Summary
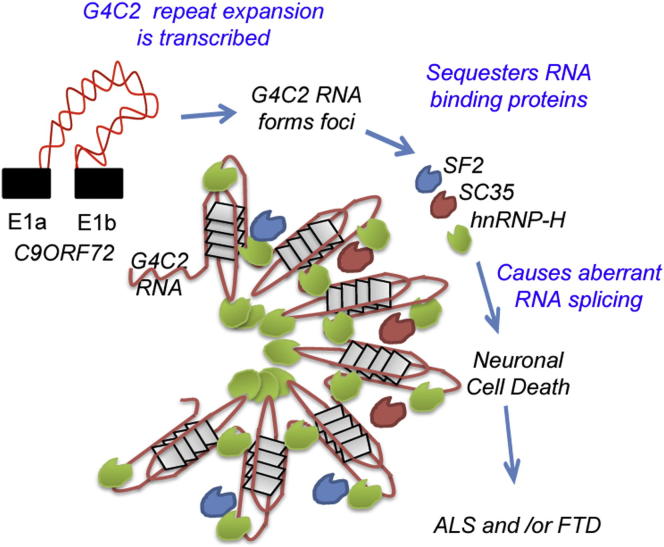
The GGGGCC (G4C2) intronic repeat expansion within C9ORF72 is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Intranuclear neuronal RNA foci have been observed in ALS and FTD tissues, suggesting that G4C2 RNA may be toxic. Here, we demonstrate that the expression of 38× and 72× G4C2 repeats form intranuclear RNA foci that initiate apoptotic cell death in neuronal cell lines and zebrafish embryos. The foci colocalize with a subset of RNA binding proteins, including SF2, SC35, and hnRNP-H in transfected cells. Only hnRNP-H binds directly to G4C2 repeats following RNA immunoprecipitation, and only hnRNP-H colocalizes with 70% of G4C2 RNA foci detected in C9ORF72 mutant ALS and FTD brain tissues. We show that expanded G4C2 repeats are potently neurotoxic and bind hnRNP-H and other RNA binding proteins. We propose that RNA toxicity and protein sequestration may disrupt RNA processing and contribute to neurodegeneration.
Graphical Abstract
Highlights
-
•
Longer G4C2 transcripts form neurotoxic RNA foci in cells and zebrafish
-
•
Longer G4C2 foci sequester RNA binding proteins hnRNP-H, SC35, and SF2
-
•
G4C2 RNA foci and hnRNP-H knockdown similarly affect TARBP2 splicing
-
•
Seventy percent of G4C2 RNA foci in C9ORF72 ALS/FTD brains colocalize with hnRNP-H
In this study, Shaw and colleagues explore mechanisms underlying toxicity of the expanded G4C2 hexanucleotide intronic repeat in C9ORF72, the most common known cause of ALS and FTD. Pathologically expanded G4C2 RNA transcripts form intranuclear foci, sequester specific RNA-binding proteins, and are potently toxic in transfected cells and zebrafish embryos. One protein, hnRNP-H, is detected in 70% of foci in C9ORF72 brain tissues, and loss of hnRNP-H leads to aberrant RNA splicing that could contribute to neurodegeneration.
Introduction
Amyotrophic lateral sclerosis (ALS) causes progressive muscle weakness and spasticity due to the degeneration of motor neurons. Frontotemporal dementia (FTD) causes changes in personality, language, and behavior due to the degeneration of neurons in frontal and temporal lobes. Both are fatal within 3–5 years of symptom onset. Multiple lines of evidence indicate that these two disorders are phenotypic variants of common pathological processes involving the deposition of TDP-43 (Neumann et al., 2006) or FUS (Vance et al., 2009). A common basis for ALS and FTD came from genetic linkage studies that identified a locus on chromosome 9p21 in familial ALS-FTD (Vance et al., 2006) and genome-wide association studies in sporadic ALS and FTD (Shatunov et al., 2010). The underlying mutation was subsequently shown to be an expansion of the GGGGCC (G4C2) repeat within intron 1 of C9ORF72 (DeJesus-Hernandez et al., 2011; Renton et al., 2011). This mutation accounts for 20%–80% of familial and 5%–15% of sporadic ALS and FTD in North American and European populations (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Smith et al., 2012). The size of the repeat in ALS and FTD cases has been estimated by Southern blotting to range between 700 and 1,600 repeats (DeJesus-Hernandez et al., 2011). The mean number of G4C2 repeats in controls is two; 95% have less than eight repeats (Smith et al., 2012).
