Abstract
Cell penetrating peptides have demonstrated potential to facilitate the cellular delivery of therapeutic molecules. Here we develop a set of 50 cell penetrating peptide based formulations with potential to deliver small interfering RNAs intracellularly. The transfection efficacy of siRNA containing lipid-like nanoparticles decorated with different peptides was evaluated both in vitro and in vivo and correlated with the peptide physical and chemical properties. In vitro, these particles were internalized primarily through macropinocytosis. When the peptides were presented to bone marrow-derived dendritic cells they induce low immunoactivation relative to control cell penetrating peptides including the antennapedia homeodomain and TAT, as quantified by the expression of activation specific surface proteins like CD80, CD86 and major histocompatibility complex class II. In vivo, peptide decorated nanoparticles primarily accumulated in the lungs and the liver. Three human peptides derived from surfactant protein B; a lung surfactant protein, orexin; a neuropeptide hormone and lactoferricin; a globular glycoprotein that exist in many physiological fluids, facilitated the in vivo delivery of siRNA and induce significant knock down (90%) of a hepatocyte expressed protein, coagulation Factor VII.
Keywords: Cell penetrating peptides, siRNA, lipid nanoparticles, computational modeling
Cell penetrating peptides (CPPs), or peptides that can facilitate entry into the cytoplasm, have been used as drug delivery vehicles for a variety of biomedical applications.1–6 In general, cell penetrating 2 or transduction domain 1 peptides possess the ability to transverse the cellular membranes and transport their cargo intracellularly. Mechanistically the CPPs are endocytosed either by fusing with lipid cell membranes following a vacuole-based endocytosis or by creating pores to the cellular membrane.7–9 Many CPPs are derived from bacterial or viral proteins or they are synthetic model peptides. Typical physichochemical features that characterize such sequences are their high positive charge and amphiphilicity.2, 3, 9 One family of therapeutics that cell penetrating peptides have been examined for use as delivery vehicles, is small interfering RNAs (siRNAs).10–13 siRNAs are oligonucleotides, 21–23 bases long, which utilize the endogenous RNA interference mechanism in order to cleave mRNAs and inhibit the translation of target proteins.14 The success of siRNAs as therapeutics in the clinic requires the development of safe and effective delivery systems.15
Our current knowledge of CPPs is focused on a limited number of sequences with cell penetrating properties, examples of which include poly-arginine or poly-lysine based peptides that contain a large number of arginines (R6–R12)16 or lysines (K6–K12),17 Tat-based peptides from the HIV-1 virus sequence (which are a combination of arginines and lysines),18 Drosophila melanogaster antennapedia homeodomain peptide or “penetratin”,19 short amphipathic peptide Pep-1 marketed as “chariot peptide”,20 and the VP22 protein based peptide from herpes simplex virus.21 Various modified, synthetic or hybrid versions of the CPPs like the MPG peptide which contains a hydrophobic domain derived from the fusion sequence of HIV glycoprotein 41 and a hydrophilic domain derived from the nuclear localization sequence of the SV40 T-antigen have also been investigated.22
Most of the CPP sequences in use today are based on non-human proteins. Non-human protein sequences may be recognized by the immune system as non-self, leading to generation of anti-CPP (and anti-drug delivery vehicle) immune responses.23 This problem is further complicated by the development of immunological memory against the CPP sequences (both B cell and T cell memory), which can lead to faster and more severe responses upon repeat dosing of the CPP based drug delivery vehicle.23, 24 These complications could potentially be addressed through the development of humanized CPP sequences. Here we describe the development of humanized CPPs for use in drug delivery. A bioinspired systematic proteomic approach is applied to identify novel peptides that can fuse or attach to a plasma membrane, internalize, and escape from the endosomes in the cytoplasm. These peptides have been evaluated for their potential as cellular delivery agents, both in vitro and in vivo.
Results
Selection of Candidate Peptides
Initially we identified amino acid sequences from peptides or protein domains that reside or interact with cellular plasma membranes, such as transmembrane proteins. We primarily considered sequences derived from the human proteome, with diverse physichochemical and structural characteristics like charge, amphiphilicity, and tertiary structure (Table 1). We collected amino acid sequences and structural information from databases including the Orientation of Proteins in Membranes (OPM),25 the PDBsum,26 and the PDBTM.27 Such databases provide information on the spatial positions of proteins with respect to the lipid membrane.
Table 1.
The amino acid sequences of the novel cell penetrating peptides along with properties such as molecular weight, isoelectric point, peptide charge at pH = 7, and the names of the proteins that the peptides are derived from.
| Sequence | MW | pI | Charge | Protein |
|---|---|---|---|---|
| 1. PSKDAFIGLM | 1244 | 6.75 | 0 | Eledoisin |
| 2.SKEWQPAQVILL | 1578 | 6.94 | 0 | Cholecystokinin |
| 3.INLKALAALAKKIL | 1645 | 10.84 | 3 | Mastoparan |
| 4.QKTVEGAGSIAAATG | 1526 | 6.94 | 0 | Alpha synuclein-1 |
| 5.CWLCRALIKRIQAMIP | 2080 | 10.12 | 2.9 | Surfactant protein B |
| 6.TLEDLRGWLRALGRASR | 2135 | 11.93 | 2 | Pleckstrin-1 |
| 7.DWLKAFYDKVAEKLKEAF | 2366 | 7.03 | 0 | Amphipathic peptide 18a |
| 8.GQRAAENRQGTLTEYCSTLMSL | 2594 | 6.43 | 0 | NCF-1 |
| 9.ICVVQDWGHHRCT | 1719 | 7.16 | 0.1 | Compstatin |
| 10.AEFLKVFLPSLLLSHLLAIGLGIYIG | 2963 | 7.74 | 0.1 | Bnip3a |
| 11.FKRLVFLPLQIVGVTLLTLAALNCLG | 2978 | 10.11 | 2 | Steryl sulfatase |
| 12.PLPDCCRQKTCSCRLYELLHGAGNHAAGILTL | 3619 | 7.86 | 1 | Orexin-a |
| 13.KQEEETHIRNEQVRQRAKECSQALSLIDIDHG | 3626 | 5.39 | −1.9 | LDL-phospholipase A2 |
| 14.APAAAAQAVAGLAPVAAEQP | 1939 | 3.3 | −1 | Alamethicin |
| 15.GSEKMSTAISVLLAQAVFLLLTSQR | 2828 | 10.09 | 1 | Acetylcholine receptor M2 |
| 16.QAPAYKKAAKKLAES | 1769 | 10.29 | 3 | Model amphipathic peptide |
| 17.GLFDIIKKIAESF | 1646 | 6.99 | 0 | Aurein 1.2 analog b |
| 18.ENFVGGCATGFKRTADGRCKPTF | 2627 | 9.11 | 1.9 | Silkworm paralytic peptide |
| 19.FASADLQGAAAAAPAAAQ | 1868 | 3.1 | −1 | Peptaibol Chrysospermin C |
| 20.GCCSYPPCFATNPDC | 1743 | 3.1 | −1.2 | alpha Conotoxin |
| 21.RRWWRF | 1172 | 12.7 | 3 | L-glutamate oxidase |
