Abstract
G protein-coupled receptors (GPCR) are integral membrane proteins that transmit signals from external stimuli to the cell interior via activation of GTP-binding proteins (G proteins) thereby mediating key sensorial, hormonal, metabolic, immunological, and neurotransmission processes. Elucidating their structure and mechanism of interaction with extracellular and intracellular binding partners is of fundamental importance and highly relevant to rational design of new effective drugs. Surface plasmon resonance (SPR) has become a method of choice for studying biomolecular interactions at interfaces because measurements take place in real-time and do not require labeling of any of the interactants. However, due to the particular challenges imposed by the high hydrophobicity of membrane proteins and the great diversity of receptor-stimulating ligands, the application of this technique to characterize interactions of GPCR is still in the developmental phase. Here we give an overview of the principle of SPR and analyze current approaches for the preparation of the sensor chip surface, capture and stabilization of GPCR, and experimental design to characterize their interaction with ligands, G proteins and specific antibodies.
Keywords: Surface plasmon resonance, G protein-coupled receptors
1. Introduction
Within the last 20 years, surface plasmon resonance (SPR) has become a method of choice for studying biomolecular interactions at interfaces. This technique could be particularly useful for studying interactions with membrane proteins like G protein-coupled membrane receptors (GPCR). They are located in the plasma membrane of cells and transmit extracellular signals elicited by light or compounds like neural transmitters, hormones, odorants to the cell interior where they activate GTP-binding proteins (G proteins). This large superfamily of heptahelical molecules comprises receptors for dopamine, serotonin, epinephrine, opioids, and cannabinoids, just to mention a few. According to various estimates 30-40% of drugs under development target GPCR. The proper function of GPCR is critical for all higher forms of life. Structural and functional studies using purified receptors are essential to understand the molecular mechanisms of their function and facilitate rational design of drugs that target a disease with high specificity [78]. It has been shown that GPCR require a fluid lipid matrix of proper composition for their function. SPR is only beginning to be adapted to the needs of membrane proteins and to GPCR in particular. The purpose of this review is (i) to give an introduction into the basics of the SPR technique, and (ii) to address the challenges of its application to GPCR.
2. Surface plasmon resonance
2.1. Phenomenon description
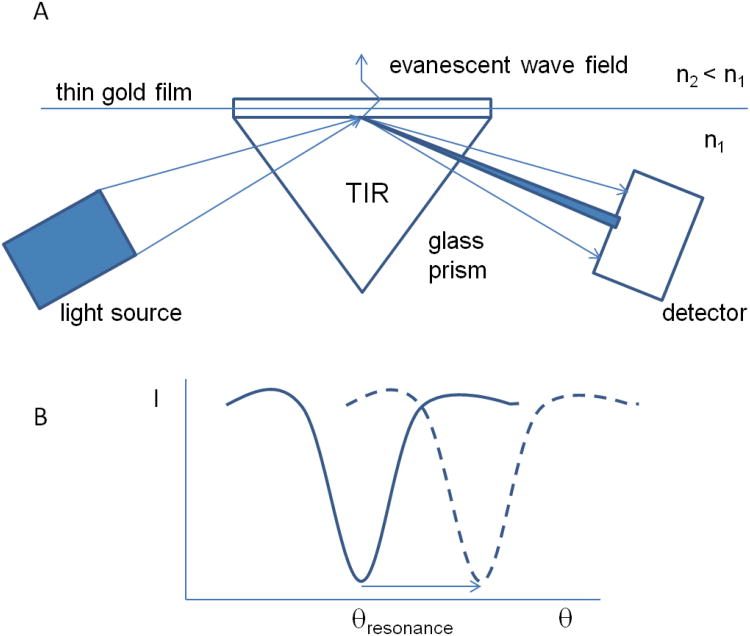
The SPR-based optical measurements do not require molecular labeling and are conducted in real time with a typical time resolution from milliseconds to seconds. The SPR effect has been known for more than a century but was interpreted as light-stimulated, collective oscillations of valence electrons (surface plasmons) in thin solid layers much later [35, 52]. For most practical applications, thin gold layers with a thickness on the order of 50 nm on the surface of a glass support are used. In a typical setup, a glass/gold interface is illuminated through the glass with a convergent beam of light that is totally reflected from the interface (Fig. 1). Plasmons are excited under a particular angle of incidence (θresonance), and excitation observed as a corresponding minimum of light intensity at a particular angle of reflection that is measured with a diode array detector. Experiments are commonly conducted with monochromatic p-polarized infrared light (for Biacore, wavelength 760 nm) generated by a light emitting diode.
Figure 1. SPR measurements.
A. Schematic presentation of the optical system to detect SPR phenomena: TIR - total internal reflection, n1 and n2 – refractive index of glass and the medium adjacent to the glass/gold interface, respectively. The amplitude of the evanescent wave field decays exponentially with distance from the surface. B. Dependence of intensity of reflected light (I) versus the angle of reflection (θ). The arrow indicates a shift in the angle of plasmon resonance (θresonance) due to the presence of macromolecules at the sensor surface.
Total internal reflection occurs upon a transition of light from a medium with higher refractive index, e.g. glass (n1=1.5-1.8), to a medium with lower refractive index, e.g. water (n2=1.333). An evanescent light wave penetrates over a short distance into the medium with the lower refractive index (about half a wavelength) (Fig. 1). The angle under which surface plasmon resonance occurs depends critically on the refractive index of the medium adjacent to the glass/gold interface. This allows to quantitatively record adsorption of biomaterials to the interface but makes experiments also very sensitive to any change in solvent composition, pH, ionic strength as well as temperature.
A comprehensive description of the physics of SPR including the theory of surface plasmons and their excitation, details of design of various SPR instrumentation and sensor surface preparation are provided e.g. in the Handbook of Surface Plasmon Resonance [34] [60] as well as in reference [27]. A simplified, qualitative description of SPR with emphasis on practical aspects of applications is provided in [19].
The number and variety of optical biosensors is steadily increasing (see a list of commercially available instruments and technologies for example in [56]). In our review, we refer mainly to Biacore instruments[1] as they have been the most frequently used in studies on GPCR. A SPR instrument for biomedical applications consists of an optical detection unit, a sensor chip, and a microfluidic liquid handling unit to deliver reactants to the sensor surface. Since the refractive index is temperature dependent, the sensor chip as well as the microfluidic handling unit must be thermostated. While optical detection and microfluidics are universal, the sensor chip surface must be specifically designed for the application.
2.2. Sensor chips and immobilization techniques
A fundamental requirement for the sensor surface is its ability to immobilize one of the interacting partners without interfering with activity. It is required that immobilization is sufficiently strong such that microfluidic flows do not result in loss of material and reproducible to be able to conduct experiments under a variety of conditions. It may also be desirable that such binding is reversible to enable repeated use of the same sensor chip.
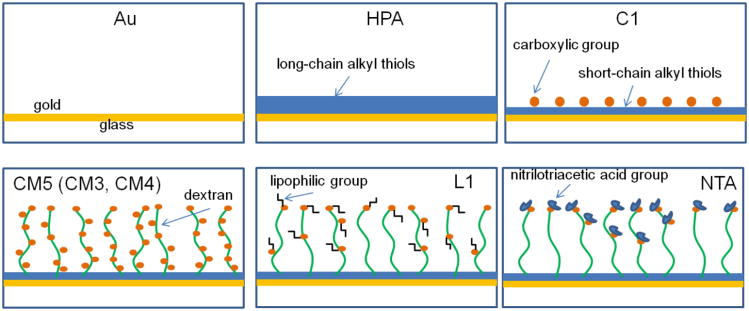
The following chip surfaces (Fig. 2) are commercially available for immobilization of interacting molecules, all of them gold coated [2]: (i) blank gold for attachment of molecules via a thiol (-SH) group (e.g. Biacore Au-chips), (ii) saturated alkyl chains of various length (e.g. Biacore HPA chips), (iii) alkyl chains of various length ending with a carboxylic group (e.g Biacore C1 chip), (iv) polysaccharide chains like dextran of various length and density, carboxy-methyl modified (e.g. Biacore CM3,4,5,7 chips), (v) polysaccharide chains like dextran with lipophilic groups attached via carboxy-methylated sites (e.g. Biacore L1-chip). (vi) chains ending with a nickel chelating nitrilotriacetic acid (NTA) group for capturing recombinant proteins with a polyhistidine tag (e.g. Biacore NTA-chips). In addition, the use of a variety of custom-made chips with properties that are specifically designed for particular classes of biomolecules has been reported [21].
Figure 2. SPR sensor chips.
Schematic presentation of commercially available sensor chips from Biacore (GE Healthcare). The 50-nm-thick gold layer is shown in yellow, saturated alkyl thiol chains of different length in blue, dextran chains of different length in green, lipophilic groups in black, red dots represent carboxylic groups attached to the surface layer with or without dextran, and blue dots are nitrilotriacetic acid groups.
These different chips enable surface immobilization of proteins by a variety of techniques such as: (i) coupling of protein thiol-, amino-, aldehyde-, or carboxyl groups to the sensor surface or to coatings, (ii) capturing of proteins via natural or engineered binding sites or tags, (iii) secondary interactions between already captured proteins and their interaction partners, (iv) interaction of the lipid matrix containing the embedded protein with lipophilic groups at the sensor surface. The coupling via formation of chemical bonds, e.g. amino coupling, is usually irreversible while capture via affinity tags or hydrophobic interactions is reversible and allows surface regeneration and re-use.
While in many reports utilizing Biacore, the immobilized interaction partner is called “ligand”, in this review, in order to avoid confusion with the more common pharmacological use of “ligand”, we reserve this term for natural and synthetic compounds that bind to specific sites on the GPCR and modulate its activity. The interacting partner captured at a surface of a sensor chip can be a small ligand, antibody or the receptor itself, depending on experimental design. The interacting partner in the mobile phase will be referred to as “analyte”.
2.3. Experimental design and data analysis
After one of the interacting partners is immobilized to the sensor chip surface and its quantity estimated as the difference in the SPR signal before and after immobilization, the binding experiment may be conducted. A change in the local refractive index measured by the SPR instrument is proportional to changes in surface mass concentrations of macromolecules up to 50 ng / mm2 [66]. The refractive index of the medium near the chip surface is also affected by the surface concentration of analyte. Therefore, the shift of the SPR angle, also called the SPR response, provides a measure of the change in total surface mass concentration under the condition that there are no other perturbing influences. For Biacore instruments, the signal is reported in resonance units (RU). Very close to the chip surface, 1 RU is equivalent to a protein surface concentration of about 1 pg/mm2 [66] or to an almost identical amount of lipid, 0.98 pg/mm2 [18]. However, the sensitivity to refractive index changes decays exponentially with increasing distance from the surface, typically to 1/3 at distances of 300-400 nm. Therefore, sensitivity may differ significantly between chips and coupling reagents that cause differences in patterns of refractive index changes as a function of distance from the chip.
The very high sensitivity of SPR to small changes in the refractive index near the surface is beneficial for detection of the capture of very low amounts of material. At the same time it can be seen as a disadvantage because it also detects minor compositional changes of analyte solutions, any kind of mechanical perturbation, changes in temperature as low as ±10-2 K and other causes of instrumental drift. Most of the unwanted perturbing influences can be eliminated by measurement of the differential response between an active surface in a primary cell and an inert but otherwise identical surface in a reference cell. Typically, the binding partners are delivered via a microfluidic handling unit that supplies the same solutes to the reference and measurement cells connected to the same microfluidic flow in series.
