Abstract
Nanoparticles are employed for delivering therapeutics into cells1,2. However, size, shape, surface chemistry and the presentation of targeting ligands on the surface of nanoparticles can affect circulation half-life and biodistribution, cell specific internalization, excretion, toxicity, and efficacy3-7. A variety of materials have been explored for delivering small interfering RNAs (siRNAs) - a therapeutic agent that suppresses the expression of targeted genes8,9. However, conventional delivery nanoparticles such as liposomes and polymeric systems are heterogeneous in size, composition and surface chemistry, and this can lead to suboptimal performance, lack of tissue specificity and potential toxicity10-12. Here, we show that self-assembled DNA tetrahedral nanoparticles with a well-defined size can deliver siRNAs into cells and silence target genes in tumours. Monodisperse nanoparticles are prepared through the self-assembly of complementary DNA strands. Because the DNA strands are easily programmable, the size of the nanoparticles and the spatial orientation and density of cancer targeting ligands (such as peptides and folate) on the nanoparticle surface can be precisely controlled. We show that at least three folate molecules per nanoparticle is required for optimal delivery of the siRNAs into cells and, gene silencing occurs only when the ligands are in the appropriate spatial orientation. In vivo, these nanoparticles showed a longer blood circulation time (t1/2 ∼ 24.2 min) than the parent siRNA (t1/2 ∼ 6 min).
Previously, self-assembled three-dimensional structures of short oligonucleotides have been explored for imaging and delivery applications13-15. In this study, we prepared oligonucleotide nanoparticles (ONPs) by programmable self-assembly of short DNA fragments and therapeutic siRNAs to develop a population of molecularly identical nanoparticles with controllable particle size and target ligand location and density. As shown in Fig. 1a, six DNA strands with complementary-overhangs at the 3′ ends can self-assemble into a tetrahedron consisting of 186 Watson-Crick base pairs. The six edges are 30 base pairs in length and the theoretical tetrahedron height is approximately 8 nm with 10 nm edges. Each edge contains a nick in the middle where the 5′ and 3′ ends of an oligonucleotide meet. The overhang at this nick is complementary to the overhang of siRNA strands. Thus six siRNAs are bound per nanoparticle (1 per each edge). Chemically modified siRNA with 2′-OMe modifications shown to significantly enhance serum stability as well as reduce immune stimulation potential was used in these experiments16. Native polyacrylamide gel electrophoresis (PAGE) analysis shows the step-wise assembly of DNA tetrahedron particles as each strand is added. A distinct band shift was observed indicating DNA assembly and yields over 95% and 98% were observed for tetrahedron formation and siRNA hybridization respectively (Fig. 1b). The tetrahedron structure was imaged by AFM in aqueous buffer and high-resolution images confirmed the presence of the three upper edges of individual tetrahedron as well as a height of ∼ 7.5 nm (Fig. 1c). Dynamic light scattering measured a hydrodynamic diameter of ∼28.6 nm with a narrow size distribution, even when ONPs were prepared at a high concentration (8 μM) (Supplementary Fig. 1).
Figure 1. Programmable self-assembly of oligonucleotide nanoparticles.
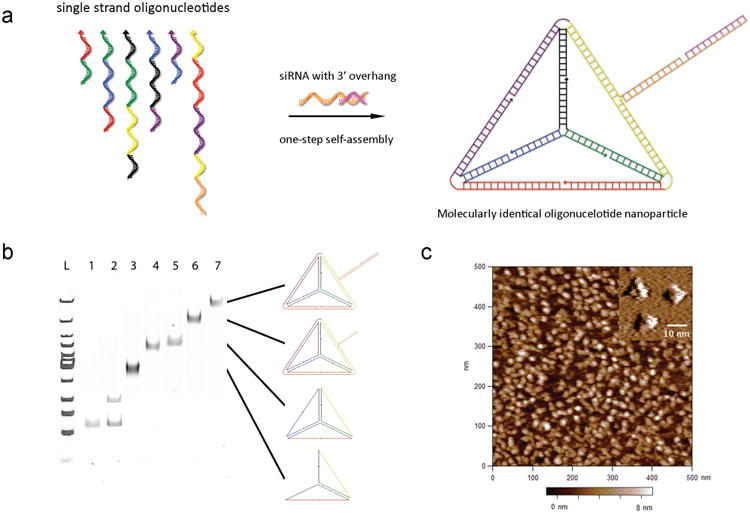
a, Schematic of DNA strands for tetrahedron formation (arrow head represents 5′ end of the nucleic acid strand and each colour corresponds to one of the six edges of the tetrahedron) and representation showing site-specific hybridization of siRNA to the self-assembled nanoparticles. b, Native PAGE analysis to verify self-assembly of DNA tetrahedron and hybridization of siRNAs to DNA core, with the presumed structures schematically drawn to the right of the gel (Lane 1: strand 1; Lane 2: strand 1 and strand 2; Lane 3: strands 1–3; Lane 4: strands 1–4; Lane 5: strands 1–5; Lane 6: strands 1–6; Lane 7: strands 1–6 and siRNAs). C, AFM image showing mono-disperse tetrahedron nanoparticles on mica. Inset indicates AFM images recorded with an ultra-sharp tip resolving the three upper edges of tetrahedral.