| 22.ENREVPPGFTALIKTLRKCKII | 2692 | 10.3 | 3 | Distinctin Antimicrobial peptide |
| 23.EPSKDAFIGLM | 1373 | 4.07 | −1 | Eledoisin |
| 24.LDKEAVYFCHLDIIW | 2030 | 4.3 | −2 | Endothelin-1b |
| 25.CELCCNPACAGC | 1352 | 3.29 | −1.2 | Enterotoxin |
| 26.HSQGTFTSDYSKYLDSRRAQDFVQWLMNT | 3647 | 7.68 | 0.1 | Glucagon |
| 27.VGALAVVVWLWLWLW | 1976 | 6.01 | 0 | Antibiotic peptide |
| 28.ICIFCCGCCHRSKCGMCCKT | 2364 | 8.1 | 2.7 | Hepcidin |
| 29.ILGKIWEGIKSLF | 1669 | 9.88 | 1 | Antimicrobial peptide ISCT |
| 30.DVPKSDQFVGLM | 1501 | 3.88 | −1 | Kassinin-1 |
| 31.FKCRRWQWRMKKLGAPSITCVRRAF | 3290 | 12.24 | 7.9 | Lactoferricin |
| 32.CRRWQWRMKKLGC | 1916 | 11.37 | 4.9 | Lactoferricin (short) |
| 33.GIGKFLHSAKKFGKAFVGEIMNS | 2467 | 10.55 | 3.1 | Magainin 2 |
| 34.INWKGIAAMAKKLL | 1722 | 10.84 | 3 | Mastoparan-X |
| 35.GIGAVLKVLTTGLPALISWIKRKRQQ | 3012 | 12.43 | 5 | Melittin |
| 36.GGAGHVPEYFVGIGTPISFYG | 2290 | 5.13 | −0.9 | Microcin J25 |
| 37.FVPIFTYGELQRMQEKERNKGQ | 2864 | 9.63 | 1 | Motilin |
| 38.NGVCCGYKLCHPC | 1562 | 7.85 | 0.9 | Mria Conotoxin |
| 39.HKTDSFVGLM | 1300 | 7.94 | 0.2 | Neurokinin-A |
| 40.DMHDFFVGLM | 1377 | 3.88 | −1.9 | Eledoisin |
| 41.RQIKIWFQNRRMKWKK | 2411 | 12.7 | 7 | Antennapedia homeodomain |
| 42.GSTLYTESRKLLRSWHLPSV | 2495 | 10.39 | 2.1 | Semliki mRNA capping enzyme NSP1 |
| 43.CSCSSLMDKECVYFCHLDIIW | 2660 | 4.3 | −2.1 | Endothelin-1 |
| 44.LDKEAVYFCHLDIIW | 2030 | 4.3 | −2 | Endothelin-1 (short) |
| 45.ICLKKWPWWPWRRCK | 2251 | 10.82 | 4.9 | Indolicidin |
| 46.GAHWGVLAGIAYFSMVGNWAK | 2400 | 9.72 | 1.1 | Hepatitis C envelope glycoprotein E1 |
| 47.KLCYLLDGILFIYGVILTALFLR | 2823 | 8.8 | 1 | CD3-zeta |
| 48.MAQDIISTIGDLVKWIIDTVNKFTKK | 3143 | 9.67 | 1 | Delta-toxin |
| 49.SGNYVLDLIYSLHKQINRGLKKIVLGWA | 3364 | 10.2 | 3 | Non-structural protein 5A |
| 50.SPDEREEWMRAIQMVANSLK | 2555 | 4.65 | −1 | RAC-beta |
We picked 50 peptide sequences derived from 46 proteins that can associate with the plasma membrane; this selection contained enough diversity of physical and chemical properties to consider for the study. We considered only short peptide so that they could be synthesized using common solid phase synthesis techniques. We sampled the sequence space to ensure that these 50 sequences exhibit diverse physicochemical characteristics like different amino acid lengths, hydrophobicities, isoelectric points and thus have diverse structural characteristics like helix or sheet propensities (Table 1). In order to compare the activity of the novel peptides relatively to a commonly used CPP, we included the antennapedia homeodomain peptide (penetratin)19 in the list of the peptides studied.
Peptide-siRNA and Nanoparticle Formulations
The peptides were conjugated to siRNA molecules in a 1:1 molar ratio using siRNA with a pyridyl disulfide group at the 3’ sense strand, separated from the siRNA sequence with an 18 carbon atom spacer. The disulfide bond of the siRNA molecule was cleaved in a tris-(2-carboxyethyl) phosphine (TCEP) resin exposing a free sulfhydryl and directly applied to the maleimide-peptide solution in dimethyl sulfoxide.
As building blocks for the nanoparticles we used a novel family of lipid-like materials, termed lipidoids.28, 29 Lipidoid-based nanoparticles can encapsulate and deliver siRNA in vitro and in vivo depending on the selection of the lipid-like building blocks.28, 29 The nanoparticles were formulated with four components 1) a lipidoid molecule, 2) cholesterol 3) a thiol modified lipid-polyethylene glycol (PEG) molecule (DSPE-PEG-SH), 4) siRNA.29–32 Initially the lipid molecule, cholesterol and PEG were dissolved in ethanol and added dropwise to a NaOAc acidic solution were electrostatic interactions drive the nanoparticle formation. After particle formation, siRNA added to the acidic solution was absorbed in the nanoparticles. Once the nanoparticles were loaded with siRNA the solution was dialyzed against DPBS to remove excess of materials and solvent. The peptides were covalently attached on the surface of the nanoparticles post-particle formation (Supplementary Figure 1B). The peptides were synthesized containing a maleimide chemical group on their amine terminus with a small 8 carbon atom PEG linker between the maleimide and the peptide (Supplementary Figure 1A). The PEG portion of the peptide increases the solubility of the hydrophobic peptides to aqueous solvents. Particle functionalization with thiols (using the DSPE-PEG-SH) on their external surface was necessary in order to achieve conjugation of the peptides on the surface of the nanoparticles post-particle formation.
Depending on their composition, nanoparticles created with lipidoids display variable activity in transfecting cells with siRNA without the presence of any ligands on their surface in vitro and in vivo.28, 29 In order to examine the activity of multivalent conjugates of the novel peptides on the surface of nanoparticles in releasing their cargo in the cytoplasm, we utilized lipidoid materials that form stable nanoparticles and encapsulate high levels of siRNA but demonstrate lower efficiency in transfecting cells alone. Specifically we selected the lipidoid C12-117 (the structure of C12-117 is displayed in Supplementary Figure 2) as a building block for nanoparticles.29 Nanoparticles composed of C12-117 do not exhibit any activity in transfecting cells with siRNA.29
Conjugation of the peptides on the surface of the nanoparticles was performed post nanoparticle formation. The peptide solutions were added after the formation of the particles and the reaction of the maleimides with the PEG thiols was performed overnight at room temperature. The reactions were monitored with MALDI (Supplementary Figure 5). To quantify the degree of siRNA entrapment in the lipidoid nanoparticles, a modified Ribogreen assay was performed.29 The entrapment efficacy was more than 80%. To monitor the stability of the formulations we quantified the size of the nanoparticles using a dynamic light scattering system (DLS). The diameter of the nanoparticles before peptide conjugation was 89.9 nm and the average diameter after peptide conjugation 96.3 nm ±5.5 nm (Supplementary Table 1). In two instances the peptide conjugation completely destabilized the particles and those peptides were not considered for further analysis. Finally we quantified the relative amount of peptide coverage on the surface of the nanoparticles. After conjugating fluorescein to the peptides and formulating the nanoparticles we quantified the amount of fluorescence emitted by the formulations relatively to the amount of fluorescence emitted if there was a complete surface coverage (Supplementary Materials and Methods). We estimated that the coverage ranged from 22% to 39% (Supplementary Table 1).
