Abstract
Glioblastoma multiforme (GBM) is an aggressive brain malignancy characterized by high heterogeneity and invasiveness. It is increasingly accepted that the refractory feature of GBM to current therapies stems from the existence of few tumorigenic cells that sustain tumor growth and spreading, the so-called glioma-initiating cells (GICs). Previous studies showed that cytokines of the bone morphogenetic protein (BMP) family induce differentiation of the GICs, and thus act as tumor suppressors. Molecular pathways that explain this behavior of BMP cytokines remain largely elusive. Here, we show that BMP signaling induces Smad-dependent expression of the transcriptional regulator Snail in a rapid and sustained manner. Consistent with its already established promigratory function in other cell types, we report that Snail silencing decreases GBM cell migration. Consequently, overexpression of Snail increases GBM invasiveness in a mouse xenograft model. Surprisingly, we found that Snail depletes the GBM capacity to form gliomaspheres in vitro and to grow tumors in vivo, both of which are important features shared by GICs. Thus Snail, acting downstream of BMP signaling, dissociates the invasive capacity of GBM cells from their tumorigenic potential.
Keywords: BMP, glioblastoma multiforme, glioma-initiating cells, invasiveness, snail
Introduction
The aggressive brain tumor glioblastoma multiforme (GBM) accounts for about 50% of all glial tumors in adults, with a median survival of 12–15 months.1, 2 GBMs are heterogeneous, invasive and vascularized with a strong hypoxic/necrotic component, making them refractory to radio- and chemotherapy.3 Besides genetic alterations that contribute to the aggressiveness of GBM,4, 5 it is evident that not all tumor cells are equally tumorigenic. Proliferative and undifferentiated tumor cell subpopulations, described as glioma-initiating cells (GICs), can contribute to cancer initiation, propagation and relapse after therapy.2, 3 Similar to adult stem cells that sustain tissue homeostasis, GICs can sustain aberrant tumor growth.6 A correlation found between tumor subtypes and stage of differentiation suggested that glioblastoma aggressiveness might be governed by processes that regulate adult brain-stem cell fate.7 Cytokines are critical regulators of adult brain-stem cell, and GIC differentiation and renewal.8
Transforming growth factor β (TGFβ) and its related bone morphogenetic proteins (BMPs) signal via serine/threonine kinase receptors, intracellular Smad proteins and kinase effector pathways that regulate gene expression.9 TGFβ helps maintaining human GICs by inducing expression of platelet-derived growth factor B, leukemia inhibitory factor (LIF) and the transcription factor Sox4 that contribute to GIC proliferation and survival.10, 11, 12 TGFβ type I receptor kinase inhibitors13 can deplete a pool of primary human GICs,14 providing a promising example of TGFβ antagonists in cancer therapy.15, 16, 17
On the other hand, BMP4 can induce astrocytic differentiation of primary human GICs, thus depleting their tumorigenicity and acting therapeutically.18 Along the same lines, a group of primary human GICs misexpresses the Polycomb regulator, EZH2, which silences epigenetically one of the BMP type I receptors, causing enhanced tumorigenicity as BMP-induced differentiation is bypassed.19 The molecular repertoire controlled by BMP signaling in GICs remains elusive.
In this study, we screened for BMP-regulated genes in GBM and focussed on the transcription factor Snail. Snail is a pro-invasive factor in carcinomas and promotes breast cancer stem cell survival.20 Consistently, we show that Snail is required for GBM cell migration and invasion, as tested by wound healing and xenograft assays, respectively. However, Snail decreases gliomasphere formation and suppresses tumor formation, suggesting a dual function of Snail in GBM that dissociates invasiveness from tumorigenic potential.
Results
Characterization of human GBM cell models
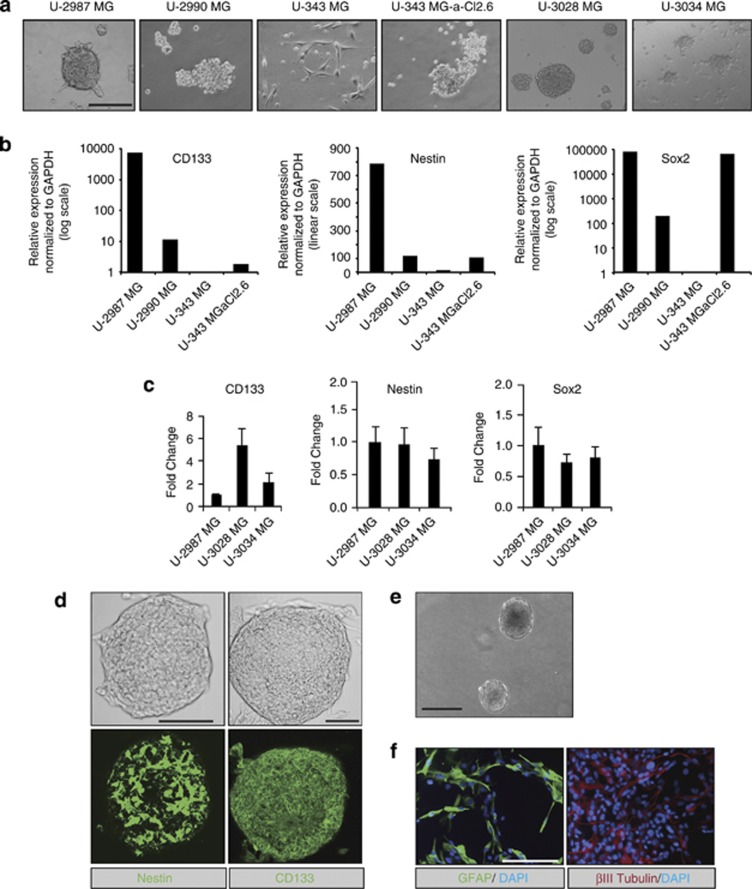
GICs are found in some GBM cell lines derived from human patients.21 To investigate human GIC responses to TGFβ and BMP, we analyzed in vitro growth and gliomasphere formation in the established cell lines (U-2987 MG, U-2990 MG, U-343 MG and U-343 MGa-Cl2.6)22 and in newly isolated patient-derived stem cell lines (U-3028 MG and U-3034 MG) explanted from glioblastomas. We found that U-2987/3028/3034 MG cells readily formed gliomaspheres, whereas the other cell lines formed unordered aggregates (U-2990/343 MG-a-Cl2.6) or sparse adherent cells (U-343 MG; Figure 1a).
Figure 1.
In vitro stem cell properties of glioblastoma cell models. (a) Primary gliomaspheres of glioblastoma cell lines grown in stem cell media (bar: 100 μm). (b, c) Real-time qRT–PCR analysis of stem cell marker expression in the glioblastoma cells normalized to the housekeeping GAPDH. Average values from triplicates with s.d.'s are plotted in this and all subsequent qRT–PCR assays; in (b), values are normalized to U-343 MG cells, whose expression is set to 1; in (c) values are normalized to U-2987 MG cells. (d) Confocal microscopy on primary U-2987 MG gliomaspheres showing phase contrast images (top) and Nestin or CD133 immunofluorescence (bottom; bar: 50 μm). (e) Phase contrast image of U-2987 MG secondary gliomaspheres (bar: 100 μm). (f) GFAP, βIII tubulin and DAPI staining of glioblastoma cells after dissociation and differentiation from individual secondary U-2987 MG gliomaspheres (bar: 100 μm).
mRNA analysis of established GIC markers showed that U-2987/3028/3034 MG cells expressed significantly high levels of the cell surface marker CD133, the transcription factor Sox2 and the intermediate filament protein Nestin, whereas the other cell lines expressed undetectable or variable levels of the markers (Figures 1b and c). Nestin and CD133 protein expression was confirmed in U-2987 MG primary gliomaspheres (Figure 1d).
The self-renewal capacity of U-2987 MG cells was verified after dissociation of primary gliomaspheres and low-density replating in stem cell media, forming secondary gliomaspheres (Figure 1e). Immunostaining for GFAP and βIII tubulin confirmed the expression of both glial and neuronal markers, respectively, which verified multipotency (Figure 1f). Thus, U-2987 MG cells fulfilled the expected features of GICs in vitro, and were used to identify new downstream targets of TGFβ and BMP.