The mechanism by which the G4C2 intronic repeats cause neurodegeneration is unknown. Decreased tissue levels of the C9ORF72 transcript implicate a loss of protein function due to haploinsufficiency (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Recent reports describe the aggregation of peptides due to repeat-associated non-ATG (RAN) translation (Mori et al., 2013b) as previously described in SCA8 (Zu et al., 2011). Antibodies against poly (Gly-Ala), (Gly-Pro), and (Gly-Arg) peptides selectively label p62-positive, TDP-43-negative neuronal inclusions that are the pathological hallmark of mutant C9ORF72 cases (Al-Sarraj et al., 2011), but evidence that peptide aggregation initiates neurodegeneration is currently lacking. The identification of intranuclear neuronal RNA foci containing G4C2 repeats in ALS and FTD tissues (DeJesus-Hernandez et al., 2011) is similar to other intronic repeat expansion disorders, including myotonic dystrophy, fragile X tremor ataxia syndrome, and several spinocerebellar ataxias (Todd and Paulson, 2010). RNA foci in myotonic dystrophy sequester and deplete muscle-blind-like protein (MBNL1), ultimately causing widespread RNA splicing abnormalities and degeneration of affected tissues (Miller et al., 2000). The fact that overexpression of MBNL1 can rescue the phenotype caused by the CTG repeats implies that loss of MBNL1 is one of the key drivers of cellular degeneration (Kanadia et al., 2003a).
In order to determine whether expanded G4C2 transcripts might be toxic and sequester RNA binding proteins, we generated G4C2 with 8×, 38×, and 72× repeats and expressed them in cell lines, primary neurons, and zebrafish embryos. Here, we demonstrate that the longer repeat lengths generate RNA foci that are toxic to neurons and bind the RNA binding proteins SF2, SC35, and hnRNP-H. hnRNP-H directly binds to G4C2 RNA and colocalizes closely with RNA foci in transfected cells and the brains of human C9ORF72 ALS and FTD cases. Our findings indicate that G4C2 repeat expansions are potently toxic, sequester RNA proteins, and may initiate neurodegeneration in mutant C9ORF72 ALS and FTD.
Results
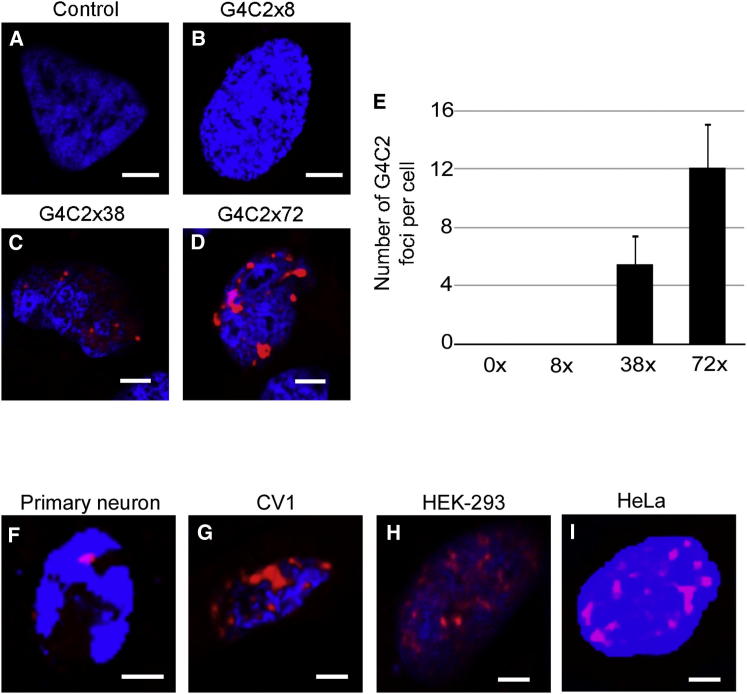
Using direct ligation we generated constructs containing 8×, 38×, and 72× G4C2 repeats, which were cloned into an untagged plasmid vector (Figures S1A–S1D). Following transfection into neuronal (SH-SY5Y) cell lines, intranuclear G4C2-positive RNA foci were detected by RNA fluorescence in situ hybridization (FISH) in all cell types expressing 38× and 72× repeats, but not in cells expressing 8× repeats (Figure 1A). The mean number of foci per cell was six for 38× repeats and 12 for 72× repeats (Figure 1E). These were expressed in a range of human nonneuronal cells (CV1, HEK, HeLa) and primary rat cortical neurons (Figures 1F–1I). The RNA foci were resistant to RNase and DNase treatments (Figures S1E and S1F), consistent with the description of G4C2 DNA repeat hairpin loops forming G quadruplexes in vitro (Fratta et al., 2012).
Figure 1.
Expanded G4C2 Repeats Form Intranuclear RNA Foci
(A–D) SH-SY5Y cells were transfected with various length G4C2 (0, 8, 38, 72 repeat) plasmids and analyzed 24 hr after transfection by RNA FISH using a Cy3-labeled (G2C4)×8 RNA probe.
(E) The mean number of G4C2 foci was counted in 50 cells.
(F–I) Primary hippocampal mouse neurons (F), CV1 (G), HEK293 (H), and HeLa (I) cells were transfected with a plasmid expressing G4C2 72× repeats; foci were detected by FISH (red) and nuclei were stained with DAPI (blue). All cell types transfected with 38× and 72× repeats showed foci (scale bar represents 3 μm).
See also Figure S1.
