can be relieved by a competitive inhibitor miRNA sponge (Ebert et al., 2007). Thus, the observed decrease in miRNA-mediated repression is not likely due to an overall increase in SG assembly or SG-PARP overexpression, but instead due to the specific function of PARP-13.1/13.2.
To further explore the role of PARP-13 in miRNA silencing, the association between PARP-13 and Ago2 were investigated (Figure 3C). Immunoprecipitation of endogenous Ago2 identified PARP-13.1 and -13.2 as binding partners mediated by mRNA. Given that the amount of PARP-13 bound to Ago2 remains unchanged in non-stress and stress conditions, it seems that other properties, such as post-translational modifications, of PARP-13 are likely altered during stress, resulting in enhanced Ago2 modification.
We therefore examined whether PARP-13 proteins are modified by pADPr. GFP-PARP-13.1 and -PARP-13.2 were immunoprecipitated from unstressed or stressed cells and probed for pADPr. Both isoforms were modified under non-stress conditions. Remarkably, PARP-13.2 exhibited a dramatic increase in modification upon stress, while PARP-13.1 modification remained roughly the same (Figure 3D). Similarly, immunoprecipitation of endogenous PARP-13 via a pan-PARP-13 antibody demonstrated an increase in pADPr modification upon stress (Figure S3A). PARP-13 modification was further confirmed by antibody 10H, which recognizes pADPr with ≥ 20 subunits (Figure S3A-B); and the pADPr staining can be eliminated by glycohydrolase ARH3 or PARG (Figure S3C), but not RNase A (data not shown). Interestingly, the amount of pADPr modification shifted among the SG-PARPs during stress conditions – similar to PARP-13.1, there was no change in PARP-12 modification, while PARP-5a and PARP-15 modifications were reduced (Figure S3D). These results suggest a possible shift in PARP activity from auto-modification to modification of stress targets including Ago1-4, TIA-1, G3BP1 and PARP-13 family members.
PARP-13 family members are pADPr-modified by other SG-PARPs
The pADPr modification attached to PARP-13 proteins are not due to auto-ADP-ribosylating activities since PARP-13.1’s PARP domain is inactive in vitro and PARP-13.2 lacks a PARP domain (Kleine et al., 2008). To determine which PARPs modify PARP-13, we examined the catalytic activity of each SG-PARP. GFP-tagged SG-PARPs were purified by immunoprecipitation from either unstressed or stressed cells (PARP-1 serves as a positive control), and the immunoprecipitates were washed twice with buffer containing high salt (450 mM NaCl) then divided into equal aliquots for ADP-ribosylating activity (Figure 4A and S4A). As expected, PARP-1 and PARP-5a demonstrated auto-poly(ADP-ribosylating) activities, as shown by the mobility shifts above their respective molecular weights (asterisks in Figure 4A). On the other hand, PARP-12 and PARP-15 exhibited mono(ADP-ribosylating) activities as demonstrated by single radioactive bands at their respective molecular weights. Consistent with their lack of active PARP domains, significant amounts of radioactivity were not associated with PARP-13.1 or PARP-13.2. Interestingly, PARP-12 and PARP-15 samples also contained single bands of radioactivity near the expected mobility of endogenous PARP-13.1 (circle) and PARP-13.2 (triangle). Similar results were observed from stress conditions (data not shown). These results suggest that PARP-12 and PARP-15 could be part of complexes that modify PARP-13 family members even in high salt conditions.
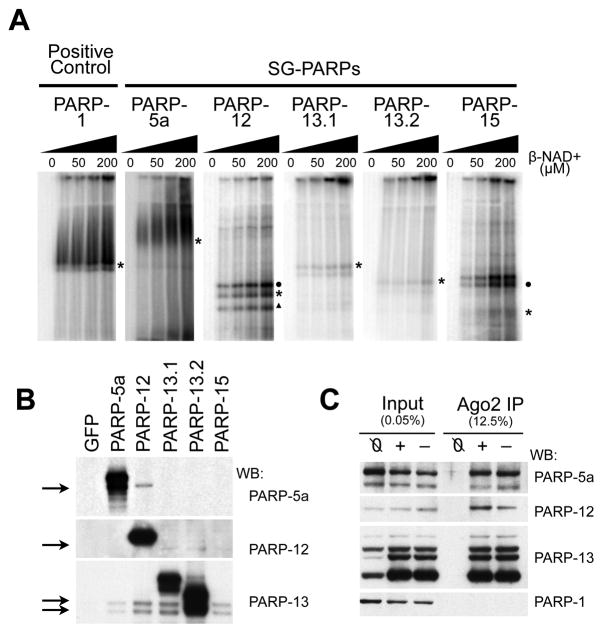
Figure 4. PARP-13 family members are poly(ADP-ribosylated) by other SG-PARPs.