Loss of U-2987 MG GIC potential by BMP7
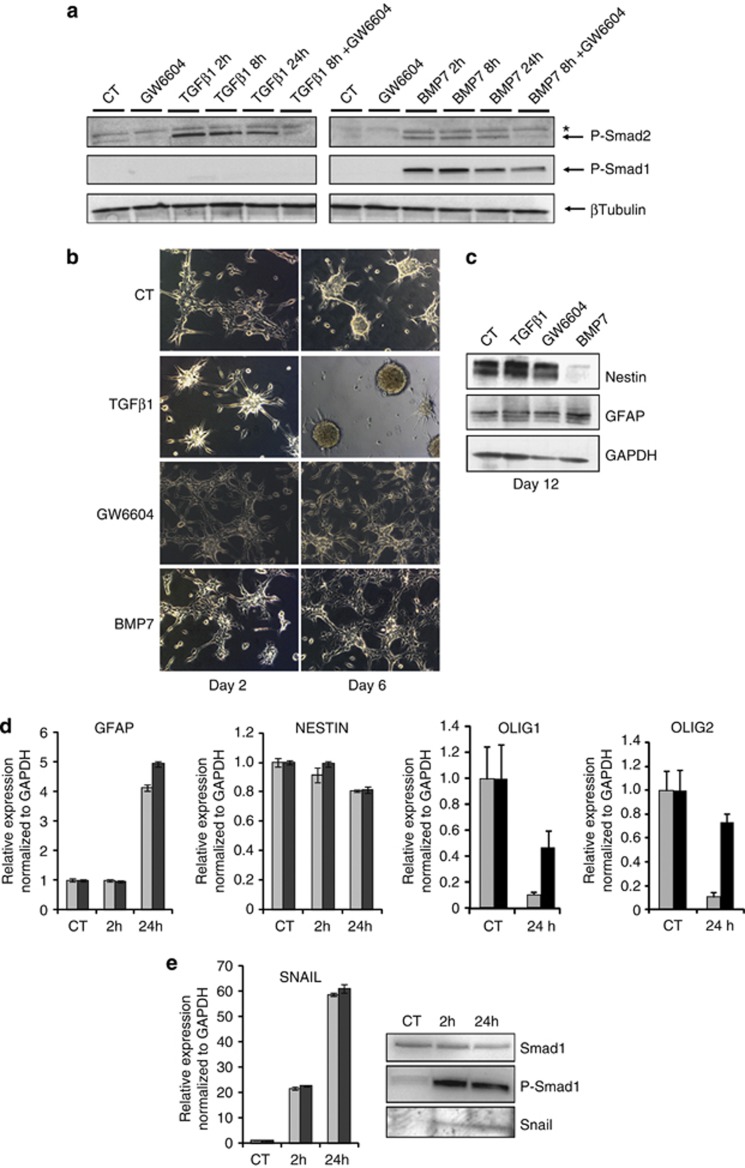
TGFβ1 promotes GIC maintenance and tumorigenic potential, whereas BMP4 induces GIC differentiation and loss of tumorigenicity.10, 11, 12, 18 We used BMP7 instead of BMP4, as BMP7 and its receptors have been involved in GBM development.23 Analysis of Smad1 and Smad2 C-terminal phosphorylation showed that TGFβ1 induced P-Smad2, but not P-Smad1. As a negative control, the TGFβ type I receptor kinase inhibitor, GW6604,24 blocked basal and TGFβ1-inducible P-Smad2 (Figure 2a, left panel). BMP7 induced P-Smad1; unexpectedly, GW6604 weakly blocked this response (Figure 2a, right panels), whereas BMP7 also induced P-Smad2, albeit much weaker than TGFβ1, and the latter was fully blocked by GW6604 (Figure 2a, right panels). Thus, TGFβ1 and BMP7 elicit their respective Smad pathways in U-2987 MG cells. Moreover, BMP7 may induce secretion of TGFβ-like ligands that activate P-Smad2 or, alternatively, BMP7 may signal via heteromeric type I receptors that activate both Smad1 and Smad2.
Figure 2.
BMP7 inhibits and TGFβ1 promotes gliomasphere formation. (a) Phospho-Smad2 and phospho-Smad1 levels in U-2987 MG cells treated with TGFβ1 or BMP7 for 2, 8 or 24 h in the presence or absence of GW6604. Protein loading controls: β tubulin; asterisk marks a non-specific protein. (b) Primary U-2987 MG gliomasphere formation upon TGFβ1, GW6604 or BMP7 treatment after 2 and 6 days in stem cell medium. (c) Western-blot of Nestin, GFAP and GAPDH expression in 12 day-old primary gliomaspheres treated with TGFβ1, GW6604 or BMP7. (d) qRT–PCR of GFAP, Nestin, Olig1 and Olig2 mRNA in U-2987 MG cells treated for 2 or 24 h with BMP7. (e) qRT–PCR of Snail mRNA in U-2987 MG cells treated as indicated; western-blot of total Smad1, P-Smad1 and Snail proteins. Control, unstimulated cells are marked as CT and two independent samples are analyzed (light and darker bars).
Primary and secondary gliomasphere assays showed that TGFβ1 induced, whereas GW6604 or BMP7 suppressed the growth of gliomaspheres (Figure 2b). The effects of GW6604 on P-Smad2 levels and gliomaspheres suggested that autocrine TGFβ helped in gliomasphere formation (Figures 2a and b). The positive effect that TGFβ1 had on gliomasphere formation could not result from increased cell proliferation, as these tumor cells, as expected, did not exhibit any robust growth responses to TGFβ1 treatment (Supplementary Figure S1a). Quantification of primary and secondary gliomaspheres confirmed the positive effect of TGFβ1 and the inhibitory effect of the GW6604 inhibitor (Supplementary Figure S1b). Stem-cell marker analysis showed that TGFβ1 induced LIF and the stem cell transcription factor Nanog, and downregulated the transcription factors Olig1 and Olig2 (Supplementary Figure S1c). Upon BMP7 treatment, cells acquired an adherent phenotype (Figure 2b) and expressed high levels of GFAP, a marker of astrocytic differentiation (Figure 2c).
To better understand the effects of TGFβ1 and BMP7 on U-2987 MG gliomasphere formation, we measured Nestin expression, which was weakly downregulated at the mRNA and more potently at the protein level by BMP7 (Figures 2c and d). We also confirmed that long-term stimulation with BMP7 induced GFAP and downregulated Olig1 and Olig2 mRNAs (Figure 2d). Olig1/2 are basic helix-loop-helix transcription factors expressed in neural progenitor cells (NPCs) that promote oligodendrocyte differentiation and potentiate GBM initiation in xenograft experiments.25, 26, 27, 28 We conclude that in U-2987 MG cells, TGFβ1 and BMP7 produce phenotypes similar to those recently established in independent GICs.
BMP7 signals via Smads to induce Snail expression in glioblastoma cells
In order to find new regulators involved in GIC responses to BMP7, we analyzed BMP7-regulated genes using Affymetrix transcriptomic profiling in U-2987 MG cells (data not shown). Human NPCs were also analyzed in order to compare GICs to normal stem cells. The details of the transcriptomic profiles will be published elsewhere.
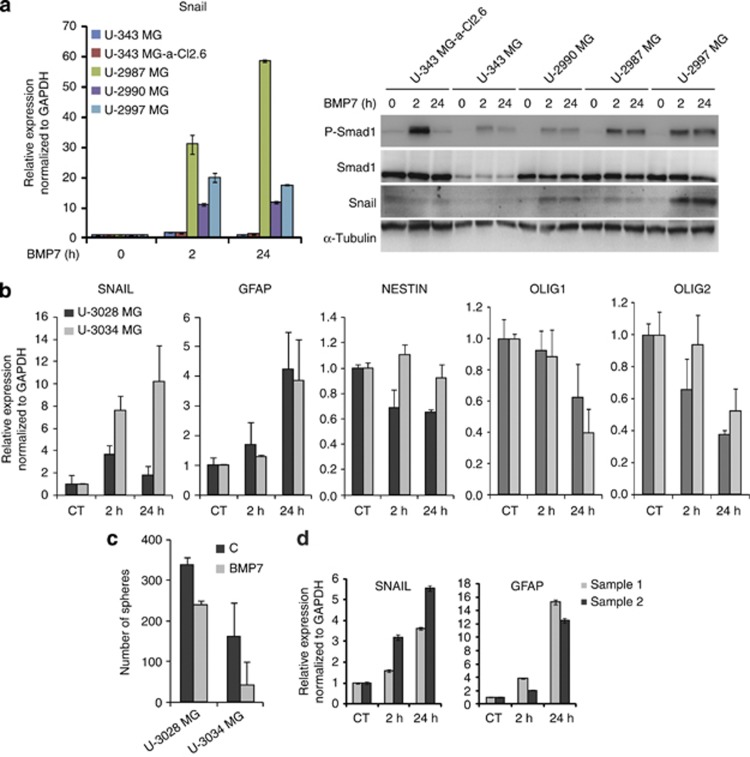
Among the BMP7-regulated genes that showed early (2 h) and sustained (24 h) response in both U-2987 MG and human NPCs was the transcription factor Snail that is an established regulator of epithelial–mesenchymal transition.29 Quantitative (q) RT-PCR and immunoblot analysis confirmed that BMP7 induced Snail mRNA and protein in U-2987 MG cells (Figure 2e). Furthermore, BMP7 induced Snail in several independent GBM lines (Figure 3a), which was verified in additional patient-derived GBMs (U-3028/3034 MG), albeit with different kinetic profiles in each cell line (Figures 3a and b). Snail upregulation correlated with GFAP upregulation and Olig1/2 downregulation, whereas a weak but significant downregulation of Nestin was measured in U-3028 MG cells (Figure 3b). As expected from the transcriptional profile and similar to U-2987 MG cells (Figure 2b), BMP7 decreased the gliomasphere-forming capacity of U-3028/3034 MG cells (Figure 3c). Gliomasphere and GIC marker screens in additional patient-derived GBM cell lines revealed an interesting pair, U-3013 MG and U-3024 MG cells (Supplementary Figures S2a and b). BMP7 failed to suppress gliomasphere formation and growth in these cells and concomitantly failed to induce Snail expression (Supplementary Figures S2c and d). It should be noted that U-3013/3024 MG cells showed robust P-Smad1 induction and other established BMP7 responses (data not shown). Finally, BMP7 induced Snail and GFAP expression in human NPCs (Figure 3d). These experiments validate the identification of Snail as a new target of BMP7 signaling in normal neural progenitors and tumor-derived GBM cells.