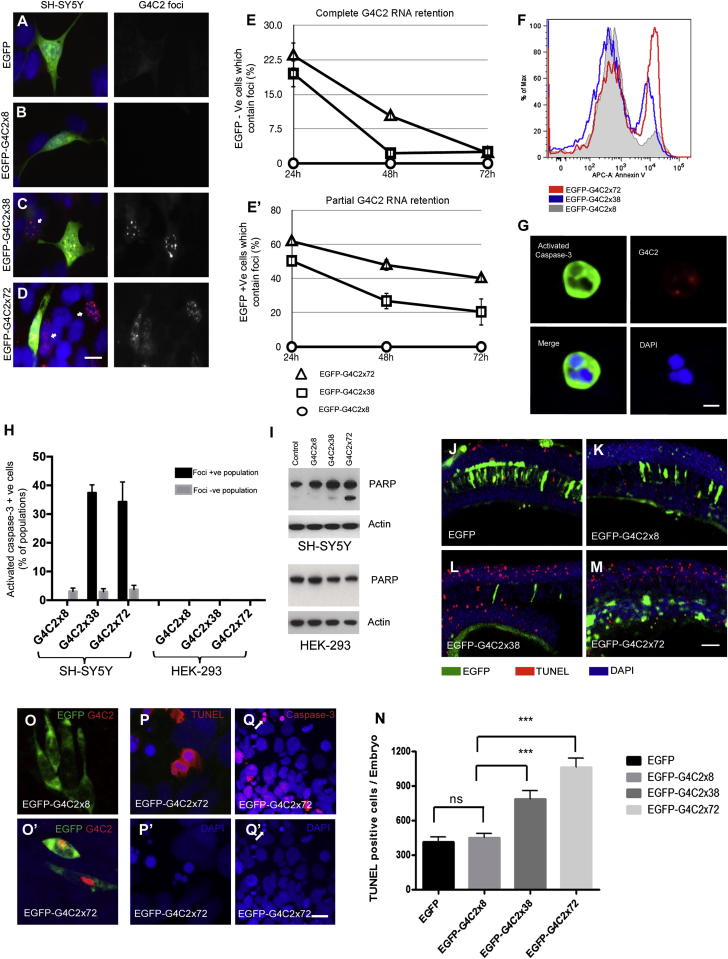
In order to determine whether the length of the G4C2 repeat affected nucleocytoplasmic trafficking of the transcript, we cloned the 8×, 38×, and 72× repeat constructs into the 3′ position of EGFP expression vectors (Figure S2A). Equivalent transcript expression was detected by RT-PCR and northern blotting for 8×, 38×, and 72× repeat vectors (Figures S2B and S2C). Again, 38× and 72×, but not 8×, repeats or EGFP alone, generated G4C2 RNA foci in SH-SY5Y cell lines (Figures 2A–2D). After 24 hr we observed that a significant proportion of cells with intranuclear RNA foci expressed little or no EGFP (19% ± 1.9% for 38× and 23% ± 1.8% for 72× repeats) (Figure 2E). The absence of EGFP in cells containing foci implies nuclear retention of mRNA containing longer repeats and that foci form shortly after transcription precluding nuclear export. In a time course experiment we observed that the number of SH-SY5Y cells bearing RNA foci decreased dramatically over 72 hr. This was most marked in foci-positive cells that were EGFP negative, such that by 72 hr there was a >90% decrease in the number of cells showing foci for 38× and 72× G4C2 repeats (Figure 2E). Cells with foci that also expressed EGFP (demonstrating nuclear export and cytoplasmic translation) showed a more modest decrease of 30% (EGFP-G4C2 38×) and 21% (EGFP-G4C2 72×), respectively (Figure 2E’).
Figure 2.
Loss of G4C2 Foci-Positive Cells Is Due to Apoptotic Cell Death in Culture and In Vivo
(A–D) SH-SY5Y cells were transfected with EGFP-tagged G4C2 constructs. G4C2 RNA foci (red) were found only in cells transfected with EGFP-G4C2 38× and EGFP-G4C2 72× repeats. Many cells with RNA foci were negative for EGFP (arrows in C and D), implying near-complete nuclear retention. Nuclei were stained with DAPI (blue) (scale bar represents 10 μm).
(E) The percentage of cells with G4C2 foci by FISH that did not express EGFP were counted at 24, 48, and 72 hr posttransfection (E), as were foci-positive cells that did express EGFP (E’). Foci-positive cells declined in number, most markedly in those showing greater nuclear retention (EGFP−). In all experiments, a total of 250 cells from three independent transfections were counted, and results are presented as mean ± SD.
(F) EGFP-tagged G4C2 plasmids were transfected into SH-SY5Y cells, stained with the early apoptosis marker Annexin V, and analyzed by FACS. Cells expressing 38× and 72× repeats showed 3- or 5-fold higher levels of Annexin V, respectively, than did cells expressing 8× repeats.
(G) G4C2 foci-positive SH-SY5Y cells were found positive for activated caspase-3 (scale bar represents 5 μm).
(H) G4C2 RNA foci-positive SH-SY5Y cells, but not HEK293 cells, express activated caspase-3. Caspase-3 activation was scored in 250 foci-positive cells per coverslip, and three coverslips were analyzed per experiment. The background level of active caspase-3 was estimated by counting the frequency of active caspase-3-positive cells in the foci-negative population.
(I) Western blot of PARP cleavage in control or G4C2-transfected SH-SY5Y and HEK293 cells. Actin was used as a loading control.