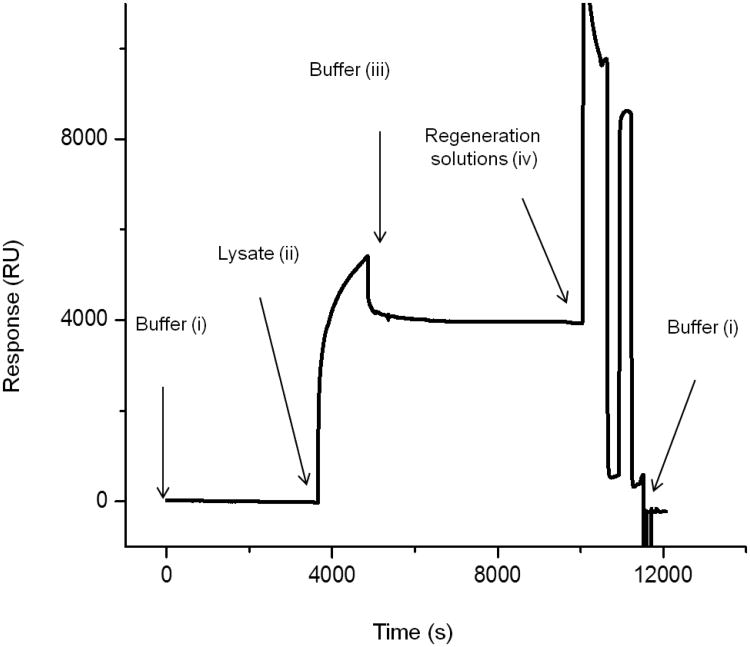
Binding of an analyte delivered by a microfluidic flow to an immobilized interaction partner at the interface results in a time-dependent SPR response called a sensorgram (Fig. 3). A typical SPR experiment consists of (i) establishment of a stable baseline in the presence of an appropriate flow of a running buffer, (ii) injection of a known volume (5- 425 μL for Biacore) of an analyte at a flow rate in the range of 1- 100 μL/min (Biacore) for a defined time of association, and (iii) injection of buffer to monitor analyte release from the surface, (iv) regeneration of the chip by flowing solutions over the surface that result in guaranteed release of analyte, if possible.
Figure 3. Sensorgram showing the capture of a GPCR followed by surface regeneration.
Recombinant Rho-tagged cannabinoid receptor type II, CB2, from cell lysate was captured to a 1D4 antibody-coated CM4 chip and the surface regenerated afterwards. Arrows show injection of lysate, buffer, and regeneration solutions, respectively; the labels i, ii, iii, and iv correspond to the typical steps of a SPR experiment as presented in section 2.3 [38].
A theoretical maximal SPR response to ligand interaction with a known amount of immobilized GPCR is easily calculated if the molecular weight of the GPCR, the ligand, and the binding stoichiometry are known. A few typical examples are shown in Table 1. They indicate that immobilization of a single layer of tightly packed GPCR at the surface of a chip may yield up to several thousand RU. An even higher response to GPCR immobilization is possible if surface capture occurs in multiple layers or in highly folded single layers. However, such packing is usually undesirable because of restrictions for access of analytes to the GPCR at the surface.
Table 1. Maximal theoretical response (RU) of a Biacore 3000 instrument with a flow cell area of 1.2 mm2 [1] for a single layer of GPCR in detergent, a lipid bilayer, or nanodiscs, and to binding of ligand.
| GPCR in | max RU1 total (GPCR+ lipid/detergent) | max RU GPCR (42 kDa) | max RU2 ligand (500 Da) |
|---|---|---|---|
| Detergent3 | 7,300 | 3,650 | 43 |
| lipid bilayer4, no GPCR | 5,000 | - | - |
| lipid bilayer, GPCR/lip. 1/70, mol/mol | 6,150 | 2,630 | 31 |
| lipid bilayer, GPCR/lip. 1/250, mol/mol | 5,340 | 930 | 11 |
| lipid bilayer, GPCR/lip. 1/500, mol/mol | 5,130 | 490 | 6 |
| lipid bilayer, GPCR/lip. 1/1000, mol/mol | 5,020 | 250 | 3 |
| Nanodiscs5 | 3,830 | 890 | 10 |
It is assumed that 1 RU is equivalent to a surface concentration of 1×10-12 g/mm2 for protein [ref 63] and 0.98×10-12 g/mm2 for lipid [ref 16].
It is assumed that the GPCR binds the analyte at a 1/1 stoichiometry. The maximal response to analyte binding is proportional to the ligand/GPCR ratio of molecular weights.
It is assumed that the GPCR has a circular lateral area of 900 Å2 [28] and that the GPCR/detergent weight ratio per micelle is 1/1.
It is assumed that the lateral area per lipid is 65 Å2 and the lipid molecular weight 800 Da.
It is assumed that the nanodiscs have a circular lateral area of 7,390 Å2 (MSP1D1) [7], contain 112 lipids and one GPCR per disc.
The binding response of an analyte is proportional to the ratio of molecular weights of analyte and immobilized partner indicating that detection of the binding of typical ligands with low molecular weight (MW = 500 Da or lower) is considerably more challenging than detection of GPCR capture at the chip surface. The requirements for baseline stability increase with decreasing amplitude of the SPR response from ligand binding. Establishment of a stable baseline is particularly challenging for membrane proteins that require the presence of detergents and/or lipids for their stability. Detergents and lipids loosely associated with the protein and interface and may respond sensitively to changes in buffer composition and the flow rate of buffer.
Special consideration should be given to design of experiments involving hydrophobic ligands like the cannabinoids. These compounds readily partition into lipid bilayers or detergent micelles making detection of their specific interaction with surface-captured GPCR by SPR technically challenging. This difficulty, however, can be resolved if the SPR detection relies on binding of larger, water-soluble molecules to the receptor-ligand complex. The analyte can be either a cognate G protein or a specific antibody recognizing a particular ligand-induced conformation of the receptor. Examples of such experimental design are presented in this review.
Under favorable conditions, SPR does not just report an interfacial interaction of an analyte with the surface but also the physico-chemical properties of this interaction. This is achieved by measuring on- and off- rates of analyte binding (kon and koff, respectively) from a set of sensorgrams detected as function of analyte concentration. The ratio of off- and on-rates yields the thermodynamic dissociation constant KD = koff/kon [3]. Measurement of on- and off- rates of analyte poses challenges due to the presence of an unstirred layer of solvent with a thickness of several microns at the chip surface. Before reaching their interacting partners at the chip, analytes must pass through this unstirred layer by diffusion which may limit binding due to mass transport restrictions [62]. It also adds additional fit parameters to the data analysis and may restrict precision of results. Diffusion rates through the unstirred layer of high molecular weight analytes are significantly lower than diffusion rates of analytes with low molecular weight. Furthermore, the thickness of the unstirred layer depends on the flow rate of the running buffer over the sample. Higher flow rates yield thinner unstirred layers which may be beneficial for data analysis but also requires larger quantities of analyte and may perturb the surface mechanically. Experiments as a function of flow rate may help to determine the influence of mass transport through the unstirred layer on analyte binding. Another approach of measuring KD is to determine the steady state response of analyte binding as a function of concentration and using the Langmuir adsorption equation for interpretation. Ideally, both approaches yield similar dissociation constants of analyte binding [63].
An additional technical issue to be aware of is that the immobilized interaction partner that binds the analyte may have an intrinsic heterogeneity of analyte binding properties as well as induced heterogeneity from immobilization on the chip surface. Methods for analysis of results in case of more complex situations have been developed [22, 68-70, 82] [ 4].
The majority of applications of SPR involve measurement of protein-protein interactions of the type antigen-antibody or peptide-receptor and only a small segment of SPR literature deals with interactions involving peripheral- and integral membrane proteins including GPCR [46, 57]. This is mainly due to difficulties with immobilization of GPCR at a solid interface at sufficiently high density while ensuring adequate stability of the proteins to allow functional studies by SPR [56]. In the following, we describe the current state of SPR studies applied to GPCR. The review analyzes reports on GPCR interaction with various binding partners and discusses prospects for novel applications of this technology to receptor research.
3. Preparation of protein samples for SPR experiments
Structurally, GPCR are comprised of an extracellular region including an N-terminus and three loops that connect transmembrane helices, the membrane region with seven transmembrane helices that span the lipid bilayer and participate in signal transduction to the interior of the cell, and the intracellular region consisting of three loops, and a C-terminal region that may contain a short amphipathic helix imbedded into the lipid-water interface. Depending on the specific receptor, binding of ligands occurs either to the N-terminal domain or to a ligand-binding pocket comprised of the transmembrane helices. The intracellular surface of the receptor is involved in interaction with cytosolic signaling partners such as G protein and arrestin [73]. SPR offers an excellent opportunity to study these interactions at the molecular level including information on binding affinity, specificity, and kinetics. Furthermore, SPR has the potential to be used in a high-throughput format for screening of drugs that target GPCR.
SPR requires substantial surface concentrations of functional GPCR either in their native plasma membrane or purified and reconstituted into lipid bilayers or any other environment that supports GPCR function. With the exception of rhodopsin, the vast majority of GPCR are present in their native tissues at low levels preventing their investigation by SPR using native membranes or isolation in a purified form via chromatographic procedures. A technically feasible alternative is overexpression of GPCR in a heterologous system such as bacterial, yeast-, insect-, or human cell lines. Critical for success are the selection of a proper expression host that allows production at acceptable yield and expense, high stability and functional activity of the GPCR, and the ability to introduce essential posttranslational modifications [78]. Cell-free expression of GPCR is also gaining attention, although the preparation of fully functional receptors by this method is still problematic [29].
Cell-based strategies for expression of functional GPCR typically produce recombinant receptor inserted into cell membranes. If the use of entire cell membranes is not practical, the expressed protein needs to be solubilized in detergent micelles and purified by one or more chromatographic steps to obtain a pure, homogenous preparation [79]. GPCR are notoriously unstable once extracted from lipid membranes [23, 59, 78], and even short exposure to detergents may induce irreversible unfolding. Furthermore, effects of detergents on solubilization, purification and preservation of the functional fold of GPCR are somewhat receptor-specific [59, 77].
Typically, SPR experiments require that the receptor remains stable at the chip surface for at least several hours, and sometimes as long as several days, preferably at ambient temperature. Several approaches to stabilize GPCR in a functional state in various environments have been developed. It was observed that non-ionic- and zwitterionic detergents are less harmful to GPCR, but optimization of solubilization conditions is often required [59]. Supplementation of micellar solutions of GPCR with strong stabilizing agents such as cholesteryl hemisuccinate (CHS) and/or strongly binding ligands, addition of glycerol, adjustment of ionic strength, and lowering of temperature may further improve stability [23, 78]. Alternative approach to GPCR stabilization involves mutagenesis to raise the temperature of protein thermal denaturation [36, 40, 71]. It is also well known that incorporation of membrane proteins into specific amphipathic environments such as proteoliposomes or amphipols [11] greatly improves protein stability compared to detergent micelles [5, 6, 37]. Another option to stabilize purified GPCR is their reconstitution into nanodiscs that are small lipoprotein particles encircled by scaffolding protein [81]. Nanodiscs have the additional advantage that they allow unrestricted access to both the N- and C-terminal faces of the lipid-embedded GPCR.