(A) in vitro ADP-ribosylating assay for PARP-1, -5a, -12, -13.1, -13.2 and -15. HeLa S3 cells were transfected with individual GFP-tagged SG-PARPs and GFP-PARP immunoprecipitates were washed twice with 450 mM NaCl then once with 150 mM NaCl. The immunoprecipitates were incubated with 0, 25, 50, 100 or 200 μM NAD+ (with 1/175 fold of P32-labeled NAD+) at 16°C for 30 min, separated on a 6% SDS-PAGE gel and visualized by autoradiography. Asterisks indicate the position of the corresponding GFP-tagged SG-PARPs; circles and triangle indicate the endogenous position of PARP-13.1 and PARP-13.2 respectively. (B) Western blots of the immunoprecipitates from panel A were probed with PARP-5a, -12 and -13 (antibodies for PARP-15 are not good for detecting endogenous protein). Immunoprecipitates from cells transfected with GFP were used as a negative control. (C) HeLa S3 cells were transfected with GFP-tagged Ago2 and cells either treated with (+) or without (−) 20 nM pateamine A for 30 min. Untransfected cells treated with 20 nM pateamine A were used as a negative control (Ø). The cytoplasmic lysates were immunoprecipitated with anti-GFP and washed thrice with cytoplasmic lysis buffer. The input and immunoprecipitates were probed with antibodies against PARP-1, -5a, -12 and -13. See also Figure S4.
To determine if SG-PARPs bind to and modify one another, the binding interactions between different PARPs were analyzed in the immunoprecipitates. Samples were analyzed either by immunoblot (Figure 4B) or mass spectrometry (LC-MS/MS; Figure S4B). Both endogenous PARP-13 isoforms were detected in immunoprecipitates from PARP-12, -13.1, -13.2 and -15 by both methods. The interaction between PARP-13.1 and PARP-13.2 confirms that both isoforms function as a complex (Law et al., 2010). In comparison, the associations of PARP-5a and both PARP-13 isoforms are relatively weak as they were only detected by immunoblot but not by mass spectrometry. However, the association with PARP-13 is specific with these PARPs as it was not observed with the negative control, GFP (Figure 4B). Interestingly, we also observed an association between PARP-12 and PARP-5a. Therefore, it seems likely that individual PARPs associate with other PARPs and account for their respective ADP-ribosylating activities in vitro and in cells.
In light of these findings, we re-examined whether Ago2 binds to other SG-PARPs. GFP-Ago2 was immunoprecipitated from either unstressed or stressed cells and probed for individual PARPs (Figure 4C). Immunoblots identified associations between GFP-Ago2 and PARP-5a and PARP-12, in addition to PARP-13, in both unstressed and stressed cells. No such association was observed for the nuclear PARP-1. Such association of Ago2 with catalytically active PARPs is consistent with the in vitro ADP-ribosylating activities observed in GFP-Ago2 immunoprecipitates (Figure S1H).
Knockdown of glycohydrolase PARG alleviates miRNA-mediated repression
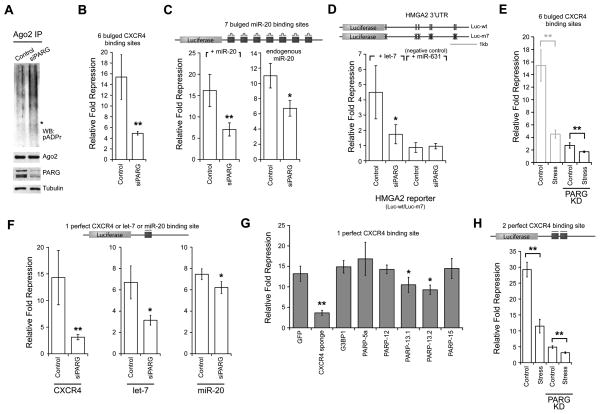
As pADPr modification is dynamically regulated by PARP and PARG activities, we next examined the effect of miRNA activity upon knocking down PARG. PARG knockdown results in an increase in pADPr modification on endogenous Ago2 (Figure 5A) and a ~5-fold decrease in miRNA-mediated repression (Figure 5B and S5A). Thus, similar to stress conditions, an inverse correlation between the pADPr modification of Ago2 and miRNA-mediated repression was observed upon PARG knockdown in non-stress conditions.
