Summary
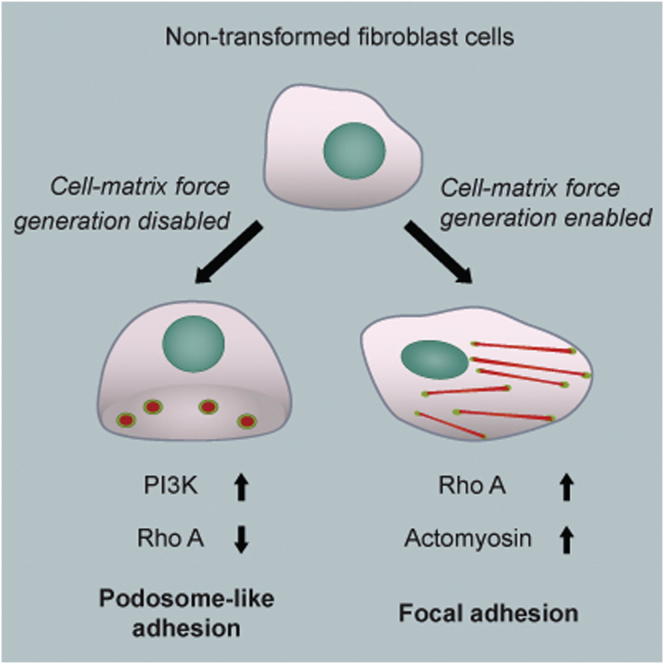
Matrix-activated integrins can form different adhesion structures. We report that nontransformed fibroblasts develop podosome-like adhesions when spread on fluid Arg-Gly-Asp peptide (RGD)-lipid surfaces, whereas they habitually form focal adhesions on rigid RGD glass surfaces. Similar to classic macrophage podosomes, the podosome-like adhesions are protrusive and characterized by doughnut-shaped RGD rings that surround characteristic core components including F-actin, N-WASP, and Arp2/Arp3. Furthermore, there are 18 podosome markers in these adhesions, though they lack matrix metalloproteinases that characterize invadopodia and podosomes of Src-transformed cells. When nontransformed cells develop force on integrin-RGD clusters by pulling RGD lipids to prefabricated rigid barriers (metal lines spaced by 1–2 μm), these podosomes fail to form and instead form focal adhesions. The formation of podosomes on fluid surfaces is mediated by local activation of phosphoinositide 3-kinase (PI3K) and the production of phosphatidylinositol-(3,4,5)-triphosphate (PIP3) in a FAK/PYK2-dependent manner. Enrichment of PIP3 precedes N-WASP activation and the recruitment of RhoA-GAP ARAP3. We propose that adhesion structures can be modulated by traction force development and that production of PIP3 stimulates podosome formation and subsequent RhoA downregulation in the absence of traction force.
Graphical Abstract
Highlights
-
•
Nontransformed fibroblasts on RGD membranes form podosome-like protrusions
-
•
Nanopatterned RGD membranes enable traction force, suppressing protrusion formation
-
•
Local activation of PI3K transforms prepodosomal-like RGD clusters
-
•
PIP3-bound RhoA GAP ARAP3 is recruited at the protrusion and downregulates RhoA-GTP
Outside-in integrin activation plays a critical role in cell adhesion and migration. Yu, Sheetz, and colleagues now report that plating cells onto fluid RGD membranes, thus inhibiting the development of traction force, causes the formation of podosome-like protrusions. The podosome-like adhesions on RGD membranes lack matrix metalloproteinases but share many other molecular components with classic podosomes in macrophages. The authors demonstrate that podosome-like protrusion formation is mediated by a PIP3-dependent pathway and is further aided by ARAP3-mediated RhoA inactivation.
Introduction
Activation of integrin receptors by extracellular ligand binding mediates the formation of cell-matrix adhesions (Miranti and Brugge, 2002). The clustering of activated integrins and integrin-associated proteins locally promotes the activation of downstream signal transduction paths leading to events such as cell migration (Huttenlocher and Horwitz, 2011), differentiation (Engler et al., 2006), and cancer metastasis (Levental et al., 2009). The recruitment of actin-binding proteins, such as talin and vinculin, provides structured linkages between integrins and the actin cytoskeleton (Vogel and Sheetz, 2006; Wehrle-Haller, 2012). While the initial clustering of integrin receptors upon binding mobile Arg-Gly-Asp (RGD) moieties is independent of traction forces (Yu et al., 2011, 2012a), contraction-mediated maturation of integrin clusters results in stable adhesion formation (Moore et al., 2010). More importantly, the physical characteristics of extracellular matrix (ECM) can initiate differential assembly of the actomyosin cytoskeletal network (Geiger et al., 2009). For example, fibroblasts on a rigid ECM substrate (100 kPa) are flat, polarized cells with actin stress fibers across the cell body. On softer but chemically identical ECM substrates (10 kPa), fibroblasts fail to polarize and exhibit fewer and less robust actin stress fibers (Prager-Khoutorsky et al., 2011). Despite numerous studies, the interplay among actin assembly, force generation, and adhesion structure remains unclear.
Podosomes and focal adhesions are both integrin-mediated multimolecular assemblies for cell adhesion (Calle et al., 2006; Geiger et al., 2001; Machesky et al., 2008). Many adherent cells, such as epithelial cells or stromal fibroblasts cultured in vitro, maintain stable adhesions to the substratum via focal adhesions, adhesion structures interconnected by an actomyosin contractile network (Cai and Sheetz, 2009; Vogel and Sheetz, 2009). On the other hand, monocytic-lineage-derived cells such as macrophages utilize an alternative structure known as a podosome as their primary adhesion machinery (Cox et al., 2012; Murphy and Courtneidge, 2011). Podosomes characteristically contain WASP, cortactin Arp2/Arp3, and actin filaments in the center (podosome core, usually 1 μm in diameter and 2 μm in height), which is surrounded by a ring of integrin and integrin-associated proteins, such as talin, vinculin, and paxillin. Alternatively, transformation of fibroblasts by constitutively active Src kinase will also drive podosome or invadopodia formation with N-WASP substituting for leukocyte-restricted WASP. N-WASP/WASP and Arp2/Arp3 are regarded as markers of podosomes as they are not seen at focal adhesions, but otherwise the two adhesive structures share many molecular components, though the spatial organization of these components is very different (Gimona et al., 2008). Although a wealth of experimental detail is now available, the underlying mechanism of podosome assembly and whether it is force dependent comparable to the situation with focal adhesions are largely unknown.
Mobile RGD ligands on nanopatterned supported lipid membranes provide a simple means to study force-mediated signal transduction events at the cell membrane and have been widely used in various cell biological investigations, such as studies of the immunological synapse (Mossman et al., 2005), ephrin-mediated cancer metastasis (Salaita et al., 2010), and force-modulated integrin adhesion (Yu et al., 2011, 2012a). Previously, we have utilized RGD-tagged lipids in supported membranes (RGD biotin bound to Cascade blue neutravidin bound to biotin lipid) with or without nanopartitioned lines to trigger integrin activation and to investigate force-dependent and independent functions during early cell spreading (Yu et al., 2011). Continuous films of RGD membranes generally exhibited long-range lateral mobility (diffusion coefficient 2 μm2/s) and were substrates with infinitesimal elasticity (Evans and Yeung, 1994; Evans and Hochmuth, 1978) (zero rigidity/shear modules, equivalent film viscosity 0.1 N-s/m2). When the long-range mobility of the RGD membrane was locally restricted by fabricating metal lines as nanopartitions within the bilayers (typically 100 nm line width and 5 nm thick with 1–4 μm line pitch, passivated by BSA or casein) (Yu and Groves, 2010; Yu et al., 2010), mobile RGD-integrin clusters assembled stable adhesions across the adjacent partitions with 1 and 2 μm, but not with 4 μm, pitch through force generation and adhesion maturation. This system was ideal for testing matrix-dependent mechanical regulation of adhesion formation.