Figure 3.
BMP7 induces Snail and astrocytic differentiation. (a) Treatment with BMP7 for 2 and 24 h in several GBM cell lines; qRT–PCR analysis of Snail mRNA (left panel), and western blot analysis (right panel) of Snail, P-Smad1, Smad1 and α tubulin, which serves as loading control. (b) qRT–PCR analysis of Snail, GFAP, Nestin, Olig1 and Olig2 mRNA in U-3028 MG and U-3034 MG cells treated with BMP7 as indicated, and presented as average of two independent experiments. (c) Primary gliomasphere formation (>50 μm) upon BMP7 treatment of U-3028 MG and U-3034 MG cells for 6 days in stem cell medium. (d) qRT–PCR of Snail and GFAP mRNA in two independent cultures (samples) of human NPCs treated with BMP7 for 2 and 24 h, respectively.
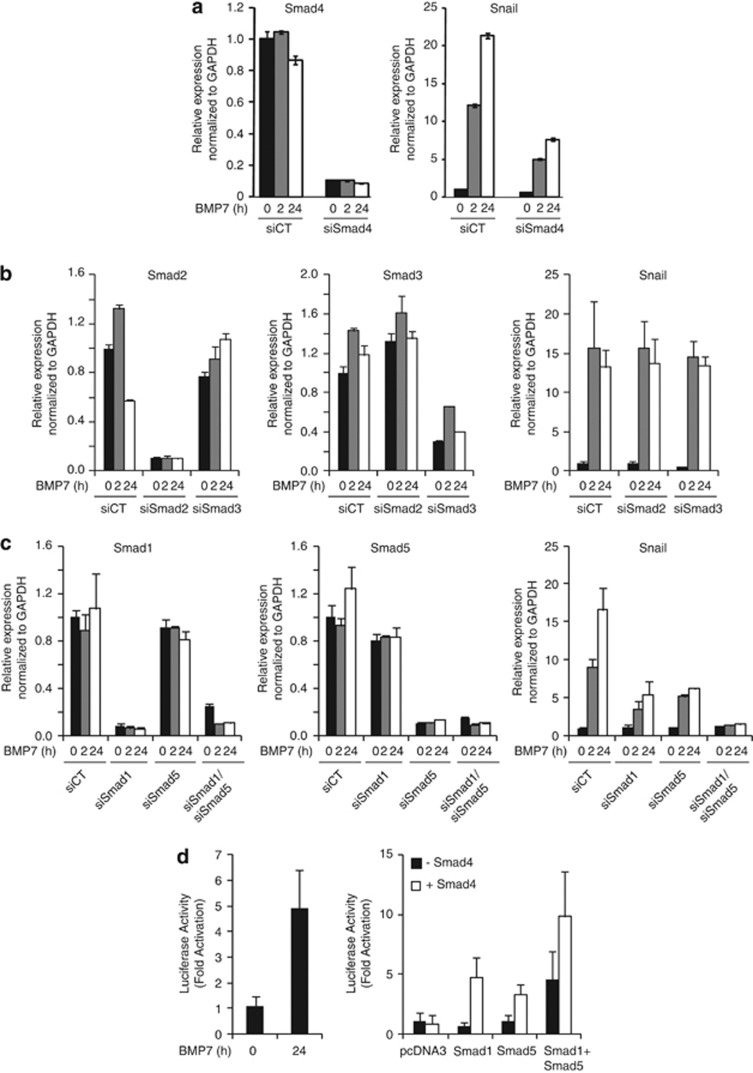
As BMP7 induced the phosphorylation of Smad1 and Smad2 (Figure 2a), we asked whether BMP7 upregulated Snail by activating TGFβ receptors. Two different chemical inhibitors of the TGFβ receptor type I kinase could not affect Snail upregulation by BMP7, whereas blocking the BMP receptor type I kinase by DMH1, a dorsomorphin derivative, completely abrogated this effect (Supplementary Figure S3). Furthermore, silencing Smad4, the common transducer of TGFβ/BMP signaling, strongly impaired BMP7-induced Snail expression (Figure 4a). Knockdown of the TGFβ receptor-activated Smads, Smad2 and Smad3 had no effect on BMP7-induced Snail expression (Figure 4b). In contrast, silencing the BMP receptor-activated Smads, Smad1 or Smad5 impaired, and their double knockdown completely abrogated Snail upregulation by BMP7 (Figure 4c). Endogenous Snail induction was corroborated by recombinant human Snail promoter studies.30 BMP7 induced Snail promoter activity; moreover, transfection of Smad1 or Smad5 further activated the Snail promoter in human hepatocarcinoma HepG2 cells, which was further enhanced by Smad4 co-transfection (Figure 4d). When Smad1, Smad5 and Smad4 were co-transfected, mimicking BMP7 stimulation, it resulted in the most efficient promoter activation (Figure 4d). We conclude that Smad1/Smad5/Smad4 transcriptionally induce Snail downstream of BMP receptors in tumor cells.
Figure 4.
Regulation of BMP7-induced Snail expression by Smads. (a–c) U-2987 MG cells were transiently transfected with different siRNAs and treated or not with BMP7 (2 and 24 h). (a) siControl or siSmad4: qRT–PCR analysis of Smad4 and Snail mRNA. (b) siControl, siSmad2 or siSmad3: qRT–PCR analysis of Smad2, Smad3 and Snail mRNA. (c) siControl, siSmad1 and/or siSmad5; qRT–PCR analysis of Smad1, Smad5 and Snail mRNA. (d) Luciferase reporter assay of a human Snail promoter construct in HepG2 cells stimulated with 30 ng/ml BMP7 for 24 h (left panel) or transiently transfected with pcDNA3, Flag-Smad1, Flag-Smad5 and/or Flag-Smad4 plasmids (right panel). Each bar represents mean±s.d. values of normalized luciferase data from triplicate samples.
Snail contributes to GFAP induction by BMP7
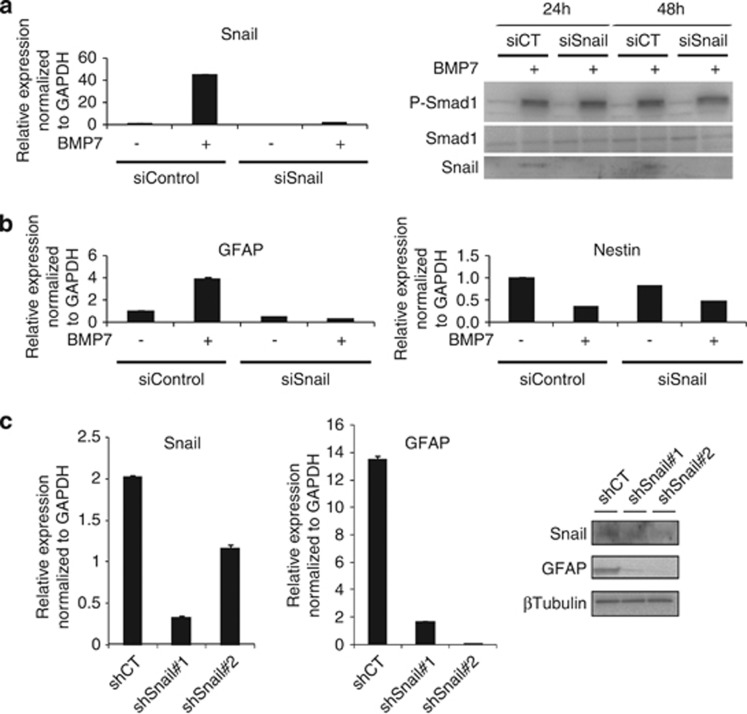
We then asked whether BMP-induced Snail regulated the GBM markers GFAP and Nestin in U-2987 MG cells. Transient knockdown of endogenous Snail did not perturb P-Smad1 levels, but inhibited the induction of GFAP by BMP7, without perturbing the downregulation of Nestin (Figures 5a and b). In two independent stable clones expressing short hairpin RNA (shRNA) targeting Snail in U-2987 MG cells (Figure 5c), the low basal Snail mRNA and protein levels were potently silenced, causing dramatic reduction of endogenous GFAP levels (Figure 5c). Thus, Snail upregulation is required for BMP7-induced GFAP expression.