(J–M) Apoptotic cell death was analyzed by TUNEL staining in zebrafish prim-5 embryos injected with plasmids mosaically expressing EGFP (J), EGFP-G4C2 8× (K), EGFP-G4C2 38× (L), or EGFP-G4C2 72× (M). The number of TUNEL-positive cells (red) increased in the embryos injected with 38× and 72× repeats (L and M) (scale bar represents 200 μm).
(N) Quantification of all embryos (n = 5) from three independent experiments is presented. Error bars show the standard error for each sample, and p values (p < 0.0001, ∗∗∗) are also determined.
(O) G4C2 RNA foci (red) were found only in zebrafish embryo cells injected with 72×, but not with 8× repeats. EGFP expression is green.
(P and Q) High-resolution images of TUNEL-positive (P and P′) and active caspse-3 (Q and Q′) from zebrafish embryo cells injected with 72× (scale bar represents 10 μm). Nuclear staining is blue.
See also Figure S2.
In order to determine whether this decline in numbers was due to toxicity induced by RNA foci, we measured the expression of the apoptosis marker Annexin V by fluorescence-activated cell sorting (FACS). FACS analysis showed that the proportion of EGFP-positive cells expressing Annexin V was 3-fold higher for 38× repeats and 5-fold higher for 72× repeats compared to 8× repeats (Figure 2F). To investigate this further, immunocytochemistry (ICC) for activated caspase-3 was performed; it showed that in SH-SY5Y cells, RNA foci dramatically enhanced the activation of caspase-3 (Figure 2G). To quantify the induction of apoptosis by G4C2 foci, we counted caspase-3-activated cells in both foci-negative and foci-positive cell populations. We found no change in the proportion of caspase-3-activated cells above background (3.2% ± 1.2%) in SH-SY5Y cells expressing 8× repeats but a dramatic increase in cells with foci from 38× (37% ± 2.7%) and 72× (34% ± 6.9%) repeats (Figure 2H). Interestingly, HEK293 cells expressing G4C2 38× or 72× showed no evidence of apoptosis despite abundant G4C2 foci (Figure 2H). Accordingly, poly(ADP-ribose) polymerase (PARP) cleavage was detected in 38× or 72× repeat transfected SH-SY5Y cells, but not in HEK293 cells (Figure 2I), suggesting that neuronal cells may be more vulnerable to apoptotic cell death triggered by G4C2 RNA foci.
We next sought to test for length-dependent G4C2 toxicity in vivo by injecting the EGFP constructs with 8×, 38×, and 72× repeats into zebrafish embryos. DNA injection leads to mosaic distribution of the DNA and expression in only a small portion of the embryonic cells. Embryos were injected at the one-cell stage and analyzed at prim-5 (approximately 24 hr postfertilization). Expression of the EGFP-G4C2 8× repeat did not increase the number of apoptotic cells above EGFP alone (Figures 2J and 2K); however, injection of the 38× or 72× G4C2 repeat constructs (Figures 2L and 2M) resulted in a significant increase in TUNEL-positive cells of 0.8- and 1.4-fold, respectively (graphically represented in Figure 2N). G4C2-positive RNA foci detected by FISH were only observed in 72× embryos, but not in 8× embryos (Figures 2O and 2O’). The majority of 72×-transfected cells that had G4C2-positive RNA foci were also active caspase-3 positive (Figure S2D). TUNEL and active caspase-3-positive cells showed nuclear condensation and fragmentation confirming apoptotic cell death (Figures 2P and 2Q). Thus, evidence from cellular and in vivo assays indicates that longer G4C2 repeats lead to the nuclear retention of transcripts, forming RNA foci that are RNase resistant and potently toxic.
