Abstract
A chemical amplifier was constructed based on enzyme-encapsulated mesoporous silica nanoparticles. By employing a supramolecular nanogate assembly that is capable of controlling the access to the encapsulated enzyme, selectivity towards substrate sizes is enabled. When an analyte molecule actuates the mechanical nanogate and exposes the enzymes, a catalytic production of fluorescent molecules is initiated. This study demonstrates a new concept of self-amplification of a chemical sensing process and can potentially increase the detection sensitivity.
In this communication we present a new concept of an autonomously amplifying chemical sensing process, where an analyte molecule actuates a mechanical nanogate and exposes enzyme amplifiers that lead to catalytic production of fluorescent molecules. Traditional enzymatic amplification, such as ELISA, requires the conjugation of an enzyme to a recognition moiety that is capable of binding to the analyte.[1–8] This binding stoichiometricly translates the concentration of the analyte to that of the enzyme. After removing the excess enzyme conjugates and adding the enzyme substrates, the enzymatic catalysis effectuates the amplified readouts. In our design, the analyte recognition process exposes the enzyme amplifiers and turns on enzyme catalyzed fluorophore generation. This first stage of the amplification causes exposure of more enzyme molecules and this cascade effect is a second amplification stage. This design eliminates washing or separation procedures and can dramatically increase the detection sensitivity. We demonstrate here an example of this type of design, which consists of two components: a mesoporous silica matrix with encapsulated enzymes in the pores, and a pH-responsive supramolecular nanogate assembly that controls the substrate access towards the enzyme. We first prove that the encapsulated enzymes retain their activity and that the nanogate enables substrate size-selectivity. Then, we show that the analyte molecule activates the nanogate, exposes the enzymes, and initiates autonomously amplified chemical sensing process.
Enzyme encapsulation in silica matrices has been widely studied.[9–20] In this study, we use mesoporous silica nanoparticles that are synthesized through a sol-gel process, where a tri-block copolymer (Pluronic P104) serves as a templating agent.[21] The resulting material exhibits pore sizes around 6.5 nm, determined from N2 adsorption-desorption measurements (Supporting Information). The pore structure provides ample space for the encapsulation of the model enzyme (Porcine Liver Esterase), which has a hydrodynamic radius of about 4 nm.[22]
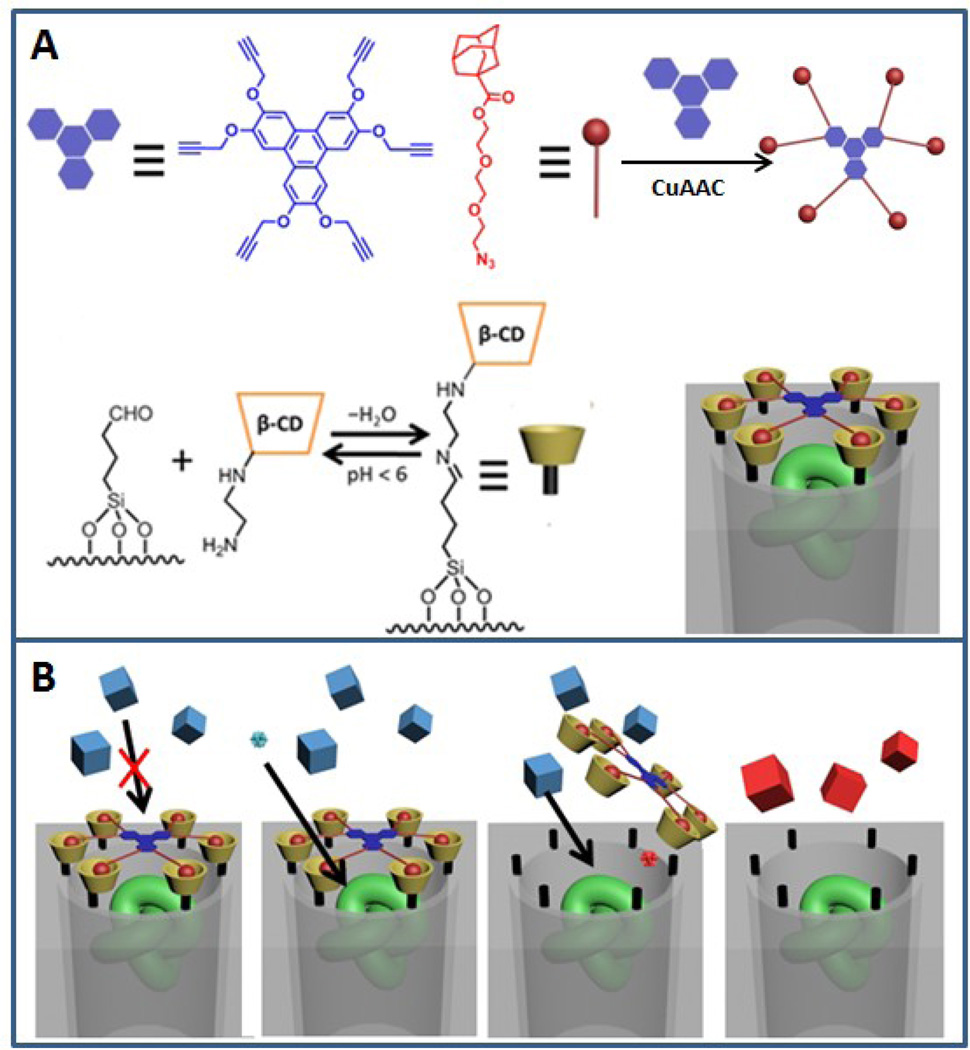