Figure 5.
BMP7 effects are caused in part by Snail. (a, b) U-2987 MG cells were transiently transfected with siControl or siSnail and treated or not with BMP7 for 24 h. (a) qRT–PCR of Snail mRNA (left panel) and western-blot of P-Smad1, total Smad1 and Snail upon 24 and 48 h of BMP7 treatment (right panel). Smad1 serves as a loading control. (b) qRT–PCR of GFAP and Nestin mRNA. (c) U-2987 MG stably transfected with pSGN-shControl (shCT) or pSGN-shSnail (shSnail no. 1 and shSnail no. 2). qRT–PCR of Snail and GFAP mRNA (left panels), and western blot of Snail and GFAP (right panel). β tubulin serves as loading control.
Snail has an impact on tumor formation and invasiveness in vivo
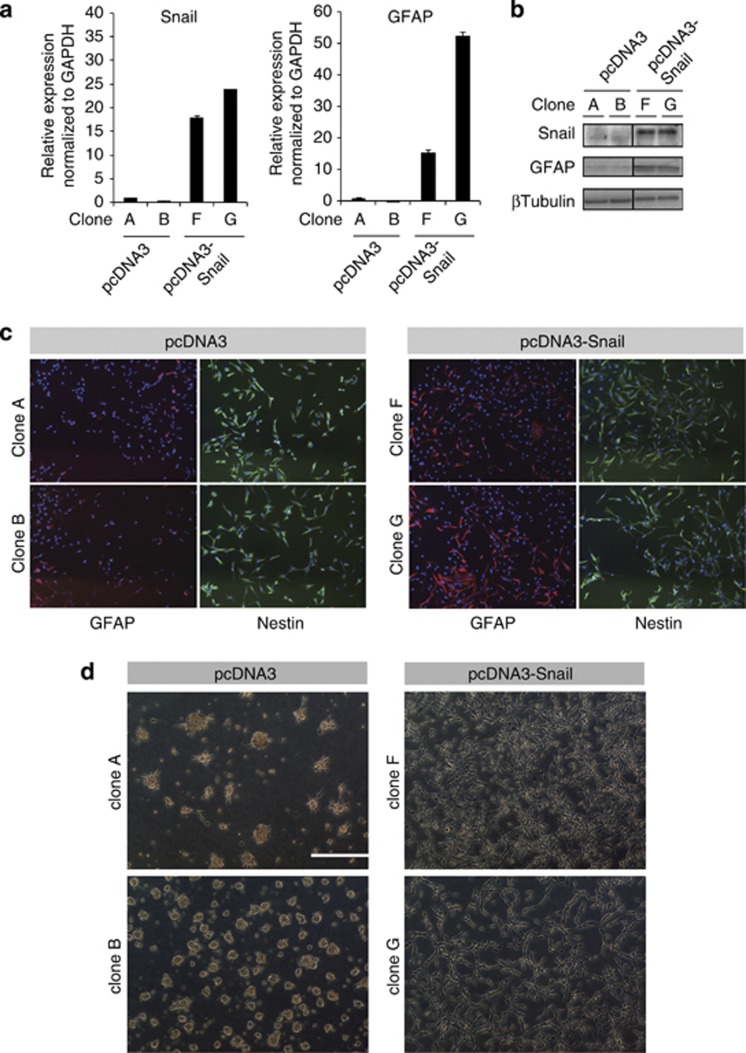
As U-2987 MG cells express low basal Snail levels, we stably expressed Snail (clones F and G) or a mock vector (clones A and B) in individual clones (Supplementary Figures S4a and b). Snail overexpression did not affect cell proliferation in serum-containing- or stem cell media (Supplementary Figures S4c and d).
Orthotopic injection of mock clones in the brain of immunocompromised mice generated tumors (Figure 6). Mock clones injected either as adherent cells or after dissociation from gliomaspheres led to similar tumor growth (Table 1). However, tumors generated from gliomaspheres were more invasive, independent of the expression level of Snail. Previous studies suggested that Snail favors cell survival.31, 32 Despite that, Snail-transfected cells induced tumor formation in 40% of the injected mice compared with 94% of the mice induced by mock clones (Table 1). In addition, mice bearing Snail-overexpressing tumors survived longer and lacked signs of illness compared with mice bearing control tumors, which developed illness and were sacrificed earlier (P<0.001; Table 1).
Figure 6.
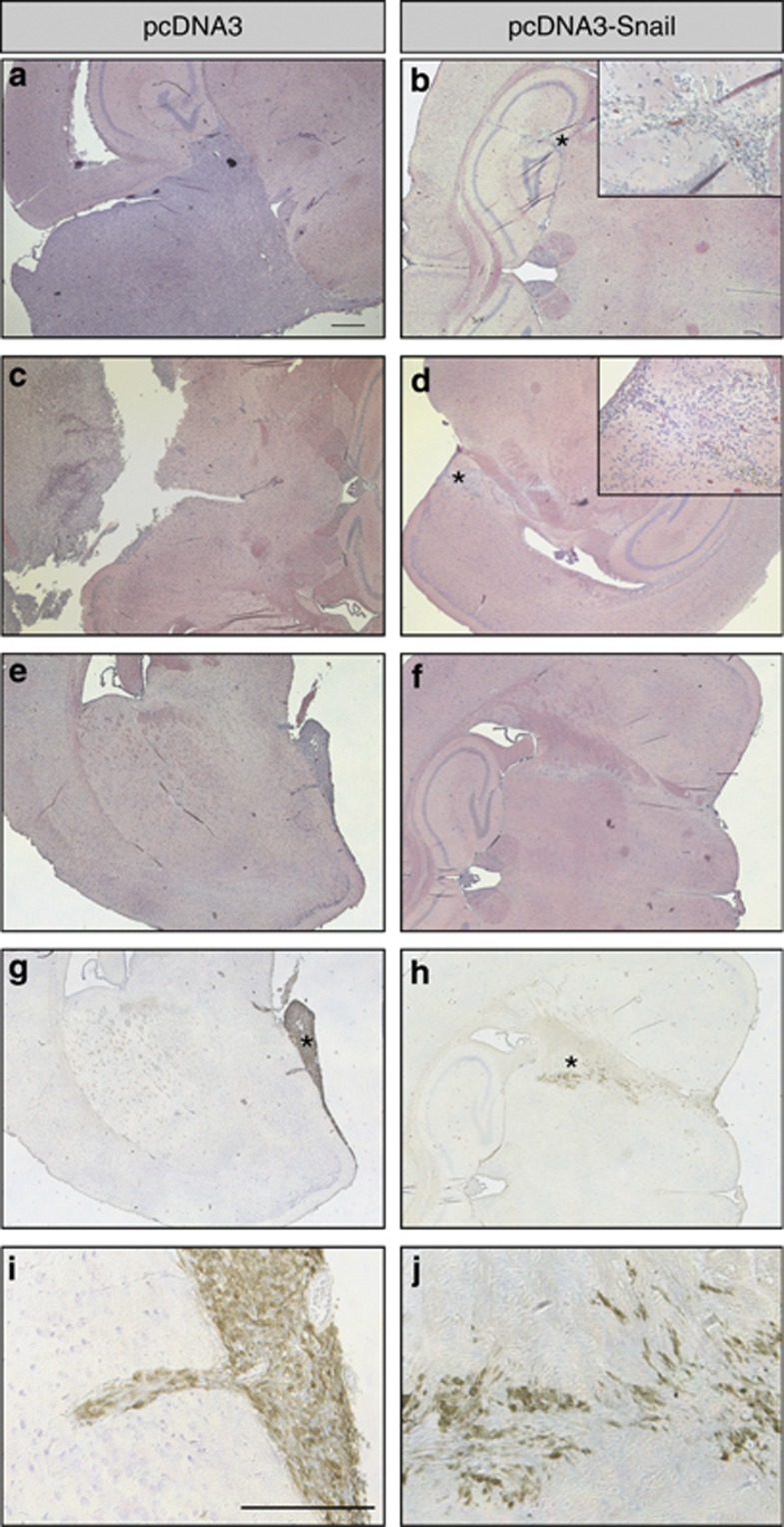
Snail decreases U-2987 MG tumorigenicity in vivo. Tumors derived after injecting adherent U-2987 MG cells (a, b) or cells dissociated from gliomaspheres (c–j). (a–f) Hematoxylin-eosin staining; (g–j) immunostaining with anti-GFP antibody. Panels g and h show the same section as panels e and f, respectively, and panels i and j are higher magnifications of panels g and h, respectively. Stars indicate tissue positions that are magnified 8 times. Panel b shows the absence of tumor development in the brain of a mouse engrafted with Snail-overexpressing cells. Panel d shows a section from an injected mouse brain that exhibited very rudimentary tumor growth. Panels e–j show sections from small but detectable tumors. Scale bar: 500 μm.