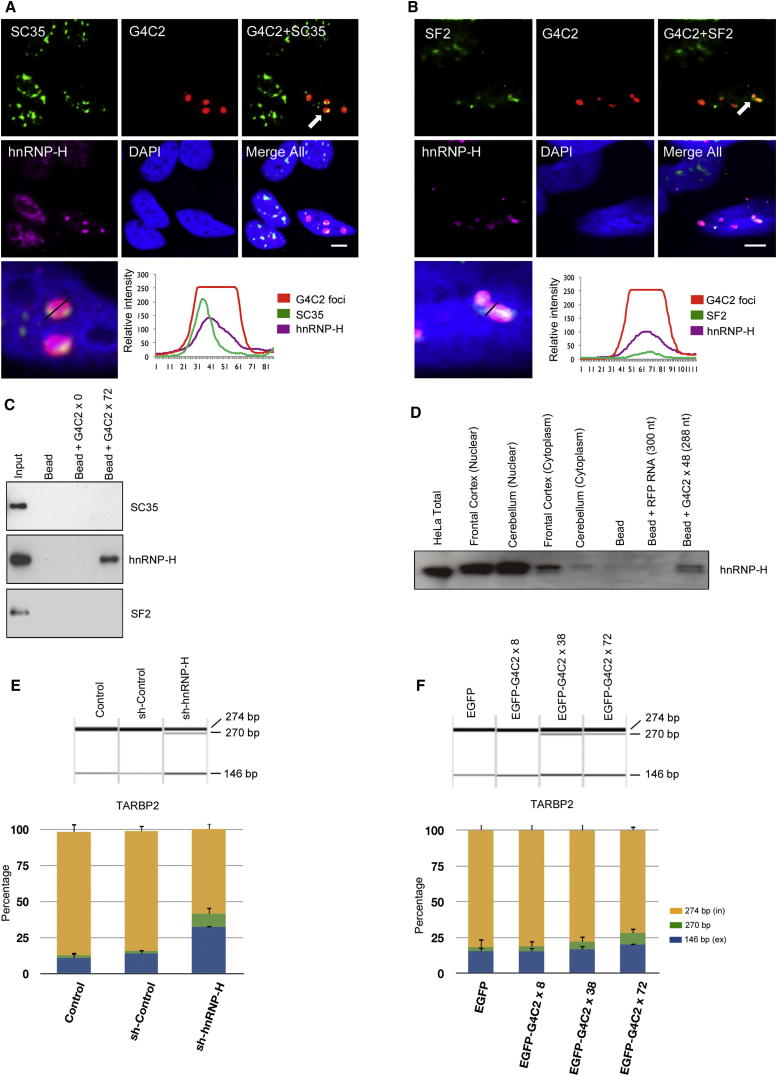
To investigate whether RNA processing proteins were bound to the G4C2 RNA foci, we probed transfected SH-SY5Y cells with antibodies to 30 different RNA-binding proteins based on their known or predicted ability to bind GC-rich sequences, including heterogeneous nuclear proteins (hnRNP) and serine-arginine (SR) proteins involved in splicing and trafficking (Tables S1 and S2). We found that antibodies to three proteins colocalized with the G4C2 RNA foci: hnRNP-H, serine-arginine-rich splicing factor 1 (SF2), and serine-arginine-rich splicing factor 2 (SC35). hnRNP-H showed the closest association with the foci with almost complete overlap, compared to the distribution of SF2 and SC35 (Figures 3A and 3B). TDP-43 was not mislocalized in any cells with RNA foci. To determine whether these proteins are physically binding to G4C2 RNA, we extracted nuclear lysate from SH-SY5Y cells, incubated it with biotin-labeled G4C2×72 RNA, and performed RNA immunoprecipitation (RIP). Western blots of the eluate were probed for all three proteins, but only hnRNP-H was detected (Figure 3C). To further validate the interaction between G4C2 repeats and hnRNP-H, we performed RIP using rat brain nuclear lysate. hnRNP-H was pulled down from rat brain lysate by G4C2×48 RNA, but not by control RNA of the same length (300 nt), implying that hnRNP-H binds specifically to G4C2 repeats (Figure 3D). Sequestration of hnRNP H by G4C2 RNA foci is expected to reduce the amount of hnRNP-H available and reduce its splicing efficiency on specific RNA targets. hnRNP-H is required for the efficient inclusion of exon 7 into the mature TARBP2 RNA (Xiao et al., 2009). We therefore compared the effect of expanded G4C2 repeats and knockdown of hnRNP-H on TARBP2 exon 7 inclusion. Lentiviral constructs expressing sh-hnRNP-H knocked down hnRNP-H with an efficiency of 95% in SH-SY5Y cells (Figure S3), which dramatically decreased the inclusion of exon 7 into TARBP2 (Figure 3E). A more modest decrease in exon 7 inclusion was also observed in SH-SY5Y cells stably transfected with 72× G4C2 plasmids (Figure 3F). Thus, the presence of G4C2 repeats has a similar effect on the splicing of TARBP2 exon 7 to hnRNP-H knockdown and is consistent with the effect of hnRNP-H sequestration by G4C2 foci.
Figure 3.
SC35, SF2, and hnRNP-H Colocalize with G4C2 Nuclear Foci, but hnRNP-H Binds to G4C2 RNA Transcripts
(A–B) SH-SY5Y cells were transfected with a plasmid expressing 72× repeats and probed 24 hr after transfection for G4C2 by FISH and immunocytochemistry (ICC). (A and B) Endogenous SC35 (A) and SF2 (B) were detected by ICC using a Dylight-488-labeled secondary antibody. hnRNP-H was simultaneously detected by ICC, using a Dylight-649-labeled secondary antibody. The intensity of endogenous SC35, SF2, hnRNP-H, and G4C2 foci were analyzed by Leica line profile tools (scale bar represents 5 μm).
(C) Biotin-labeled G4C2×72 RNA transcripts were synthesized and used for RNA pull-down of SH-SH5Y lysates; G4C2×0 transcripts were used as control. Partial colocalization of SC35 and SF2 is seen with the RNA foci, whereas hnRNP-H shows very close colocalization. Only hnRNP-H coprecipitated with G4C2×72 RNA.
(D) Biotin aptamer-labeled G4C2×48 RNA transcripts were synthesized and used for RNA pull-down of rat brain lysates (Supplemental Experimental Procedures); RNA of equivalent length to G4C2×48 (300 nt) was used as a control. hnRNP-H coprecipitated with G4C2×48 RNA.