Various signal transduction pathways can regulate cell-matrix adhesions. Anionic phospholipids, such as phosphatidylinositol (4,5)-bisphosphate (PIP2) and phosphatidylinositol (3,4,5)-triphosphate (PIP3) are dynamically regulated in plasma membranes (McLaughlin et al., 2002; Xu et al., 2003). Local enrichment of these negatively charged lipids can initiate N-WASP/WASP-mediated actin polymerization at plasma membranes (Papayannopoulos et al., 2005; Pollitt and Insall, 2009). Class IA phosphoinositide 3-kinase (PI3K) is composed of two subunits, p85 (regulatory) and p110 (catalytic), that phosphorylate PIP2 to generate PIP3 (Vanhaesebroeck et al., 2012), raising local PIP3 concentrations. On the other hand, PTEN dephosphorylates PIP3 to PIP2, decreasing PIP3 concentrations. While the biochemical interaction of p85beta and focal adhesion kinase (FAK) has been reported previously (Chen et al., 1996), N-WASP and Arp2/Arp3 actin polymerization complexes are absent at tension-loaded focal adhesions. Here, we report that spatiotemporal recruitment of PI3K and local enrichment of PIP3 at integrin-mediated adhesion sites on traction-force free RGD membranes play an important role in differential signal transduction leading to podosome formation.
Results
Formation of Podosome-like Adhesion Follows Initial Integrin Clustering of RGD Lipids
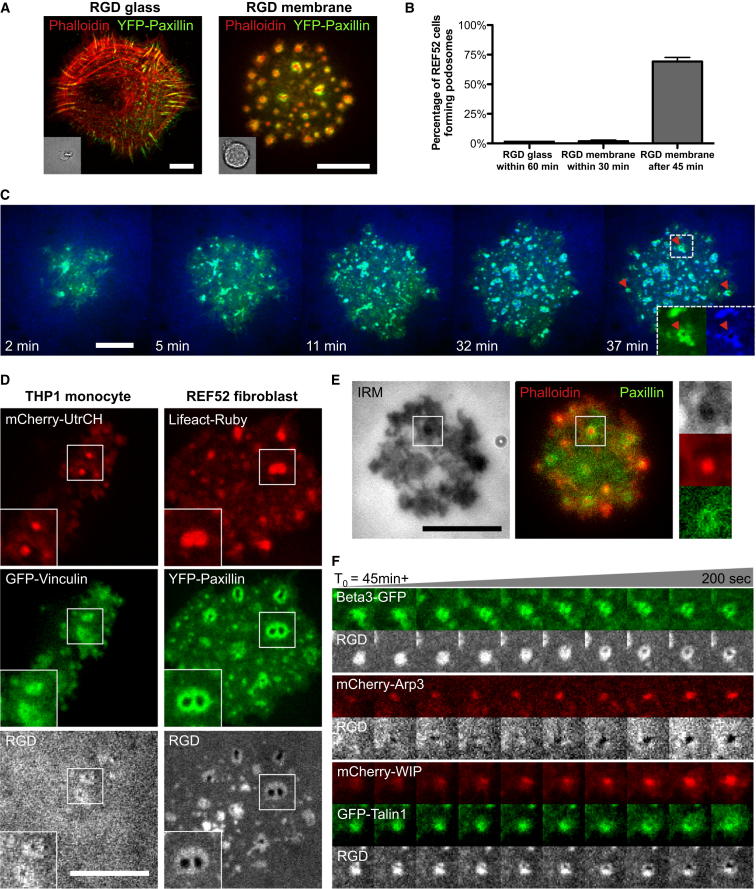
Although THP1 monocytic cells treated with transforming growth factor β1 have been used as a model system to study podosomes on regular matrix-coated substrates (Monypenny et al., 2011), fibroblasts generally do not form podosomes on matrix-coated substrates unless transformed by Src (Oikawa et al., 2008; Tarone et al., 1985). It was therefore surprising to see that nontransformed fibroblasts (RPTPα+/+ mouse embryonic fibroblasts and REF52 rat fibroblasts) formed podosome-like adhesions when plated on freely diffusive RGD lipids (Figure 1A) without artificially elevated Src activity. About 70% of REF52 fibroblasts developed podosome-like adhesions after 45 min of initial adhesion (Figure 1B; total of 321 cells in four experiments), while the same cells seeded on immobilized RGD-coated glass consistently formed classic focal adhesions (Figure 1A). In parallel, we also observed podosome formation in THP1 cells on RGD-supported bilayers (Figure 1D; Movie S1).
Figure 1.
Podosome Formation on RGD Membrane
(A) REF52 fibroblast forms regular focal adhesion on RGD glass but developed podosomes on RGD membrane after 45 min of initial adhesion.
(B) Percentage of REF52 fibroblast cells forming regular adhesion and podosomes on RGD-coated glass or RGD membrane. A total 321 cells in four experiments were used.
(C) Transition from initial RGD-integrin clusters to podosomes in REF52 fibroblast adhered on RGD membrane (Movie S2). Inset: podosomes (red arrowheads) were identified by ring formations of both RGD and YFP paxillin. (D) THP1 monocytes and REF52 fibroblasts both formed podosomes on RGD membrane. The dense actin core is surrounded by adhesion proteins, such as paxillin, vinculin, and RGD-integrin clusters. The center of the podosome ring is depleted from RGD.
(E) Interference reflection microscopy reveals tight contacts at the protrusive podosome core in REF52 fibroblast cells. Inset (top to bottom): RICM, CF594 phalloidin (F-actin staining), and YFP paxillin.
(F) Development of podosomes (4 × 4 μm2 each frame) in REF52 fibroblast cells. Integrin β3, talin, and RGD clusters are reorganized to form the podosome ring. Arp3 and WIP are enriched at the podosome core.
Error estimates are SEM. The scale bar represents 10 μm.
To understand the development of podosome-like adhesions in fibroblast cells, we examined the process of adhesion formation on RGD membranes. We found that cells assembled RGD-integrin clusters during the early adhesion process, as we described previously (Yu et al., 2011, 2012a). Similar to the case of focal complex and focal adhesion formation, the activated RGD-integrin receptors promptly recruited various integrin-binding proteins such as talin and paxillin and nucleated micrometer-sized clusters as nascent adhesion structures. However, RGD-integrin clusters on continuous RGD lipid bilayers were not interconnected by actin stress fibers and developed into podosomes after 45 min of initial adhesion (Figure 1C; Movie S2). The formation of podosome-like adhesions on RGD membranes was characterized by actin filament assembly in the podosome core at the center of individual RGD-integrin clusters (Figure 1D). Integrin-associated proteins, such as talin, paxillin, and vinculin, were consequently repartitioned into the ring structure (podosome ring) surrounding the actin core (Figures 1D and 1F).
Podosome-like Adhesions Have the Same Components as Classic Podosomes
To better identify the molecular organizations of podosome-like adhesion on RGD membranes, we rigorously examined more than 20 different molecular components (Table S1) that have been identified previously (Linder and Kopp, 2005). We found that podosome-like adhesions shared identical components in classic podosomes in macrophages. Therefore, we refer to podosome-like adhesions on RGD membranes as “podosomes” in the rest of this article. Podosome cores were enriched in F-actin and other characteristic molecular markers, such as Arp2/Arp3, WIP, and N-WASP, that were absent in classic focal adhesions (Figures 1F and S2A). The metalloproteinase, MMP-14 (MT1-MMP), was not enriched at podosomes of nontransformed fibroblasts or THP1 macrophages (Figure S1B). However, Src-transformed fibroblasts exhibited a high level of MMP-14 at invadopodia or long-lasting podosomes (Figure S1B), as previously reported (Wu et al., 2005). In addition, Tks5, a key adaptor protein in invadopodia formation, was not enriched at podosomes in nontransformed fibroblast on RGD membranes (Figure S1C).
Podosomes Are Dynamic Structures Formed by Arp2/Arp3-Mediated Actin Polymerization
Intense F-actin polymerization within RGD-integrin clusters as visualized by LifeAct was a signature of podosome formation at prepodosomal RGD clusters, and Arp2/3 was enriched at podosome cores (Figure 1F). The Arp2/Arp3 inhibitor CK-666 (Nolen et al., 2009) effectively abolished podosome formation (100 μM, 28 cells in two experiments; Figure S1). Formins, such as mDia1, DAAM1, and FHOD1, were not enriched at the podosomes (Figure S2A). SMIFH2 has been shown to inhibit mDia1- and mDia2-mediated actin polymerization in vitro and in vivo (Rizvi et al., 2009). However, SMIFH2 formin inhibitor did not suppress podosome formation in RPTPα+/+ mouse embryonic fibroblasts (Figure S2B). Thus, the actin cores of these podosomes were assembled by Arp2/Arp3-dependent actin polymerization and were not dependent on formin activity. Myosin-II, visualized by myosin regulatory light chain (MLC) was not enriched at podosomes. The disassembly of podosomes correlated with sparse recruitment of MLC around the dissociating actin core (Movie S3; Figure S4C) and a return to prepodosomal RGD-integrin clusters. The transition between podosome and adhesion clusters often repeated multiple times. Unlike long-lasting invadopodia (stable for hours), each podosome on RGD membranes exhibited a lifespan of 2–20 min.