In order to successfully encapsulate the model enzyme as well as to enable the substrate-size selectivity towards the enzymatic activity, a previously-established pH-responsive supramolecular nanogate assembly was constructed at the pore opening.[23] This nanogate consists of two components: β-cyclodextrin molecules that are grafted onto the silica surface through imine bonds, and an adamantane cluster molecule with a star-like structure that forms an inclusion complex with the cyclodextrin moieties. (Figure 1A) When the derivatized mesoporous silica is exposed to the adamantane cluster, the clusters that bind over the pore openings block the pore entrances and only allow molecules with small enough sizes to diffuse in and out of the pores. The pH-sensitive imine bonds are cleaved under acidic conditions[24] and the cyclodextrin molecules together with the included adamantane cluster move away from the pore opening and allow molecules with larger sizes to freely diffuse through. When the analyte molecule recognition generates acid, the acid can activate the nanogate and open the access towards the enzyme. This will facilitate the enzymatic catalysis for larger substrate molecules and provide chemical amplification. (Figure 1B)
Figure 1.
A) Assembly of the nanogate-based amplifier. The capping agent is synthesized using a Cu catalyzed azid-alkyne coupling reaction. The green knot represents the enzyme molecule that is trapped in the pore. B) The mechanism of the self-amplifying chemical sensing process. The size selectivity of the nanogate enables separation of the substrate (blue cubes) and the enzyme. A small analyte molecule (blue ball) actuates the nanogate, which opens the access to the enzyme and fluorescent products are generated catalytically.
In this study, the cyclodextrin-modified silica nanoparticles were loaded with porcine liver esterase (PLE) and then capped with the adamantane cluster molecules. Enzymes that were not successfully capped by the nanogate were removed through extensively washing with Polyethylene glycol (MW 2000).[25] Two PLE substrates with different sizes (4-acetoxycinnamic acid, ACA, and 5-carboxyfluorescein diacetate, CFDA) were chosen to serve as model substrate molecules. These substrates exhibit no fluorescence before undergoing hydrolysis reactions, but the resulting products are strongly fluorescent. Moreover, the thermal hydrolysis process of these substrates in solution occurs at a significantly slower rate without the enzymatic catalysis. A continuous-monitoring fluorescence spectroscopy method was employed to monitor the hydrolysis of the substrate. In those experiments, a substrate solution and the enzyme-encapsulated silica nanoparticles were stirred at room temperature and an excitation beam was introduced to excite the fluorescent product molecules. The corresponding fluorescence intensity was recorded over the course of the experiment.