(E) Splicing assay of TARBP2 exon 7 shows that G4C2 RNA sequestration of hnRNP-H impairs RNA processing. Comparison of hnRNP H knockdown and control SH-SY5Y cells validates that hnRNP H promotes inclusion of TARBP2 exon 7.
(F) Analysis of TARBP2 exon 7 splicing in SH-SY5Y cells stably transfected with C9ORF72 repeats. Inclusion of TARBP2 exon 7 is decreased in cells expressing G4C2 ×72 but is not affected in cells expressing lower repeat numbers. PCR products including (in) or excluding (ex) the regulated alternative exon are marked on the right. Average quantification values of exon inclusion (yellow) and exclusion (blue) are shown. Error bars show SD of three replicates. To test significance, the ratio between exon 7 inclusion and exclusion was calculated and tested by one-way ANOVA and Tukey’s honestly significant difference test.
See also Figure S3.
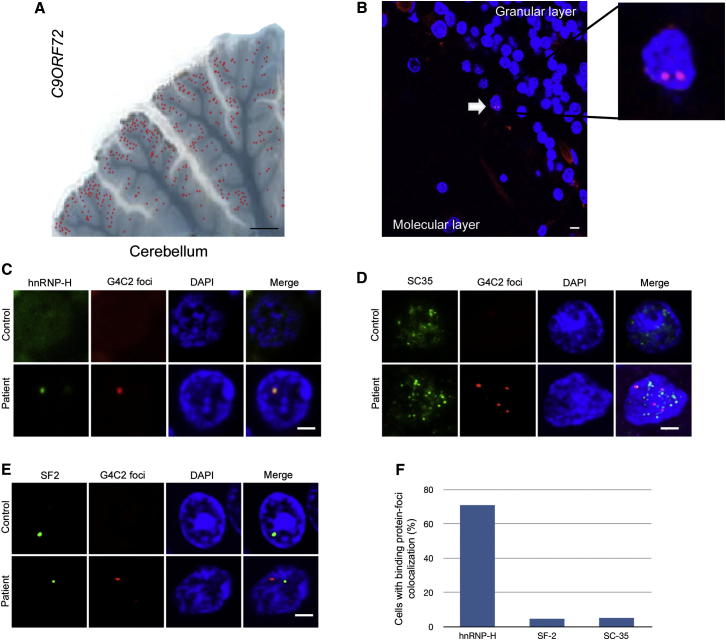
Having determined that three RNA binding proteins colocalize with G4C2 RNA foci in cells, we sought a similar association in human tissues. We optimized our FISH protocol (adapted from DeJesus-Hernandez et al. [2011]) and observed abundant RNA foci in the cerebellum and less abundant foci in the frontal cortex and temporal lobes of ALS and FTD patients carrying the C9ORF72 expansion mutation (confirmed by repeat primed PCR and Southern blot, n = 5) that are absent from controls (n = 5) (Figure S4; Table S3). Discrete intranuclear neuronal RNA foci were larger in the cerebellum (∼500 nm) compared to the cortex (∼200 nm) and were most frequent in neurons adjacent to Purkinje cells (Figures 4A and 4B). By combining FISH for RNA foci and immunofluoresence for the foci-associated RNA binding proteins identified in transfected cells, we demonstrated that SF2 and SC35 rarely colocalized with G4C2 RNA in cerebellum, being present in <5% of foci, whereas hnRNP-H showed striking colocalization overlapping almost with ∼70% of foci (Figures 4C–4F). We have therefore confirmed the presence of RNA foci in ALS and FTD patients with pathological G4C2 intronic expansions in C9ORF72 and demonstrated that they bind RNA-binding proteins, being most closely associated with hnRNP-H.
Figure 4.
Intranuclear Neuronal RNA Foci in C9ORF72 Mutant ALS and FTD Brain Tissues Colocalize Very Closely with hnRNP-H
(A) Image of mutant C9ORF72 patient cerebellum overlaid with the location of neurons containing G4C2 RNA foci (red dots) (scale bar represents 4 mm).
(B) G4C2 RNA foci-positive neurons (white arrow) were observed between the granular and molecular layer of the cerebellum (scale bar represents 10 μm).
(C–E) FISH and ICC were performed for hnRNP-H (C), SC35 (D), and SF2 (E), with a G4C2 mutation-negative ALS case used as control (scale bar represents 3 μm).
(F) The percentage of foci that colocalized with hnRNP-H, SF2, and SC-35 were counted (n = 50 cells). Of the three RNA binding proteins that colocalized with foci in transfected cells, only hnRNP-H shows a striking degree of overlap for 70% of all foci in the cerebellum.
See also Figure S4.
Discussion
The G4C2 expansion mutation in C9ORF72 is the most common gene defect associated with ALS and FTD in the European population. Several mechanistic hypotheses are emerging, including the loss of the C9ORF72 encoded protein, RNA-mediated toxicity (DeJesus-Hernandez et al., 2011; Renton et al., 2011), and toxic dipeptides from repeat-associated non-ATG (RAN) translation (Mori et al., 2013b). Our results provide experimental evidence that RNA containing longer G4C2 repeats causes cellular toxicity in a length-dependent manner in transfected cells and in vivo and may contribute to neurodegeneration in C9ORF72-related ALS and FTD.