Table 1. Tumor formation in xenografted immunodeficient mice.
| Cell clone | Culture condition | No. of cells injected | No. of injected mice | No. of tumors | Average survival (weeks) | % of developed tumors |
|---|---|---|---|---|---|---|
| pcDNA3 (clone A) | Adherent | 1 × 106 | 9 | 9 | 8.87 | 100 |
| GFP-pcDNA3 (clone A) | 3 | 3 | 8.24 | 100 | ||
| Sum | 12 | 12 | 8.71 | 100 | ||
| pcDNA3 (clone A) | Spheres | 1 × 105 | 5 | 4 | 8.63 | 80 |
| GFP-pcDNA3 (clone A) | 11 | 10 | 10.38 | 91 | ||
| Sum | 16 | 14 | 9.51 | 88 | ||
| pcDNA3-Snail (clone F) | Adherent | 1 × 106 | 9 | 4 | 12.00 | 44 |
| GFP-pcDNA3-Snail (clone F) | 3 | 1 | 12.00 | 33 | ||
| Sum | 12 | 5 | 12.00 | 42 | ||
| pcDNA3-Snail (clone F) | Spheres | 1 × 105 | 6 | 3 | 12.00 | 50 |
| GFP-pcDNA3-Snail (clone F) | 10 | 3 | 12.00 | 30 | ||
| Sum | 16 | 6 | 12.00 | 38 |
Significance on the average survival:
P-value (adherent pcDNA3 versus pcDNA3-Snail): 2.8645E-08.
P-value (gliomasphere pcDNA3 versus pcDNA3-Snail): 3.03226E-05.
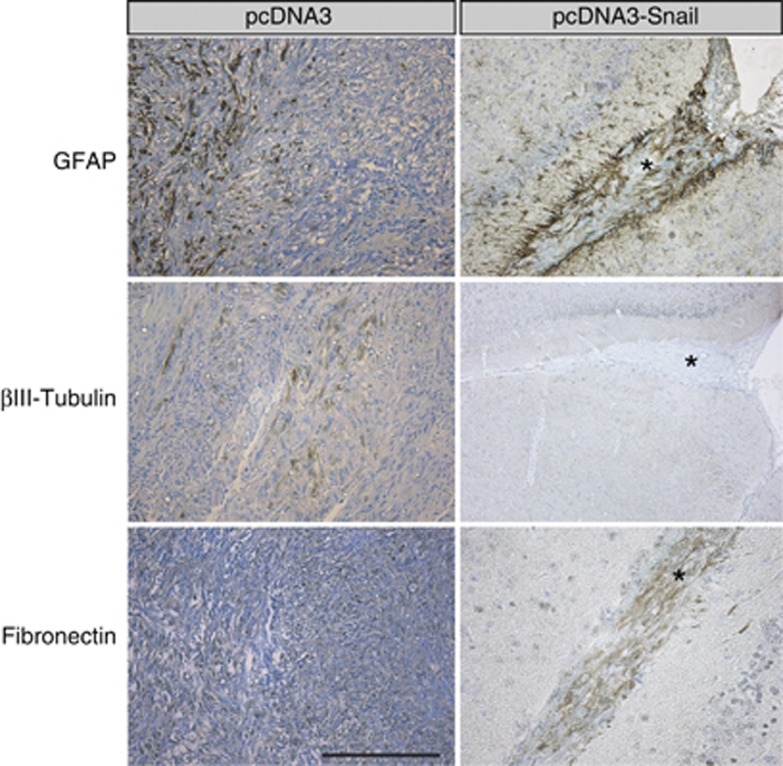
Brain histochemical analysis showed that invasive Snail-overexpressing GBM cells were distinguishable from host cells by the high levels of activated, phosphorylated Akt kinase (Supplementary Figure S5). Mock- and Snail-overexpressing GBMs formed invasive tumors, however, Snail-overexpressing GBMs had a more scattered pattern, migrating deeper into the brain, whereas mock-GBMs invaded as cell sheets, showing less spreading into the host tissue (Figures 6a–d). To facilitate the identification of Snail-expressing tumor cells within the mouse brain, we co-transfected GFP and again observed that Snail-expressing tumor cells spread in a patterned manner from the site of injection towards the brain center (Figures 6e–j). In contrast, GFP-tagged mock clones generated large tumor masses with less cell scattering (Figures 6e, g and i). Finally, whereas mock-transfected GBMs had a heterogeneous differentiation profile, expressing astrocytic GFAP, neuronal βIII tubulin and mesenchymal fibronectin (Figure 7, left panels), Snail-expressing GBMs completely lacked expression of the neuronal marker βIII tubulin (Figure 7, right panels). These results suggest that Snail promotes astrocytic differentiation, leading to less heterogeneous and smaller tumors while increasing tumor scattering within the brain.
Figure 7.
Cell-specific marker expression in U-2987 MG invading tumor cells in vivo. Immunostaining for GFAP, βIII tubulin and fibronectin on tumor sections from mice injected with pcDNA3 or pcDNA3-Snail transfected U-2987 MG cells dissociated from gliomaspheres. Note the widespread tumor cell staining in the control tumors (left panels) versus the limited, streak-like formation of the invasive Snail-expressing tumors (right panels, stars). Scale bar: 200 μm.
Snail promotes cell migration and suppresses gliomasphere formation
To better understand the effects of Snail in vivo, we analyzed tumor cell migration and gliomasphere formation in vitro. Whereas Snail induces cell migration of carcinoma cells,20 its effect on GBM cell migration is unclear. Scratch assays with Snail-knockdown clones compared with control clones (shCT) showed significant reduction in wound closure (Supplementary Figure S6), which reflected the degree of Snail knockdown (stronger in shSnail no. 1 and weaker in shSnail no. 2) (Figure 5c). Thus, under in vitro culture conditions, Snail promoted GBM cell migration. Furthermore, stimulation with BMP7 enhanced shCT cell migration, however, no significant effect by BMP7 was seen after Snail knockdown (Supplementary Figure S6). Importantly, the effect of BMP7 on migration did not result from increased cell proliferation (Supplementary Figure S1a).
Prior to gliomasphere assays, we analyzed GFAP and Nestin levels by comparing side-by-side cells with low Snail (mock clones A and B) to cells with high Snail (Snail clones F and G) (Figures 8a–c). High Snail expression positively correlated with high GFAP levels. Immunostaining also revealed a strong increase in GFAP-positive cell numbers in the presence of high Snail without effects on Nestin expression (Figure 8c). Moreover, Snail overexpression resulted in reduced Olig1/2 expression (Supplementary Figure S7). Strikingly, Snail-transfected cells failed to form gliomaspheres compared with mock cells (Figure 8d). Snail-transfected cells became adherent and resembled parental BMP7-stimulated cells (Figure 2b). Therefore, in vitro, Snail impairs the potential of GBM cells to form gliomaspheres in a manner similar to BMP7. Snail overexpression only partially phenocopied the BMP7 response of GBM cells, as BMP7 downregulated Nestin expression (Figure 2c), but Snail did not (Figure 8c).
Figure 8.