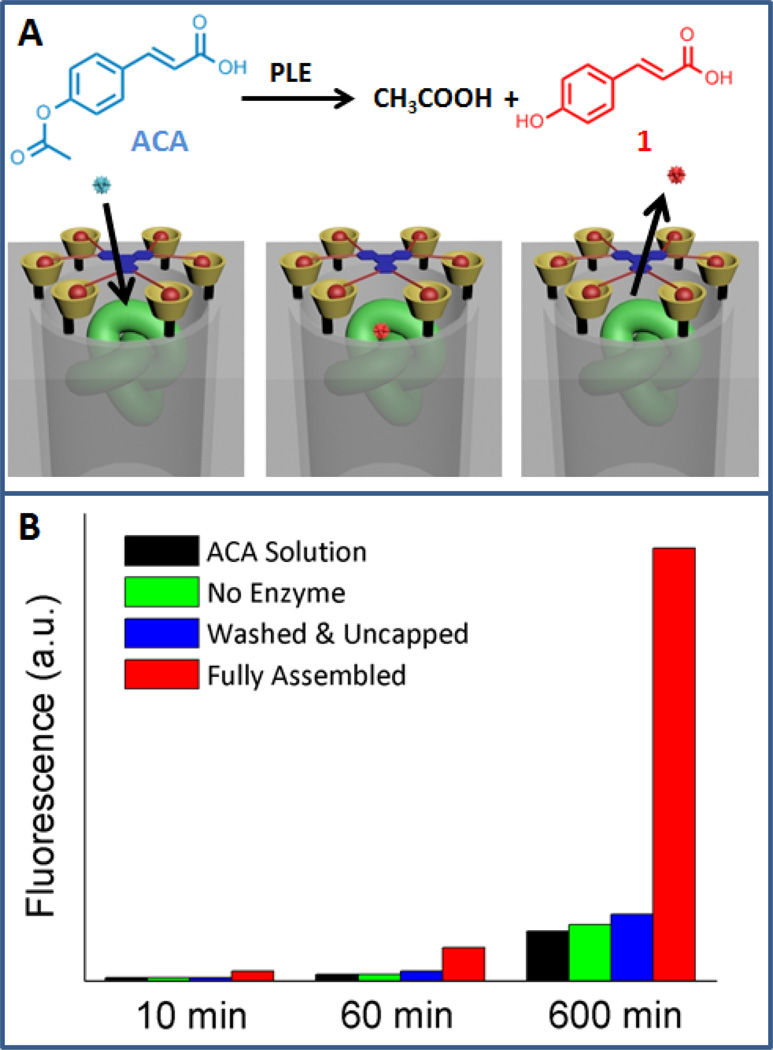
The operation of the amplifier was investigated in two stages. First, the analyte recognition step of the chemical amplifier was studied. 4-acetoxycinnamic acid (ACA) molecules are small enough to pass through the nanogate.[26] After they interact with the enzymes in the mesopores, the corresponding fluorescent product molecules diffuse out and are detected. As shown in Figure 2, the fluorescence intensity of the solution dramatically increases over time in the presence of the enzyme-encapsulated nanoparticles, indicating that the ACA molecules pass through the nanogate and interact with the enzyme (red bars). These results also prove that the enzymes remain their activity when trapped in the mesopores. When the silica nanoparticles are not loaded with the enzyme, the hydrolysis rate (green bars) is the same as that of the thermal hydrolysis of the ACA molecules in the buffer solution (black bars). In another control experiment, the nanoparticles are loaded with enzyme, but without a capping agent. In this case, the loaded enzymes were extracted by ethylene glycol during the washing step. As a result, there is no enzymatic hydrolysis activity (blue bars). These experiments prove that the ACA analyte is recognized and processed by the encapsulated enzymes. However, this recognition process is stoichiometric, i.e. the fluorescence intensity is directly correlated with the amount of ACA molecules in the solution, and no amplification is involved.
Figure 2.