While many commercially available SPR biosensors are quite suitable for measuring the kinetics of binding of water-soluble analytes to a target immobilized on a derivatized gold surface, it is challenging to use them for functional studies of membrane proteins. The close proximity of integral membrane proteins to a solid interface requires special surface properties not to perturb their biological activity. Furthermore, in particular detection of smaller ligands requires very high sensitivity that can only be achieved if the receptor molecules are captured on the surface at high density (see Tab. 1). This, in turn, is quite challenging for the low-abundant GPCR. Perhaps, the only exception is represented by the rhodopsin-containing rod outer segment discs that can be used for surface immobilization without further enrichment of the receptor. For the recombinant GPCR expressed in heterologous hosts, preparation of vesicles derived from expression host membranes, solubilization in detergents, or reconstitution into liposomes and nanodiscs prior to surface capture is required. Capture of the detergent-solubilized target receptor can be performed either directly from the crude extract, provided that the interaction with the capture molecule at the surface of the sensor chip is sufficiently strong and specific, or after chromatographic purification of the GPCR and consecutive binding to the chip surface. The surface capturing of nanodiscs can be performed via engineered tags either on the scaffolding protein or on the receptor.
Membrane vesicles containing high concentrations of receptor can be captured on the sensor chip as well. However, GPCR are typically oriented in a random manner in membrane preparations or reconstituted into liposomes [42, 50] which may render a significant fraction of receptors inaccessible for analytes.
Another approach to capture GPCR at the surface of sensor chips is the formation of planar membranes that are tethered to the surface or held at the surface by tethered membrane proteins. In order to accommodate transmembrane proteins with large cytosolic or extracellular domains, tethered spacers have been developed that keep the lipid bilayer at a certain distance from the surface. That is achieved e.g. by using thiophospholipids that carry a triethylenglycol spacer unit to capture membranes and membrane proteins [25]. Also formation of self-assembled monolayers using hydroxyl- and cholesteryl-terminated thiols to capture lipid bilayers was reported [20]. Immobilization of membrane proteins can be achieved either by incorporation of protein from micellar solution into a pre-formed membrane at the chip surface or by attachment of the protein to the chip with consecutive formation of the surrounding membrane. On-chip reconstitution of detergent-solubilized GPCR was reviewed by Cooper [17].
In addition, the membrane-embedded protein can be captured via hydrophobic interaction with lipophilic chains on the sensor surface [37]. This preserves the lateral mobility of receptor molecules which may be critical for studying interactions that involve formation of receptor dimers or oligomers [62]. Surface capture can also be performed via specific interaction of protein tags, labels or epitopes with the sensor chip.
4. SPR studies of GPCR interactions
While the majority of applications involve surface capture of the target GPCR on a sensor chip for interaction with analytes, there are also a few examples where the GPCR itself was used as analyte. Methods for surface capture of GPCR can be separated into two categories to produce either randomly or uniformly oriented protein on the sensor surface (Table 2). Random orientation may be desirable when GPCR-containing vesicles are immobilized via a tag on the receptor since it guarantees that there are always receptor molecules at the surface with their N-terminal- or C-terminal side available for interaction with analytes. In contrast, a uniform orientation of receptor molecules allows controlling access to epitopes on the receptor, e.g. for quantitative studies of ligand-, antibody- and G protein interaction.
Table 2. Experimental strategies for the application of SPR to the study of GPCR interactions.
| Orientation | Environment | Surface | Capturing method | Fig | Protein | Source | Functional test6 | Ref |
|---|---|---|---|---|---|---|---|---|
| GPCR captured in random orientation | GPCR in planar membrane | Phospholipid bilayer (custom made) | Via hydrophobic interactions | 4.A | Rho | Native, not purified, solubilized in detergents | Interaction with G-protein upon activation | [58] |
| Lipid monolayer (custom made) | Via hydrophobic interactions | 4.B | Rho | Native, purified, solubilized in detergents and supplemented with phospholipids | Interaction with G-protein upon activation | [25] | ||
| Lipid monolayer (custom made) | Via hydrophobic interactions | 4.C | CCR5 | Recombinant, enriched membrane vesicles | Ligand binding | [53] | ||
| L1 chip | Via hydrophobic interaction of lipids | 4.D | Rho | Native, not purified membrane vesicles | None | [41] | ||
| L1 chip | Via hydrophobic interaction of lipids | 4.D | OR | Recombinant, not purified membrane vesicles (nanosomes) | Interaction with G-protein upon activation | [74, 76] | ||
| L1 chip | Amine coupling of GPCR followed by formation of lipid bilayer | 4.E | Rho | Native, purified, solubilized in detergents | Interaction with G-protein upon activation | [31] | ||
| GPCR in vesicle | Biotinylated antibody on SA coated chip | Via C-terminal epitope | 4.F | Rho | Native, not purified membrane vesicles | None | [41] | |
| Antibody on custom made surface | Via N- and C-terminal epitope | 4.F,G | OR | Recombinant, not purified membrane vesicles (nanosomes) | Interaction with G-protein upon activation | [8, 75] | ||
| Antibody on custom made surface | Via C-terminal epitope | 4.F | CCR5 | Recombinant, enriched membrane vesicles | Ligand binding | [65] | ||
| Antibody on CM5 chip surface | Via internal and C-terminal epitope | 4.F | α2 AR | Recombinant, not purified membrane vesicles | Ligand binding | [64] | ||
| Ni-loaded NTA chip | Via C-terminal his-tag | 4.F | StaR (A2AR) | Recombinant, not purified membrane vesicles | Ligand binding | [15] | ||
| GPCR captured in uniform orientation | GPCR in planar membrane | SA on custom made surface | Via biotin label on GPCR carbohydrates | 4.H | Rho | Native, not purified membrane/detergent mixed micelles | Interaction with G-protein upon activation | [9] |
| Con A on CM5 chip | Via GPCR carbohydrates | 4.H | Rho | Native, not purified membrane/detergent mixed micelles | Ligand binding and interaction with G-protein upon activation | [13] | ||
| Antibody on modified L1 chip | Via C-terminal epitope followed by formation of lipid bilayer | 4.I | CXCR4 CCR5 | Recombinant, not purified, solubilized in detergents | Ligand and conformation-specific antibody binding | [67] | ||
| GPCR in detergents | Con A or antibody on CM5 chip | Via carbohydrates, N- or C-terminal epitope | 4.J,K | Rho | Native, not purified or recombinant, enriched, solubilized in detergents | Interaction with G-protein upon activation | [32, 33] | |
| Antibody on modified L1 or CM5 chip | Via C-terminal epitope | 4.J | CXCR4CCR5 | Recombinant, not purified, solubilized in detergents | Ligand and conformation-specific antibody binding | [67] | ||
| Antibody on CM4 chip | Via C-terminal epitope | 4.J | CXCR4 CCR5 | Recombinant, purified or not, solubilized in detergents | Ligand or conformation-specific antibody binding | [44, 45, 47, 48, 55] | ||
| Antibody on CM4 chip | Via C-terminal epitope | 4.J | OR | Recombinant, not purified, solubilized in detergents | Ligand binding | [16, 30] | ||
| Antibody on CM4 chip | Via C-terminal epitope | 4.J | CB2 | Recombinant, purified or not, solubilized in detergents | Ligand binding followed by radioassay7 | [38] | ||
| Avidin or antibody on custom made surface | Via biotin on N-terminal region or via N-terminal epitope | 4.J,K | β2AR | Recombinant, purified, solubilized in detergents | Conformational change upon ligand binding followed by fluorescence7 | [49] | ||
| Antibody on CM5 chip | Via internal and C-terminal epitope | 4.J | α2 AR | Recombinant, not purified, solubilized in detergents | Ligand binding | [64] | ||
| Ni loaded NTA on custom made surface | Via C-terminal his-tag | 4.J | CB2 | Recombinant, purified, solubilized in detergents | None | [72] | ||
| Ni loaded NTA chip | Via C-terminal his-tag | 4.J | StaR (A2AR) | Recombinant, purified or not, solubilized in detergents | Ligand binding | [15, 83] | ||
| Ni-loaded NTA chip | Via His-tag followed by amine coupling | 4.L | StaRs (A2AR, β1AR) | Recombinant, purified, solubilized in detergents | Ligand binding | [54] | ||
| GPCR in nanodisc | Antibody on CM5 chip | Via C-terminal epitope on GPCR | 4.M | CCR5 | Recombinant, membrane fraction solubilized in detergents, reconstituted into nanodiscs | Ligand and conformation-specific antibody binding | [81] | |
| GPCR in virus particle | C1 and CM3 chips | Amine coupling of GPCR-containing virus particle | - | CXCR4 CCR5 | Recombinant, virus particles | Conformation-specific antibody binding | [26] | |
| GPCR on cell surface | Gold layered chip (Moritex) | Cells grown on the chip surface | - | mAChRDR BLT1 PAFR | Recombinant, whole cells | Whole cell response upon stimulation with lligand measured by SPR | [12] | |
| GPCR as SPR analyte | Ligand captured on SA coated chip | - | - | NTS-1R | Recombinant, purified, solubilized in detergents | Ligand binding | [24] | |
| Ligand immobilized on CM5 chip | - | - | PTHR | Recombinant, purified, solubilized in detergents | Ligand binding | [39] | ||
| Ligand immobilized on CM5 chip | - | - | GLP-1R | Recombinant, purified, solubilized in detergents | Ligand binding | [61] |
Functional tests performed on captured GPCR by SPR or indicated technique
No SPR involved in the assay
4.1. Studies on GPCR captured in random orientation
A random orientation of receptor molecules on the sensor surface results either from non-specific attachment of the receptor to the surface, e.g. by amine coupling or incorporation into pre-formed bilayers, or from random orientation of protein molecules in a captured proteoliposome or native membrane vesicle.
4.1.1. GPCR in planar membranes
Rhodopsin
Since bovine rhodopsin can be conveniently isolated at high concentration from rod outer segment discs (ROS) of bovine retinae, it was the most frequently investigated GPCR until the recombinant expression and stabilization techniques for these receptors became more readily available. Salamon et al. [58] reported capturing of rhodopsin in a lipid bilayer. The procedure involved formation of a phosphatidylcholine (PC) bilayer on the silver layer of an SPR chip. Incorporation of bovine rhodopsin into the lipid layer was achieved by addition of a solution of rhodopsin/octyl glucoside (OG)/lipid mixed micelles that was diluted below the critical micelle concentration (CMC) of the detergent OG in the aqueous compartment of the SPR cell (Fig. 4 A). Similarly, successful reconstitution of purified rhodopsin from rhodopsin/OG/egg-PC mixed micelles into micrometer-sized alternating regions of pure, fluid phospholipid bilayers separated by bilayers composed of an outer phospholipid leaflet on a gold-attached inner thiolipid was reported (Fig. 4 B) [25]. In both cases, the ability of rhodopsin to bind transducin in the dark and to dissociate from it in the presence of GTP upon illumination with light was recorded by SPR demonstrating functional reconstitution of rhodopsin on the biosensor surface.