Figure 5. PARG knockdown alleviates miRNA-mediated repression and miRNA-directed cleavage.
(A) pADPr modification levels of endogenous Ago2 in HeLa S3 cells transfected with 25 nM control siRNA or siPARG for 48 hr. Asterisk indicates where Ago2 migrated. Shown are western blots for Ago2, PARG and tubulin. (B) 293T cells were transfected with 25 nM control siRNA or siPARG for 72 hr. Relative fold repression was measured as in Figure 3A; n = 3. (C) PARG knockdown effect observed in luciferase reporter with 7 artificial miR-20 binding sites. The relative fold repression was calculated by the amount of expression of the construct normalized to a construct with all binding sites mutated at their seed positions. The assay was tested with exogenous addition of miR-20 (left) or with endogenous miR-20 (right); n = 4. (D) PARG knockdown effect observed in luciferase reporter with endogenous HMGA2 3′UTR. The relative fold repression is calculated by the amount of expression by the wild-type construct (Luc-wt) normalized to the mutant construct Luc-m7; n = 4. (E) siPARG-transfected cells were either treated with or without 30 nM pateamine A for 2 hr (right). As a comparison, part of Figure 3A is reproduced here on the left to show cells transfected with a control siRNA. (F) The effect of PARG knockdown on miRNA-directed cleavage was examined for luciferase construct with 1 perfect siCXCR4, let-7 or miR-20 binding site; n = 4 in each case. (G) The effect of SG-PARP overexpression on miRNA-directed cleavage assay as in panel F; n = 5. (H) The effect of stress on miRNA-directed cleavage was tested with a luciferase reporter with 2 perfect binding sites for siCXCR4 using the same transfection conditions and drug treatment as in Panel E; n = 3. For panels B-H, error bars indicate SD; paired t-test p < 0.05 (*) and < 0.01 (**). See also Figure S5.
The effect of PARG knockdown on miRNA activity in non-stress conditions was further examined with other constructs. Using a luciferase construct with 7 miR-20 binding sites, the expression was reduced ~15 fold upon addition of exogenous miR-20 but such repression was reduced by half upon PARG knockdown (Figure 5C). Given that miR-20 is expressed in 293T cells (Landgraf et al., 2007), the repression mediated by endogenous miR-20 was examined. Under this condition, ~11 fold repression was observed and such repression was reduced 1.8-fold in siPARG-transfected cells (Figure 5C). Similarly, miRNA-mediated repression was examined with a luciferase construct (Luc-wt) fused with the endogenous HMGA2 3′UTR, which contains 7 let-7 binding sites (Mayr et al., 2007) (Figure 5D). As a control, a luciferase (Luc-m7) construct with each miRNA-binding site mutated was used to normalize expression. Upon addition of exogenous let-7, the wild-type construct was repressed by 4.5 fold compared to the mutant. On the other hand, there is no such repression upon addition of miR-631, which does not bind anywhere in the HMGA2 3′UTR. Similar to other constructs, the let-7-mediated repression is reduced by half upon PARG knockdown.
Since both PARG knockdown and stress can alleviate miRNA-mediated silencing individually, we asked whether the combination of both results in further relief in miRNA-mediated silencing (Figure 5E and S5B). Indeed, a significant further reduction of miRNA-mediated repression was observed upon stress in siPARG-treated cells, though the magnitude is less than multiplicative. While stress resulted in a ~3.4-fold repression in control cells, only ~1.6-fold repression was observed upon stress in siPARG-treated cells. Given that stress and PARG knockdown did not synergistically attenuate miRNA silencing, it is likely that the stress pathway involved in modulating the miRNA-mediated silencing is not independent of the pathway that regulates pADPr level via PARG.