A) Illustration of an ACA molecule passing through the nanogate resulting in catalytic production of a fluorescent product. B) Normalized fluorescence intensity for compound 1 (hydrolyzed ACA) in four different samples: the fully assembled nanogate system with encapsulated enzymes, the control showing that all the enzymes in the uncapped nanoparticles were removed through washing with PEG solution, the no enzyme control where no enzyme was loaded into the pore, and the ACA solution by itself.
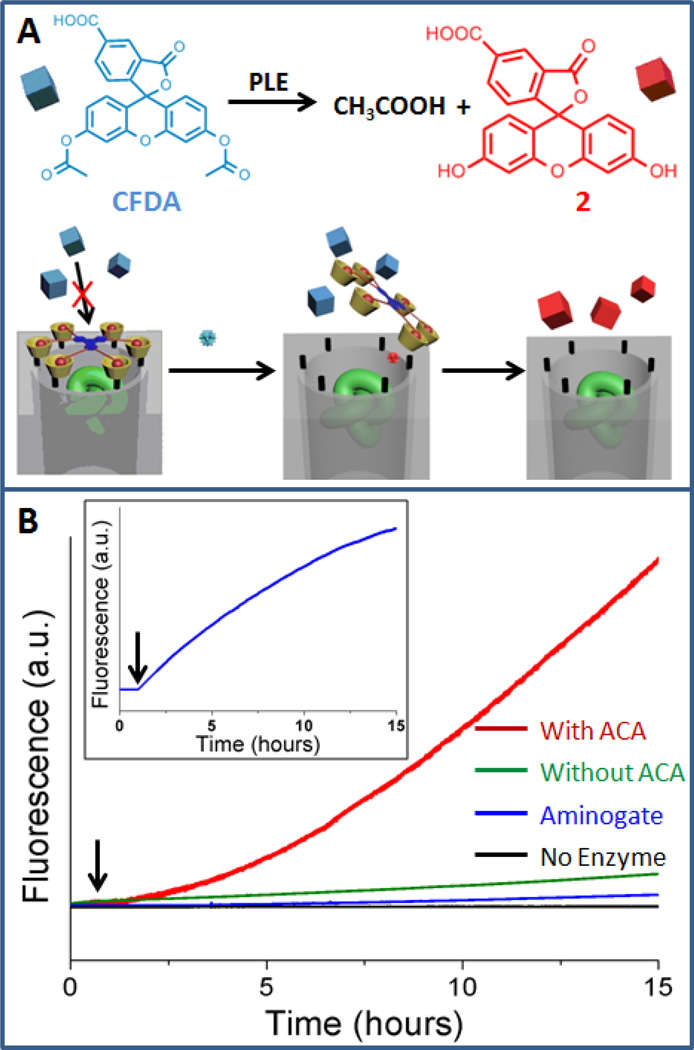
The amplification steps were demonstrated using a second enzyme substrate, 5-carboxyfluorescein diacetate (CFDA), and utilizing the design features of the nanogate. The analyte recognition process triggers the amplification of the fluorescence readout. Unlike the ACA molecules, the CFDA molecules are too big to freely enter the gated pore (Figure 3A). As a result, the enzymatic production of the fluorescent compound 2 (hydrolyzed CFDA) is prohibited by the nanogate. As shown in Figure 3B, in the presence of the enzyme-encapsulated nanoparticles, the fluorescence intensity of the compound 2 does not exhibit much increase over a long period (green curve). This result proves that the size-selectivity of the nanogates prohibits catalytic hydrolysis of CFDA.[23] In contrast, when a small amount of ACA is introduced to the solution, the fluorescence intensity increases over time (Figure 3B, red curve). As demonstrated above, the ACA molecules enter the pore and are recognized by the esterase. This analyte recognition generates acetic acid and therefore actuates the nanogate (Supporting Information). When the access to the enzymes occurs, the CFDA molecules diffuse into the pore and a large amount of fluorescent product molecules (compound 2) are generated. At this stage, the fluorescence intensity readout for the compound 2 is no longer stoichiometricly correlated with the amount of the analyte (ACA molecules) and this serves as a signal amplification process. Moreover, the fluorescence intensity of the compound 2 (Figure 3B, red curve) exhibits a self-accelerating feature, while that of the compound 1 shows a typical enzymatic kinetics (Figure 3B, insert). This positive curvature can be explained by the cascade effect of this system. Because the hydrolysis of CFDA also generates acetic acid, the catalytic process in one mesopore can activate nanogates on other neighboring pores and can open the access to more enzymes. When the initial amount of CFDA molecules in the solution is high, this process functions as a chain reaction and provides additional amplification features to the system.