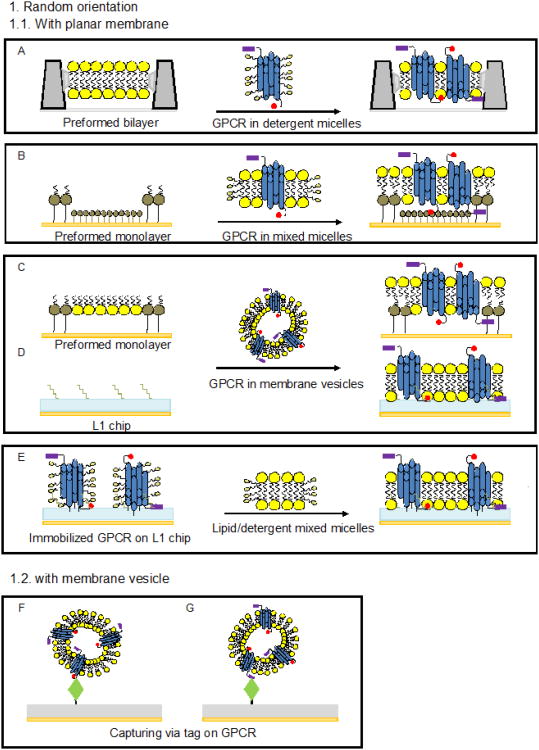
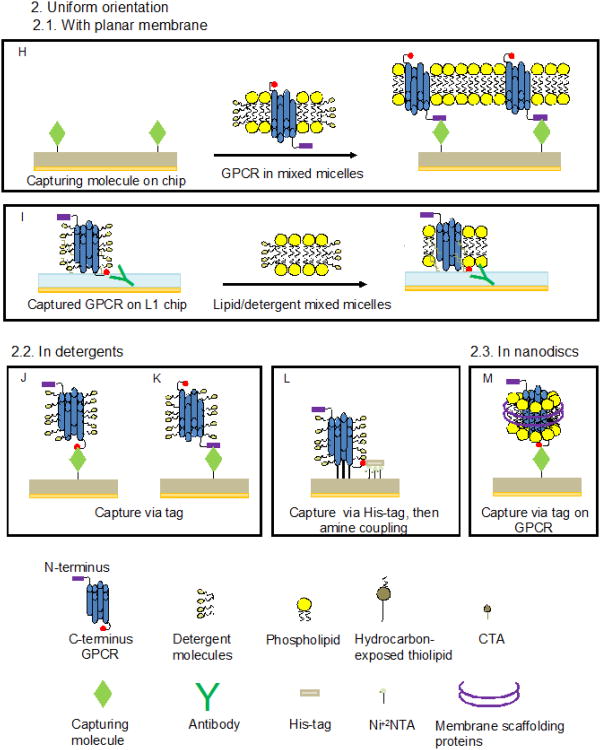
Figure 4. Methods of surface capture of GPCR for SPR studies.
A: Formation of a GPCR-containing lipid bilayer on a sensor chip by providing the GPCR in detergent micelles to a pre-formed supported PC bilayer [58]. B: Formation of a GPCR-containing lipid bilayer on a sensor chip by providing the GPCR in PC/detergent mixed micelles to a patterned organic monolayer consisting of alternating regions of carboxyl-exposed thiols (CTA) and hydrocarbon-exposed thiolipids [25]. C: Capture of GPCR-containing membrane vesicles onto a phospholipid/thiohexa (ethylene oxide) octadecane (THEO-C18) mixed monolayer on a gold surface [53]. D: Capture of GPCR-containing membrane vesicles onto a L1 sensor chip [41, 74, 76]. E: Immobilization of a GPCR in detergent micelles by amine coupling on a L1 chip followed by injection of lipid/detergent mixed micelles and formation of a lipid bilayer [31]. F and G: Capture of GPCR-containing membrane vesicles via tags or epitopes. Capture methods take advantage of the following interactions: Rho-tag/1D4 antibody [41, 65], c-Myc/antibody [8, 75], HA-tag or GPCR internal epitope/antibody [64] his-tag/NiNTA [15]. H: Capture of a GPCR delivered in detergent-solubilized membranes onto a functionalized surface. Examples include the use of a GPCR biotinylated at N-terminal carbohydrates and a patterned monolayer consisting of ω-hydroy-undecanethiol (HTA) and biotinylatedthiols to which streptavidin was attached [9] or a non-labeled GPCR attached via carbohydrates to an immobilized lectin [13]. I: Capture of a GPCR in detergent micelles via a C-terminal tag on an antibody-coated L1 chip followed by injection of lipid/detergent mixed micelles and formation of a lipid bilayer [67]. J and K: Capture of a GPCR in detergent micelles onto a functionalized surface. Capture methods were based on the following interactions: N-terminal cahrbohydrates of the GPCR/Concanavalin A [32, 33], N- and C-terminal GPCR epitopes/antibody [32], GPCR internal epitope or HA-tag/antibody [64], biotinylated N-terminal carbohydrates of the GPCR/streptavidin or FLAG-tag/antibody [49], Rho-tag/1D4 antibody [16, 32, 38, 44, 45, 47, 48, 55, 67], His-tag/anti polyhistidine antibody [30], or His-tag/NiNTA [15, 72, 83]. L: Capture of a GPCR in detergent micelles via C-terminal His-tag onto a NTA chip followed by amine coupling [54]. M: Capture of GPCR-containing nanodiscs via a C-terminal Rho-tag on the GPCR onto an antibody-coated surface [81].
Another approach to surface capture was demonstrated in [31], where rhodopsin was immobilized by amine coupling to the hydrophobic surface of a L1 sensor chip at a concentration equivalent to 4,000 RU followed by formation of a lipid bilayer (Fig. 4 E). The ability of the receptor to bind α transducin was preserved; the release of G protein was demonstrated upon photoactivation of rhodopsin in the presence of GTP. In another approach using a L1 chip, rhodopsin at a concentration of 30 μg/mL was directly captured from mildly sonicated ROS at a surface density equivalent to 4,000 RU (Fig. 4.D). The presence of rhodopsin on the surface was confirmed by binding of a specific antibody. However, questions regarding the structure of rhodopsin-containing bilayer fragments at the surface and the functional state of the GPCR remained unanswered [41].
Recombinant GPCR
For human olfactory receptor OR 17-40 several strategies for capturing the receptor on a chip surface in an environment of native lipids were developed. The OR 17-40 and its cognate Gα- subunit (Gαolf) were co-expressed in membranes of the yeast Saccharomyses cerevisiae. The Biacore L1 sensor was used to attach receptor-containing membrane vesicles (nanosomes) to the sensor surface resulting in formation of planar membranes containing randomly oriented receptor at a surface density of ∼ 3,000 RU (Fig. 4D) [76]. Due to the low molecular weight and hydrophobicity of odorant ligands, their binding could not be directly monitored by SPR. However, the desorption of Gαolf triggered by the ligand helional in the presence of GTPγS was measured quite accurately. Using the same experimental setup, a specific interaction of the odorant binding protein to the receptor and its release by specific ligands was demonstrated and the values of koff at 25° C for two binding sites (1.5 × 10-4 s-1 and 3.1 × 10-6 s-1) were reported [74].
Capture of recombinant human CCR5 chemokine receptor in membrane bilayers onto a custom designed hydrophobic surface (Fig. 4 C) was reported [53]. By using monoclonal antibodies against either the N- or C-termini of the receptor, the presence of randomly oriented receptor molecules in the bilayer was established. The receptor retained ligand binding competence upon surface immobilization as demonstrated by chemokine binding.
4.1.2. GPCR in membrane vesicles
Rhodopsin in ROS prepared as described in section 4.1.1. [41] was also captured onto an antibody-coated sensor chip taking advantage of the corresponding C-terminal nanopeptide epitope on the receptor (Rho-tag, Fig. 4 F). The presence of rhodopsin on the surface was confirmed by binding of antibody 1D4, the same as used for capturing. However, the functional state of immobilized receptor was not tested.
Other examples of capture of GPCR-containing membrane vesicles, described in the following paragraphs, were reported for the recombinant olfactory receptor 17-40 expressed in yeast nanosomes [75] [8], chemokine receptor CCR5 produced in COS-1 cell membranes [65], the α2 adrenergic receptor [64], and StaR adenosine A2A receptor [15], both expressed in insect cell membranes.
A comparison of strategies to capture olfactory receptor (OR) nanosomes onto different functionalized surfaces was performed in [75]. Both, specific attachment via uniformly immobilized antibodies against N-terminal c-Myc and C-terminal HA protein tags (Fig. 4 F and G), or non-specific adsorption of the receptor-containing vesicles were investigated. Best quantitative capture of functional receptor was observed when the OR was specifically attached via the C-terminal tag [75]. The capture method was further advanced by using the N-terminal c-Myc tag on the receptor interacting with an anti- c-Mycantibody randomly adsorbed on a gold surface [8]. Only an estimated 11% of surface-attached antibodies were properly oriented to interact with the OR resulting in biofilms with lower thickness and higher porosity than those obtained by using uniformly oriented antibodies for OR captur. The low density of receptor-containing nanosomes on the sensor surface gave rise to a measurable increase in odorant sensitivity which was attributed by the authors to a better access of GTPγS to the receptor-bound Gαolf. It was also reported that the nanosomes with a diameter of 50 nm, containing up to ten receptor molecules per vesicle, flattened out due to collapse of the particle upon capture at the chip surface but did not merge into a continuous bilayer.
In all studies on OR 17-40, whether in closed membrane vesicles or in planar membranes, the detected desorption response of Gαolf triggered by odorant binding was of low magnitude (∼ 20 RU) indicating potential problems with delivery of odorant ligands to the binding site on the receptor or low receptor/Gα density on the surface. However, these biofilms generated signals that were reproducible, ligand-specific, and, therefore, useful to assess functionally relevant binding of various interaction partners to OR.
Vesicles containing chemokine receptor CCR5 were specifically captured via a C-terminal Rho-tag on the receptor by the above mentioned 1D4 monoclonal antibody immobilized on a functionalized gold surface (Fig. 4 F) [65]. Binding of antibodies and a chemokine ligand to the captured receptor confirmed the presence of ligand binding-competent receptor on the sensor surface. Furthermore, α2 adrenergic receptor membranes were captured onto an antibody-coated CM5 chip via a C-terminal HA affinity tag or a GPCR internal epitope on the receptor [64], and adenosine A2A receptor vesicles were captured via a receptor C-terminal polyhistidine tag onto a Ni+2-loaded NTA chip [15]. Both surface-immobilized receptors retained the ability to bind ligands.
4.2. GPCR captured in uniform orientation
Surface capture of GPCR in a desired orientation can be achieved by taking advantage of a naturally occurring carbohydrate moiety, binding epitopes, or engineered affinity tags like Rho-tag, Flag-tag, c-Myc-tag, poly-histidine tag, or biotin tag placed at either the N- or C-terminus of the receptor. SPR studies on surface-attached GPCR can be performed (i) using detergent-containing buffers, (ii) upon formation of a lipid bilayer around the captured receptor, or (iii) after reconstitution of the GPCR into nanodiscs. The advantage of the nanodiscs over bilayers is that both sides of the captured receptor are potentially accessible for interaction with the binding partners. Furthermore, nanodiscs allow selection for receptor monomers or dimers which could be important for understanding mechanisms of activation of cognate G protein.