PARG knockdown, PARP-13 overexpression or stress alleviates miRNA-directed cleavage
Apart from inhibiting translation or accelerating mRNA decay, miRNAs can also induce mRNA cleavage when the miRNA binds to its mRNA target in a perfectly complementary manner (e.g. Yekta et al., 2004). The effect of PARG knockdown on the miRNA-directed cleavage was next examined. First, a luciferase construct with a perfectly complementary binding site for siCXCR4 or let-7 was tested (Figure 5F and S5C). Upon addition of siCXCR4 or let-7, expression is reduced 15-fold or 7-fold, respectively. As in the case of the constructs with bulged configuration for siRNA/miRNA, this repression is relieved upon PARG knockdown (4.5 fold for siCXCR4 and 2-fold for let-7). Such relief in repression upon PARG knockdown was also examined for endogenous miR-20 and the repression was reduced ~1.2 fold upon PARG knockdown (Figure 5F). Since Ago2 is the only member of the Argonaute family that is capable of mediating miRNA-directed cleavage, these statistically significant decreases in repression must reflect inhibition of a complex containing this factor.
Given that the relief of miRNA-mediated translational inhibition/mRNA decay can result from overexpression of specific PARPs or stress, their effects on miRNA-directed cleavage were also examined. Overexpression of PARP-13.1/-13.2 reduced the level of miRNA-mediated directed cleavage 1.3/1.4 fold and 1.6/1.7 fold in the reporter construct containing 1 and 2 perfect siCXCR4 binding site(s), respectively (Figure 5G and S5D). Thus, both major miRNA-mediated processes involve PARP-13 family members. Similarly, stress reduced the repression ~2.5 fold using the reporter containing 2 perfect siCXCR4 binding sites (Figure 5H and S5E). When stress and siPARG treatment were combined, the repression was reduced to ~1.6-fold (Figure 5H and S5F). Thus, similar to miRNA-mediated silencing (Figure 5E), the reduced magnitude observed upon stress in siPARG-treated cells, as compared to untreated cells, is likely because the stress pathway involved in modulating the miRNA-directed cleavage overlaps with the pathway that regulates pADPr level via PARG.
Discussion
pADPr regulates post-transcriptional gene expression in the cytoplasm
We report a previously uncharacterized function for pADPr – cytoplasmic post-transcriptional regulation of mRNA. pADPr regulates miRNA silencing and catalyzes the assembly of microscopically visible SG structures. This post-transcriptional regulation occurs in the interphase cytoplasm, further extending the function of pADPr outside the nucleus. Our findings help explain several intriguing observations regarding pADPr function in the cytosol. First, pADPr synthesis and hydrolysis activities are enriched in post-nuclear, post-mitochondrial fractions and in the free mRNP fractions (Elkaim et al., 1983; Thomassin et al., 1985). Free mRNP fractions are enriched in factors that regulate translation and decay of mRNAs, many of which are SG components. Second, our data indicate potential functions for the two cytoplasmic isoforms of PARG, which together are more active than the single nuclear isoform (Meyer-Ficca et al., 2004). Third, large amounts of RNA-binding proteins were associated with pADPr in our previous analyses and by global proteomic analyses; some of them are SG components, including G3BP1 and PABP (Chang et al., 2009; Gagne et al., 2008).
Here we report that specific cytoplasmic RNA-binding proteins, Ago2, G3BP1 and TIA-1, are pADPr-modified dependent on the presence of their RNA-binding domains. Each protein is increasingly modified by pADPr upon stress and enriched in SGs (though not all components, e.g. PABP, are modified). Perhaps one general function of pADPr is to recruit RNA-binding proteins to specific locations, such as SGs, thus functioning as a scaffold for protein recruitment. This scaffold function in the cytoplasm is conceptually similar to the role pADPr plays in other complexes/structures: the mitotic spindle where pADPr recruits spindle pole proteins (Chang et al., 2009; Chang et al., 2005); Cajal bodies, a nuclear organelle enriched in nucleic acid binding proteins that can bind pADPr (Kotova et al., 2009); or at DNA damage sites, pADPr recruits nucleic acid binding proteins for chromatin remodelling and DNA repair (Ahel et al., 2009). However, in contrast to these examples involving the activation of a single PARP, we identified multiple PARPs in SGs from distinct subfamilies: Tankyrase PARP-5a; RNA-binding PARP-12 and PARP-13 isoforms; and PARP-14 and PARP-15 that contain pADPr-binding macro-domains. These pADPr-synthesizing activities along with PARG99 and PARG102 isoforms likely regulate the local pADPr concentration that determines the assembly and maintenance of SGs.