Cellular toxicity was associated with the nuclear retention of transcripts containing 38× and 72× repeats and the appearance of RNA foci. By placing the repeats 3′ to EGFP we were able to monitor nuclear export and show that transfected neuroblastoma cells, but not HEK cells, declined in number over time because of apoptotic cell death. This was most marked in those showing the greatest nuclear retention and foci, indicating cell type and neuron-specific toxicity of longer G4C2 repeats. These data suggest that similar to trinucleotide diseases (Hirth, 2010), cellular toxicity of hexanucleotide repeat expansion directly correlates with G4C2 repeat length, indicating that the longer the repeat, the more toxic its nature.
In order to explore the sequestration hypothesis, we screened antibodies to 30 RNA binding proteins in G4C2 72×-transfected neuroblastoma cells and demonstrated colocalization of G4C2 RNA foci with SF2, SC35, and, most strikingly, hnRNP-H. Overexpression of 72× G4C2 and knockdown of hnRNP-H had a similar effect on the splicing of a target RNA TARBP2, consistent with the sequestration of hnRNP-H. Using biotinylated 72× RNA, we were only able to pull down hnRNP-H, indicating that is able to interact with G4C2 RNA. On probing human cerebellar ALS and FTD tissues, we detected a striking colocalization of hnRNP-H with 70% of RNA foci. We did not detect changes in the distribution or colocalization with RNA foci in human tissues for pur-α (Xu et al., 2013) or hnRNP-A3 (Mori et al., 2013a) to suggest sequestration, as had previously been postulated. Our findings suggest that the sequestration of hnRNP-H and other RNA binding proteins may play a mechanistic role in neurodegeneration associated with the C9ORF72 mutation.
Our observation that longer stretches of G4C2 RNA form neurotoxic foci and bind specific RNA binding proteins is similar to other intronic microsatellite expansion disorders (Todd and Paulson, 2010). Expanded CUG (DM1) and CCUG (DM2) intronic repeats generate RNA foci in cells, in animal models, and in patients with myotonic dystrophy. The foci bind and deplete muscle-blind-like proteins (MBNL1, 2, and 3), causing widespread RNA splicing abnormalities and degeneration of affected tissues (Miller et al., 2000). The fact that MBNL1 knockout mice develop key aspects of human DM and that overexpression of MBNL1 in Drosophila and mice can rescue the phenotype caused by the CTG repeats implies that loss of MBNL1 is one of the key drivers of cellular degeneration (Kanadia et al., 2003b).
hnRNP-H binds strongly to G-runs in intronic sequences to enhance exon skipping (Xiao et al., 2009) and G-rich RNA quadruplexes (Millevoi et al., 2012). hnRNP-H binding is known to inhibit the nuclear export of RNA containing expanded CUG repeats associated with myotonic dystrophy type 1 (DM1) (Kim et al., 2005). It is a component of the splicing enhancer complex that activates alternative splicing of c-src in neurons (Chou et al., 1999). Thus, the nuclear retention and aggregation of G4C2 RNA may be enhanced by hnRNP-H binding, which could in turn facilitate further G4C2 RNA binding in a positive feedback loop to generate large RNase-resistant foci. This would result in the sequestration of hnRNP-H itself, other RNA binding proteins, and multiple RNA transcripts, leading to significant dysregulation of RNA processing and toxicity. We did not observe TDP-43 binding to G4C2 repeats or mislocalization in our cellular models as has been described in C9ORF72 mutant ALS and FTD cases, and it is not yet clear whether TDP-43 misaccumulation is mechanistic in these cases. The observation that RAN translation generates dipeptides in C9ORF72 mutant cases is of great interest (Mori et al., 2013b). Recent evidence has emerged that RAN polyglycine peptides are present in cellular and Drosophila models of fragile X tremor ataxia syndrome and in patients carrying the CGG expansion and that cell death could be prevented by the suppression of RAN translation (Todd et al., 2013). Toxicity due to RNA foci and RAN translation are not mutually exclusive, and their relative contribution to repeat-associated neurodegenerative disorders requires further investigation.
Experimental Procedures
Detailed Experimental Procedures are described in the Supplemental Experimental Procedures.
Patient Samples
All cases were provided by the Medical Research Council (MRC) London Neurodegenerative Diseases Brain Bank (Institute of Psychiatry, King’s College London) and were collected and distributed in accordance with local and national research ethics committee approvals. See Table S3 for case details.
FISH with Immunofluoroscence
Cy3-labeled G2C4 ×8 RNA probes were synthesized by Integrated DNA Technologies and used as antisense probes. Cells were fixed in 4% PFA for 15 min and washed with PBS. Then cells were incubated with 70% ethanol overnight (16 hr) at 4°C. The next day, the cells were rehydrated in PBS for 15 min and permeabilized with 0.1% Triton X-100 for 5 min. Following this, the coverslips were incubated in prehybridization solution (40% formamide, 2×SSC) for 15 min. Probes were used at working solutions of 5 ng/μl and diluted in hybridization buffer (40% formamide, 1 mg/ml tRNA, 10% dextran sulfate, 2×SSC). Following overnight hybridization at 48°C, coverslips were washed three times in 2X SSC at room temperature for 15 min each. These were then washed with PBS three times for 15 min each. Coverslips were treated with RNase A/T1 mix (Fermentas, 1:50,000) for 5 min at room temperature and washed with PBS three times for 15 min each. For double immunostaining with FISH, coverslips were blocked in 5% donkey serum for 1 hr, and then cells were incubated with primary antibodies (Table S2), and the appropriate secondary antibodies were used. DAPI (Sigma) was used for counterstaining. Leica confocal SP systems were used for high-resolution imaging.