It has been suggested that the optimal particle size for a cancer targeting nanodelivery carrier is 20–100 nm3,4,17. Nanoparticles with a diameter above 20 nm avoid renal clearance, which is a typical occurrence for monomeric siRNA, and enhance delivery to certain tumour types through the enhanced permeability and retention effect (EPR)18,19. In theory, ONPs are large enough to avoid renal filtration (>20 nm) but small enough to penetrate through the leaky vasculatures in a tumour region, bind to cell surface receptors, and facilitate intracellular uptake, while reducing reticuloendothelial system (RES)-mediated clearance. Because the ONPs are molecularly defined, they exist as a single uniform population in terms of size and shape. This is distinctly different from traditional cationic delivery carriers that can exist in a range of shapes and sizes. The nucleic acid composition of the ONPs allows precise spatial control of all decorating ligands via hybridization and means that the siRNA component of the particle can be varied.
To ascertain whether ONPs can provide effective targeted delivery of siRNA to human cancer cells, we conjugated various cancer-targeting ligands from peptides to small molecules. It is hypothesized that intracellular delivery of the ONPs will be promoted by active targeting of specific surface receptors on cancer cells20,21. Among the 28 different targeting ligands tested here, folic acid (FA) conjugated ONPs exhibited the greatest gene silencing, over 50 % reduction of firefly luciferase expression in Hela cells in a dose-dependent manner (Fig. 2a). A few cationic peptides (Hph-1 and Penetratin) also showed the reduction of firefly luciferase expression, however the structures of the ONPs modified with these ligands were less stable causing unwanted aggregation due to the charge interaction between the cationic peptides and anionic nucleic acids. For folate-mediated gene silencing, folate receptor overexpressing KB cells (expressing green fluorescent protein (GFP)) were also tested to confirm specificity of targeted gene silencing. Our data showed over 60 % reduction of GFP expression in KB cells at a 35 nM dose of FA conjugated ONPs carrying siRNA targeting GFP (Supplement Fig. 2).
Figure 2. In vitro screening and gene silencing using ONPs.

a, Screening of tumour-targeting ligands using ONPs in a luciferase silencing assay in HeLa cells (Control: ONPs without targeting ligands, LF: Lipofectamine RNAiMax, peptide abbreviations are described in supplementary information). b, GFP gene silencing efficiency varies with folic acid (FA) density on ONPs (n=4, siRNA concentration=35 nM). *p<0.003, **p<0.001 as compared with the control; ns=not significant. c, The structure and function relationship of the orientation of ligand (bullet shapes on end of siRNA strands) and the efficiency of gene silencing for ONPs (n=4, siRNA concentration=35 nM, set A: FA on 1, 2, and 3a; set B: FA on 1, 2, and 3b; set C: FA on 1, 2, and 3c). *p<0.018 **p<0.019 as compared with the control; ns=not significant. d, Automated confocal analysis of intracellular uptake of ONPs with different FA orientations (n=8).