pADPr regulates miRNA function in the interphase cytoplasm
miRNA targets are preferentially expressed relative to total protein synthesis under three conditions – stress, PARP-13 overexpression and PARG knockdown. Two of these conditions, PARP-13 overexpression and stress trigger SG assembly. Yet this apparent correlation is paradoxical since SGs are not sites of active translation as they do not contain 60S ribosomes (Anderson and Kedersha, 2008). Therefore any preferential translation of miRNA targets probably occurs outside SGs in the cytoplasm. Several results suggest that the two phenomena are likely to be co-incidental events that are not necessarily mechanistically linked. For example, overexpression of G3BP1, a known inducer of SG assembly, does not result in the relief of miRNA silencing, whereas PARG knockdown results in the relief of miRNA silencing in the presence or absence of SG formation. This is not surprising as the majority of Ago/miRNA complexes are located in the diffuse cytoplasm; only 5% are localized in SGs upon stress and such pool is rapidly exchanging with the cytoplasm (Leung et al., 2006). Thus, the relief of miRNA silencing as a result of poly(ADP-ribosylation) likely occurs in the diffuse cytoplasm.
At what step does pADPr modulate miRNA silencing? Given that the miRNA silencing is alleviated upon increase in pADPr modification level for both endogenous miRNAs and exogenously added siRNA, pADPr likely regulates a step downstream of miRNA processing in the cytoplasm. Consistent with this, the expression levels of nearly all (>99%) miRNAs examined using miRNA microarray remained unchanged upon PARG knockdown (data not shown). In addition, the relief of miRNA silencing was observed in constructs that can be cleaved through perfectly complementary sites and silenced through partially complementary sites. Thus, pADPr likely regulates miRNA function at a step upstream of the direct activity of the Argonaute complex.
Here we propose that the accessibility of Argonaute/miRNA complex to its target mRNA is affected by an increase in local pADPr modification on multiple proteins that bind to the target, resulting in the relief of miRNA silencing (Figure 6). Those proteins that are modified by pADPr include all Argonaute members and PARP-5a, -12, -13.1 and -13.2, among which Ago1-4 and PARP-13.2 are increasingly modified upon stress when miRNA silencing is relieved. Consistent with the importance of poly(ADP-ribosylation), relief of miRNA silencing was observed upon PARG knockdown when pADPr modification on proteins is generally increased. Such high concentration of negatively-charged pADPr modification near the sites of miRNA:mRNA-binding likely disrupts the electrostatic interaction between similarly charged miRNA and mRNA. Alternatively, the sizeable pADPr modification might cause steric hindrance to prohibit effective miRNA silencing. Recently, it has been shown that in vitro addition of pADPr inhibits the RNA-binding ability of a Drosophila heterogeneous nuclear ribonucleoprotein (Ji and Tulin, 2009). Given that pADPr also exhibited binding affinity to RNA-binding proteins (Chang et al., 2009; Gagne et al., 2008), one function of pADPr could be to regulate the binding of RNAs to RNA-binding proteins.
Figure 6. A working model: A high local concentration of pADPr at miRNA complex results in relief of miRNA silencing.
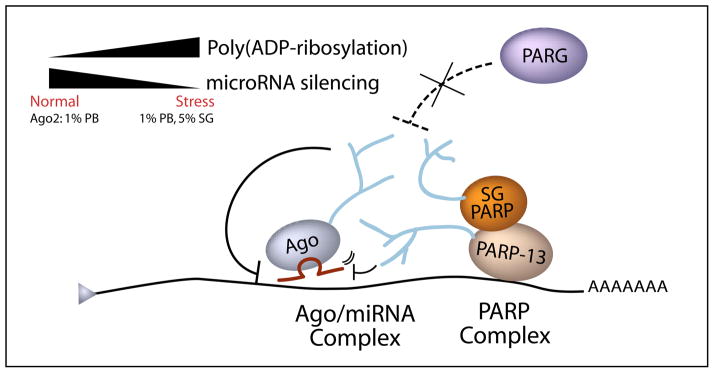
Upon stress, multiple proteins including all Argonaute family members and PARP-13.1/2 complex are increasingly modified by pADPr. Such increase in poly(ADP-ribosylation) during stress could be due to increase in PARP activity and/or decrease in PARG activity (dotted line). High concentration of pADPr near the Argonaute/miRNA complex might disrupt electrostatic interaction or cause steric hindrance for effective miRNA silencing. Similar relief of miRNA silencing is also observed upon overexpression of PARP-13 or, conversely, upon knockdown of PARG.