4.2.1. Planar membranes
Rhodopsin
In order to capture bovine rhodopsin in a uniform orientation, ROS membranes were solubilized in detergent, and carbohydrates attached at the N-terminal end were specifically biotinylated. This facilitated interaction of rhodopsin with streptavidin immobilized on a mixed self-assembled monolayer (SAM) and created micropatterns of receptor molecules with the intracellular domain pointing away from the surface (Fig. 4 H) [9]. The removal of detergent upon dilution generated a supported lipid bilayer used for studies of binding of α transducin. Activity of the immobilized, dark adapted rhodopsin was monitored by inducing desorption of G protein by a flash of light performed in the presence of GTP. Binding of 11-cis- or 9-cis retinal by opsin was evaluated, and the KD value at ambient temperature for binding of 11-cis retinal to opsin determined to be 130 nM.
In a somewhat simplified approach, rhodopsin was directly captured from solubilized ROS membranes via its N-terminal carbohydrate moiety on a CM5 sensor coated with covalently attached concanavalin A (ConA) lectin (Fig. 4 H) [13, 51]. While a continuous planar membrane was not formed in this case, lipids were still present on the chip surface as membrane patches. Using this strategy, capture levels of 600-1,000 RU for the GPCR and 400-700 RU for cognate G proteins were achieved. The captured rhodopsin retained activity as a catalyst for GTP exchange on G protein subunits and was stable for several days at ambient temperature. The study of the interaction of rhodopsin with G protein subunits reveled a relatively low affinity to Gαt and Gβ1γ1 when binding to these subunits was performed separately (KD at ambient temperature ∼ 300 nM) and much tighter binding when the heterotrimeric G protein was provided in the assay.
Chemokine receptors
Detergent-solubilized chemokine receptors CXCR4 and CCR5, which are essential mediators of the immune response and entry receptors for HIV, were successfully captured onto a L1 chip coated with 1D4 antibody followed by reconstitution of a lipid bilayer around the captured receptor (Fig. 4 I). The binding of a native chemokine ligand of CXCR4 (stromal cell-derived 1α) and of conformation-specific antibodies to the reconstituted receptor was demonstrated [67]. Capturing without membrane formation was also attempted (see below). The results suggest that both detergent-solubilized as well as membrane-reconstituted receptors preserve the ability to bind a specific ligand.
4.2.2. GPCR in detergent
Capture via carbohydrate moiety
Native rhodopsin solubilized in CHAPS from ROS disc preparations was captured onto a concavaline A-coated CM5 chip via its N-terminal-located carbohydrate chains [33] (Fig. 4 K). The binding of Gt to photoactivated rhodopsin in the absence of nucleotides was characterized. A KD value of ∼ 13.6 nM was determined for the heterotrimeric G protein at 25 °C [33]. However, only about 6-9% of available binding sites were occupied by G protein upon light-activation of the receptor at maximal G protein concentration. The same authors studied alternative capturing strategies for the native rhodopsin (Fig. 4 J and K) and established better capturing of the receptor via its C-terminal rather than N-terminal epitope [32].
Capture via Rho-tag
Purified recombinant rhodopsin, generated by expression of opsin in COS-1 cells and reconstituted with 9-cis retinal, was solubilized in CHAPS and captured via its C-terminal epitope onto a 1D-4 antibody-coated sensor chip surface (Fig. 4 J) [32]. The recombinant receptor was able to bind Gt upon light exposure and to dissociate from it in the presence of GTP. While the affinity of this interaction at 25° C (KD = 9.1 nM) was similar to the one determined for native rhodopsin [33], the rates of association and dissociation were 3-4 times lower at the same temperature. Furthermore, regeneration of rhodopsin activity using 9-cis retinal took significantly longer to complete [9]. This suggests that some structural features of rhodopsin might be affected in the absence of a proper lipid bilayer.
The capture of chemokine receptors CCR5 and CXCR4 via a C-terminal Rho-tag was studied as well [43]. Stenlund et al. [67] established a method for capturing of functional receptor from crude cell lysate extracts on two different antibody coated surfaces, a CM5 chip (Fig. 4 J) and a hydrazide-modified L1 sensor chip (Fig. 4 I). Carboxymethylated dextran chips (mostly CM4) were further used to explore detergent solubilization and crystallization conditions for chemokine receptors, to develop purification strategies, and to study interaction with HIV envelope glycoprotein gp120, ligands, and small-molecule inhibitors. Functionality of the captured receptors was investigated by ligand binding [44, 45, 67] and interaction with antibodies that recognize structural epitopes in the extracellular N-terminal domain of the receptors [47, 48, 55, 67].
Similar to the chemokine receptors, the successful capture of C-terminal Rho-tag modified olfactory receptor OR 17-4 expressed in HEK293S cells [16] was demonstrated (Fig. 4 J). The receptor solubilized from a crude cell extract with detergent was captured on the 1D4-antibody-coated surface of a CM4 sensor chip at a surface density of 2,800 RU. In addition, capture of the same receptor via a poly-histidine tag onto an anti-His antibody-coated CM4 chip resulted in a surface coverage equivalent to 4,000 RU [30]. Such high densities of capture enabled direct measurement of binding of small odorant ligands: undecanal (170 Da), lilial (204 Da) and floralozone (190 Da). While specific- and dose-dependent binding was shown and ligand binding affinities estimated to be in the low μM-range at ambient temperature, the magnitude of these responses did not exceed 8 RU. The low water solubility and low molecular weight of the ligands in combination with a relatively low affinity for the receptor render this SPR-based ligand binding assays very challenging. On the other hand, a quantitative capture of receptor molecules with a uniformly accessible extracellular surface makes the detection of the ligand binding event by SPR feasible.
Cannabinoid receptor type II (CB2) was expressed in E. coli cells as a C-terminal fusion with the thioredoxin A (TrxA) of E. coli and a Rho-tag, and captured either directly from crude cell extracts (as a N-terminal fusion with maltose-binding protein (MBP)) or after purification and removal of expression tags, at a density of about 3,000 RU. The accessibility of eptitops on the receptor and affinity at 25 °C, (KD ∼2.2 nM) of a monoclonal antibody against CB2 were determined. The kinetics of removal of the N-terminal MBP from the fusion by action of specific TEV protease was studied on a surface-captured MBP-CB2 fusion protein by SPR [38].
Capture via FLAG-tag
The N-terminal FLAG-tag was used to capture the β2-adrenergic receptor on a custom-made surface in an oriented fashion (Fig. 4 K) [49]. Special attention was paid to avoiding surface-induced denaturation of receptor by introducing a spacer between the metal surface and the GPCR. The receptor was delivered to the surface solubilized in dodecyl maltoside (DDM) micelles and ligand binding competence upon immobilization was confirmed in real-time by fluorescence microscopy to monitor the conformational changes of the receptor in response to binding of an agonist. While it remains to be seen if ligand binding can also be measured by SPR, this approach is promising since formation of a lipid bilayer that may limit access to one surface of the receptor is not required for its functional immobilization.
Capture via HA-tag
The use of the N-terminal HA-tag to capture the α2-adrenergic receptor in an oriented fashion on the surface of a CM5 sensor chip coated with anti-HA monoclonal antibody was reported (Fig. 4 J) [64]. Receptor expressed in insect Sf9 cells was captured after solubilization in DDM and Big CHAP detergents. Binding of several low molecular weight ligands to the receptor was detected generating binding responses ranging from 1-2 to ∼37 RU.
Capture via His-tag
Capture of the recombinant human cannabinoid receptor CB2 expressed in E. coli and purified by affinity chromatography was performed using a C-terminal decahistidine tag [80] (Fig. 4 J). A custom-modified gold surface covered with a polyethylene oxide SAM with controlled density of Ni+2-NTA functional groups was used. The receptor was stable when delivered to the surface in buffers containing DDM, CHAPS, the cholesterol derivative CHS, glycerol and a high affinity cannabinoid ligand CP-55,940, at 8°C. The surface could be regenerated at mild conditions by supplementing the running buffer with high concentrations of imidazole. At ambient temperature, partial inactivation and irreversible precipitation of the receptor on the chip surface occurred [72].
The use of the C-terminal His-tag for receptor capture was demonstrated for surface immobilization of the thermostabilized A2A adenosine receptor (StaR) produced in Trichoplusia ni (Tni) cells. Purified receptor molecules were deposited onto a Ni+2-charged NTA chip, either reconstituted into membranes (section 4.1.2.) or solubilized in detergents at densities equivalent to 10,000 or 7,000 RUs, respectively (Fig. 4 J) [15]. Functional activity was established by binding of the ligand xanthine amine congenerer (XAC). The full kinetic analysis of ligand binding for five antagonists with molecular weights in the range from 286-428 Da, a kinetic screening of antagonists binding, and a binding screen of ligand fragments selected from a compound library (MW 136-194 Da) were successfully performed demonstrating the viability of SPR for characterizing the binding properties of StaR. The capturing strategy was further adapted to map the ligand binding pocket on the receptor [83]. The new method, called “biophysical mapping” involved single amino acid substitutions on the stabilized receptor at positions likely involved in interaction with ligands. Receptor variants were then solubilized from HEK-293T cell membranes, captured onto the sensor surface, and the binding of each of them to a set of ligands tested at 10°C. GPCR capturing levels were between 4,000-10,000 RU while ligand binding responses were below 40 RU, sometimes as low as 2 RU.
In another study [54], His-tagged StaR A2A R and β1AR proteins were immobilized by a hybrid capture-coupling technique consisting of a mild pre-concentration of receptors via the His-tag on the Ni-NTA surface and subsequent amine coupling to the pre-activated surface (Fig. 4 L) resulting in stable protein layers. The binding of eight low molecular weight antagonists ranging in affinity from 2.8 nM to 84 μM at ambient temperature was characterized by SPR.
4.2.3. GPCR in nanodiscs
CCR5 with a C-terminal Rho-tag incorporated into nanodiscs was captured onto a CM5 sensor chip surface via 1D4 antibody interaction (Fig. 4 M) [81]. The stability of the receptor reconstituted into nanodiscs or solubilized in detergent micelles was evaluated by monitoring SPR binding responses of a conformation-sensitive antibody. While CCR5 in nanodiscs yielded a stable SPR signal for up to 24 hours at 4°C, the receptor was rather unstable in 1% DDM with a half-life of less than 2 hours. Dose-dependent binding of a chemokine MIP-1α monomeric variant (MW 12 kDa) containing three amino acid substitutions to the nanodisc-imbedded receptor yielded a KD=5 μM at ambient temperature. However, the SPR response with amplitude of less than 10 RU was rather low. Previously reported values of IC50 obtained from competition binding experiments [10] and KD values obtained by saturation binding [14], both using radiolabeled ligand, yielded affinities in the subnanomolar range, suggesting that optimization of the procedure to measure interaction of molecules with nanodisc-reconstituted proteins is required.