Snail impairs gliomasphere formation in vitro. (a, b) Snail and GFAP expression in U-2987 MG cells stably transfected with pcDNA3-Snail (clones F and G) or the empty vector (clones A and B) by qRT–PCR (a), and western blot (b), respectively. (c) Immunofluorescence staining of GFAP (red), Nestin (green) and DAPI (blue) in pcDNA3 or pcDNA3-Snail transfected U-2987 MG cells. (d) Primary gliomasphere formation capacity of U-2987 MG cells stably transfected with pcDNA3-Snail or the empty vector (scale bar: 400 μm).
Discussion
Autocrine TGFβ favors growth, invasiveness and angiogenesis of GBM.17 Blocking TGFβ signaling is therefore advantageous as a complementary treatment for GBM, and phase I/II clinical trials evaluate this clinical approach.15 Mechanistically, TGFβ induces GBM tumor growth via Smad-mediated transcriptional induction of Sox4, platelet-derived growth factor-B and LIF, the latter two cytokines supporting GIC survival and tumor expansion.10, 11, 12 Consistently, we found that TGFβ increased the ability of GICs to self-renew in low-density assays and induced the expression of genes involved in self-renewal, such as LIF and Nanog (Figure 2, Supplementary Figure S1). On the other hand, BMP members suppress GBM tumorigenesis possibly by driving GICs towards astrocytic differentiation,18 suggesting that BMP ligands could also act therapeutically. Consistently, we found that BMP7 impaired gliomasphere formation and enhanced GFAP expression (Figures 2 and 3).
By interrogating the roles of BMP in modulating the in vitro stem-like cell characteristics of human GBM cells, we identified the transcription factor Snail as a new molecular factor downstream of BMP–Smad signaling (Figures 2 and 3). BMP7 induced Snail expression via Smad1, Smad5 and Smad4, independently of the TGFβ receptor-activated Smads, Smad2 and Smad3, even though BMP7 could promote Smad2 C-terminal phosphorylation (Figures 2 and 4). Moreover, Snail was required for BMP7-induced GFAP expression (Figure 5b). Snail expression has been reported in some GBMs,33, 34 whereas its contribution to GBM tumorigenicity remains unknown. Snail expression in glioblastoma cells promoted migration in vitro and enhanced tumor cell scattering within the recipient brain in vivo (Figure 6, Supplementary Figure S6), which is reminiscent of its role in neural crest migration during embryonic development,35 and consistent with the effect of Snail2/Slug, a closely related transcription factor, in inducing glioblastoma cell migration and invasiveness.36 In vitro, Snail was required for both basal and BMP7-induced migratory capacity (Supplementary Figure S6). This finding deserves deeper investigation in order to explain the molecular circuit that is driven by Snail to promote GBM invasiveness.
Moreover, Snail-overexpressing cells in vivo showed limited pluripotency, as they expressed stronger astrocytic and mesenchymal markers, whereas lacking neuronal marker expression (Figure 7). Along the same lines, BMP4 can induce astrocytic differentiation and potent migration of the normal neural stem cells.37 Although Snail-overexpressing cells showed high phospho-Akt levels during brain invasion (Supplementary Figure S5), which might confer a survival advantage to the tumor, we reproducibly found that Snail minimized the extent of tumorigenicity (Figure 6, Table 1). We cannot fully exclude the possibility that Snail-expressing cells within the mouse brain might have a different late-onset impact on GBM outgrowth, if allowed to remain beyond 12 weeks. However, the antitumorigenic effect of Snail is compatible with the established tumor-suppressor actions of BMPs in GBM18 and the suppression of gliomasphere formation by Snail (Figure 8). In agreement with this model, GBMs that showed relative resistance to BMP-induced gliomasphere suppression also failed to induce Snail (Supplementary Figure S2). Thus, one way by which BMPs may counteract GBM tumorigenicity is by Snail induction. It has also been described that suppression of GBM tumorigenesis by BMP7 requires downregulation of transcription factor Olig2.25 In agreement, we have found that BMP7 downregulated Olig1/2 expression, an effect also seen in Snail-overexpressing cells (Figures 2 and 3, Supplementary Figure S7). It would be interesting to analyze whether repression of Olig1/2 is required for the invasive activity of Snail.
Snail induction correlated with GFAP upregulation and GIC differentiation (Figures 2, 3, 5, 6, 7, 8). Both Snail and BMP7 induced GFAP, however, only BMP7 but not Snail, downregulated Nestin (Figures 2, 3, 8). It is therefore interesting to define the subset of BMP responses in GBM cells that depend on the direct activity of Snail. A transcriptomic screen that we have performed aims at addressing this question in the context of GICs. GFAP expression is known to be controlled by cytokines such as LIF and also by BMPs via a mechanism that depends on crosstalk between Stat3 and Smad1.38, 39 As Snail is an established transcriptional repressor,29 it is likely that Snail indirectly regulates GFAP expression, possibly by repressing other neuroectodermal genes, which directly regulate GFAP transcription. This notion is supported by our observation that Snail was unable to regulate a 2 kbp promoter fragment of the GFAP gene (data not shown). The GFAP promoter contains E-box sequences whose functional importance remains unknown.40
Thus, our work opens the exciting possibility that comparative analysis of invasive properties of glioblastomas and carcinomas may provide new clues into tumor cell behavior. It is likely that Snail is a central factor in these processes independent from the specific cell type of origin that the tumor evolved from. This work represents one of the few examples where the interlinked capacity of GBMs to grow and invade is dissociated. Moreover, it supports the notion that both invasiveness and tumorigenicity need to be targeted by new therapies against brain cancer.
Materials and methods
Cell culture, cytokines, antibodies and plasmids
The new human glioblastoma stem cell lines, U-3013/3024/3028/3034 MG, were established as follows: human GBM grade IV biopsies were obtained in accordance with the protocol approved by the Uppsala ethical review board (2007/353), and were graded by neuropathologist Irina Alafuzoff, Uppsala University Hospital, according to the World Health Organization guidelines. Tumor biopsies were minced (1 mm × 1 mm bits) and digested by 1:1 ratio of Accutase (eBioscience, San Diego, CA, USA)/TrypLE (Invitrogen, Life Technologies Europe BV, Stockholm, Sweden), at 37 °C for 15 min and triturated through 18 and 21 g needles five times. Dissociated cells were resuspended in DMEM/F12 Glutamax (GIBCO, Life Technologies Europe BV, Stockholm, Sweden) and Neurobasal medium (GIBCO) mixed in 1:1 ratio, with the addition of 1% B27 (Invitrogen, Life Technologies Europe BV), 0.5% N2 (Invitrogen, Life Technologies Europe BV), 1% penicillin/streptomycin (Sigma-Aldrich Sweden AB, Stockholm, Sweden), 10 ng/ml EGF and FGF2 (PeproTech, EC Ltd, London, UK), and plated at 100 000 cells/ml. After primary sphere formation, spheres were seeded onto poly-ornithine/laminin-coated dishes and cultured as adherent cells as described.41 U-3013/3024/3028/3034 MG cells were used at passages 15–30. GBM cells U-343 MG, U-343 MGa-Cl2:6, U-2990 MG and U-2987 MG were previously described,22 and were maintained in MEM, 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Primary human NPCs were obtained from Lonza Ltd, Basel, Switzerland (NHNP, PT-2599/5F0029) and were cultured per manufacturer's instructions. Human hepatocarcinoma HepG2 cells were cultured in DMEM, 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin, as described.42
Glioblastoma cells in regular or stem cell media were treated with TGFβ1, BMP7 or with the inhibitors GW6604, SB505124 and DMH1 (dorsomorphin homolog 1) after 8–24 h starvation in a medium without serum or in the continuous presence of stem cell medium. DMSO served as a vehicle. Recombinant human TGFβ1 was from (PeproTech, EC Ltd.) or from Biosource International, Inc (Camarillo, CA, USA), and recombinant human BMP7 was a gift from K. Sampath (Curis, Inc., Boston, MA, USA).
The antibodies used are: rabbit anti-Nestin, (Chemicon International Inc., Temecula, CA, USA), rabbit anti-GFAP (Dako Sweden AB, Stockholm, Sweden), goat anti-Snail, mouse anti-β tubulin and mouse anti-GAPDH (Santa Cruz Biotechnologies Inc., Heidelberg, Germany), mouse anti-βIII tubulin (Abcam, Cambridge, UK), mouse anti-CD133 (Myltenyi Biotec Norden AB, Lund, Sweden), rabbit anti-P-Smad2 (home-made, Uppsala, Sweden), rabbit anti-P-Smad1, rabbit anti-Smad1, rabbit anti-pAkt (Cell Signaling Technology Inc., Danvers, MA, USA) and rabbit anti-Fibronectin (Sigma).