In addition to optimal particle size and cancer specificity for intracellular siRNA delivery, ONPs allow full control of spatial orientation of ligands as well as the density of ligands on the nanoparticle surface. Since the geometry of the ONPs is well defined, it is possible to investigate correlations of ligand density and orientation with gene silencing. To evaluate the importance of spatial orientation for the targeting ligand, six different overhang sequences were designed for siRNA hybridization and the number of hybridized siRNAs was varied as well as the density and location of the folate. We can specifically control the number of siRNAs on the core DNA tetrahedron from one to six (Supplement Fig. 3). By using both folic acid conjugated and non-conjugated siRNAs, the level of GFP gene silencing was investigated with various numbers of folate ligands, while maintaining the same number of siRNAs on each nanoparticle (Fig. 2b). Our results indicate that minimum of three folate ligands are required to achieve GFP gene silencing. Interestingly, more than three ligands on one particle did not improve the gene silencing efficiency. Importantly, the orientation/location of the ligands dramatically affected the gene silencing (Fig. 2c). When three folic acids are decorated on the tetrahedron such that the local density is maximized (three folate encompass a face or vertex of the tetrahedron: example location 1, 2, 3a or 1, 2, 3c in Fig. 2c, respectively), GFP silencing was observed. However, when three folic acids were located with greater distance from each other on the tetrahedron, such that the local density is lower (locations 1, 2, 3b), we did not observe any GFP silencing. Interestingly, our confocal study revealed that the intracellular uptake of these nanoparticles were similar, regardless of folic acid location, however potent gene silencing activity was only observed for nanoparticles with the appropriate spatial orientation of folic acid (Fig. 2d). It is conceivable that, with a specific orientation of FA, the higher local ligand density may influence the intracellular trafficking pathway of nanoparticles through the cells and corresponding gene silencing. Although it is believed that the density and location of ligands can greatly influence the nanoparticle/cell membrane interactions as well as the intracellular uptake pathways, this phenomenon is extremely difficult to prove by conventional nanoparticles due to the lack of precise control of ligand density and orientation on a single nanoparticle22,23. To our knowledge, this is the first in vitro evaluation of ligand density and orientation that can alter the function of a nanoparticle-delivery system.
To verify in vivo delivery of ONPs, the pharmacokinetic (PK) profile and organ bio-distribution were investigated in nude mice bearing KB xenograft tumours. Cy5-labeled ONPs with folate ligands (3 nmol of siRNA, ∼ 2.0 mg/kg) (6 FA ligands per ONP) were systemically delivered via tail-vein injection and the in vivo behavior was quantitatively measured over the course of 24 h by fluorescence molecular tomography fused with computed tomography (FMT-CT)24,25. Co-registration of FMT and CT provided high-resolution fluorescence images of the tumour targeting by ONPs and the distribution of nanoparticles in five major organs. As shown in Fig. 3a, ONPs primarily accumulated in tumour and kidney, with little accumulation in other organs such as liver, spleen, lung, and heart. Ex vivo fluorescence images at 12 h post-injection also correspond well with the biodistribution data. A reconstructed 3D FMT-CT image of a tumour bearing mouse revealed the accumulation of ONPs in the tumour region as early as 25 min post-injection (Fig. 3b). In addition, the blood half-life data indicated that ONPs have a longer blood circulation time (t1/2 ∼ 24.2 min) compared to the parent siRNA (t1/2 ∼ 6 min) (Supplementary Fig. 4)26.
Figure 3.
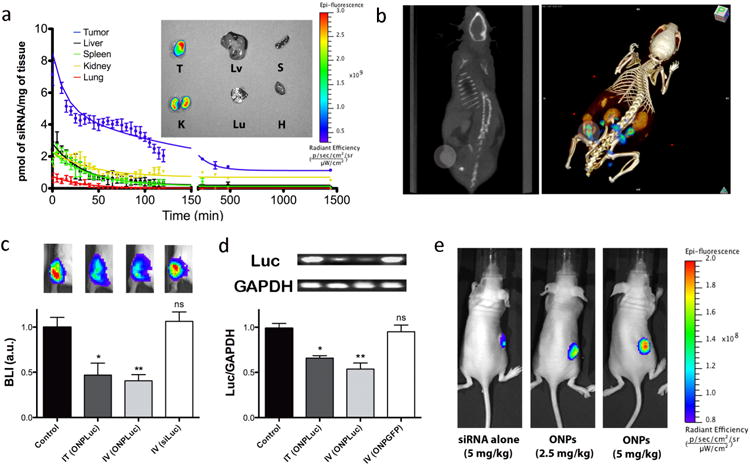