FISH for Human Tissue
Sections from frontal and temporal cortices and cerebellum were provided as 10% formalin fixed and paraffin-embedded blocks. To perform FISH, paraffin was removed with xylene, and sections were rehydrated in an ethanol series (100%, 95%, 70%) for 3 min per step. These slides were incubated in 0.3% Sudan black for 5 min and washed with water. Next, slides were treated with proteinase-K (20 μg/ml, TBS [pH 7.4]) in a 37°C water bath for 20 min. Next, slides were treated with ice-cold 20% acetic acid in 1× TBS for 2 min and then incubated in prehybridization buffer (40% formamide, 2×SSC) for a further 15 min. Probes were used at working solutions of 5 ng/μl and diluted in hybridization buffer (40× formamide, 1 mg/ml tRNA, 10% dextran sulfate, 2×SSC). Following overnight hybridization at 48°C, slides were washed three times in 2X SSC at room temperature for 15 min each. These were then washed with PBS three times for 15 min each.
Statistics
Statistical analysis was carried out using GraphPad Prism 5. For one-way ANOVA, Bonferroni post hoc tests were used to compare the abundance of TUNEL-stained cells in zebrafish. Data are presented as mean ± SEM. Statistical significance was considered at p < 0.0001 (∗∗∗).
Acknowledgments
This project was funded by a Strategic Grant Award from The Wellcome Trust and Medical Research Council (MRC) (089701), the Motor Neuron Disease Association, the American ALS Association, Heaton-Ellis Trust, an MRC grant (G0900688), the National Institutes of Health Research Biomedical Research Centre for Mental Health at the South London and Maudsley National Health Service Foundation Trust, the Psychiatry Research Trust, and the European Community’s Seventh Framework Programme (FP7/2007-2013) under the grant agreement number 259867. J.A. is supported by the Boehringer Ingelheim Fonds.
Published: November 27, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes Supplemental Experimental Procedures, four figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.10.049.
Supplemental Information
References
- Al-Sarraj S., King A., Troakes C., Smith B., Maekawa S., Bodi I., Rogelj B., Al-Chalabi A., Hortobágyi T., Shaw C.E. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- Chou M.Y., Rooke N., Turck C.W., Black D.L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P., Mizielinska S., Nicoll A.J., Zloh M., Fisher E.M., Parkinson G., Isaacs A.M. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth F. Drosophila melanogaster in the study of human neurodegeneration. CNS Neurol. Disord. Drug Targets. 2010;9:504–523. doi: 10.2174/187152710791556104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia R.N., Johnstone K.A., Mankodi A., Lungu C., Thornton C.A., Esson D., Timmers A.M., Hauswirth W.W., Swanson M.S. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia R.N., Urbinati C.R., Crusselle V.J., Luo D., Lee Y.J., Harrison J.K., Oh S.P., Swanson M.S. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr. Patterns. 2003;3:459–462. doi: 10.1016/s1567-133x(03)00064-4. [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Langlois M.-A., Lee K.-B., Riggs A.D., Puymirat J., Rossi J.J. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33:3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S., Moine H., Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA. 2012;3:495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- Mori K., Lammich S., Mackenzie I.R., Forné I., Zilow S., Kretzschmar H., Edbauer D., Janssens J., Kleinberger G., Cruts M. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125:413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- Mori K., Weng S.M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A., Cruts M., Van Broeckhoven C. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatunov A., Mok K., Newhouse S., Weale M.E., Smith B., Vance C., Johnson L., Veldink J.H., van Es M.A., van den Berg L.H. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9:986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B.N., Newhouse S., Shatunov A., Vance C., Topp S., Johnson L., Miller J., Lee Y., Troakes C., Scott K.M. The C9ORF72 expansion mutation is a common cause of ALS+/–FTD in Europe and has a single founder. Eur. J. Hum. Genet. 2012;21:102–108. doi: 10.1038/ejhg.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd P.K., Paulson H.L. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd P.K., Oh S.Y., Krans A., He F., Sellier C., Frazer M., Renoux A.J., Chen K.C., Scaglione K.M., Basrur V. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C., Al-Chalabi A., Ruddy D., Smith B.N., Hu X., Sreedharan J., Siddique T., Schelhaas H.J., Kusters B., Troost D. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129:868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Wang Z., Jang M., Nutiu R., Wang E.T., Burge C.B. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat. Struct. Mol. Biol. 2009;16:1094–1100. doi: 10.1038/nsmb.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Poidevin M., Li X., Li Y., Shu L., Nelson D.L., Li H., Hales C.M., Gearing M., Wingo T.S. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl. Acad. Sci. USA. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.