Figure 3.
A) Illustration of the operation of the amplifier. B) Normalized fluorescence profiles of hydrolyzed CFDA in the presence of enzyme-encapsulated silica nanoparticles in different experiments: with the addition of ACA analyte (red curve), no ACA (green curve), no enzyme in the particles (black curve), and the amino-version of the nanogate (blue curve). Insert: the corresponding fluorescence profile of the hydrolyzed analyte for the experiment with ACA. The arrows indicate when ACA was added.
In order to further prove the mechanism of this chemical amplifier, three control experiments were conducted. In the case where the nanoparticles were not loaded with the enzymes, negligible change of the fluorescence intensity was observed. This proves that there are no side reaction involving the ACA and the CFDA molecules. In the second experiment, the acid-responsive imine groups in the nanogate assembly were reduced to amines, which disabled the acid-responsive feature.[27] When the amino version of the nanogates were employed to trap the enzymes, negligible change of the fluorescence intensity was observed (Figure 3B, blue curve). This was because the acid generated from the analyte recognition could not activate the nanogate and thus the access for the CFDA molecules to the enzyme remained blocked. This result proves that the pH-responsiveness is critical for the operation of this system and the nanogate must be mechanically actuated to initiate the self amplifying chemical sensing process. In the third experiment, the nanoparticles were removed after the initiation of amplification process, and after removal the solution fluorescence stopped increasing in absence of the nanoparticles. This experiment demonstrates that the amplification does not result from the enzymes escaping from the pores into the solution and accelerating the hydrolysis (Supporting Information).
In summary, a new concept of autonomously amplifying chemical sensing process was introduced. The specific example based on the enzyme-encapsulated mesoporous silica nanoparticles and the pH-responsive size-selective supermolecular nanogates is proof of principle. The nanogate assembly allows the coexistence of the enzyme substrates and the enzymes and prevents their interaction before the system is activated. An analyte molecule can mechanically actuate the nanogate and opens the access to the enzyme. Fluorescent molecules are catalytically produced to generate amplified readout. Because the product of this process can also activate the nanogate, this system operates in a cascaded manner and provides self-acceleration and additional signal amplification.
Supplementary Material
ACKNOWLEDGMENT
The work was supported by NIH RO1 133697 CA. The authors thank Juyao Dong, Matt Kiesz and Angela Hwang for their assistance with the experiments.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The synthetic procedures, the characterizations of the nanoparticles, the fluorescence spectrum of ACA and CFDA, the fluorescence profiles of the acetic acid induced nanogate opening and the control experiment where the nanoparticles were removed from the solution, this material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Clark MF, Adams AN. J. Gen. Virol. 1977;34:475–483. doi: 10.1099/0022-1317-34-3-475. [DOI] [PubMed] [Google Scholar]
- 2.Wilson GS, Hu Y. Chem. Rev. 2000;100:2693–2704. doi: 10.1021/cr990003y. [DOI] [PubMed] [Google Scholar]