In vivo PK and gene silencing in tumour xenograft mouse model. a, PK of ONPs in KB tumour-bearing mice and ex vivo fluorescence image of five major organs and tumour at 12h post-injection (T: Tumour, Lv: Liver, S: Spleen, K: Kidney, Lu: Lung, H: Heart). High level of siRNA accumulation occurs in tumor tissue. b, Tumour specific accumulation of ONPs as determined by FMT-CT after 25 min post-injection (left: CT scan, right: 3D FMT-CT). Circular region indicates the location of tumour xenograft. Warmer colour in tumour region represents the site-specific accumulation of Cy5 labeled ONPs. c, In vivo luciferase silencing in KB tumour xenografts (n=7, BLI: Bioluminescence Intensity, Control: PBS injection, IT: intratumoural, IV: intravenous, ONPLuc: ONPs with Folate conjugated anti-luciferase siRNA, siLuc: Folate conjugated anti-luciferase siRNA). siRNA concentration=2.5mg/kg. *p<0.138, **p<0.002 as compared with the control, ns=not significant. Four Image boxes shown above the bar graph represent the live BLI image of mice from each group. Warmer colour indicates strong BLI in tumour xenografts. d, Quantitative analysis of luciferase mRNA degradation in KB tumours 2 days after ONP injection (n=3, Luc: Firefly luciferase gene, ONPGFP: ONPs with Folate conjugated anti-GFP siRNA). siRNA concentration=2.5mg/kg. GAPDH is used as house keeping gene. Luciferase mRNA level is expressed as a ratio to GAPDH (Luc/GAPDH). *p<0.05, **p<0.03 as compared with the control, ns=not significant. e, In vivo live fluorescence images showing dose responsive accumulation of ONPs in KB tumours compared to folic acid conjugated siRNA (siRNA alone); animals were treated by systemic injection (n=3) and images are representative of each group.
In order to assess the therapeutic potential of ONPs as cationic-free gene delivery carriers, we conducted in vivo gene silencing of firefly luciferase expressing KB xenografts. Folic acid conjugated ONPs with anti-luciferase siRNA were administrated at a dose of 2.5 mg/kg of siRNA into mice either by tail-vein injection or intra-tumour injection. Silencing was evaluated 48 h post-injection by measuring bioluminescent intensity in the tumour, (Fig. 3c). Both tail vein and intra-tumour injections resulted in approximately 60 % decrease in bioluminescent intensity. When mice were injected in either mode of administration with folic acid conjugated anti-luciferase siRNAs (not assembled into nanoparticles), no decrease in bioluminescent intensity was observed. This result corresponds well with our in vitro silencing experiment, confirming that the ONPs are a critical factor for successful gene silencing. In addition, measurement of the firefly luciferase mRNA level in tumour cells strongly supported the target specific mRNA cleavage by both systemic and local delivery of ONPs (Fig. 3d). A dose dependent study revealed that tumour specific accumulation of ONPs was achieved by systemic delivery (Fig 3e) and the IC50 for luciferase silencing was estimated to be approximately 1.8 mg/kg (Supplementary Fig. 5a). Finally, the immune response of ONPs was monitored by measuring the IFN-α levels in blood samples 6 h post-injection and there was no significant increase in IFN-α as compared to levels in untreated mice (Supplementary Fig. 5b).
This study has demonstrated that six single-stranded DNA fragments, and six double-stranded siRNAs, can self-assemble in a one-step reaction to generate DNA/siRNA tetrahedral nanoparticles for targeted in vivo delivery. The overhang design on the DNA strands allowed specific hybridization of complementary siRNA sequences, and provided full control over spatial orientation of siRNA and the locations and density of cancer targeting ligands. The ONPs can be modified with different tumour targeting ligands by simple conjugation chemistry, extending the use of these nanoparticles for the treatment of various cancers. We observed robust gene silencing via both intra-tumour and systemic injection of ONPs in KB xenograft tumours without any detectable immune response. We believe these particles may also have utility in the treatment of other tissues through modification of size and ligand type.
Method Summary
Preparation of self-assembled DNA/siRNA nanoparticles (ONPs)
Six single-stranded DNAs from IDT (Chicago, IL) and 6 double strand siRNAs from Alnylam (Cambridge, MA) were annealed to prepare ONPs. DNA strands (final strand concentration 3.3 μM each) were mixed in equal molar ratio in annealing buffer containing 5 mM Mg2+ and a 6-fold molar excess of siRNA strands were added to the solution. The solution was heated to 90°C for 2 min and rapidly cooled to 4°C to generate ONPs (3.3 μM particle concentration). For folic acid conjugated ONPs, folic acid conjugated luciferase or GFP siRNA (Alnylam) were used. All siRNAs were chemically modified with site-specific 2′-OMe chemistry to avoid immune response activation and to improve nuclease resistance26.