4.2.4. Virus- and cell-based SPR
For the study of low-affinity interactions, a quantitative immobilization of one of the binding partners is desirable. High surface densities of chemokine receptors in a native lipid environment were achieved by immobilizing virus particles expressing recombinant CXCR4 and CCR5 receptors by amine coupling on the surface of C1 and CM3 biosensors [26]. Receptor function was tested using specific, conformational-sensitive antibodies. Furthermore, the thermodynamic dissociation constant for interaction with gp120 at 25° C was estimated to be 506 nM.
A SPR-based approach to measure whole cell responses triggered by ligands and mediated by GPCR was reported [12]. Chinese hamster ovary (CHO) cells stably expressing either muscarinic acetylcholine receptor M2 (mAChR M2), dopamine receptor (DR D2), leukotriene B4 receptor (BLT1) or platelet-activating factor receptor (PAFR) in addition to endogenous GPCR were grown on the gold surface of a SPR sensor chip. A dose-dependent SPR response was observed after stimulation with cognate ligands, and the EC50 values determined. The SPR response was attributed to density changes at the sensor chip surface resulting from cytoskeletal rearrangements upon agonist binding to the GPCR. While the study presented an interesting attempt to characterize the in-vivo response of several recombinant GPCR to various stimuli, this approach did not allow accurate measurements of the affinity of interaction of a GPCR with binding partners. The system could be potentially useful for screening of libraries of compounds that modulate GPCR signaling in an environment that closely resembles native tissues.
4.3. GPCR as an analyte
Studies of ligand-GPCR interaction can also be performed using a target receptor as a “soluble analyte” while ligands are attached to the surface of the sensor chip. An example of this approach was reported by Michalke et al. [39] using the recombinant human parathyroid hormone receptor (huPTH1R). The GPCR was expressed in E. coli as inclusion bodies, solubilized and refolded in detergent micelles, and its affinity to surface-immobilized parathyroid hormone (PTH) studied by SPR. The 4.1 kDa PTH peptide ligand was covalently attached to a CM5 chip by amine coupling. Immobilization of the ligand resulted in an interfacial binding of the refolded receptor allowing rapid regeneration of the chip surface by flushing with SDS solution. Following a similar strategy, binding of the neurotensin receptor to a ligand-coated SA chip (Biacore) and of the glucagon-like peptide-1 receptor to a captured cysteinylated agonist were reported in [24] and [61], respectively.
The use of GPCR as an analyte has the advantage of generating a very high SPR response to GPCR binding. The challenge is the need for development of surface-tethered ligands with high affinity for the GPCR. This is easier to achieve for hydrophilic ligands such as water soluble peptides than for small hydrophobic ligands. The same technology is used for selective adsorption of functional GPCR molecules to the packing material of columns for receptor purification. A disadvantage for functional studies could be that tethered ligands may not have the same affinity for GPCR as their untethered counterpart.
5. Conclusions
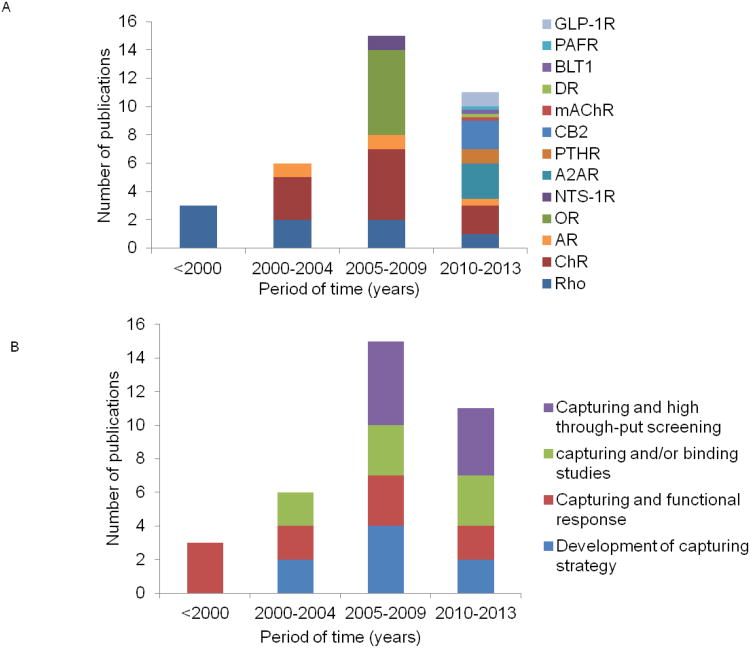
This review illustrates that SPR was successfully applied to study GPCR isolated from native tissues (rhodopsin) or produced by recombinant expression. Since the introduction of SPR methodology in early 1990s, there has been a steady increase in the number of published papers on applications to GPCR (Fig. 5). We are aware of SPR experiments with 13 different types of GPCR. However, SPR studies using rhodopsin and chemokine receptors represent about half of published reports. The mechanism of activation of G protein was studied for two GPCR only, the olfactory receptor and rhodopsin. While the majority of early SPR studies focused on bovine rhodopsin, more recently the fraction of papers on a variety of recombinant receptors increased. This trend parallels the increase in the number of reported high resolution structures of GPCR and is the result of significant technical advances in expression, structure-preserving purification and stabilization of these receptors.
Figure 5. Analysis of SPR publications on GPCR.
A, Timeline of publications on SPR applied to the study of GPCR. The receptors analyzed include rhodopsin (Rho) [9, 13, 25, 31-33, 41, 58], chemokine receptors CCR5 and CXCR4 (ChR) [26, 44, 45, 47, 48, 53, 55, 65, 67, 81], β1, β2 and α2 adrenergic receptors (AR)[49, 54, 64], olfactory receptors (OR) [8, 16, 30, 74-76], neurotensin receptor (NTS-1R)[24], adenosine receptor (AA2R)[15, 54, 83], parathyroid hormone receptor (PTHR)[39], glucagon-like peptide-1 receptor (GLP-1R)[61], cannabinoid receptor type II (CB2) [38, 72], muscarinic acetylcholine receptor (mAChR), dopamine receptor (DR), leukotrien B(4) receptor (BLT1), and platelet-activating factor receptor (PAFR)[12]. B, SPR studies of GPCR over time. Publications were grouped according to the main focus of the reported study. Some of the papers with main focus on development of strategies for capture of GPCR may include confirmation of the functional state of GPCR performed by SPR [8, 32, 53, 65, 75] or by another method [49] and some of them do not [41, 72]. Studies on capture of GPCR and investigation of a functional response include [9, 12, 13, 25, 31, 33, 58, 74, 76], capture and/or binding studies with ligands or antibodies [16, 24, 26, 30, 38, 39, 61, 67, 81], and high throughput screening [15, 44, 45, 47, 48, 54, 55, 83].
The applications covered investigation of (i) mechanistic aspects of GPCR activation by measuring the turnover of G protein triggered by an inducer (light or small ligands), (ii) characterization of binding partners such as antibodies, ligands, or allosteric modulators, and (iii) high-throughput screening of buffers to optimize solubilization and crystallization conditions, as well as screening for binding of ligands and ligand fragments. Earlier publications focused on the development of procedures to capture GPCR on SPR chips while more recent papers explore application of SPR to study interaction with binding partners and to high-throughput screening of ligands and conditions for analyte binding. Considering the high cost of sensor chips for SPR, it is highly desirable that the surface can be regenerated and re-used. In many instances this requirement is met by using affinity tag-antibody interactions that also ensure high stability of the SPR signal over long injection times in addition to surface regeneration. Non-specific surface attachment of membrane vesicles containing GPCR is less desirable since it usually does not generate a steady SPR response.
The intrinsic instability of GPCR in detergent micelles complicates SPR studies and makes preservation of the functional structure of receptors on a sensor chip a particularly challenging task. To assess the functional state of the receptor captured to the sensor chip surface, binding of G protein, ligands, and of conformation-specific antibodies is performed. Measurement of the cellular response upon treatment with ligands that bind to specific receptors expressed by the cell was also reported. The quantitative measurement of G protein binding is quite challenging and depends on several parameters such as the functional state of the receptor, accessibility of the relevant epitopes to ligand and G protein, and the presence of a lipid bilayer. A direct, quantitative measurement of small ligand binding can be performed reliably with (i) hydrophilic ligands such as peptides or hormones, and (ii) on receptors captured at the surface at sufficiently high density. Hydrophobic ligands, on the other hand, tend to partition into the lipid matrix or the micelles which makes SPR determination of their specific binding to the receptor extremely challenging. Binding of conformation-specific antibodies may, in principle, deliver more accurate, quantitative results on the functional state of the receptor provided that an antibody that selectively recognizes a functional fold of the receptor is available.
In addition to ligand binding, studies of the turnover of G proteins on a ligand-activated receptor are very informative. They not only confirm the ability of the receptor to specifically interact with ligands but report on the entire cascade of conformational changes that lead to activation and release of G protein. It is desirable to conduct experiments on receptors imbedded into lipid bilayers since the lipid matrix stabilizes the GPCR and provides a native-like environment for interaction with G protein. Such experiments do not necessarily require a uniform orientation of captured receptor molecules as long as there is a sufficient fraction of receptor molecules with an accessible C-terminal surface and unrestricted access to the ligand binding site. However, the scenario in which the captured GPCR is randomly oriented on the chip surface may present problems for reproducibility of experiments and for quantification of the SPR response. On the other hand, a uniform orientation of lipid-imbedded receptors on the chip surface, either in nanodiscs or in tethered planar bilayers generates a more native-like environment with higher surface density of quantifiable receptor that is properly oriented for interaction. Experiments on oriented GPCR in single lipid bilayers are likely to be more challenging than experiments on nanodiscs because in bilayers only the intra- or extracellular surface of the receptor are exposed to analytes. In this regard, the use of hydrophobic ligands is potentially an advantage since they readily partition into the lipid matrix and may access the binding site on the receptor from the lipid matrix, see e.g.[61, 83]. While bilayers attached to the surface via tethered lipids may present a good experimental setup since they allow lateral mobility of imbedded proteins, they require custom designed SAM at the chip surface that may not be readily available to most users.
Compared to other methods for conducting functional studies on GPCR, e.g. ligand binding studies on GPCR-containing cell membrane preparations using a radioactive assay or G protein-activation studies, application of SPR has the advantage of potentially producing quantitative data on analyte binding that are difficult to measure by other methods, e.g. rates of analyte binding and release, but are of high relevance for pharmacological applications. Furthermore, SPR experiments are easily converted to a high-throughput format for automatic screening of a large set of analytes. The challenges with application of SPR are the necessity of achieving reproducible immobilization of GPCR at high concentration at the chip surface in an environment close to natural conditions such that function is not impaired.
Regarding stability of GPCR and proper function, the stable and physiologically relevant environment provided by lipid bilayers has many advantages over detergent-micelles. Therefore, the use of supported lipid bilayers is attractive for application of SPR to GPCR. The recently developed nanodisc technology still needs further testing in SPR applications to ensure sufficient surface density of captured nanodiscs and to account for possible artifacts in receptor function caused by the presence of scaffolding protein. However the method holds promise for SPR applications to GPCR since it potentially allows uniform orientation of surface-captured receptor molecules while maintaining unobstructed access to both intra- and extra-cellular surfaces of the receptor.
Acknowledgments
I.G. was supported the intramural research program of the National Institute of Biomedical Imaging and Bioengineering, NIH, and S. L-H, A.Y. and K.G. were supported by the intramural research program of the National Institute on Alcohol Abuse and Alcoholism, NIH.