In-vitro Silencing Activity and Endocytosis Mechanisms of the Peptide Decorated Nanoparticles
Initially we screened formulations’ ability to transfect cells with siRNA in vitro using a dual luciferase expressing HeLa cell line.29–31 Dual luciferase HeLa cells have been transformed to stably express firefly and renilla luciferase, both of which emit a different luminescent signal. When firefly luciferase siRNA is used, specific silencing induces reduction in only firefly luciferase, without affecting renilla luciferase levels. Reduced expression of renilla luciferase results from non-specific silencing typically associated with cytoxicity. The value of the firefly luciferase luminescent signal is scaled to the value of the renilla luciferase and this ratio corresponds to the knockdown of luciferase in viable cells. We consider formulations with renilla luciferase signal >75% relatively to untreated controls. We have chosen to use the particular cell line, as we have shown in previous studies in our lab, that HeLa cells is the optimal in vitro system for predicting the transfection efficacy of the lipidoid nanomaterials in vivo.
siRNA-peptide (1:1) conjugates or nanoparticles decorated with peptides were added to cells in culture. Cell culture supernatants were replaced with fresh medium 4 hours post addition of formulations. In both cases the medium contained 10% serum. Luminescence from cells was measured 3 days post addition of formulations to cells. Similarly to previous observations, the 1:1 siRNA to peptide formulations exhibit only modest delivery efficacy. The maximum knockdown was approximately 30% at the highest applied concentration of 500 ng of RNA per well, in 200 μl of cell medium or 2.5 μg/ml siRNA (0.5 μg/ml peptide) (Figure 1A and Supplementary Figure 3). Given this result, we hypothesized that delivery would require additional functionality. At these concentrations with stoichiometric conjugation, the final siRNA concentration would have very low efficacy and be impractical.
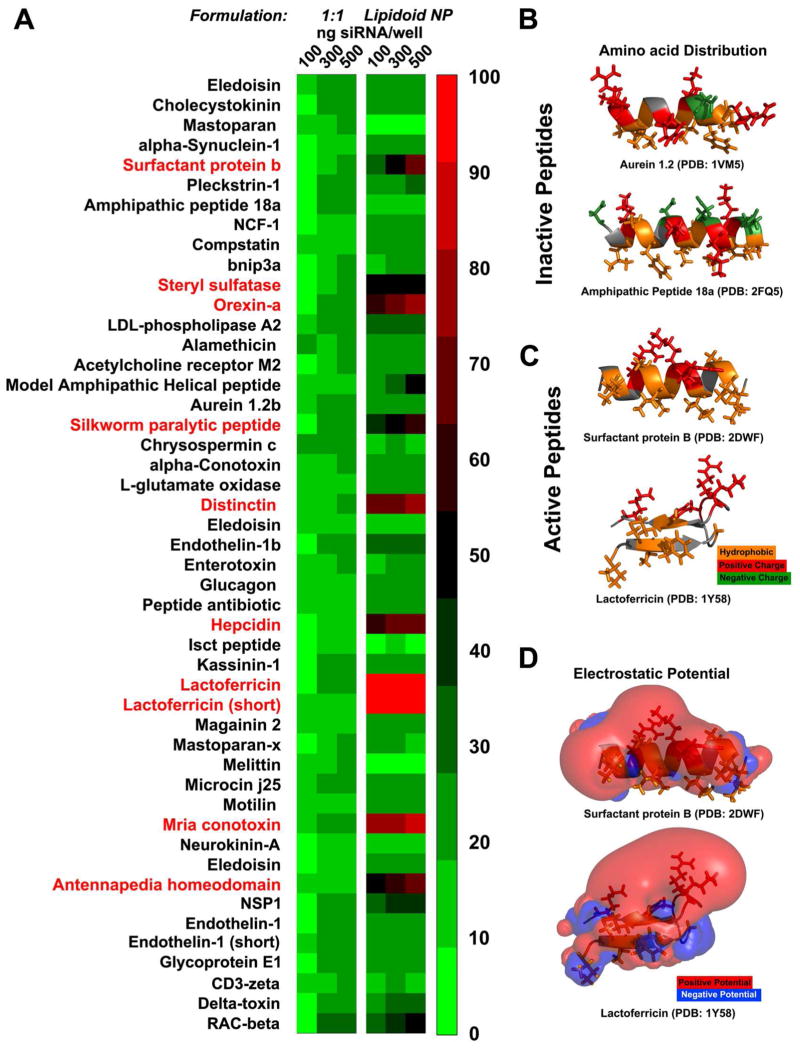
Figure 1. In vitro.
studies of the novel cell penetrating peptides. (A) The 1:1 peptide:siRNA conjugates (left panel) exhibit lower efficacy than the peptide decorated nanoparticles (right panel) at knocking down luciferase in dual HeLa cells. Ten of the peptides (red) exhibited efficacy greater than 60% reduction in the luciferase expression (n=5). (B, C and D) Structure-function correlations of the peptides’ activity. The electrostatic and hydrophobic properties of two inactive and two active peptides can correlate with their activity. The electrostatic potential is visualized as isosurfaces at +1kT/e for positive potential (red) and −1kT/e for negative (blue). Hydrophobic amino acids are represented with orange color while the positively charged with red and the negatively charged with green. In the case of the inactive peptides negatively charged amino acids disrupt the sequence of positively charged residues. For example the peptides derived from aurein 1.2 47, and amphipathic peptide 18a 48 (top and bottom). In the case of active peptides the positively charged amino acids cluster thus creating a surface of positive electrostatic potential that covers almost half of the volume around the peptide allowing the hydrophobic residues to reside in a neutral surface. For example the peptides derived from surfactant protein B 49 (top) and lactoferricin 50 (bottom) are positively charged and only the former is amphiphilic.
Unlike the 1:1 siRNA to peptide formulations, the peptide decorated nanoparticles exhibited higher activity at similar siRNA concentrations (Figure 1A). When conjugated on the surface of nanoparticles, 10 peptides exhibited greater than 60% knockdown of luciferase expression in the dual HeLa cells (Figure 1A). Nanoparticles formulated with the C12-117 lipidoid alone have no activity in transfecting cells with siRNA (Figure 1A). The luciferase expression also followed a quantitative dose response at increasing dosages of peptide-decorated siRNA-containing nanoparticles (Figure 1B and Supplementary Figure 4). The peptide conjugated nanoparticle which exhibited the strongest silencing of the luciferase expression was the lactoferricin peptide formulation. We utilized two lactoferricin peptides; a 25 amino acid peptide (FKCRRWQWRMKKLGAPSITCVRRAF) and a truncated 13 amino acid peptide (CRRWQWRMKKLGC). Lactoferricin is a peptide derived from lactoferrin, a secretory protein present in a variety of secretory fluids like milk and saliva.33, 34 Both of these peptides induce 95% knockdown at 500 ng of RNA per well of nanoparticle formulations (2.5 μg/ml). Similar to lactoferricin, surfactant protein B a lung surfactant,35 induced 86% knockdown of luciferase expression at 500 ng of siRNA per well. A number of peptides also showed 70% knockdown efficiency (Figure 1A). The lowest amount of siRNA complexed in nanoparticles tested 100 ng/well or 30 nM of siRNA, a typical concentration of the molecule tested in vitro, yielded a significant luciferase knockdown 60–70% for the top peptide hits.