In vitro testing of ONPs
HeLa cells, modified to stably express both firefly and Renilla luciferase genes, were utilized for in vitro screening of ONPs. ONPs with different ligands were applied to 1.5 × 104 HeLa cells in medium containing serum. Firefly luciferase silencing was assessed 24 h post-transfection using a Dual-Glo Luciferase Assay kit (Promega, Madison, WI). Transfections were performed in quadruplicate. siRNA transfected into cells using Lipofectamine RNAiMax (Invitrogen) was used as a positive control (Fig. 2). For folate receptor targeted GFP gene silencing, GFP expressing KB cells were used. The level of GFP silencing was evaluated at 24 h post-transfection using a BD FACSCalibur (Franklin Lakes, NJ).
siRNA delivery in mice using ONPs
All animal experiments were conducted using institutionally approved protocols. Luc-KB tumour bearing female BALB/c nude mice (Charles River Laboratories, Wilmington, MA) received tail vein or intra-tumoural injections of either PBS (negative control) or ONPs containing anti-Luc siRNA diluted in PBS (n=7 per each group, siRNA concentration=2.5mg/kg). Two days post-injection, bioluminescence intensity (BLI) in KB tumours was measured by a IVIS Lumina imaging system (Hopkinton, MA).
Full Methods and any associated references are available in the online version of the paper at www.nature.com/naturenanotechnology.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (EB000244), Center for Cancer Nanotechnology Excellence (U54 CA151884), Alnylam Pharmaceuticals, and the National Research Foundation of Korea (NRF-2011-357-D00063). The authors thank J. Hong and C. Hong for figure drawing, and J.B. Lee and A. Schroeder for helpful discussion.
Footnotes
Author contributions: H.L., A.L.J. and D.G.A planned the experiments, H.L., A.P., K.L., A.S., W.Q., C.Z., J.S.D., A.L.J. and J.T. conducted the experiments, H.L., A.L.J., Y.C., K.L., E.D.K., M.N., R.L., and D.G.A. analyzed the data, and H.L., A.L.J., M.M., and D.G.A. wrote the paper.
Additional information: Supplementary information accompanies this paper at www.nature.com/naturenanotechnology.
References
- 1.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 2.Peer D, et al. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 3.Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 4.Choi HS, et al. Design consideration for tumor-targeted nanoparticles. Nat Nanotechnol. 2011;5:42–47. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nature Biotech. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 6.De Jong WH, Borm P. Drug delivery and nanoparticles: Applications and hazards. Int J Nanomedicine. 2008;3:133–149. doi: 10.2147/ijn.s596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akinc A, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 9.Bumcrot D, Manoharan M, Koteliansky V, Sah DWY. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nature Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61:850–862. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 12.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Bhatia D, et al. A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat Communications. 2011;2:339. doi: 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- 14.Walsh AS, Yin H, Erben CM, Wood MJA, Tuberfield AJ. DNA cage delivery to mammalian cells. ACS Nano. 2011;5:5427–5432. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 15.Keum JW, Ahn JH, Bermudez H. Design, assembly, and activity of antisense DNA nanostructures. Small. 2011;7(24):3529–3535. doi: 10.1002/smll.201101804. [DOI] [PubMed] [Google Scholar]
- 16.Gaglione M, Messere A. Recent progress in chemically modified siRNAs. Mini Rev Med Chem. 2010;10:578–595. doi: 10.2174/138955710791384036. [DOI] [PubMed] [Google Scholar]
- 17.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 18.Allen TM, Cullis PR. Drug delivery systems: entering the mainstream. Science. 2004;303:1818–1822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 19.Mok H, Lee SH, Park JW, Park TG. Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nature Mater. 2010;9:272–278. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- 20.Song E, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nature Biotech. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 21.Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16:163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarapore P, Shu Y, Guo P, Ho SM. Application of Phi29 motor pRNA for targeted therapeutic delivery of siRNA silencing metallothionein-IIA and surviving in ovarian cancers. Mol Ther. 2011;19:386–394. doi: 10.1038/mt.2010.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manz BN, et al. T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc Natl Acad Sci USA. 2011;108:9089–9094. doi: 10.1073/pnas.1018771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nahrendorf M, et al. Hybrid PET-optical imaging using targeted probes. Proc Natl Acad Sci USA. 2010;107:7910–7915. doi: 10.1073/pnas.0915163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leuschner F, et al. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat Biotechnol. 2011;29:1005–1010. doi: 10.1038/nbt.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