Abbreviations
- GPCR
G protein-coupled receptors
- SPR
surface-plasmon resonance
- TIR
total internal reflection
- SAM
self-assembled monolayers
- CMC
critical micelle concentration
- DDM
dodecyl maltoside
- OG
octyl glucopyranoside
- CHS
cholesteryl hemisuccinate
- PC
phosphatidylcholine
- ROS
rod outer segment discs
- GTP
guanosine triphosphate
- ConA
concanavalin A
- SA
streptavidin
- Rho-peptide
C-terminal nanopeptide of bovine rhodopsin
- HA
Human influenza hemagglutinin
- Rho
rhodopsin
- OR
olfactory receptor
- ChR
chemokine receptors
- AR
adrenergic receptors
- NTS-1R
neurotensin receptor
- AA2R
adenosine receptor
- PTHR parathyroid hormone receptor
CB2, cannabinoid receptor type II
- mAChR
muscarinic acetylcholine receptor
- DR
dopamine receptor
- BLT1
leukotrien B(4) receptor
- PAFR
platelet-activating factor receptor
- GLP-1R
glucagon-like peptide-1 receptor
Footnotes
Authors declare no competing interests.
References
- 1.Biacore 3000 Instrument handbook. GE Healthcare; Uppsala: 2007. [Google Scholar]
- 2.Biacore sensor surface handbook. GE Healthcare; Uppsala: 2007. [Google Scholar]
- 3.BIAevaluation software handbook. GE Healthcare; Uppsala: 2007. [Google Scholar]
- 4.Altschuh D, Bjorkelund H, Strandgard J, Choulier L, Malmqvist M, Andersson K. Deciphering complex protein interaction kinetics using Interaction Map. Biochem Biophys Res Commun. 2012;428:74–79. doi: 10.1016/j.bbrc.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 6.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayburt TH, Sligar SG. Membrane protein assembly into nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benilova I, Chegel VI, Ushenin YV, Vidic J, Soldatkin AP, Martelet C, Pajot E, Jaffrezic-Renault N. Stimulation of human olfactory receptor 17-40 with odorants probed by surface plasmon resonance. Eur Biophys J. 2008;7:807–814. doi: 10.1007/s00249-008-0272-5. [DOI] [PubMed] [Google Scholar]
- 9.Bieri C, Ernst OP, Heyse S, Hofmann KP, Vogel H. Micropatterned immobilization of a G protein-coupled receptor and direct detection of G protein activation. Nat Biotechnol. 1999;17:1105–1108. doi: 10.1038/15090. [DOI] [PubMed] [Google Scholar]
- 10.Blanpain C, Migeotte I, Lee B, Vakili J, Doranz BJ, Govaerts C, Vassart G, Doms RW, Parmentier M. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood. 1999;94:1899–1905. [PubMed] [Google Scholar]
- 11.Charvolin D, Perez JB, Rouviere F, Giusti F, Bazzacco P, Abdine A, Rappaport F, Martinez KL, Popot JL. The use of amphipols as universal molecular adapters to immobilize membrane proteins onto solid supports. Proc Natl Acad Sci U S A. 2009;106:405–410. doi: 10.1073/pnas.0807132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen K, Obinata H, Izumi T. Detection of G protein-coupled receptor-mediated cellular response involved in cytoskeletal rearrangement using surface plasmon resonance. Biosens Bioelectron. 2010;25:1675–1680. doi: 10.1016/j.bios.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Clark WA, Jian X, Chen L, Northup JK. Independent and synergistic interaction of retinal G-protein subunits with bovine rhodopsin measured by surface plasmon resonance. Biochem J. 2001;358:389–397. doi: 10.1042/0264-6021:3580389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colin P, Benureau Y, Staropoli I, Wang Y, Gonzalez N, Alcami J, Hartley O, Brelot A, Arenzana-Seisdedos F, Lagane B. HIV-1 exploits CCR5 conformational heterogeneity to escape inhibition by chemokines. Proc Natl Acad Sci U S A. 2013;110:9475–9480. doi: 10.1073/pnas.1222205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Congreve M, Rich RL, Myszka DG, Figaroa F, Siegal G, Marshall FH. Fragment screening of stabilized G-protein-coupled receptors using biophysical methods. Method Enzymol. 2011;493:115–136. doi: 10.1016/B978-0-12-381274-2.00005-4. [DOI] [PubMed] [Google Scholar]
- 16.Cook BL, Steuerwald D, Kaiser L, Graveland-Bikker J, Vanberghem M, Berke AP, Herlihy K, Pick H, Vogel H, Zhang S. Large-scale production and study of a synthetic G protein-coupled receptor: human olfactory receptor 17-4. Proc Natl Acad Sci U S A. 2009;106:11925–11930. doi: 10.1073/pnas.0811089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper MA. Advances in membrane receptor screening and analysis. J Mol Recognit. 2004;17:286–315. doi: 10.1002/jmr.675. [DOI] [PubMed] [Google Scholar]
- 18.Cooper MA, Try AC, Carroll J, Ellar DJ, Williams DH. Surface plasmon resonance analysis at a supported lipid monolayer. Biochim Biophys Acta. 1998;1373:101–111. doi: 10.1016/s0005-2736(98)00091-1. [DOI] [PubMed] [Google Scholar]
- 19.De Mol NJ, Fisher MJE. Surface Plasmon Resonance: methods and protocols. In: De Mol NJ, Fisher MJE, editors. Method Mol Biol. Vol. 627. Springer; New York: 2010. [Google Scholar]
- 20.Fruh V, AP IJ, Siegal G. How to catch a membrane protein in action: a review of functional membrane protein immobilization strategies and their applications. Chem Rev. 2010;111:640–656. doi: 10.1021/cr900088s. [DOI] [PubMed] [Google Scholar]
- 21.Gedig ET. Surface Chemistry in SPR Technology. In: Schasfoort RBM, Tudos AJAJ, editors. Handbook of surface plasmon resonance. Roy Soc Chem; Cambridge: 2008. pp. 173–218. [Google Scholar]
- 22.Gorshkova II, Svitel J, Razjouyan F, Schuck P. Bayesian analysis of heterogeneity in the distribution of binding properties of immobilized surface sites. Langmuir. 2008;24:11577–11586. doi: 10.1021/la801186w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grisshammer R. Understanding recombinant expression of membrane proteins. Curr Opin Biotechnol. 2006;17:337–340. doi: 10.1016/j.copbio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Harding PJ, Hadingham TC, McDonnell JM, Watts A. Direct analysis of a GPCR-agonist interaction by surface plasmon resonance. Eur Biophys J. 2006;35:709–712. doi: 10.1007/s00249-006-0070-x. [DOI] [PubMed] [Google Scholar]
- 25.Heyse S, Ernst OP, Dienes Z, Hofmann KP, Vogel H. Incorporation of rhodopsin in laterally structured supported membranes: observation of transducin activation with spatially and time-resolved surface plasmon resonance. Biochemistry. 1998;37:507–522. doi: 10.1021/bi971564r. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman TL, Canziani G, Jia L, Rucker J, Doms RW. A biosensor assay for studying ligand-membrane receptor interactions: binding of antibodies and HIV-1 Env to chemokine receptors. Proc Natl Acad Sci U S A. 2000;97:11215–11220. doi: 10.1073/pnas.190274097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homola J. Surface Plasmon Resonance Based Sensors. In: Wolfbeis OS, editor. Chem Sens Biosens. Vol. 4. Springer; Berlin Heidelberg: 2006. [Google Scholar]
- 28.Huber T, Botelho AV, Beyer K, Brown MF. Membrane model for the G-protein-coupled receptor rhodopsin: hydrophobic interface and dynamical structure. Biophys J. 2004;86:2078–2100. doi: 10.1016/S0006-3495(04)74268-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kai L, Roos C, Haberstock S, Proverbio D, Ma Y, Junge F, Karbyshev M, Dotsch V, Bernhard F. Systems for the cell-free synthesis of proteins. Method Mol Biol. 2012;800:201–225. doi: 10.1007/978-1-61779-349-3_14. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser L, Graveland-Bikker J, Steuerwald D, Vanberghem M, Herlihy K, Zhang S. Efficient cell-free production of olfactory receptors: detergent optimization, structure, and ligand binding analyses. Proc Natl Acad Sci U S A. 2008;105:15726–15731. doi: 10.1073/pnas.0804766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlsson OP, Lofas S. Flow-mediated on-surface reconstitution of G-protein coupled receptors for applications in surface plasmon resonance biosensors. Anal Biochem. 2002;300:132–138. doi: 10.1006/abio.2001.5428. [DOI] [PubMed] [Google Scholar]
- 32.Komolov KE, Aguila M, Toledo D, Manyosa J, Garriga P, Koch KW. On-chip photoactivation of heterologously expressed rhodopsin allows kinetic analysis of G-protein signaling by surface plasmon resonance spectroscopy. Anal Bioanal Chem. 2010;397:2967–2976. doi: 10.1007/s00216-010-3876-4. [DOI] [PubMed] [Google Scholar]
- 33.Komolov KE, Senin II, Philippov PP, Koch KW. Surface plasmon resonance study of G protein/receptor coupling in a lipid bilayer-free system. Anal Chem. 2006;78:1228–1234. doi: 10.1021/ac051629t. [DOI] [PubMed] [Google Scholar]
- 34.Kooyman RPH. Physics of Surface Plasmon Resonance. In: Schasfoort RBM, Tudos AJ, editors. Handbook of Surface Plasmon resonance. Roy Soc Chem; Cambridge: 2008. pp. 15–34. [Google Scholar]
- 35.Kretschmann E, Raether H. Radiative decay of non radiative surface plasmons excited by light. Z Naturforsch Pt A. 1968;A 23:2135–2136. [Google Scholar]
- 36.Lebon G, Bennett K, Jazayeri A, Tate CG. Thermostabilisation of an agonist-bound conformation of the human adenosine A(2A) receptor. J Mol Biol. 2011;409:298–310. doi: 10.1016/j.jmb.2011.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques. 2006;40:601–602, 604, 606. doi: 10.2144/000112169. passim. [DOI] [PubMed] [Google Scholar]
- 38.Locatelli-Hoops S, Gorshkova I, Gawrisch K, Yeliseev AA. Expression, surface immobilization, and characterization of functional recombinant cannabinoid receptor CB2. Biochim Biophys Acta-Proteins and Proteomics. 2013 doi: 10.1016/j.bbapap.2013.06.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michalke K, Huyghe C, Lichiere J, Graviere ME, Siponen M, Sciara G, Lepaul I, Wagner R, Magg C, Rudolph R, Cambillau C, Desmyter A. Mammalian G protein-coupled receptor expression in Escherichia coli: II. Refolding and biophysical characterization of mouse cannabinoid receptor 1 and human parathyroid hormone receptor 1. Anal Biochem. 2010;401:74–80. doi: 10.1016/j.ab.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Miller JL, Tate CG. Engineering an ultra-thermostable beta(1)-adrenoceptor. J Mol Biol. 2011;413:628–638. doi: 10.1016/j.jmb.2011.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minic J, Grosclaude J, Aioun J, Persuy MA, Gorojankina T, Salesse R, Pajot-Augy E, Hou YX, Helali S, Jaffrezic-Renault N, Bessueille F, Errachid A, Gomila G, Ruiz O, Samitier J. Immobilization of native membrane-bound rhodopsin on biosensor surfaces. Biochim Biophys Acta-Gen Subjects. 2005;1724:324–332. doi: 10.1016/j.bbagen.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell DC, Kibelbek J, Litman BJ. Effect of phosphorylation on receptor conformation: the metarhodopsin I in equilibrium with metarhodopsin II equilibrium in multiply phosphorylated rhodopsin. Biochemistry. 1992;31:8107–8111. doi: 10.1021/bi00150a001. [DOI] [PubMed] [Google Scholar]