The problem of the uptake of such formulations is a complex question. There is a more than 20 year ongoing debate on the pathways and mechanisms that cell penetrating peptides follow in order to get endocytosed. In addition, lipidoid nanoparticles by themselves also follow distinct endocytosis pathways. Thus the combination of cell penetrating peptides with lipidoid nanoparticles generates a very complex combinatorial problem that we have just started delineating. To elucidate the mechanisms of endocytosis we utilized inhibitors of endocytosis which include dynasore (inhibits dynamin dependent clathrin and caveolae-mediated endocytosis), cytochalasin D (inhibits macropinocytosis) and geinstein (caveolae mediated endocytosis).36–38 We incubated HeLa cells with Alexa 647 labeled nanoparticles decorated with CPPs in the presence of these inhibitors. Cells were then analyzed using an automated spinning disk confocal microscope. Interestingly cytochalasin-D appeared to concentrate the nanoparticles on the periphery of the cells independent of the CPPs attached to it. Dynasore or geinstein however did not appear to inhibit the nanoparticles uptake. Thus, macropinocytosis appears to be the predominant pathway for internalization of CPP-nanoparticles. (Figure 2A). These results also support previous reports that CPPs may induce macropinocytosis to gain entry inside cells.39
Figure 2.
Endocytosis mechanistic studies of the peptide decorated nanoparticles and immunostimulation of the peptides to mouse derived dendritic cells. (A) We administered nanoparticle formulations in combination with various endocytosis inhibitors and studied the alteration in their endocytosis patterns. Such inhibitors included dynasore a dynamin inhibitor affecting clathrin mediated endocytosis, cytochalasin D that inhibits actin polymerization thus affecting receptor mediated endocytosis and genistein that inhibits caveolae-mediated endocytosis. (B) Purified bone marrow derived dendritic cells were exposed to 30 μg/ml of peptide solutions for 3 days. The expression of dendritic cell activation markers including CD40, CD80, CD86, MHC class I and II was quantified with FACS. Primarily CD80, CD86 and MHC class II were upregulated. The plot contains 2 peaks of cells given that all of the cells considered do not contain the surface markers studied. The expression (as quantified by the geometric mean of the expression curves) of these 3 markers on the cells is scaled as 100% representing the expression induced by LPS treatment and 0% the PBS treatment and plotted the results as a heatmap. The short lactoferricin derived peptide exhibited the least activation of the dendritic cells relatively to well established CPPs like the antenappedia homeodomain and TAT.
Structure-Function Correlations
Most of the 50 peptides studied are positively charged though only a fraction of those were effective at delivering siRNA intracellularly when conjugated on the surface of nanoparticles (Figure 1A). We first investigated whether there was a specific spatial distribution of the positively or negatively charged amino acids within the peptide sequence relative to the hydrophobic residues. In order to study the distribution of the amino acids within the peptide sequence in the context of their tertiary structure we modeled their three dimensional structures using information from the Protein Data Bank.40 For each structure we color coded charge and hydrophobicity information for each amino acid; with yellow we represented the hydrophobic, with red the positively charged and with green the negatively charged amino acids (Figure 1B, 1C and 1D). We repeated the color coding on the 10 most active and the 10 least active peptides as measured in the HeLa luciferase assay. We observed that active peptides did not have any negatively charged amino acids in their sequences (Figure 1B). In the case of inactive peptides the inclusion of negatively charged amino acids in between the positively charged amino acids, even if the positive charges are in stoichiometric excess, seemed to disrupt their effectiveness (Figure 1C).
Next we calculated and visualized the electrostatic potential of the peptides and correlated it to the hydrophobicity patterns in their sequences. We solved the Poisson-Boltzmann equation 41 describing electrostatic interactions between molecular solutes in buffer solutions (Supplementary Materials and Methods). We display two examples describing the electrostatic potential of surfactant protein B and lactoferricin (Figure 1D) as isosurfaces for positive potential (red) and for negative (blue). In the case of the active peptides we observed that the positive charge clusters at specific locations within the tertiary structure of the peptide (Figure 1D). Although most of the peptides that were active are amphiphilic, amphiphilicity was not a requirement for the effectiveness; peptides like lactoferricin have a tertiary structure composed of a beta hairpin with positively charged amino acids in between antiparallel beta sheets. Those amino acids contributed similarly to what we have observed in all the examples creating a defined volume of positive charge relatively to a neutral volume where hydrophobic amino acids exist.
Immunostimulation, In vivo Biodistribution and Activity Studies
Ideally, a therapeutic delivery peptide should possess minimal immunostimulation; allowing for prolonged and repeated delivery of the drug without anti-peptide antibodies and associated immune responses. In order to evaluate the immunostimulation properties of the most potent peptides that we identified, we incubated different concentrations of the peptide-formulations with mouse-derived bone marrow-derived dendritic cells (BMDCs) and quantified the expression of markers which correlate with activation of the cells in the presence of immunostimulating materials.42 After incubating BDMCs for 18 hrs with 30 μg/ml of the peptides derived from surfactant protein B, orexin and the lactoferricin protein in addition to two well established CPPs derived from the antennapedia homeodomain protein and TAT, we quantified the expression of activation specific markers including CD40, CD80, CD86 as well as the major histocompatibility complexes class I and II (MHC-I and MHC-II) on the surface of the peptideexposed CD11c+ BDMCs using flow cytometry (Figure 2B). As a positive control we included lipopolysacharide (LPS). We calculated the geometric mean of the antigen expression distribution and scaled the data so that 100% is the activation induced by LPS. Among the 5 tested surface markers, CD80, CD86 and MHC-II were upregulated upon presentation of all the stimulatory peptides. Of the 5 tested peptides, lactoferricin derived short peptide caused the least cell activation (Figure 2B).
To evaluate the potential of these peptides for in vivo application we studied the biodistribution of nanoparticle formulations in C57BL/6 mice using two non-invasive in vivo imaging platforms; a fluorescent trans-illumination system tracking nanoparticles containing near infrared fluorophores 43 and an x-ray micro computed tomography system 44 for studying the biodistribution of nanoparticles containing x-ray contrast agents.