Human Flag-tagged Smad1, Smad4 and Smad5 were expressed under the control of pcDNA3, as described.42 The human Snail promoter luciferase plasmid was provided by A García-Herreros (IMIM, Barcelona, Spain).30
Cell growth, proliferation and migration assays
Cells growing in regular media were counted in a Beckman Coulter (Beckman Coulter AB, Bromma, Sweden) counter after each passage (every 3–4 days) over a 2-week period. Growth was measured by MTS assay (CellTiter-96 Aqueous-One-Solution Assay, Promega Biotech AB, Stockholm, Sweden). Proliferation in stem cell media was determined by measuring nuclear 5′-ethynyl-2′-deoxyuridine (EdU) incorporation. Briefly, cells seeded over poly-lysine/laminine-coated chamber slides (Beckton Dickinson AB, Stockholm, Sweden) for 24 h to allow for adhesion, were fed EdU for 1.5 h and EdU-positive cells were recorded using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen).
Wound healing assays were performed by scratching confluent cell monolayers, followed by immediate stimulation with BMP7 (30 ng/ml) in the presence of 3% FBS; wound size was monitored 48 h later.
Gliomasphere and differentiation analysis
Primary gliomaspheres were formed by plating 80 000 GBM cells per well in stem cell medium: MEM, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 25 ng/ml EGF, 25 ng/ml bFGF and B27 complement (Invitrogen). Medium was refreshed every 2–3 days, and gliomasphere numbers and size were recorded; gliomaspheres were then collected for RNA or protein extraction, or adhered onto poly-L-lysine/laminin-coated slides and maintained undifferentiated in stem cell medium for 6 days before immunostaining.
GBM self-renewal capacity was assessed by dissociating primary gliomaspheres and plating the cells at low/clonogenic density (1000 cells per well). For dissociation into single cell suspension, primary gliomaspheres were triturated by pipetting in 37 °C PBS/50 U/ml collagenase 50 times, and passaging through a 40-μm strainer. Secondary gliomaspheres formed in dilution assays and their number was recorded.
For differentiation assays, secondary gliomaspheres adhered onto poly-L-lysine/laminin-coated slides and differentiated in the presence of 1% FBS-containing MEM. Six days after plating, cells were immunostained to monitor marker expression.
Orthotopic intracranial injections
U-2987 MG cells stably transfected with either pcDNA3 or pcDNA3-Snail with or without pcDNA3-GFP, were collected after trypsinization of adherent cultures grown in FBS-containing media or after dissociation of gliomaspheres grown in stem cell media in non-adherent plates. Then, 106 cells from adherent cultures or 105 cells from gliomaspheres were resuspended in 5 μl of FBS-free media and injected in the right brain hemisphere of newborn immunocompromised mice (2–3 days after birth) using a Hamilton syringe according to the ethical permit. Mice were followed up in terms of health and tumor development. Mice were sacrificed 12 weeks after injection if no clinical signs became apparent and their brains were dissected, dehydrated and embedded in paraffin for histological analysis and tumor identification. The number of tumor-bearing mice was scored along with the time of their survival. All mouse xenograft experiments were performed in accordance with the local animal ethics committee after permission granted to the Department of Immunology, Genetics and Pathology, Uppsala University (responsible Bengt Westermark).
Transfections and RNAi
The empty vectors pcDNA3 and pSuperior-Neo-GFP (pSGN/shControl), and the vector containing Snail cDNA (pcDNA3-Snail) or the small hairpin RNA against Snail (shSnail) were previously described;43, 44 they were transfected into U-2987 MG cells using the calcium phosphate coprecipitation method (pcDNA3 and pcDNA3-Snail) or Fugene-HD (Promega Biotech AB) (pSGN and shSnail), and clones were obtained after selection with antibiotics. Stable clones expressing pcDNA3 and pcDNA3-Snail were transfected with pcDNA3-GFP using Fugene-HD, and GFP-positive cells were sorted by FACS, 1 week after selection with antibiotics. Transient transfection of HepG2 cells with the Snail luciferase reporter and Smad expression vectors was performed using the calcium phosphate coprecipitation method, as described.44
ON-TARGETplus SMARTpool siRNA specific for human Snail, Smad1, Smad2, Smad3, Smad4, Smad5, non-target siRNA control and transfection reagents were from Dharmacon Thermo Fischer Scientific, Lafayette, CO, USA.
Histology, immunostaining and immunobloting
For immunocytochemistry, gliomaspheres or adherent cells were washed twice with PBS containing Ca2+ and Mg2+, and either fixed in 3% paraformaldehyde and permeabilized with 0.3% Triton X-100, or fixed in ice-cold methanol. The slides were blocked in 3% goat serum and 0.5% glycine, incubated with primary antibody, briefly washed three times, incubated in the dark with the secondary antibody either conjugated to Alexa-543 or Alexa-488 and stained with either DAPI or propidium iodide to visualize the nuclei before mounting in fluoromount (SouthernBiotech, Birmingham, AL, USA).
Imaging was performed under a fluorescent microscope (Leica Microsystems AB, Kista, Sweden) or a Zeiss Axiovert 200 M confocal microscope equipped with LSM 510 laser with a 63 × /0.75 objective lens and photographing at ambient temperature in the presence of immersion oil, with a Hamamatsu C4742-95 CCD digital camera and software QED Camera Plugin v1.1.6 (QED Imaging Inc., Pittsburgh, PA, USA) and Volocity (PerkinElmer Inc., Waltham, MA, USA). Phase contrast images were obtained either with a Zeiss confocal microscope or with an inverted Zeiss AxioVision microscope and AxioCam camera. Histological analysis was done using a Leica Qwin Series microscope with a Leica DFC camera. Image file size was reduced with Adobe Photoshop CS2.
Immunoblot analysis of GBM protein extracts using the antibodies listed above was performed as described.45
Real-time qRT–PCR analysis
Total cellular RNA was purified with the RNeasy kit (Qiagen AB, Sollentuna, Sweden) and cDNA was synthesized from 0.5 μg of RNA using the iQ kit (Bio-Rad Laboratories AB, Solna, Sweden). qRT–PCR conditions were as described45 with forward (F) and reverse (R) primers used: CD133, F5′-ACCCAACATCATCCCTGTTCTT-3′/R5′-AGCTCTTCAAGGTGCTGTTCATG-3′ Nestin, F5′-AGCCCTGACCACTCCAGTTTAG-3′/R5′-CCCTCTATGGCTGTTTCTTTCTCT-3′ Sox2, F5′-ACACCCTGATCTGGCATGGA-3′/R5′-GGCTGTTGCCTGGCTTCTC-3′ Snail, F5′- CACTATGCCGCGCTCTTTC-3′/R5′-GCTGGAAGGTAAACTCTGGATTAGA-3′ GFAP, F5′-TGCGGCTCGATCAACTCA-3′/R5′-GTTGGTTTCATCCTGGAGCTTCT-3′ POU5F1/Oct4, F5′-AGTGCCCGAAACCCACACT-3′/R5′-CTTCTGGCGCCGGTTACA-3′ Nanog, F5′-TGCCTCACACGGAGACTGTCT-3′/R5′-AGTGGGTTGTTTGCCTTTGG-3′ LIF, F5′-TGAACCAGATCAGGAGCCAACT-3′/R5′-CCCCCTGGGCTGTGTAATAG-3′ OLIG1, F5′-AGAAAGTGCTCGCTCTCACC-3′/R5′-GCAAGGCGCCCTAAGTTCTA-3′ OLIG2, F5′-CTTCAAGTCATCCTCGTCCAG-3′/R5′-TGTTGATCTTGAGACGCAGC-3′ Smad1 F5′-GTCTGCATCAATCCCTACCAC-3′/R5′-GCTCATTTTGTCCTAAGTTACGG-3′ Smad2, F5′-TGGCTGGCACCCTGCAACAG-3′/R5′-TGCCTTCGGTATTCTGCTCCCCA-3′ Smad3-F5′-GCAATATTCCAGAGACCCCACC-3′/R5′-TAGGTTTGGAGAACCTGCGTCC-3′ Smad4, F5′-CATCCTGGACATTACTGGCCA-3′/R5′-CCTACCTGAACGTCCATTTCAA-3′ Smad5, F5′-TTCCCTTATCTCCAAACAGCC-3′/R5′-CATCAGGTGGCATATAGGCAG-3′ GAPDH, F5′-GGAGTCAACGGATTTGGTCGTA-3′/R5′-GGCAACAATATCCACTTTACCA-5′.