Previously, interference reflection microscopy (IRM) was used to highlight close-contact zones at the cell-matrix interface (Holt et al., 2008), and the podosome core indeed showed close contact to the supporting substrate (Figure 1E). The tight contact revealed by destructive interference between two closely apposed interfaces indicated that the podosomes were protrusive (Evans et al., 2003; Monypenny et al., 2011). Intriguingly, the intensity of RGD fluorescence at the podosome core was diminished and immediately recovered after podosome disassembly (Figure S3B), which indicated that either the cell protrusive force was pushing out the RGD ligands and/or was creating a void in the bilayer. When bilayer continuity was tested with fluorescent lipid probes, such as Texas red-labeled 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (Texas red DHPE), doped into the supported bilayer, the fluorophores remained evenly distributed at the podosome core (Figure S3A), suggesting that the podosomes were indeed protrusive.
Along with Arp2/Arp3, cortactin, and WIP accumulation (Figure S2A), RGD depletion from the core was another unique phenomenon observed during the transformation from prepodosomal RGD clusters to podosomes. In more than 20 independent experiments with both macrophages and nontransformed fibroblasts, we rigorously examined and confirmed that F-actin polymerization and RGD depletion were always concurrent. While prepodosomal RGD clusters can be in an arbitrary ring shape, we always verified podosome formation by classic podosome markers and RGD depletion inside the RGD ring during live cell imaging (Table S1; Figure 3B). Thus, the podosomes in fibroblasts on RGD membranes exhibit the normal protrusive behavior, morphology, and dynamics of podosomes in macrophages.
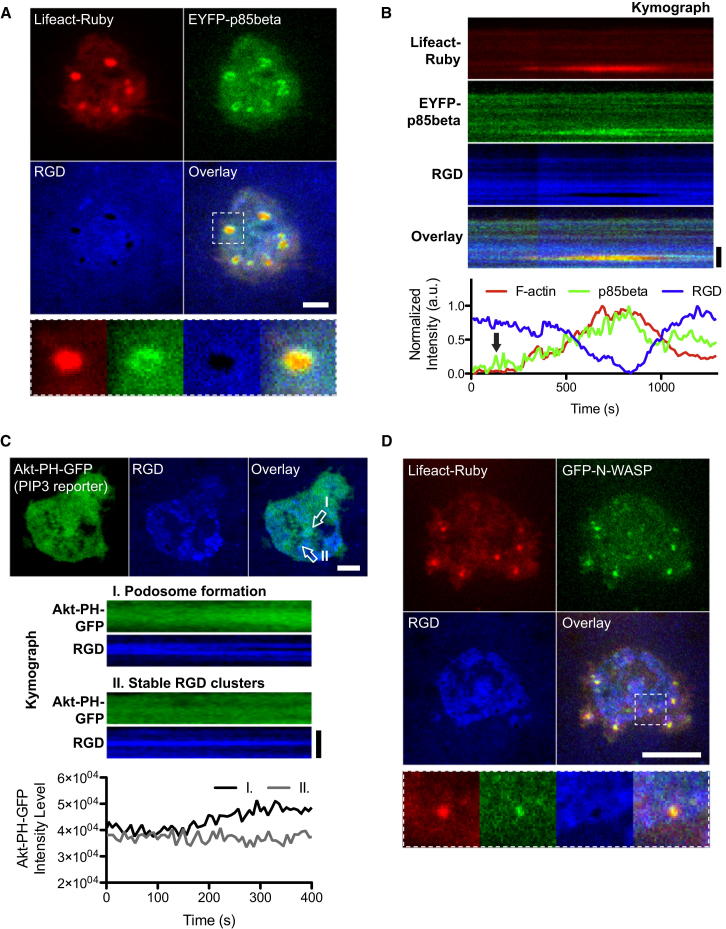
Figure 3.
Dynamical p85beta Recruitment and Increased PIP3 Level Turn RGD Clusters into Podosomes
(A) Class IA PI3K regulatory subunit p85beta was recruited at podosomes.
(B) Increased recruitment of p85beta at RGD clusters preceded F-actin polymerization. Spatial-temporal recruitment of F-actin and p85beta were analyzed by kymographs and intensity-time plot. p85beta was recruited at a subset of RGD clusters that subsequently turned into podosomes. As dot-like F-actin started to polymerize podosome core, p85beta reorganized from podosome core to podosome ring (Movie S5).
(C) Local enrichment of PIP3 during podosome formation. PIP3 levels were monitored by Akt-PH. The PIP3 level increased during the void formation within the RGD cluster (zone I) as a result of podosome formation on RGD membranes. The PIP3 level remains unaltered in stable RGD clusters (zone II).
(D) N-WASP was recruited at podosome cores. The scale bar represents 5 μm.
Force Generation in Nanopartitioned RGD Membranes Suppresses Podosome Formation
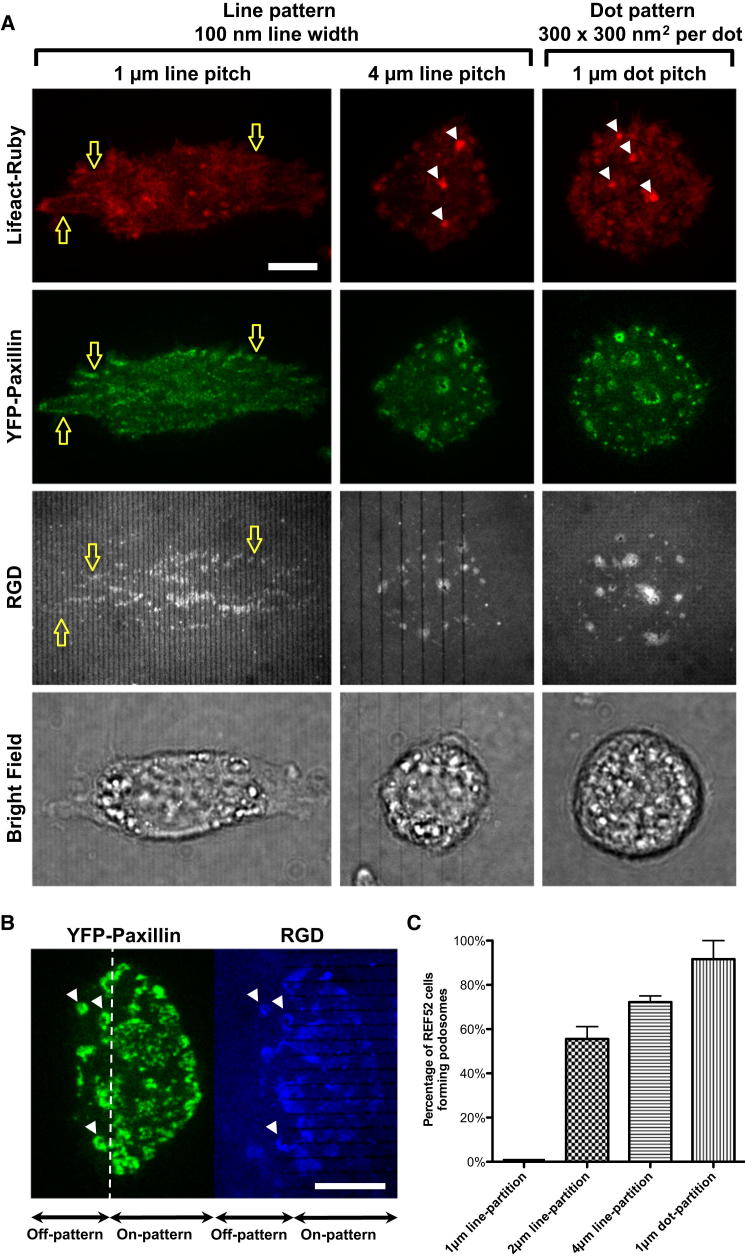
To test whether or not force on adhesions would affect podosome formation, we spread cells on nanopartitioned RGD bilayers where it was previously shown that cells would generate force on adhesions and stabilize them (Yu et al., 2011). The RGD bilayers were partitioned by nanofabricated metal lines (typically 100nm line width and 5nm in height) that provided passive resistance to adhesion movement. Previously, we demonstrated that the line pitch of partitioning barriers on an RGD membrane was inversely related to cell adhesion area. A smaller line pitch between barriers provided a higher density of barriers to RGD-integrin cluster movement and cells spread over larger areas. Cells formed focal adhesions with stress fibers, but they did not form podosomes when plated on nanopartitioned RGD membranes with a 1 μm line pitch (10% surface density). The immobilized RGD-integrin clusters were linear and were linked by actin fibers (hollow arrows in Figure 2A) after 60 min of initial adhesion. Furthermore, when cells were plated on nanopartitioned RGD membranes with a 4 μm line pitch that did not support force generation, podosome formation was restored. At the beginning, cells nucleated RGD clusters adjacent to nanopatterned lines with a 4 μm pitch, but there were no actin stress fibers between RGD clusters and cells had a smaller spread area. After 60 min of adhesion, these RGD clusters were also converted to podosomes (white arrowheads in Figure 2A). Interestingly, when RGD membranes were partitioned by dot arrays (300 × 300 nm2 metal areas with 1 μm pitch, also 9% of surface density) that provided no spatial confinement of RGD ligands, cells failed to develop force-stabilized RGD-integrin clusters and formed podosomes (Figure 2A). With increasing distance between the membrane partitions, fewer force-stabilized adhesion sites were nucleated, and podosome formation consequently increased (Figure 2C; total of 47 cells in three experiments). More surprisingly, when a single cell adhered to both a continuous and a partitioned RGD membrane, podosomes formed only on the continuous region and did not form at the partitioning lines (Figure 2B; Movie S4). Thus, we suggest that force generation by contraction to the lines produced a local signal that suppressed podosome formation within spatially restricted regions.