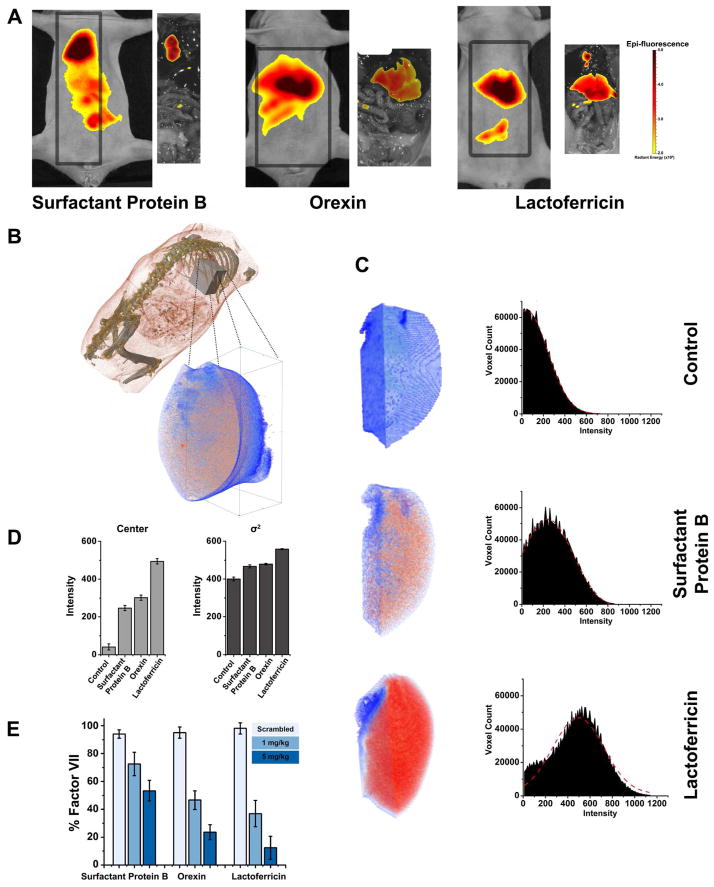
For the in vivo fluorescent imaging the lipidoid nanoparticles were formulated using a near infrared dye conjugated (Cy5.5) siRNA. Non-invasive tracking of siRNA-nanoparticles using this fluorescent dye is facile as there is little near infrared fluorescence contrast generated by most tissues thus minimizing tissue autofluorescence.45 We studied nanoparticle formulations decorated with surfactant protein B, orexin and lactoferricin. Six hours post i.v. injection of nanoparticle formulations, at a concentration of 5 mg/kg, animals were imaged using an IVIS Spectrum imaging system (Figure 3A). Most of the formulations were localized in the liver and lungs of the animals. In order to verify the localization of the nanoparticles we dissected the animals post mortem and imaged them again using the IVIS system (Figure 3A).
Figure 3. In vivo.
studies of the novel cell penetrating peptides. (A) and (B) Biodistribution studies of the peptide decorated nanoparticles. In (A) we utilized a near infrared dye (Cy5.5) conjugated siRNA to formulate the nanoparticles and delivered them via tail vein injection. The animals were imaged with an IVIS Spectrum imaging system 6 hours post injection. The nanoparticles are primarily accumulated in the lungs and livers of the animals. In (B) we conjugated the siRNA to an iodine containing hormone, thyroxine, and injected the formulations via tail vain. We imaged the animals using a micro CT system. In order to quantify the relative amounts of each formulation we considered as a region of interest the left lobe of the liver below the heart after the diaphragm (indicated using grey cube in the figure). (C) Image intensity quantified after averaging the intensity in a 3x3x3 voxel volume and fitted to a Gaussian distribution. (D) We utilized the characteristics of the distributions, like the center and the variance of the distribution, in order to compare the nanoparticle accumulation in the tissue. In an example of three variants of decorated nanoparticles with surfactant protein B, orexin and lactoferricin peptides, the latter exhibited the highest accumulation in the liver with the other two also exhibiting high accumulation. (E) Mice were injected with peptide decorated nanoparticles containing siRNA against a liver synthesized blood circulating protein, Factor VII. Three days post injection, amount of Factor VII in the blood was measured using Factor VII-specific ELISA. In all of the cases n=5 and p<0.01. Interestingly the nanoparticle activity correlated with the biosdistribution as quantified with the contrast agent based analysis. Nanoparticles decorated with scrambled peptide sequences, at 5 mg/kg siRNA concentration, had no activity in knocking down Factor VII.
Although Cy5.5 containing nanoparticles could provide evidence on the biodistribution of the formulations, we could not achieve detailed spatial representation of their localization. Furthermore, the fluorescent based technique did not allow us to quantify the amount of the nanoparticles localized in the liver. This was primarily attributed to the signal variability from batch to batch, or even due to different positioning of the animals in the imaging system. To circumvent such limitations inherent to fluorescent based measurements we utilized an x-ray based micro CT system that provided detailed quantification of the signal distribution. In order to utilize nanoparticle formulations we developed a bioconjugate molecule that contains iodines and can be used as a contrast agent for nanoparticle based microtomography applications. We utilized a human thyroid hormone, thyroxine,46 which contains 4 iodine atoms as well as a primary amine that was used as conjugation point to siRNA molecules (Supplementary Figure 1B).
We injected peptide decorated nanoparticles nanoparticles containing the thyroxine conjugated siRNA (20 mg/kg) and imaged the animals using a GE Healthcare eXplore Locus micro CT system 6 hours post injection. We then process the 3D imaged from the micro CT. Initially we selected within the animal a region of interest with similar volume and same anatomical location among different samples. For all the animals we considered the same liver region that spans in the left lateral lobe of the tissue, below the heart following the diaphragm, keeping the volume of interest the same. We then averaged the intensity data using a sampling volume of 3x3x3 voxels to reduce the background signal noise, quantified the distribution of the signal intensity from the siRNA conjugated contrast agent and fitted the data to a Gaussian distribution. We utilized the center of the signal intensity distribution as a metric for accumulation; the higher the value the more intense the signal from the tissue. In the control case where we injected nanoparticles without any contrast agent conjugated siRNA the center of the Gaussian distribution of the signal intensity is close to zero. Nanoparticles containing siRNA-conjugated lactoferricin demonstrated highest signal, meaning the highest among the samples accumulation in the liver, over 60%, as quantified using the center of the intensity Gaussian distribution (Figure 3C and D). Formulations with other peptides like surfactant protein B and orexin also exhibited high signal intensities relative to the control (particles with no peptides on their surface).
Having established that the nanoparticle formulations primarily accumulate in the liver, we further examined the potency of the formulations at silencing a liver specific protein. As a target for silencing we selected Factor VII, one of the clotting factors that participate in the coagulation cascade. Factor VII is produced solely by the liver and has the shortest half-life of all the circulating coagulation factors thus provides a robust indicator for the effectiveness of the formulations. We formulated peptide decorated nanoparticles with Factor VII-specific siRNA and delivered the formulations to mice via tail vein injection. For each formulation we injected 5 mice at 2 different siRNA concentrations; 1 mg/kg and 5 mg/kg. Three days post-injection we collected blood from the animals and quantified the amounts of circulating Factor VII using a chromogenic enzyme-linked immunosorbent assay (ELISA). We tested surfactant protein B, orexin and lactoferricin. Out of the 3 peptides the one that exhibited the highest potency at reducing the expression of Factor VII, by up to 80% at 5 mg/kg, was lactoferricin (Figure 3E). At the same concentration surfactant protein B and orexin reduced the Factor VII expression by 50% and 70% respectively. Scrambled control sequences of the aforementioned peptides (Surfactant Protein B: MLRIAIRAWLCIPQCK, Orexin: LRGYEGHRCGILALLDLACTKHANCLPCTQPS, Lactoferricin: KRCRGWRQKMCLW) did not exhibit any activity in decreasing Factor VII expression (Figure 3E). Furthermore a C12-227 lipidoid formulation containing only Factor VII siRNA without any peptide conjugates has been shown not to exhibit any in vivo transfection efficacy.29 The weight of the animals remained the same throughout the experiment indicating low systemic toxicity of the formulations. Interestingly the nanoparticle efficacy followed the biodistribution trend as quantified with the contrast agent conjugated siRNA using the micro CT analysis. Presumably such analysis can be used for predicting the efficacy of in vivo formulations.
Conclusions
Membrane associated proteins have evolved to associate with cellular plasma membranes. Other proteins, including some of bacterial or viral origin, can transverse the plasma membrane and enable the host to enter the target cell. Within these protein sequences, small peptide fragments are responsible for such activity.12 In our study, we sought to develop protein fragments for intracellular deliver of small interfering RNAs. Although for the last 30 years there have been many examples of utilizing such protein fragments, called cell penetrating or transduction domain peptides for intracellular delivery,1–3, 9 to our knowledge there has been no systematic examination of the human proteome for new sequences that exhibit such properties.