Promoter reporter assays
The human Snail promoter was co-transfected with reporter plasmid pCMV-βGal for normalization, and expression vectors pcDNA3 (control), Flag-Smad1, Flag-Smad5 and/or Flag-Smad4 in HepG2 cells. Cells were stimulated with 30 ng/ml BMP7 for 24 h. The enhanced luciferase assay kit from BD Pharmingen (Beckton Dickinson AB, Stockholm, Sweden) was used. Normalized promoter activity data are plotted in bar graphs representing mean±s.d. from triplicate samples. Each independent experiment was repeated at least twice.
Statistical analysis
Statistical analysis was done using excel and P values <0.05 were considered significant. qRT–PCR, wound healing and mouse survival data were tested for statistical significance using the two-sided Student's t-test. The Fisher's exact test was applied for comparing the incidence of tumor formation in mouse xenograft experiments. All quantitative data are presented as mean±s.d. estimated from triplicate or more determinations per experiment.
Acknowledgments
We thank: J Grawé, Uppsala University FACS facility; M Kastemar for tumor sampling; I Alafuzoff for tumor pathology; A Åhgren, M Vanlandewijck for tumor immunohistochemistry; H Göransson-Kultima, A Andersson, Uppsala University Microarray facility; A Östman (Karolinska Institute) for encouragement; M Brenner, A Cano for reagents; K Sampath (Curis, Inc,) for BMP7; T Gahman and A Shiau (Ludwig Institute for Cancer Research, San Diego, USA) for GW6604 and DMH1 inhibitor synthesis; S Thuault, E-J Tan, K Kahata and other group members for discussion. Funding of the authors' work was provided by the Atlantic Philanthropies/Ludwig Institute for Cancer Research Clinical Discovery Program, the Göran Gustafsson Foundation and the Swedish Cancer Society.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon RE, Rich JN. Glioblastoma stem cells: a neuropathologist's view. J Oncol. 2011;2011:397195. doi: 10.1155/2011/397195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Nagane M, Huang HJ, Cavenee WK. Advances in the molecular genetics of gliomas. Curr Opin Oncol. 1997;9:215–222. doi: 10.1097/00001622-199709030-00001. [DOI] [PubMed] [Google Scholar]
- Westermark B, Nistér M. Molecular genetics of human glioma. Curr Opin Oncol. 1995;7:220–225. doi: 10.1097/00001622-199505000-00005. [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ, Rich JN. Making a tumour's bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7:733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the Mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin C-H. The regulation of TGFβ signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- Bruna A, Darken RS, Rojo F, Ocana A, Penuelas S, Arias A, et al. High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Penuelas S, Anido J, Prieto-Sanchez RM, Folch G, Barba I, Cuartas I, et al. TGF-β increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Hjelmeland MD, Hjelmeland AB, Sathornsumetee S, Reese ED, Herbstreith MH, Laping NJ, et al. SB-431542, a small molecule transforming growth factor-β-receptor antagonist, inhibits human glioma cell line proliferation and motility. Mol Cancer Ther. 2004;3:737–745. [PubMed] [Google Scholar]
- Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, et al. TGF-β receptor inhibitors target the CD44high/Id1high glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Bogdahn U, Hau P, Stockhammer G, Venkataramana NK, Mahapatra AK, Suri A, et al. Targeted therapy for high-grade glioma with the TGF-β2 inhibitor trabedersen: results of a randomized and controlled phase IIb study. Neuro Oncol. 2011;13:132–142. doi: 10.1093/neuonc/noq142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Miyazono K. TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- Rich JN. The role of transforming growth factor-β in primary brain tumors. Front Biosci. 2003;8:e245–e260. doi: 10.2741/992. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Lottaz C, Beier D, Meyer K, Kumar P, Hermann A, Schwarz J, et al. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70:2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- Ferletta M, Caglayan D, Mokvist L, Jiang Y, Kastemar M, Uhrbom L, et al. Forced expression of Sox21 inhibits Sox2 and induces apoptosis in human glioma cells. Int J Cancer. 2011;129:45–60. doi: 10.1002/ijc.25647. [DOI] [PubMed] [Google Scholar]
- Yamada N, Kato M, ten Dijke P, Yamashita H, Sampath TK, Heldin CH, et al. Bone morphogenetic protein type IB receptor is progressively expressed in malignant glioma tumours. Br J Cancer. 1996;73:624–629. doi: 10.1038/bjc.1996.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gouville AC, Boullay V, Krysa G, Pilot J, Brusq JM, Loriolle F, et al. Inhibition of TGF-β signaling by an ALK5 inhibitor protects rats from dimethylnitrosamine-induced liver fibrosis. Br J Pharmacol. 2005;145:166–177. doi: 10.1038/sj.bjp.0706172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirasani SR, Sternjak A, Wend P, Momma S, Campos B, Herrmann IM, et al. Bone morphogenetic protein-7 release from endogenous neural precursor cells suppresses the tumourigenicity of stem-like glioblastoma cells. Brain. 2010;133:1961–1972. doi: 10.1093/brain/awq128. [DOI] [PubMed] [Google Scholar]
- Maire CL, Wegener A, Kerninon C, Nait Oumesmar B. Gain-of-function of Olig transcription factors enhances oligodendrogenesis and myelination. Stem Cells. 2010;28:1611–1622. doi: 10.1002/stem.480. [DOI] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Tate CM, Pallini R, Ricci-Vitiani L, Dowless M, Shiyanova T, D'Alessandris GQ, et al. A BMP7 variant inhibits the tumorigenic potential of glioblastoma stem-like cells. Cell Death Differ. 2012;19:1644–1654. doi: 10.1038/cdd.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, A Cano. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype. Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Barberà MJ, Puig I, Dominguez D, Julien-Grille S, Guaita-Esteruelas S, Peiró S, et al. Regulation of Snail transcription during epithelial to mesenchymal transition of tumor cells. Oncogene. 2004;23:7345–7354. doi: 10.1038/sj.onc.1207990. [DOI] [PubMed] [Google Scholar]
- Franco DL, Mainez J, Vega S, Sancho P, Murillo MM, de Frutos CA, et al. Snail1 suppresses TGF-β-induced apoptosis and is sufficient to trigger EMT in hepatocytes. J Cell Sci. 2010;123:3467–3477. doi: 10.1242/jcs.068692. [DOI] [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung J, Cho BK, Kim YS, Snail ParkSH. and Cox-2 expressions are associated with WHO tumor grade and survival rate of patients with gliomas. Neuropathology. 2010;30:224–231. doi: 10.1111/j.1440-1789.2009.01072.x. [DOI] [PubMed] [Google Scholar]
- Sun M, Song L, Zhou T, Gillespie GY, Jope RS. The role of DDX3 in regulating Snail. Biochim Biophys Acta. 2011;1813:438–447. doi: 10.1016/j.bbamcr.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- Yang HW, Menon LG, Black PM, Carroll RS, Johnson MD. SNAI2/Slug promotes growth and invasion in human gliomas. BMC Cancer. 2010;10:301. doi: 10.1186/1471-2407-10-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Kaduwal S, Yang DH, Choi KY. Bone morphogenetic protein 4 stimulates attachment of neurospheres and astrogenesis of neural stem cells in neurospheres via phosphatidylinositol 3 kinase-mediated upregulation of N-cadherin. Neuroscience. 2010;170:8–15. doi: 10.1016/j.neuroscience.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Abematsu M, Mori H, Yanagisawa M, Kagawa T, Nakashima K, et al. Potentiation of astrogliogenesis by STAT3-mediated activation of bone morphogenetic protein-Smad signaling in neural stem cells. Mol Cell Biol. 2007;27:4931–4937. doi: 10.1128/MCB.02435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Lee Y, Messing A, Su M, Brenner M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia. 2008;56:481–493. doi: 10.1002/glia.20622. [DOI] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Kurisaki K, Kurisaki A, Valcourt U, Terentiev AA, Pardali K, ten Dijke P, et al. Nuclear factor YY1 inhibits transforming growth factor β- and bone morphogenetic protein-induced cell differentiation. Mol Cell Biol. 2003;23:4494–4510. doi: 10.1128/MCB.23.13.4494-4510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmeda D, Jorda M, Peinado H, Fabra A, Cano A. Snail silencing effectively suppresses tumour growth and invasiveness. Oncogene. 2007;26:1862–1874. doi: 10.1038/sj.onc.1209997. [DOI] [PubMed] [Google Scholar]
- Thuault S, Tan EJ, Peinado H, Cano A, Heldin C-H, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283:33437–33446. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowanetz M, Lonn P, Vanlandewijck M, Kowanetz K, Heldin C-H, Moustakas A. TGFβ induces SIK to negatively regulate type I receptor kinase signaling. J Cell Biol. 2008;182:655–662. doi: 10.1083/jcb.200804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.