- 43.Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 44.Navratilova I, Besnard J, Hopkins AL. Screening for GPCR ligands using surface plasmon resonance. ACS Med Chem Lett. 2011;2:549–554. doi: 10.1021/ml2000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navratilova I, Dioszegi M, Myszka DG. Analyzing ligand and small molecule binding activity of solubilized GPCRs using biosensor technology. Anal Biochem. 2006;355:132–139. doi: 10.1016/j.ab.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 46.Navratilova I, Myszka DG, Rich RL. Probing membrane protein interactions with real-time biosensor technology. In: Pebay-Peyroula E, editor. Biophysical analysis of membrane proteins. Wiley-VCH; Weinheim: 2008. pp. 121–140. [Google Scholar]
- 47.Navratilova I, Pancera M, Wyatt RT, Myszka DG. A biosensor-based approach toward purification and crystallization of G protein-coupled receptors. Anal Biochem. 2006;353:278–283. doi: 10.1016/j.ab.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 48.Navratilova I, Sodroski J, Myszka DG. Solubilization, stabilization, and purification of chemokine receptors using biosensor technology. Anal Biochem. 2005;339:271–281. doi: 10.1016/j.ab.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 49.Neumann L, Wohland T, Whelan RJ, Zare RN, Kobilka BK. Functional immobilization of a ligand-activated G-protein-coupled receptor. ChemBio Chem. 2002;3:993–998. doi: 10.1002/1439-7633(20021004)3:10<993::AID-CBIC993>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 50.Niu SL, Doctrow B, Mitchell DC. Rhodopsin activity varies in proteoliposomes prepared by different techniques. Biochemistry. 2009;48:156–163. doi: 10.1021/bi801835s. [DOI] [PubMed] [Google Scholar]
- 51.Northup J. Measuring rhodopsin-G-protein interactions by surface plasmon resonance. Method Mol Biol. 2004;261:93–112. doi: 10.1385/1-59259-762-9:093. [DOI] [PubMed] [Google Scholar]
- 52.Otto A. Excitation of nonradiative surface plasma waves in silver by method of frustrated total reflection. Z Phys. 1968;216:398–410. [Google Scholar]
- 53.Rao NM, Silin V, Ridge KD, Woodward JT, Plant AL. Cell membrane hybrid bilayers containing the G-protein-coupled receptor CCR5. Anal Biochem. 2002;307:117–130. doi: 10.1016/s0003-2697(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 54.Rich RL, Errey J, Marshall F, Myszka DG. Biacore analysis with stabilized G-protein-coupled receptors. Anal Biochem. 2011;409:267–272. doi: 10.1016/j.ab.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rich RL, Miles AR, Gale BK, Myszka DG. Detergent screening of a G-protein-coupled receptor using serial and array biosensor technologies. Anal Biochem. 2009;386:98–104. doi: 10.1016/j.ab.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rich RL, Myszka DG. Grading the commercial optical biosensor literature-class of 2008: ‘The mighty binders’. J Mol Recognit. 2010;23:1–64. doi: 10.1002/jmr.1004. [DOI] [PubMed] [Google Scholar]
- 57.Rich RL, Myszka DG. Survey of the 2009 commercial optical biosensor literature. J Mol Recognit. 2011;24:892–914. doi: 10.1002/jmr.1138. [DOI] [PubMed] [Google Scholar]
- 58.Salamon Z, Wang Y, Soulages JL, Brown MF, Tollin G. Surface plasmon resonance spectroscopy studies of membrane proteins: Transducin binding and activation by rhodopsin monitored in thin membrane films. Biophys J. 1996;71:83–294. doi: 10.1016/S0006-3495(96)79224-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarramegn V, Muller I, Milon A, Talmont F. Recombinant G protein-coupled receptors from expression to renaturation: a challenge towards structure. Cell Mol Life Sci. 2006;63:1149–1164. doi: 10.1007/s00018-005-5557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schasfoort RBM, McWhirter A. SPR Instrumentation. In: Schasfoort RBM, Tudos AJ, editors. Handbook of Surface Plasmon resonance. Roy Soc Chem; Cambridge: 2008. pp. 35–80. [Google Scholar]
- 61.Schroder-Tittmann K, Bosse-Doenecke E, Reedtz-Runge S, Ihling C, Sinz A, Tittmann K, Rudolph R. Recombinant expression, in vitro refolding, and biophysical characterization of the human glucagon-like peptide-1 receptor. Biochemistry. 2010;49:7956–7965. doi: 10.1021/bi101159s. [DOI] [PubMed] [Google Scholar]
- 62.Schuck P. Use of surface plasmon resonance to probe the equilibrium and dynamic aspects of interactions between biological macromolecules. Annu Rev Biophys Biomol Struct. 1997;26:541–566. doi: 10.1146/annurev.biophys.26.1.541. [DOI] [PubMed] [Google Scholar]
- 63.Schuck P, Minton AP. Kinetic analysis of biosensor data: elementary tests for self-consistency. Trends Biochem Sci. 1996;21:458–460. doi: 10.1016/s0968-0004(96)20025-8. [DOI] [PubMed] [Google Scholar]
- 64.Sen S, Jaakola VP, Pirila P, Finel M, Goldman A. Functional studies with membrane-bound and detergent-solubilized alpha(2)-adrenergic receptors expressed in Sf9 cells. Biochim Biophys Acta-Biomembranes. 2005;1712:62–70. doi: 10.1016/j.bbamem.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 65.Silin VI, Karlik EA, Ridge KD, Vanderah DJ. Development of surface-based assays for transmembrane proteins: selective immobilization of functional CCR5, a G protein-coupled receptor. Anal Biochem. 2006;349:247–253. doi: 10.1016/j.ab.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 66.Stenberg E, Persson B, Roos H, Urbaniczky C. Quantitative-determination of surface concentration of protein with surface-plasmon resonance using radiolabeled proteins. J Colloid Interface Sci. 1991;143:513–526. [Google Scholar]
- 67.Stenlund P, Babcock GJ, Sodroski J, Myszka DG. Capture and reconstitution of G protein-coupled receptors on a biosensor surface. Anal Biochem. 2003;316:243–250. doi: 10.1016/s0003-2697(03)00046-0. [DOI] [PubMed] [Google Scholar]
- 68.Sundberg EJ, Andersen PS, Gorshkova II, Schuck P. Surface plasmon resonance biosensing in the study of ternary systems of interacting proteins. In: Schuck P, editor. Protein interactions: Biophysical approaches for the study of complex reversible systems. Springer; New York: 2007. pp. 97–141. [Google Scholar]
- 69.Svitel J, Balbo A, Mariuzza RA, Gonzales NR, Schuck P. Combined affinity and rate constant distributions of ligand populations from experimental surface binding kinetics and equilibria. Biophys J. 2003;84:4062–4077. doi: 10.1016/S0006-3495(03)75132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Svitel J, Boukari H, Van Ryk D, Willson RC, Schuck P. Probing the functional heterogeneity of surface binding sites by analysis of experimental binding traces and the effect of mass transport limitation. Biophys J. 2007;92:1742–1758. doi: 10.1529/biophysj.106.094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tate CG, Schertler GF. Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 72.Vaish A, Silin V, Walker ML, Steffens KL, Krueger S, Yeliseev AA, Gawrisch K, Vanderah DJ. A generalized strategy for immobilizing uniformly oriented membrane proteins at solid interfaces. Chem Commun. 2013;49:2685–2687. doi: 10.1039/c3cc00077j. [DOI] [PubMed] [Google Scholar]
- 73.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 74.Vidic J, Grosclaude J, Monnerie R, Persuy MA, Badonnel K, Baly C, Caillol M, Briand L, Salesse R, Pajot-Augy E. On a chip demonstration of a functional role for Odorant Binding Protein in the preservation of olfactory receptor activity at high odorant concentration. Lab Chip. 2008;8:678–688. doi: 10.1039/b717724k. [DOI] [PubMed] [Google Scholar]
- 75.Vidic J, Pla-Roca M, Grosclaude J, Persuy MA, Monnerie R, Caballero D, Errachid A, Hou Y, Jaffrezic-Renault N, Salesse R, Pajot-Augy E, Samitier J. Gold surface functionalization and patterning for specific immobilization of olfactory receptors carried by nanosomes. Anal Chem. 2007;79:3280–3290. doi: 10.1021/ac061774m. [DOI] [PubMed] [Google Scholar]
- 76.Vidic JM, Grosclaude J, Persuy MA, Aioun J, Salesse R, Pajot-Augy E. Quantitative assessment of olfactory receptors activity in immobilized nanosomes: a novel concept for bioelectronic nose. Lab Chip. 2006;6:1026–1032. doi: 10.1039/b603189g. [DOI] [PubMed] [Google Scholar]
- 77.Vukoti K, Kimura T, Macke L, Gawrisch K, Yeliseev A. Stabilization of functional recombinant cannabinoid receptor CB(2) in detergent micelles and lipid bilayers. Plos One. 2012;7:e46290. doi: 10.1371/journal.pone.0046290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yeliseev A, Vukoti K. Expression of G-protein-coupled receptors. In: Robinson AS, editor. Production of membrane proteins. Wiley-VCH; Weinheim: 2011. pp. 219–248. [Google Scholar]
- 79.Yeliseev A, Zoubak L, Gawrisch K. Use of dual affinity tags for expression and purification of functional peripheral cannabinoid receptor. Protein Expr Purif. 2007;53:153–163. doi: 10.1016/j.pep.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yeliseev AA, Wong KK, Soubias O, Gawrisch K. Expression of human peripheral cannabinoid receptor for structural studies. Protein Sci. 2005;14:2638–2653. doi: 10.1110/ps.051550305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshiura C, Kofuku Y, Ueda T, Mase Y, Yokogawa M, Osawa M, Terashima Y, Matsushima K, Shimada I. NMR analyses of the interaction between CCR5 and its ligand using functional reconstitution of CCR5 in lipid bilayers. J Am Chem Soc. 2010;132:6768–6777. doi: 10.1021/ja100830f. [DOI] [PubMed] [Google Scholar]
- 82.Zhao H, Gorshkova II, Fu GL, Schuck P. A comparison of binding surfaces for SPR biosensing using an antibody-antigen system and affinity distribution analysis. Methods. 2013;59:328–335. doi: 10.1016/j.ymeth.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhukov A, Andrews SP, Errey JC, Robertson N, Tehan B, Mason JS, Marshall FH, Weir M, Congreve M. Biophysical mapping of the adenosine A2A receptor. J Med Chem. 2011;54:4312–4323. doi: 10.1021/jm2003798. [DOI] [PMC free article] [PubMed] [Google Scholar]