With the goal of identifying new cell penetrating peptides, we sampled the membrane associated protein sequence proteome and extracted the amino acid sequences responsible for the membrane attachment. We synthesized these sequences, as short peptides, with modifications to allow the chemoselective attachment to siRNA or facilitate the conjugation on the surface of nanoparticles that contain siRNA and tested the activity of the formulations in transfecting cells in vitro and in vivo. To identify the internalization mechanisms of these constructs, we investigated the involvement of various endocytosis pathways including: dynamin-mediated, clathrin-mediated, caveolae-mediated endocytosis and macropinocytosis. We observed that nanoparticle formulations that contain the peptides decorated on the surface of the particles primarily follow macropinocytosis routes.
Common properties among the already known cell penetrating peptides are their high positive charge and amphiphilicity which creates a propensity for the peptides to attach on a lipid membrane. In order to understand the effects of charge and amphiphilicity on peptide function we correlated the patterns of those two properties within the amino acid sequences with the peptide activity. Within the sequences of the active peptides the positively charged amino acids create a surface of positive electrostatic potential though allowing the existence of the hydrophobic amino acids in an electrostatically neutral zone. Furthermore the positively charged are usually clustered around the hydrophobic residues and no negatively charged amino acids disrupt this sequence (Figure 1C).
Furthermore peptide decorated nanoparticles exhibited low immunostimulatory properties relatively to established CPPs like antennapedia homeodomain and TAT (Figure 2B). Given the potency of the peptide decorated nanoparticles in transfecting cells in vitro and their low immunostimulatory effects, we tested the activity of such formulations in vivo. First we created near-infrared fluorescent and x-ray contrast bioconjugates of the siRNA molecules in order to formulate and monitor the biodistribution of the nanoparticles in different tissues in real time. Following intravenous injection, the nanoparticles primarily accumulated in the lungs and liver (Figure 3). Later we tested the activity of the nanoparticle formulations in reducing the expression of a hepatocyte synthesized protein, Factor VII. Three human peptide formulations exhibited significant activity at reducing the expression of Factor VII; surfactant protein B — a lung surfactant protein, orexin — a neuropeptide hormone and lactoferricin — a globular glycoprotein that exists in many physiological fluids. At 5 mg/kg the peptide decorated nanoparticles significantly reduced the expression of Factor VII 90% for the lactoferricin and 80% for the orexin derived peptide (Figure 3E). Furthermore the effectiveness of the nanoparticles at knocking down FVII was consistent with the biodistribution profile as quantified with micro CT image analysis (Figure 3).
Materials and Methods
Peptide-siRNA and Nanoparticle Formulations
The peptides were synthesized using solid phase synthesis methods at the Biopolymers Core Facility in the Koch Institute for Integrative Cancer research at MIT. The peptides were initially conjugated to siRNA molecules in a 1:1 molar ratio by utilizing a pyridyl disulfide group at the 3’ sense strand of the siRNA, with an 18 carbon atom spacer in between. The disulfide bond of the siRNA molecule was cleaved in a tris-(2-carboxyethyl) phosphine (TCEP) resin (Pierce/Thermo Scientific, Rockford, IL) and directly applied to the peptide solution in dimethyl sulfoxide. The nanoparticles were formulated with C12-117 lipidoid, cholesterol (MW 387, Sigma-Aldrich, St. Louis, MO) and a thiol modified lipid-PEG, DSPE-PEG-thiol from Nanocs (PEG MW 2000 and MW 3400, Boston, MA). For the in vivo formulations we used PEG MW 3400 Da. Stock solutions of lipidoid, cholesterol, and PEG were created in 200 proof ethanol at 100, 20, and 100 mg/mL concentrations, respectively. Components were combined to yield weight fractions of 52:20:28. Ethanol mixture was then added dropwise to 200 mM sodium acetate buffer (pH 5) (Sigma-Aldrich, St. Louis, MO) while stirring to spontaneously form empty liposomes. siRNA at a concentration of 10 mg/mL in 50 mM sodium acetate was added to empty lipidoid nanoparticles at a weight ratio of 10:1 total lipids:siRNA. The peptide solutions were added after the formation of the particles and the reaction of the maleimides with the thiols was performed in DPBS overnight at room temperature. For the conjugation reaction we utilized a 5 fold molar excess of the peptide relative to the PEG molecules (detailed description in Supplementary Materials and Methods).
In Vitro siRNA Transfection Assay
HeLa cells expressing Firefly and Renilla luciferase were seeded in an opaque white 96-well plate (Corning-Costar, Corning, NY) in 15,000 cells/well density. Cells were incubated with 100, 300 and 500 ng of siRNA/well either at 1:1 peptide:siRNA conjugates or in nanoparticle formulations for 3 days at 37 °C in a 5% CO2 tissue incubator. The Firefly and Renilla luciferase activity was quantified using the Dual-Glo Luciferase Assay System (Promega, Madison, WI) using the manufacturer’s instructions (detailed description in Supplementary Materials and Methods).
High throughput confocal microscopy
HeLa cells were seeded at 15,000 cells per well in black 96-well plates (Greiner Bio One, Stonehouse, UK). Cells were pre-incubated for 1 hr in the presence of either dynasore (100μM), cytochalasin D (10 μM) and geinstein (50 μM). Nanoparticles decorated with CPPs were exposed to the cells in the presence or absence of these endocytic inhibitors for 1 hour. The cells were washed, fixed in 4% paraformaldehyde and counterstained counterstained in PBS containing Hoescht (2 μg/ml) for nuclei identification (Invitrogen/Life Technologies, Grand Island, NY). Imaging was performed with an OPERA automated spinning disk confocal microscope (Perkin Elmer, Waltham, MA) with a 40X objective. The same defined pattern of 20 fields from each well was acquired to eliminate bias and provide a statistically significant number of cells for analysis. A representative image from each treatment is presented.
Peptide immunostimulatory Studies
Recombinant murine Granulocyte/Macrophage Colony-Stimulating Factor (GM-CSF) was purchased from Peprotech (Rocky Hill, NJ). CD11c-FITC, purified CD16/32, CD40-APC, CD80-APC, CD86-APC, MHCI-APC, MHCII-APC antibodies were purchased from Biolegend (San Diego, CA). Bone marrow cells, flushed from femur and tibia of C57BL/6 female mice were treated with ACK lysing buffer, washed twice, and cultured in 6-well plated in 4 mL of RPMI supplemented with 10% heat inactivated FBS, 2-mercaptoethanol, 1% Pen/strep, gentamicin and 40 ng/mL GM-CSF. On day 6, BMDCs were harvested and transferred to a 12-well plate. Aliquots of peptide solutions (20 μl) with the corresponding peptide concentration were added, and cells were mixed by pipetting. Plates were incubated at 37 °C and 5% CO2 for 18 hrs, after which the cells were centrifuged, washed and incubated with antibodies (CD11c-FITC, purified CD16/32 and CD40-APC, CD80-APC, CD86-APC, MHCI-APC or MHCII-APC) in 50 μL of FACS buffer (PBS supplemeted with 4% FBS and 0.2% sodium azide) for 20 min at 4°C. Cells were washed twice and analyzed by flow cytometry.