Figure 2.
RGD Membrane with Dense Partitioning Barriers Prevents Podosome Formation
(A) Nanopatterned lines (100 nm line width, with 1 to 4 μm line pitch) were prefabricated on glass substrate before RGD membrane deposition. The cell formed regular adhesion and actin stress fibers (white hollow arrows) on line-partitioned RGD membrane with 1 μm pitch. However, podosome formation (white arrowheads) remained when the cell adheres on RGD membrane with a 4-μm-pitch line partition, as well as 1-μm-pitch dot arrays (each dot area: 300 × 300 nm2).
(B) When a single cell adhered to both a continuous and a partitioned RGD membrane, podosomes (white arrowheads) formed only on the continuous region and did not form between the partitioning lines (Movie S4).
(C) Percentage of REF52 fibroblast cells forming podosomes when adhering on various patterned RGD membranes. Denser line partitions in RGD membranes result in less podosome formation. A total of 47 cells in three experiments were used.
Error estimates are SEM. The scale bar represents 10 μm.
Recruitment of p85beta Precedes Podosome Formation and Local Enrichment of PIP3
To determine what factors might be involved in stimulating podosome formation in the absence of force, we looked at Src kinase activity and PIP3 formation. After inhibition of Src by PP2 (10–20 μm, 2 hr), podosomes still formed, but at only 40% of the frequency of control cells (Figure S6A; total of 104 cells in three experiments). In the case of PIP3 formation, we found the localized recruitment of class 1A PI3K regulatory subunit p85beta at prepodosomal RGD clusters preceded actin assembly and appeared to trigger podosome formation (Figures 3A and 3B, arrow). During the initial phase of podosome formation, EYFP-p85beta was initially recruited to a subset of preexisting integrin-RGD clusters (Figure S4A) and then it expanded to the podosome rings (Figure 3B; Movie S5). In parallel, we monitored the time-dependence of PIP3 production by measuring the level of Akt-PH binding and we measured a marked increase in Akt-PH binding during the transition from RGD clusters to podosomes (Figure 3C). In addition, N-WASP was recruited at podosome cores (Figure 3D).
To determine if local enrichment of PIP3 triggered podosome formation, we inhibited PI3K activity by Wortmannin (100 nM). Fibroblasts can still develop initial RGD clusters after PI3K inhibition, but podosome formation on RGD membranes was blocked (Figures 4C and S6C; total of 108 cells in four experiments). While p85beta was also found at regular focal adhesions (Figure S5A), there were no significant changes in local PIP3 levels at the adhesions (Figure S5B), and N-WASP was not recruited at focal adhesions (Figure S5C). Thus, we suggest the recruitment of class IA PI3K caused the rise in PIP3 levels that led to F-actin assembly in the transformation of prepodosomal RGD-integrin clusters to podosomes on traction-force-free RGD membranes.
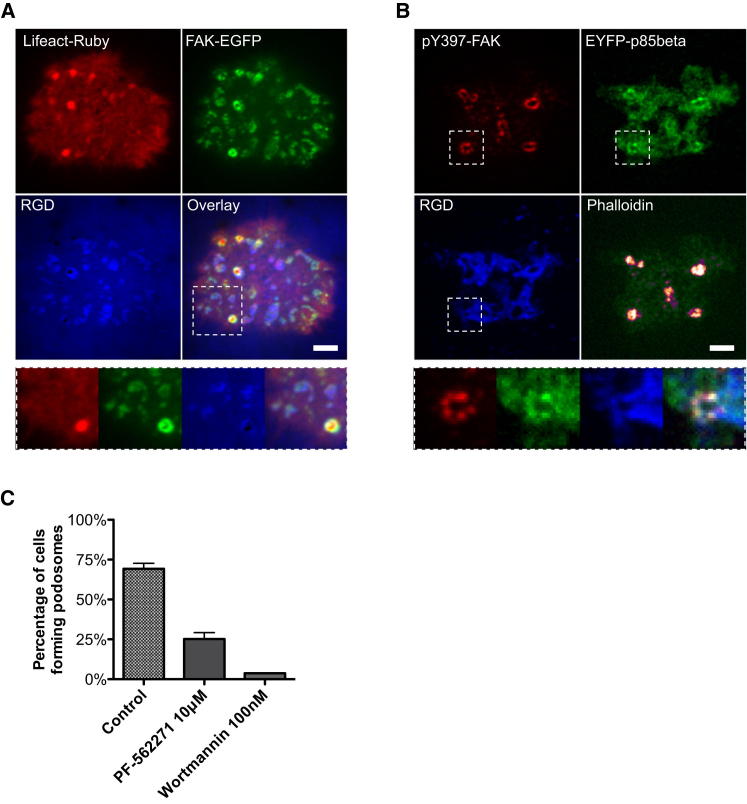
Figure 4.
Autophosphorylation of FAK Recruits p85beta at RGD Clusters
(A) FAK was recruited at RGD clusters and was more enriched at podosome rings.
(B) p85beta and pY397 FAK were both enriched at podosome rings.
(C) Podosome formation was suppressed by the FAK/Pyk2 inhibitor PF-562271, which blocked the autophosphorylation of FAK and Pyk2. The PI3K inhibitor Wortmannin also effectively abolished podosome formation. Error estimates are SEM. The scale bar represents 5 μm.
In terms of the degradation of PIP3 and podosome disassembly, PTEN was found at podosomes, but it only appeared after p85beta. It was often found above the focal plane of RGD-integrin clusters and decreased as F-actin disassembled (Figure S4B). In addition, the level of PIP3 decreased with the disassembly of the F-actin core. This suggests that a sustained level of PIP3 is necessary for maintenance of the F-actin core.
Tyrosine Autophosphorylation of Both FAK and PYK2 Regulates p85beta Recruitment
As p85beta is known to bind to substrates with phosphotyrosines via SH2 domains (Songyang et al., 1993), we looked for possible tyrosine kinases that may have been involved. Classical PI3K activation often involved autophosphorylation of receptor tyrosine kinase (RTK), as well as focal adhesion kinase (FAK) (Chen et al., 1996). After testing a number of RTK inhibitors that did not block podosome formation, we tested the dual FAK and Pyk2 kinase inhibitor PF-562271 (Roberts et al., 2008) and found that it efficiently suppressed podosome formation on RGD membranes. We found that FAK was recruited to RGD-integrin clusters (Figure 4A) and Y397 of FAK was autophosphorylated and colocalized with EYFP-p85beta at podosomes (Figure 4B). When FAK/Pyk2 autophosphorylation was inhibited by PF-562271 (10 μM, 4 hr pretreated), fibroblasts can still form initial RGD clusters, but p85beta recruitment was suppressed. Only 25% of fibroblasts were able to form podosomes on RGD membranes (Figures 4C and S6D; total of 139 cells in three experiments). Thus, we suggest autophosphorylation of FAK (Y397) and Pyk2 (Y407) recruited p85beta that triggered local enrichment of PIP3 at prepodosomal RGD clusters.