One interesting observation from this study is that overexpression of PARP-13 family members affects both miRNA-mediated repression and miRNA-directed cleavage, yet these PARPs are not catalytically active. Instead, our data suggest that PARP-5a, PARP-12 and PARP-15 are likely the source of pADPr modification, given that (1) PARP-5a has demonstrated poly(ADP-ribosylating) activities and PARP-12 and PARP-15 mono(ADP-ribosylating) activities, (2) PARP-5a and PARP-12, but not PARP-1, associate with Ago2, and (3) all of these PARPs associate with endogenous PARP-13 family members. We note that the association of an inactive PARP with active PARPs resembles the case of the receptor tyrosine kinase erbB-3, which, though itself has no active kinase domain, can mediate signalling through heterodimerization with active EGF family kinases like Her2 (Holbro et al., 2003). Perhaps, due to their ability to bind mRNA, the function of the inactive PARP-13 isoforms is to anchor the activity of the catalytically active PARPs to the mRNP complex. This might partly explain why the mRNA-binding domain is required for pADPr modification of Ago2.
In conclusion, our data point to two functions for pADPr in the cytoplasm. At SGs, pADPr modulates the assembly and maintenance of an mRNP-enriched structure. At submicroscopic miRNP complexes, pADPr relieves miRNA-mediated repression and miRNA-directed cleavage under stress conditions. These cytoplasmic functions are likely mediated through the concerted activities of catalytically inactive and mADPr- and pADPr-synthesizing PARPs. Such cross-subfamily mechanism of pADPr synthesis suggests that pADPr polymerization could be more complex than previously thought. Given that other post-transcriptional factors such as G3BP1 and TIA-1 are increasingly modified by pADPr upon stress and that these modifications depend on the presence of an RNA-binding domain, it is likely that pADPr is a key regulator of mRNA functions in the interphase cytoplasm.
Experimental Procedures
Immunofluorescence
Following SG induction, coverslips were rinsed twice in PBS and either extracted for 30 sec with buffer A, fixed for 15 min in 4% paraformaldehyde in buffer B, or fixed in ice cold methanol for 5 min, followed by slow rehydration with PBS. Coverslips were incubated with 1° antibodies for 45 min and 2° antibodies for 35 min. See Extended Experimental Procedures for buffer receipes.
Immunoprecipitation
8×107 HeLa S3 cells were transfected with GFP-PARPs or RNA-binding proteins for 48 hr using 293fectin, such that the expression did not induce visible SGs. Cells were either treated with or without 20 nM pateamine A or 250 μM Arsenite for 30 min. At 10 min pre-stress, latruculin B was added to 0.5 mg/ml. Cells were lysed with cytoplasmic lysis buffer C. Lysate was spun at 18,407 g for 10 min and a final concentration of 10 mg/ml cytochalasin B and 25 mM nocodazole was added to the supernatant. The supernatants were incubated with anti-GFP and Protein A beads for 90 min. The immunoprecipitates were washed for 10 min with buffer C, then twice in buffer C containing 300 mM NaCl and again with buffer C. Beads were then eluted with sample buffer and heated for 10 min at 70°C. For endogenous Ago2 modification, anti-Ago2 antibody was pre-incubated with Protein A beads. The supernatant was made by spinning the lysate at 2,300 g or 18,407 g for 10 min and the beads were washed 3 × 5 min with buffer C before eluting with sample buffer. See Extended Experimental Procedures for buffer receipes.
miRNA reporter assay
24 hr after transfection, cells were either untreated or stressed with 30 nM pateamine A, 1 μM hippuristanol or 250 μM arsenite for 2 hr before lysis for luciferase assay. Firefly and Renilla luciferase signals were measured using the Dual Luciferase reporter assay system (Promega). Shown are representative luciferase assay results from 3–4 replicates, as indicated in the figure legend, in each experimental condition and each condition has been independently examined two to four times.