In Vivo Fluorescence Imaging
For the in vivo fluorescence imaging studies we used Cy5.5-labeled siRNA (Alnylam, Cambridge, MA) to formulate the nanoparticles. Female SKH1 hairless mice were injected via the tail vein with either PBS (negative control) or Cy5.5-labeled siRNA containing nanoparticles. Imaging was performed 2 hours post particle delivery, with an IVIS Spectrum system (PerkinElmer, Waltham, MA). Excitation and emission wavelengths used for Cy5.5 imaging on the IVIS were 675 nm and 694 nm, respectively.
In Vivo Micro Computed Tomography
Human thyroxine (Sigma-Aldrich, St. Louis, MO) was conjugated to siRNA molecules in a 1:1 molar ratio. Thyroxine, contains 4 iodine atoms as well as a primary amine and a carboxylic acid. A hetero-bifunctional crosslinker N-[γ-maleimido butyryloxy] succinimide ester (GMBS) (Pierce/Thermo Scientific, Rockford, IL) that contains an N-hydroxysuccinimide ester (NHS) and a maleimide reacted with thyroxine’s primary amine in the presence of triethylamine in DPBS containing 5mM EDTA as a conjugation buffer. For this conjugation we used 10:1:1 molar ratio of GMBS, thyroxine and triethylamine respectively following the manufacturer’s instructions.
To perform micro CT imaging studies we used an eXplore CT120-whole mouse MicroCT (GE Healthcare, Woburn, MA) in the Applied Therapeutics and Whole Animal Imaging Core facility at Koch Institute at MIT. The animal were injected the nanoparticle formulations at 20mg/kg of thyroxine conjugated siRNA and 2 hours post injection they were imaged. 220 projection images were acquired over a rotation of 192° with an exposure time of 48 ms at 70 kVp and 32 mA. The images were reconstructed with MicroView (GE Healthcare, Woburn, MA) and post image processing was performed (detailed description in Supplementary Materials and Methods).
In Vivo Factor VII Silencing
C57BL/6 mice (Charles River Labs, Wilmington, MA) were used for in vivo siRNA silencing experiments. Factor VII 2’-O-methyl sugar modified siRNA (Alnylam, Cambridge, MA) was used to prevent activation of the Toll-like receptor 7 immune response and confer enzymatic degradation of the siRNA in vivo. Peptide decorated nanoparticles were formulated using the aforementioned protocol using the 2’-Omethyl sugar modified Factor VII siRNA. 3 days post injection the animals were sacrificed and blood was collected via cardiac puncture in K2EDTA vacuum blood collection serum tube (BD, Franklin Lakes, NJ). Serum levels of Factor VII were analyzed using a Biophen FVII assay kit (Aniara, Mason, OH) (detailed description in Supplementary Materials and Methods).
Supplementary Material
Acknowledgments
The authors are grateful to Drs. Abigail Lytton-Jean, Arturo Vegas and Daniel Siegwart for constructive discussion, suggestions and their help on chemical conjugation techniques and editing of the manuscript. This work was supported by Alnylam Therapeutics, the National Heart, Lung, and Blood Institute, National Institutes of Health, as a Program of Excellence in Nanotechnology (PEN) Award (Contract # HHSN268201000045C) and the NIH 1R01CA132091 Grant.
Footnotes
Supporting Information Available: One Table: Supplementary Table 1; Average nanoparticle size and zeta potential as measured with a ZetaPALS system and % relative peptide coverage on the surface of nanoparticles. Five Figures: Supplementary Figure 1A; Chemoselective chemistries utilized for the conjugation of the peptides on the surface of nanoparticles or directly to siRNA. Supplementary Figure 1B: Schematic of the procedure followed to formulate the lipid nanoparticles. Supplementary Figure 2; The chemical structure of the lipidoid C12-117. Supplementary Figure 3; In vitro efficacy studies of the novel cell penetrating peptides. Supplementary Figure 4; In vitro dose response studies of the novel cell penetrating peptides. Supplementary Figure 5; Maldi MS spectra of the cell penetrating peptide-PEG cojugates. Supplementary Materials and Methods. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Joliot A, Prochiantz A. Transduction Peptides: from Technology to Physiology. Nat Cell Biol. 2004;6:189–196. doi: 10.1038/ncb0304-189. [DOI] [PubMed] [Google Scholar]
- 2.Snyder EL, Dowdy SF. Cell Penetrating Peptides in Drug Delivery. Pharm Res. 2004;21:389–393. doi: 10.1023/B:PHAM.0000019289.61978.f5. [DOI] [PubMed] [Google Scholar]
- 3.Torchilin VP. Recent Approaches to Intracellular Delivery of Drugs and DNA and Organelle Targeting. Annu Rev Biomed Eng. 2006;8:343–375. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- 4.Ren Y, Hauert S, Lo JH, Bhatia SN. Identification and Characterization of Receptor-Specific Peptides for siRNA Delivery. ACS Nano. 2012;6:8620–8631. doi: 10.1021/nn301975s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson ES, Whitney MA, Friedman B, Aguilera TA, Crisp JL, Baik FM, Jiang T, Baird SM, Tsimikas S, Tsien RY, et al. In vivo Fluorescence Imaging of Atherosclerotic Plaques with Activatable Cell-Penetrating Peptides Targeting Thrombin Activity. Integr Biol. 2012;4:595–605. doi: 10.1039/c2ib00161f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng CJ, Saltzman WM. Enhanced siRNA Delivery Into Cells by Exploiting the Synergy Between Targeting Ligands and Cell-Penetrating Peptides. Biomaterials. 2011;32:6194–6203. doi: 10.1016/j.biomaterials.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drin G, Cottin S, Blanc E, Rees AR, Temsamani J. Studies on the Internalization Mechanism of Cationic Cell-Penetrating Peptides. J Biol Chem. 2003;278:31192–31201. doi: 10.1074/jbc.M303938200. [DOI] [PubMed] [Google Scholar]
- 8.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. Cell-penetrating peptides. A Reevaluation of the Mechanism of Cellular Uptake. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 9.Zorko M, Langel U. Cell-Penetrating Peptides: Mechanism and Kinetics of Cargo Delivery. Adv Drug Deliv Rev. 2005;57:529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Hoyer J, Neundorf I. Peptide Vectors for the Nonviral Delivery of Nucleic Acids. Acc Chem Res. 2012;45:1048–1056. doi: 10.1021/ar2002304. [DOI] [PubMed] [Google Scholar]
- 11.Meade BR, Dowdy SF. Exogenous siRNA Delivery Using Peptide Transduction Domains/Cell Penetrating Peptides. Adv Drug Deliv Rev. 2007;59:134–140. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Haas AK, Maisel D, Adelmann J, von Schwerin C, Kahnt I, Brinkmann U. Human-Protein-Derived Peptides for Intracellular Delivery of Biomolecules. Biochem J. 2012;442:583–593. doi: 10.1042/BJ20111973. [DOI] [PubMed] [Google Scholar]