RhoA-GTP Levels Are Decreased upon Cell Adhesion to Mobile RGD Membranes
Since artificially upregulated RhoA-GTP levels and cellular contractility abolished podosome formation (Schramp et al., 2008; van Helden et al., 2008), we decided to determine if reduced RhoA-GTP was also correlated with podosome formation. Inhibiting Rho-associated protein kinase (ROCK) activity (Y-27632, 10 μM) or downregulating myosin-II-mediated contractility (blebbistatin, 50 μM) did not affect podosome formation on RGD membranes (Figure S6B; total of 60 and 59 cells examined in four experiments, respectively). In contrast, artificially upregulating cellular contractility by the RhoA agonist lysophosphatidic acid (LPA, 40 μM) or expressing a constitutively active RhoA-Q63L mutant effectively inhibited podosome formation (Figure S6B; total of 62 and 87 cells examined in four experiments, respectively). Furthermore, we utilized a fluorescence resonance energy transfer (FRET)-based RhoA biosensor (Pertz et al., 2006) to measure RhoA activity when cells adhered to different substrates (Figure 5B). REF52 cells plated on the mobile RGD membrane had a significantly lower FRET efficiency (0.45 ± 0.01 SEM, n = 30; Figure 5C) than on RGD-coated glass (0.73 ± 0.02 SEM, n = 25; Figure 5C; p value < 0.0001, two-sample t test, two-tailed). Thus, low levels of RhoA-GTP seem to be important for podosome formation.
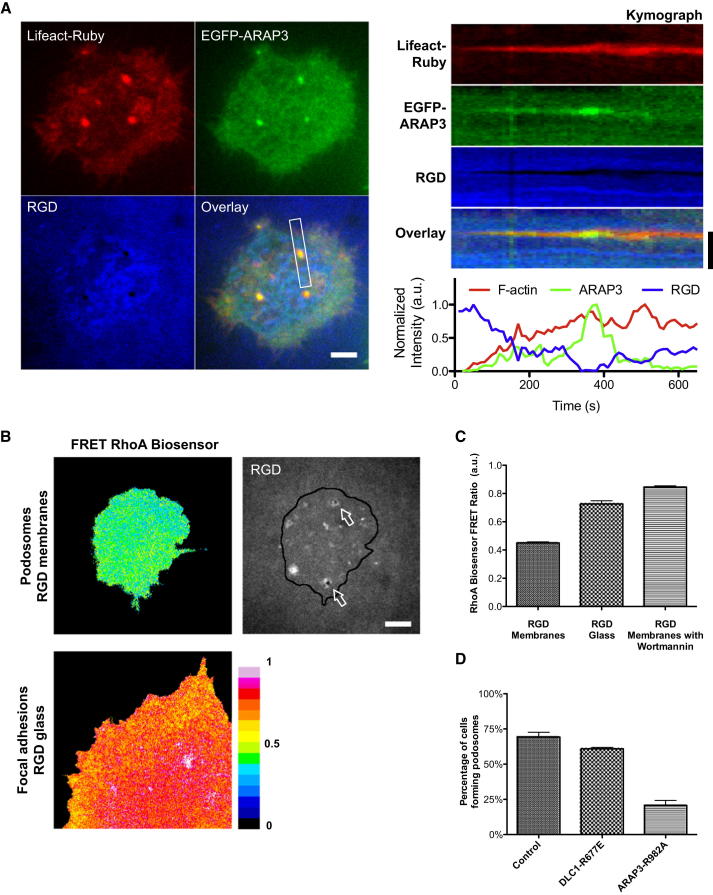
Figure 5.
Low RhoA-GTP Levels Accompany Podosome Formation
(A) ARAP3, a PIP3-bound RhoA GAP, was recruited at the podosome core. Recruitment of ARAP3 increased as the F-actin podosome core developed and then decreased before F-actin dissembled. Spatial-temporal recruitment of F-actin and ARAP3 were analyzed by kymographs and intensity time plot.
(B) Color-coded heatmap images of RhoA activity in vivo. RhoA activity measured by a FRET-based RhoA biosensor indicated lower RhoA-GTP levels when REF52 fibroblast cells adhered and formed podosomes on RGD membranes. Higher RhoA-GTP levels were measured as cells adhered and form focal adhesion on RGD-coated glass.
(C) Comparison of RhoA activity via FRET efficiency between cells adhered on RGD membranes (0.45 ± 0.01 SEM, n = 30) and RGD glass (0.73 ± 0.02 SEM, n = 25); p value < 0.0001 (two-sample t test, two tailed). When PI3K was inhibited by Wortmannin, the FRET efficiency of the RhoA biosensor became high (0.85 ± 0.01 SEM, n = 29), even when cells adhered to the mobile RGD membrane.
(D) Differential effects of overexpressing two catalytic-dead RhoA GAP mutants, DLC1-R677E and ARAP3-R982A. Podosome formation was suppressed to 25% in the case of ARAP3-R982A but unaltered in the case of DLC1-R677E. Error estimates are SEM. The scale bar represents 5 μm.
To further investigate RhoA regulation, we tested two RhoA GTPase-activating proteins (GAPs), DLC1 and ARAP3, which localized to podosomes. While DLC1 was recruited at both focal adhesion and podosome rings through tensin (Schramp et al., 2008), ARAP3 has been shown to bind to PIP3 at plasma membranes (Krugmann et al., 2002, 2004). Indeed, we found that ARAP3 was recruited at PIP3-enriched podosome cores after F-actin core formation (Figure 5A; Movie S6). However, ARAP3 was not recruited to focal adhesions (Figure S5D). We tested whether DLC1 or ARAP3 played important roles in podosome formation by transiently overexpressing catalytic-dead RhoA GAP mutants DLC1-R677E and ARAP3-R982A, respectively. Podosome formation was suppressed to 25% in the case of ARAP3-R982A (Figure 5D), but not significantly disrupted in the case of DLC1-R677E (total of 149 and 114 cells in three experiments, respectively). When PI3K was inhibited, the FRET efficiency of the RhoA biosensor increased significantly (0.85 ± 0.01 SEM, n = 29), even when cells adhered to the mobile RGD membrane (Figure 5C), and podosome formation was suppressed. These observations indicate that the RhoA-GTP level is inversely correlated with podosome formation and that recruitment of ARAP3 and possibly other RhoA-GAPs by local enrichment of PIP3 provided a positive feedback to downregulate cellular RhoA level in podosome-forming cells.
Discussion
In these studies, we have demonstrated for that plating cells onto RGD ligands linked to fluid lipid bilayers caused the formation of integrin-based podosome-like adhesions. Surprisingly, such a response was characteristic not only of cells that produce podosomes under normal culture conditions, such as the macrophage line used here, but also of fibroblasts, which do not produce podosomes when plated onto “normal” rigid substrates. Although large traction forces mediated by RhoA are important for focal adhesion maturation in fibroblasts, there is no evidence that they are major determinants in podosome formation. Notably, macrophages do not develop large traction forces on matrix-coated substrates (Féréol et al., 2009) or mature focal adhesions, but they do form podosomes. The implications are that the components needed to form podosomes are present in fibroblasts and immune cells, but the combination of cell contractility and matrix mechanics plays the critical role in determining which type of adhesion is formed.
The podosome-like adhesions that form in the absence of force in nontransformed fibroblasts are indistinguishable from podosomes in macrophages in terms of morphology, components (Table S1), and protrusive dynamics (Figure 1E). However, their physiological functions, such as chemotaxis and endocytosis/exocytosis, need to be further examined. Spatial depletion of mobile RGD ligands and destructive interference at podosomes by IRM indicated that there was active protrusion of the podosome core (Figures 1E and S3B). While ligands on supported membranes were freely diffusive, the observed spatial exclusion of RGD-neutravidin (5.4 ± 5.8 nm footprint in the x-y dimension; Hendrickson et al., 1989) at the podosome core could not be explained by a simple repartitioning effect from ligation with integrin receptor (8 ± 12 nm footprint in the x-y dimension; Nermut et al., 1988). Nevertheless, supported lipid membrane remains evenly distributed at the podosome core (Figure S3A). The vertical force required to physically penetrate a lipid membrane via biomimetic stealth probes (200 nm in diameter) has been reported as 58 nN (Almquist and Melosh, 2010). We conclude that the protrusive force at podosomes was less than needed to penetrate the membrane but sufficient to block diffusion of RGD-neutravidin into the contact region.