Supplementary Material
Highlights.
Upon stress, poly(ADP-ribose) is enriched in stress granules in the cytoplasm.
Specific PARPs and PARG isoforms localize to and regulate stress granule integrity.
Poly(ADP-ribose) modifies cytoplasmic proteins, including miRNA-binding Argonautes.
Overexpression of PARP-13 or knockdown of PARG attenuates miRNA-mediated silencing.
Acknowledgments
We thank Drs. Tanuma, Urana, J. Tazi, P. Anderson, N. Kedersha and J. Pelletier for kind gifts of reagents, A. West, T. Lai and E. Vasile for technical support, A. Young, M. Ebert, J. Wilusz, P. Boutz for comments, M. Lindstrom for illustrations. A.K.L.L was a special fellow of Leukemia and Lymphoma Society. P.C. is a Rita Allen Foundation Scholar, a Kimmel Foundation for Cancer Research scholar and a Howard S. and Linda B. Stern Career Development Professor. This work was supported by RO1-CA133404 and PO1-CA42063 to P.A.S. and partially by Cancer Center Support (core) grant P30-CA14051.
Footnotes
Supplementary Information 5 supplementary figures and 1 supplementary movie accompanies the paper.
References
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha NL. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Habermacher R, Martine U, Closs E, Filipowicz W. Relief of microRNA-Mediated Translational Repression in Human Cells Subjected to Stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Chang P, Coughlin M, Mitchison TJ. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol Biol Cell. 2009;20:4575–4585. doi: 10.1091/mbc.E09-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Chang W, Dynek JN, Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J. 2005;391:177–184. doi: 10.1042/BJ20050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkaim R, Thomassin H, Niedergang C, Egly JM, Kempf J, Mandel P. Adenosine diphosphate ribosyltransferase and protein acceptors associated with cytoplasmic free messenger ribonucleoprotein particles. Biochimie. 1983;65:653–659. doi: 10.1016/s0300-9084(84)80029-2. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. 2009;37:3501–3513. doi: 10.1093/nar/gkp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczynski P, Kutok JL, Li C, Mitra J, Aguiar RC, Shipp MA. BAL1 and BBAP are regulated by a gamma interferon-responsive bidirectional promoter and are overexpressed in diffuse large B-cell lymphomas with a prominent inflammatory infiltrate. Mol Cell Biol. 2006;26:5348–5359. doi: 10.1128/MCB.02351-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, 3rd, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci U S A. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamitsu H, Hoshino H, Okada H, Miwa M, Momoi H, Sugimura T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984;23:3771–3777. doi: 10.1021/bi00311a032. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. The Journal of Cell Biology. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Kotova E, Jarnik M, Tulin AV. Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet. 2009;5:e1000387. doi: 10.1371/journal.pgen.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law LM, Albin OR, Carroll JW, Jones CT, Rice CM, Macdonald MR. Identification of a dominant negative inhibitor of human zinc finger antiviral protein reveals a functional endogenous pool and critical homotypic interactions. J Virol. 2010;84:4504–4512. doi: 10.1128/JVI.02018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chen G, Ji X, Gao G. ZAP is a CRM1-dependent nucleocytoplasmic shuttling protein. Biochem Biophys Res Commun. 2004;321:517–523. doi: 10.1016/j.bbrc.2004.06.174. [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel S, Wang Y, Lenobel R, Korner R, Hsiao HH, Urlaub H, Patel D, Meister G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbodio JI, Lodish HF, Chi NW. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase) Biochem J. 2002;361:451–459. doi: 10.1042/0264-6021:3610451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci. 1999;112(Pt 21):3649–3656. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- Thomassin H, Niedergang C, Mandel P. Characterization of the poly(ADP-ribose) polymerase associated with free cytoplasmic mRNA-protein particles. Biochem Biophys Res Commun. 1985;133:654–661. doi: 10.1016/0006-291x(85)90955-6. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, Vervoorts J, Walsemann G, Grotzinger J, Kremmer E, et al. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24:1982–1993. doi: 10.1038/sj.onc.1208410. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.