- 13.Nam HY, Kim J, Kim S, Yockman JW, Kim SW, Bull DA. Cell Penetrating Peptide Conjugated Bioreducible Polymer for siRNA Delivery. Biomaterials. 2011;32:5213–5222. doi: 10.1016/j.biomaterials.2011.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitehead KA, Langer R, Anderson DG. Knocking Down Barriers: Advances in siRNA Delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in Humans from Systemically Administered siRNA via Targeted Nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. Polyarginine Enters Cells More Efficiently than Other Polycationic Homopolymers. J Pept Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 17.Haensler J, Szoka FC., Jr Polyamidoamine Cascade Polymers Mediate Efficient Transfection of Cells in Culture. Bioconjug Chem. 1993;4:372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 18.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo Protein Transduction: Delivery of a Biologically Active Protein into the Mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 19.Derossi D, Joliot AH, Chassaing G, Prochiantz A. The Third Helix of the Antennapedia Homeodomain Translocates Through Biological Membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 20.Morris MC, Depollier J, Mery J, Heitz F, Divita G. A Peptide Carrier for the Delivery of Biologically Active Proteins into Mammalian Cells. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 21.Elliott G, O'Hare P. Intercellular Trafficking and Protein Delivery by a Herpesvirus Structural Protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 22.Morris MC, Vidal P, Chaloin L, Heitz F, Divita G. A New Peptide Vector for Efficient Delivery of Oligonucleotides into Mammalian Cells. Nucleic Acids Res. 1997;25:2730–2736. doi: 10.1093/nar/25.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Groot AS, Scott DW. Immunogenicity of Protein Therapeutics. Trends Immunol. 2007;28:482–490. doi: 10.1016/j.it.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Chauhan A, Tikoo A, Kapur AK, Singh M. The Taming of the Cell Penetrating Domain of the HIV Tat: Myths and Realities. J Control Release. 2007;117:148–162. doi: 10.1016/j.jconrel.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of Proteins in Membranes Database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski RA, Hutchinson EG, Michie AD, Wallace AC, Jones ML, Thornton JM. PDBsum: a Web-Based Database of Summaries and Analyses of all PDB Structures. Trends Biochem Sci. 1997;22:488–490. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- 27.Tusnady GE, Dosztanyi Z, Simon I. Transmembrane Proteins in the Protein Data Bank: Identification and Classification. Bioinformatics. 2004;20:2964–2972. doi: 10.1093/bioinformatics/bth340. [DOI] [PubMed] [Google Scholar]
- 28.Akinc A, Goldberg M, Qin J, Dorkin JR, Gamba-Vitalo C, Maier M, Jayaprakash KN, Jayaraman M, Rajeev KG, Manoharan M, et al. Development of Lipidoid-siRNA Formulations for Systemic Delivery to the Liver. Mol Ther. 2009;17:872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, et al. Lipid-Like Materials for Low-Dose, in vivo Gene Silencing. Proc Natl A Sci USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegwart DJ, Whitehead KA, Nuhn L, Sahay G, Cheng H, Jiang S, Ma M, Lytton-Jean A, Vegas A, Fenton P, et al. Combinatorial Synthesis of Chemically Diverse Core-Shell Nanoparticles for Intracellular Delivery. Proc Natl A Sci USA. 2011;108:12996–13001. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehead KA, Sahay G, Li GZ, Love KT, Alabi CA, Ma M, Zurenko C, Querbes W, Langer RS, Anderson DG. Synergistic Silencing: Combinations of Lipid-Like Materials for Efficacious siRNA Delivery. Mol Ther. 2011;19:1688–1694. doi: 10.1038/mt.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahon KP, Love KT, Whitehead KA, Qin J, Akinc A, Leshchiner E, Leshchiner I, Langer R, Anderson DG. Combinatorial Approach to Determine Functional Group Effects on Lipidoid-Mediated siRNA Delivery. Bioconjugate Chem. 2010;21:1448–1454. doi: 10.1021/bc100041r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duchardt F, Ruttekolk IR, Verdurmen WP, Lortat-Jacob H, Burck J, Hufnagel H, Fischer R, van den Heuvel M, Lowik DW, Vuister GW, et al. A Cell-Penetrating Peptide Derived from Human Lactoferrin with Conformation-Dependent Uptake Efficiency. J Biol Chem. 2009;284:36099–36108. doi: 10.1074/jbc.M109.036426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K, Tomita M. Identification of the Bactericidal Domain of Lactoferrin. Biochim Biophys Acta. 1992;1121:130–136. doi: 10.1016/0167-4838(92)90346-f. [DOI] [PubMed] [Google Scholar]
- 35.Cochrane CG, Revak SD. Pulmonary Surfactant Protein B (SP-B): Structure-Function Relationships. Science. 1991;254:566–568. doi: 10.1126/science.1948032. [DOI] [PubMed] [Google Scholar]
- 36.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a Cell-Permeable Inhibitor of Dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Schliwa M. Action of cytochalasin D on cytoskeletal networks. J Cell Biol. 1982;92:79–91. doi: 10.1083/jcb.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parton RG, Joggerst B, Simons K. Regulated Internalization of Caveolae. J Cell Biol. 1994;127:1199–1215. doi: 10.1083/jcb.127.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA Fusogenic Peptide Enhances Escape of TAT-Fusion Proteins After Lipid Raft Macropinocytosis. Nat Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a Computer-Based Archival File for Macromolecular Structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 41.Honig B, Nicholls A. Classical Electrostatics in Biology and Chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 42.Lipscomb MF, Masten BJ. Dendritic Cells: Immune Regulators in Health and Disease. Physiol Rev. 2002;82:97–130. doi: 10.1152/physrev.00023.2001. [DOI] [PubMed] [Google Scholar]
- 43.Weissleder R, Tung CH, Mahmood U, Bogdanov A., Jr In vivo Imaging of Tumors with Protease-Activated Near-Infrared Fluorescent Probes. Nat Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 44.Rabin O, Manuel Perez J, Grimm J, Wojtkiewicz G, Weissleder R. An X-Ray Computed Tomography Imaging Agent Based on Long-Circulating Bismuth Sulphide Nanoparticles. Nat Mater. 2006;5:118–122. doi: 10.1038/nmat1571. [DOI] [PubMed] [Google Scholar]
- 45.Sevick-Muraca EM. Translation of Near-Infrared Fluorescence Imaging Technologies: Emerging Clinical Applications. Annu Rev Med. 2012;63:217–231. doi: 10.1146/annurev-med-070910-083323. [DOI] [PubMed] [Google Scholar]
- 46.Taurog A. Thyroid Peroxidase and Thyroxine Biosynthesis. Recent Prog Horm Res. 1970;26:189–247. doi: 10.1016/b978-0-12-571126-5.50009-1. [DOI] [PubMed] [Google Scholar]
- 47.Wang G, Li Y, Li X. Correlation of Three-Dimensional Structures with the Antibacterial Activity of a Group of Peptides Designed Based on a Nontoxic Bacterial Membrane Anchor. J Biol Chem. 2005;280:5803–5811. doi: 10.1074/jbc.M410116200. [DOI] [PubMed] [Google Scholar]
- 48.Mishra VK, Anantharamaiah GM, Segrest JP, Palgunachari MN, Chaddha M, Sham SW, Krishna NR. Association of a Model Class A (Apolipoprotein) Amphipathic Alpha Helical Peptide with Lipid: High Resolution NMR Studies of Peptide-Lipid Discoidal Complexes. J Biol Chem. 2006;281:6511–6519. doi: 10.1074/jbc.M511475200. [DOI] [PubMed] [Google Scholar]
- 49.Sarker M, Waring AJ, Walther FJ, Keough KM, Booth V. Structure of Mini-B, a Functional Fragment of Surfactant Protein B, in Detergent Micelles. Biochemistry. 2007;46:11047–11056. doi: 10.1021/bi7011756. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen LT, Schibli DJ, Vogel HJ. Structural Studies and Model Membrane Interactions of Two Peptides Derived from Bovine Lactoferricin. J Pept Sci. 2005;11:379–389. doi: 10.1002/psc.629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.