Invadopodia or long-lived stable podosomes (more than 30 min) have been widely investigated in Src-transformed cells (induction of constitutively activated Src kinase) and invasive cancer cell lines (Huveneers et al., 2008; Oikawa et al., 2008). However, podosome-like adhesions on RGD membranes and invadopodia in Src-transformed cells are different in both dynamics and molecular components (Table S1). Constitutively activated Src causes hyperactivation of various downstream targets, such as ARHGEF5 RhoA-GEF (Kuroiwa et al., 2011), phosphorylation of Tks5/Grb2 complexes (Oikawa et al., 2008), and MMP-14 secretion (Poincloux et al., 2009; Wu et al., 2005; Yu et al., 2012b). Notably, Tks5 (Figure S1C) is not enriched at podosomes in nontransformed fibroblast on RGD membranes. In addition, we have examined the potential recruitment of MMP-14 and found that it was present at invadopodia in invasive cancer cells and Src-transformed fibroblasts. However, in nontransformed fibroblasts, most of the MMP-14 remained in endocytic vesicles and there was only a weak recruitment of MMP-14 around the podosomes (Figure S1B). This is all consistent with the hypothesis that podosomes formed in the absence of force are aided by but do not require Src activity.
Likewise, diaphanous-related formins are required for invadopodia formation in invasive MDA-MB-231 breast adenocarcinoma cells (Lizárraga et al., 2009). However, when we treated nontransformed fibroblasts and THP1-differentated macrophages with the formin inhibitor SMIFH2, we still observed podosome formation at a similar density to control cells. Podosomes and invadopodia share many molecular components, but most likely not all. We suggest that podosomes on RGD membranes indeed differ from long-lived invadopodia in Src-transformed cells or invasive cancer cells in their lifespan, formin involvement, Tks5 recruitment, and MMP-14 secretion.
Our data suggest that conventional traction force development and myosin-II activities are dispensable in podosome formation. Mobile RGD membranes with nanopartitioning lines provide a unique platform to examine the force-regulated adhesion structure transformation (Figure 2A). As cells adhere to RGD membranes, initial integrin activation results in RGD clustering without traction force. With dense line-partitioned RGD membranes, such as with 1 or 2 μm line pitch, forces can be generated on RGD-liganded integrins to form traction-force loaded adhesions (Figure 2A) after activation of initial spreading. Because the local contraction units are unable to span the 4 μm spacing (Ghassemi et al., 2012; Yu et al., 2011), the podosomes form as well on the larger line spacing as on continuous bilayers. In addition, myosin-II is not recruited during podosome formation on RGD membrane (Figure S4C). Thus, our data indicate that podosome formation requires minimum traction force development and that the local force generation between RGD-integrin adhesion clusters at dense line-partitioned RGD membranes inhibits podosome formation. When the same cell covers both the 2 μm pitch lines and a continuous membrane (Figure 2B), there is a remarkably local formation of focal adhesions at the lines while podosomes form over continuous membrane regions, indicating that the effect of contractility is local and may involve spatial contact signals.
Although we do not fully know how local contractions are translated into inhibition of podosome formation, we find that peak production of PIP3 is the key upstream event to trigger the transformation from prepodosomal integrin-RGD clusters to podosomes (Figure 6). Class IA PI3K regulatory subunit p85beta is first recruited at activated integrins through binding to autophosphorylated FAK and possibly Pyk2, and local production of PIP3 is observed by an increased level of Akt-PH-GFP. Inhibition of FAK and Pyk2 by PF-562271 (Roberts et al., 2008) effectively block podosome formation. Indeed, autophosphorylation site Y397 of FAK can bind p85beta (Chen et al., 1996), and Pyk2 has the same sequence (Y-A-E-I) at its tyrosine autophosphorylation site, Y402. In the case of fibroblasts, the activation of Pyk2 appears to be best correlated with podosome formation on RGD lipids, since FAK−/− cells and cells treated with a FAK-specific inhibitor form podosomes normally (C.-H.Y. and M.P.S., unpublished data). Pyk2 also plays an important role in podosome formation in osteoclasts (Gil-Henn et al., 2007), but we cannot rule out another kinase because there could be off-target inhibition of other kinases by PF-562271. In addition, FAK can phosphorylate N-WASP and promote actin polymerization, and inhibition of FAK kinase activity suppresses N-WASP activity (Tang et al., 2013). However, N-WASP can still be phosphorylated by other kinases, such as Src family kinases (Dovas and Cox, 2010) or Abl kinase (Burton et al., 2005). While N-WASP could be activated by other kinases, N-WASP may fail to be recruited at RGD clusters without local enrichment of PIP3. We suggest that inhibition of FAK and Pyk2 autophosphorylation provides a mechanism to abolish local production of PIP3 by perturbing p85/PI3K association. As expected, inhibition of PI3K also suppresses podosome formation (Figure 4C).
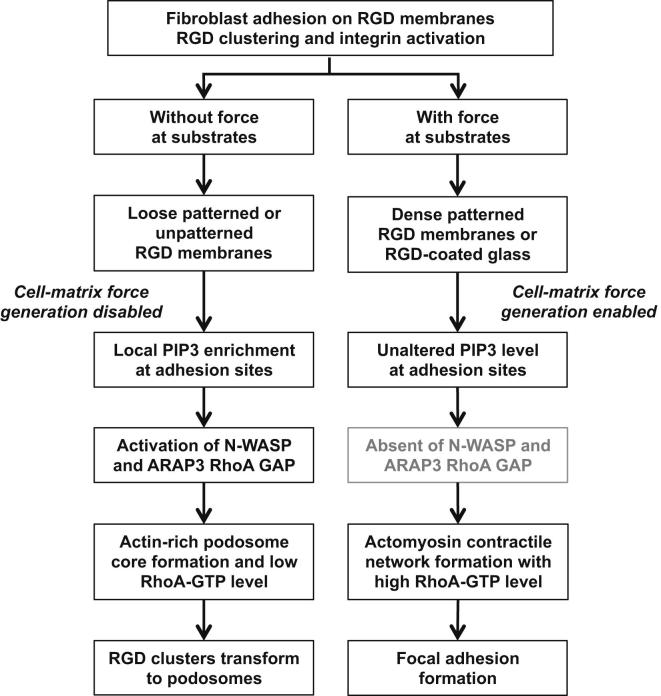
Figure 6.
Summary of Force-Mediated Adhesion Transformation Pathway
Early RGD-integrin activation triggered actomyosin contraction. When substratum provides traction force (1 μm line-pitch RGD membranes), cells form classic focal adhesions. When substratum provides no traction force, RGD-integrin clusters can turn into podosomes after 45 min of initial adhesion. Local enrichment of PIP3 by PI3K activation at the prepodosomal RGD cluster triggers N-WASP and Arp2/Arp3-mediated actin polymerization that initiated podosome formation. Recruitment of ARAP3 via local enrichment of PIP3 serves as a positive-feedback mechanism to downregulate RhoA-GTP in podosome-forming cells.
Another protein that binds to PIP3 is PTEN, and it dephosphorylates PIP3, thereby causing the loss of actin polymerizing proteins. It has previously been shown that PTEN is present in and regulates podosome/invadopodia formation (Hoshino et al., 2012; Poon et al., 2010). PTEN associates with podosomes after the actin core is formed and contributes to the disassembly of the podosome F-actin core. Thus, it seems that the cycle of podosome formation and disassembly is primarily dependent upon the local levels of PIP3 on plasma membranes. This can explain the regional differences in podosome formation we see in single cells (Figure 2B; Movie S4) through slower two-dimensional diffusion of PIP3 lipids rather than fast diffusive cytosolic signals.
A factor that could contribute indirectly to the formation of podosomes is RhoA activity. Using the FRET-based RhoA biosensor, lower RhoA-GTP levels are observed when cells develop podosomes, and pharmaceutically activating RhoA-mediated contractility using LPA is seen to abolish podosome formation on RGD membranes, in agreement with previous reports (Schramp et al., 2008; van Helden et al., 2008). More than 70 Rho GAPs have been identified in eukaryotes (Tcherkezian and Lamarche-Vane, 2007), and it remains unclear how RhoA activities are differentially regulated during adhesion formation. While DLC1 is linked to downregulation of RhoA in Src-transformed cells (Schramp et al., 2008), we find that the PIP3-binding protein ARAP3 is another RhoA-regulating factor recruited at podosome cores. ARAP3 contains both RhoA GAP and Arf6 GAP domains, and the RhoA GAP function of ARAP3 is activated by Rap-GTP (Krugmann et al., 2002, 2004). ARAP3’s Arf6 GAP function in vivo, however, is still under investigation (Gambardella et al., 2011). Inhibition of PI3K upregulates RhoA-GTP and cellular contractility (Krugmann et al., 2004; Orlova et al., 2007), and our RhoA biosensor measurements also agree with previous findings (Figure 5C). Using overexpressed catalytic-dead RhoA GAP mutants, we find ARAP3-R982A moderately suppresses podosome formation, while DLC1-R677E has no significant effect. However, ARAP3 is recruited largely after podosomes are formed. Recruitment of ARAP3 provides a positive-feedback mechanism to downregulate RhoA-GTP. Thus, our results indicate that manipulations of traction force development at integrin-matrix clusters can serve as a mechanical signal to modulate adhesion phenotype switching and RhoA activities.