- 3.Liu G, Wan Y, Gau V, Zhang J, Wang L, Song S, Fan C. J. Am. Chem. Soc. 2008;130:6820–6825. doi: 10.1021/ja800554t. [DOI] [PubMed] [Google Scholar]
- 4.Tan Y, Hoffman RM. Nature Protocols. 2008;3:1388–1394. doi: 10.1038/nprot.2008.117. [DOI] [PubMed] [Google Scholar]
- 5.Mannerstedt K, Jansson AM, Weadge J, Hindsgaul O. Angew. Chem. Int. Ed. 2010;49:8173–8176. doi: 10.1002/anie.201003435. [DOI] [PubMed] [Google Scholar]
- 6.Miranda OR, Li X, Garcia-Gonzalez L, Zhu Z-J, Yan B, Uwe UHF, Rotello VM. J. Am. Chem. Soc. 2011;133:9650–9653. doi: 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He H, Xu X, Wu H, Jin Y. Adv. Mater. 2012;24:1736–1740. doi: 10.1002/adma.201104678. [DOI] [PubMed] [Google Scholar]
- 8.Miranda OR, Chen H-T, You C-C, Mortenson DE, Yang X-C, Bunz UHF, Rotello VM. J. Am. Chem. Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivnitski D, Artyushkova K, Rincon RA, Atanassov P, Luckarift HR, Johnson GR. Small. 2008;4:357–364. doi: 10.1002/smll.200700725. [DOI] [PubMed] [Google Scholar]
- 10.Luckarift HR, Spain JC, Naik RR, Stone MO. Nat. Biotech. 2004;22:211–213. doi: 10.1038/nbt931. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Caruso F. Chem. Mater. 2005;17:953–961. [Google Scholar]
- 12.Takahashi H, Li B, Sasaki T, Miyazaki C, Kajino T, Inagaki S. Chem. Mater. 2000;12:3301–3305. [Google Scholar]
- 13.Yamanaka SA, Dunn B, Valentine JS, Zink JI. J. Am. Chem. Soc. 1995;117:9095–9096. [Google Scholar]
- 14.Dave BC, Miller JM, Dunn B, Valentine JS, Zink JI. J. Sol-Gel Sci. Tech. 1997;8:629–634. [Google Scholar]
- 15.Dunn B, Zink JI. Acc. Chem. Res. 2007;40:747–755. doi: 10.1021/ar700033b. [DOI] [PubMed] [Google Scholar]
- 16.Tarn D, Ashley CE, Xue M, Carnes EC, Zink JI, Brinker CJ. Acc. Chem. Res. 2013;46:792–801. doi: 10.1021/ar3000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avnir D, Braun S, Lev O, Ottolenghi M. Chem. Mater. 2004;6:1605–1614. [Google Scholar]
- 18.Frenkel-Mullerad H, Avnir D. J. Am. Chem. Soc. 2005;127:8077–8081. doi: 10.1021/ja0507719. [DOI] [PubMed] [Google Scholar]
- 19.Williams AK, Hupp JT. J. Am. Chem. Soc. 1998;120:4366–4371. [Google Scholar]
- 20.Wada A, Tamaru S, Ikeda M, Hamachi I. J. Am. Chem. Soc. 2009;131:5321–5330. doi: 10.1021/ja900500j. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Sun X, Zhang G, Trewyn BG, Slowing II, Lin VS-Y. ACS Nano. 2011;5:1366–1375. doi: 10.1021/nn103077k. [DOI] [PubMed] [Google Scholar]
- 22.Junge W, Krisch K. Eur. J. Biochem. 1974;43:379–389. doi: 10.1111/j.1432-1033.1974.tb03424.x. [DOI] [PubMed] [Google Scholar]
- 23.Xue M, Cao D, Stoddart JF, Zink JI. Nanoscale. 2012;4:7569–7574. doi: 10.1039/c2nr32170j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillebrenner H, Buyukserin F, Kang M, Mota MO, Stewart JD, Martin CR. J. Am. Chem. Soc. 2006;128:4236–4237. doi: 10.1021/ja058455h. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Lu D, Austin R, Agarwal A, Mueller LJ, Liu Z, Wu J, Feng P. Langmuir. 2007;23:5735–5739. doi: 10.1021/la063507h. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Li Z, Gaines JW, Zhao Y-L, Cao D, Bozdemir OA, Ambrogio MW, Frasconi M, Botros YY, Zink JI, Stoddart JF. Angew. Chem. Int. Ed. 2012;51:5460–5465. doi: 10.1002/anie.201107960. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y-L, Li Z, Kabehie S, Botros YY, Stoddart JF, Zink JI. J. Am. Chem. Soc. 2010;132:13016–13025. doi: 10.1021/ja105371u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.