In conclusion, we suggest that the development of podosomes as adhesion structures implicates the absence of traction forces between integrin receptors and matrix ligands. Lack of traction forces at activated RGD-integrin clusters results in spatial-temporal recruitment of p85beta and local enrichment of PIP3, which is not observed in force-loaded focal adhesions. This PIP3-dependent pathway of podosome formation does not require the induction of constitutively activated Src kinase and is further aided by the inactivation of RhoA by PIP3-mediated recruitment of ARAP3. We suggest that local contractions may directly inhibit podosome formation while facilitating focal adhesion formation through a block of the PIP3-dependent pathway. The transformation between prepodosomal RGD-integrin clusters and podosomes is a remarkable example of mechanosensing through cell-adhesion processes. The reorganization of adhesion structures triggered by changing microenvironments has become an emerging theme of adaptive regulation in cellular signaling. Force and matrix ligand and integrin composition are all critical factors regulating adhesion phenotype and turnover.
Experimental Procedures
Cell Culture and Fluorescent Fusion Proteins
Nontransformed RPTPα+/+ mouse embryonic fibroblasts (Su et al., 1999), rat embryonic fibroblast (REF52), and THP-1 (human monocytic leukemia cells) were used in this study. Detailed information regarding cell culture, transfection protocol, the plasmids of fluorescent fusion proteins, and microscopy methods can be found in the Supplemental Experimental Procedures.
RGD-Supported Lipid Bilayer Membranes
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (16:0 biotinyl-Cap-PE) were purchased from Avanti Polar Lipids. The lipids (0.2 mol% of biotinyl-Cap-PE and 99.8 mol% of DOPC) were mixed with an equal volume of 1× PBS and then pipetted onto cleaned glass substrates for the self-assembly processes. A total of 0.1 μg/ml of Cascade blue neutravidin (Life Technologies) or DyLight 680 neutravidin (Thermo Fisher Scientific) was added onto supported lipid membranes, followed by 1 μg/ml of biotinylated RGD, cyclo (Arg-Gly-Asp-D-Phe-Lys[Biotin-PEG-PEG]; Peptides International). Detailed information regarding lipid preparation and membrane functionalization can be found in the Supplemental Experimental Procedures.
Nanopatterned Glass Substrate
Nanoimprint lithography was utilized to fabricate the physical barriers on glass substrates, and detailed preparation methods were previously described (Yu et al., 2011). In brief, a silicon-based imprint mold was fabricated by electron-beam lithography and anisotropic etching processes. First, Coverglasses (Warner Instruments) were cleaned by Piranha solution (sulfuric acid and hydrogen peroxide, mixed in 3:1 ratio) for 15 min, rinsed with deionized water, and then spin-coated with UV-curable imprint polymers. Patterns were then transferred from the mold to the glass by high-pressure stamping the imprint mold onto the polymer-coated coverglass and curing the polymer by UV exposure. After demolding, oxygen plasma etching was used to extend imprinted trenches vertically to the surface of the coverglass. A thin chromium metal layer was deposited onto the exposed glass surface by thermal evaporation. The chromium on imprinted polymers was removed by resist lift-off processing. Typically, metal lines were 100 nm in width and 5 nm in height with a gap distance ranging from 1 to 4 μm. The density of the metal lines remains constant and is about 10% per μm2.
Author Contributions
C.-H.Y. and N.B.M.R. conducted and analyzed most of the experiments. A.K. and K.L.H. assisted in sample preparations. C.-H.Y., G.E.J., A.D.B., and M.P.S. supervised this study and prepared the manuscript.
Acknowledgments
C.-H.Y. acknowledges support from National Science Council of Taiwan (grant NSC98-2917-I-564-165). N.B.M.R. is funded by a joint National University of Singapore and King’s College studentship. G.E.J. is supported by the Medical Research Council, UK (G1100041) and a generous provision of a visiting professorship to the Mechanobiology Institute, Singapore.
Published: November 27, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental information includes Supplemental Experimental Procedures, six figures, one table, and six movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.10.040.
Contributor Information
Cheng-han Yu, Email: chyu@nus.edu.sg.
Michael P. Sheetz, Email: ms2001@columbia.edu.
Supplemental Information
Actin at podosome core was visualized by mCherry-UtrCH. Both GFP-vinculin and RGD were recruited at podosome rings. The scale bar represents 10 μm.
Podosomes (red arrows) were identified by ring formations of both RGD and YFP-paxillin in the REF52 fibroblast. The scale bar represents 10 μm.
Actin at the podosome core was visualized by LifeAct-ruby in RPTPα+/+ mouse fibroblast cells. MLC-GFP was recruited around the dissociating actin core during the disassembly of podosomes. The scale bar represents 10 μm.
When a single cell adhered to both a continuous and a partitioned RGD membrane, podosomes formed only on the continuous region and did not form between the partitioning lines. RGD membrane with dense nanopartitions impeded podosome formation. Podosomes (red arrows) were identified by ring formations of both RGD and YFP-paxillin in the REF52 fibroblast. The scale bar includes 10 μm.
Class IA PI3K regulatory subunit p85beta was first recruited at prepodosomal RGD clusters, which was followed by F-actin polymerization in the podosome core. p85beta was subsequently reorganized into podosome rings. The scale bar represents 10 μm.
PIP3-bound RhoA-GAP ARAP3 was recruited at podosome cores during F-actin polymerization. The scale bar represents 10 μm.
References
- Almquist B.D., Melosh N.A. Fusion of biomimetic stealth probes into lipid bilayer cores. Proc. Natl. Acad. Sci. USA. 2010;107:5815–5820. doi: 10.1073/pnas.0909250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton E.A., Oliver T.N., Pendergast A.M. Abl kinases regulate actin comet tail elongation via an N-WASP-dependent pathway. Mol. Cell. Biol. 2005;25:8834–8843. doi: 10.1128/MCB.25.20.8834-8843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Sheetz M.P. Force propagation across cells: mechanical coherence of dynamic cytoskeletons. Curr. Opin. Cell Biol. 2009;21:47–50. doi: 10.1016/j.ceb.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle Y., Burns S., Thrasher A.J., Jones G.E. The leukocyte podosome. Eur. J. Cell Biol. 2006;85:151–157. doi: 10.1016/j.ejcb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Chen H.C., Appeddu P.A., Isoda H., Guan J.L. Phosphorylation of tyrosine 397 in focal adhesion kinase is required for binding phosphatidylinositol 3-kinase. J. Biol. Chem. 1996;271:26329–26334. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- Cox S., Rosten E., Monypenny J., Jovanovic-Talisman T., Burnette D.T., Lippincott-Schwartz J., Jones G.E., Heintzmann R. Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat. Methods. 2012;9:195–200. doi: 10.1038/nmeth.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovas A., Cox D. Regulation of WASp by phosphorylation: Activation or other functions? Commun. Integr. Biol. 2010;3:101–105. doi: 10.4161/cib.3.2.10759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Evans E., Yeung A. Hidden dynamics in rapid changes of bilayer shape. Chem. Phys. Lipids. 1994;73:39–56. [Google Scholar]
- Evans E.A., Hochmuth R.M. Mechanochemical properties of membranes. In: Felix B., Arnost K., editors. Current Topics in Membranes and Transport. Academic Press; New York: 1978. pp. 1–64. [Google Scholar]
- Evans J.G., Correia I., Krasavina O., Watson N., Matsudaira P. Macrophage podosomes assemble at the leading lamella by growth and fragmentation. J. Cell Biol. 2003;161:697–705. doi: 10.1083/jcb.200212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Féréol S., Fodil R., Laurent V.M., Balland M., Louis B., Pelle G., Hénon S., Planus E., Isabey D. Prestress and adhesion site dynamics control cell sensitivity to extracellular stiffness. Biophys. J. 2009;96:2009–2022. doi: 10.1016/j.bpj.2008.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambardella L., Anderson K.E., Nussbaum C., Segonds-Pichon A., Margarido T., Norton L., Ludwig T., Sperandio M., Hawkins P.T., Stephens L., Vermeren S. The GTPase-activating protein ARAP3 regulates chemotaxis and adhesion-dependent processes in neutrophils. Blood. 2011;118:1087–1098. doi: 10.1182/blood-2010-10-312959. [DOI] [PubMed] [Google Scholar]
- Geiger B., Bershadsky A., Pankov R., Yamada K.M. Transmembrane crosstalk between the extracellular matrix—cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- Geiger B., Spatz J.P., Bershadsky A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- Ghassemi S., Meacci G., Liu S., Gondarenko A.A., Mathur A., Roca-Cusachs P., Sheetz M.P., Hone J. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc. Natl. Acad. Sci. USA. 2012;109:5328–5333. doi: 10.1073/pnas.1119886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Henn H., Destaing O., Sims N.A., Aoki K., Alles N., Neff L., Sanjay A., Bruzzaniti A., De Camilli P., Baron R., Schlessinger J. Defective microtubule-dependent podosome organization in osteoclasts leads to increased bone density in Pyk2(-/-) mice. J. Cell Biol. 2007;178:1053–1064. doi: 10.1083/jcb.200701148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M., Buccione R., Courtneidge S.A., Linder S. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 2008;20:235–241. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Hendrickson W.A., Pähler A., Smith J.L., Satow Y., Merritt E.A., Phizackerley R.P. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc. Natl. Acad. Sci. USA. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt M.R., Calle Y., Sutton D.H., Critchley D.R., Jones G.E., Dunn G.A. Quantifying cell-matrix adhesion dynamics in living cells using interference reflection microscopy. J. Microsc. 2008;232:73–81. doi: 10.1111/j.1365-2818.2008.02069.x. [DOI] [PubMed] [Google Scholar]
- Hoshino D., Jourquin J., Emmons S.W., Miller T., Goldgof M., Costello K., Tyson D.R., Brown B., Lu Y., Prasad N.K. Network analysis of the focal adhesion to invadopodia transition identifies a PI3K-PKCα invasive signaling axis. Sci. Signal. 2012;5:ra66. doi: 10.1126/scisignal.2002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher A., Horwitz A.R. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S., Arslan S., van de Water B., Sonnenberg A., Danen E.H. Integrins uncouple Src-induced morphological and oncogenic transformation. J. Biol. Chem. 2008;283:13243–13251. doi: 10.1074/jbc.M800927200. [DOI] [PubMed] [Google Scholar]
- Krugmann S., Anderson K.E., Ridley S.H., Risso N., McGregor A., Coadwell J., Davidson K., Eguinoa A., Ellson C.D., Lipp P. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol. Cell. 2002;9:95–108. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- Krugmann S., Williams R., Stephens L., Hawkins P.T. ARAP3 is a PI3K- and rap-regulated GAP for RhoA. Curr. Biol. 2004;14:1380–1384. doi: 10.1016/j.cub.2004.07.058. [DOI] [PubMed] [Google Scholar]
- Kuroiwa M., Oneyama C., Nada S., Okada M. The guanine nucleotide exchange factor Arhgef5 plays crucial roles in Src-induced podosome formation. J. Cell Sci. 2011;124:1726–1738. doi: 10.1242/jcs.080291. [DOI] [PubMed] [Google Scholar]
- Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F., Csiszar K., Giaccia A., Weninger W. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S., Kopp P. Podosomes at a glance. J. Cell Sci. 2005;118:2079–2082. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- Lizárraga F., Poincloux R., Romao M., Montagnac G., Le Dez G., Bonne I., Rigaill G., Raposo G., Chavrier P. Diaphanous-related formins are required for invadopodia formation and invasion of breast tumor cells. Cancer Res. 2009;69:2792–2800. doi: 10.1158/0008-5472.CAN-08-3709. [DOI] [PubMed] [Google Scholar]
- Machesky L., Jurdic P., Hinz B. Grab, stick, pull and digest: the functional diversity of actin-associated matrix-adhesion structures. Workshop on Invadopodia, Podosomes and Focal Adhesions in Tissue Invasion. EMBO Rep. 2008;9:139–143. doi: 10.1038/sj.embor.7401162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S., Wang J., Gambhir A., Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- Miranti C.K., Brugge J.S. Sensing the environment: a historical perspective on integrin signal transduction. Nat. Cell Biol. 2002;4:E83–E90. doi: 10.1038/ncb0402-e83. [DOI] [PubMed] [Google Scholar]
- Monypenny J., Chou H.C., Bañón-Rodríguez I., Thrasher A.J., Antón I.M., Jones G.E., Calle Y. Role of WASP in cell polarity and podosome dynamics of myeloid cells. Eur. J. Cell Biol. 2011;90:198–204. doi: 10.1016/j.ejcb.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S.W., Roca-Cusachs P., Sheetz M.P. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev. Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman K.D., Campi G., Groves J.T., Dustin M.L. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- Murphy D.A., Courtneidge S.A. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nermut M.V., Green N.M., Eason P., Yamada S.S., Yamada K.M. Electron microscopy and structural model of human fibronectin receptor. EMBO J. 1988;7:4093–4099. doi: 10.1002/j.1460-2075.1988.tb03303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., Pollard T.D. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Itoh T., Takenawa T. Sequential signals toward podosome formation in NIH-src cells. J. Cell Biol. 2008;182:157–169. doi: 10.1083/jcb.200801042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova I., Silver L., Gallo G. Regulation of actomyosin contractility by PI3K in sensory axons. Dev. Neurobiol. 2007;67:1843–1851. doi: 10.1002/dneu.20558. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V., Co C., Prehoda K.E., Snapper S., Taunton J., Lim W.A. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol. Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R.L., Hahn K.M. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- Poincloux R., Lizárraga F., Chavrier P. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 2009;122:3015–3024. doi: 10.1242/jcs.034561. [DOI] [PubMed] [Google Scholar]
- Pollitt A.Y., Insall R.H. WASP and SCAR/WAVE proteins: the drivers of actin assembly. J. Cell Sci. 2009;122:2575–2578. doi: 10.1242/jcs.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon J.S., Eves R., Mak A.S. Both lipid- and protein-phosphatase activities of PTEN contribute to the p53-PTEN anti-invasion pathway. Cell Cycle. 2010;9:4450–4454. doi: 10.4161/cc.9.22.13936. [DOI] [PubMed] [Google Scholar]
- Prager-Khoutorsky M., Lichtenstein A., Krishnan R., Rajendran K., Mayo A., Kam Z., Geiger B., Bershadsky A.D. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., Kovar D.R. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts W.G., Ung E., Whalen P., Cooper B., Hulford C., Autry C., Richter D., Emerson E., Lin J., Kath J. Antitumor activity and pharmacology of a selective focal adhesion kinase inhibitor, PF-562,271. Cancer Res. 2008;68:1935–1944. doi: 10.1158/0008-5472.CAN-07-5155. [DOI] [PubMed] [Google Scholar]
- Salaita K., Nair P.M., Petit R.S., Neve R.M., Das D., Gray J.W., Groves J.T. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science. 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramp M., Ying O., Kim T.Y., Martin G.S. ERK5 promotes Src-induced podosome formation by limiting Rho activation. J. Cell Biol. 2008;181:1195–1210. doi: 10.1083/jcb.200801078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z., Shoelson S.E., Chaudhuri M., Gish G., Pawson T., Haser W.G., King F., Roberts T., Ratnofsky S., Lechleider R.J. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- Su J., Muranjan M., Sap J. Receptor protein tyrosine phosphatase alpha activates Src-family kinases and controls integrin-mediated responses in fibroblasts. Curr. Biol. 1999;9:505–511. doi: 10.1016/s0960-9822(99)80234-6. [DOI] [PubMed] [Google Scholar]
- Tang H., Li A., Bi J., Veltman D.M., Zech T., Spence H.J., Yu X., Timpson P., Insall R.H., Frame M.C., Machesky L.M. Loss of Scar/WAVE complex promotes N-WASP- and FAK-dependent invasion. Curr. Biol. 2013;23:107–117. doi: 10.1016/j.cub.2012.11.059. [DOI] [PubMed] [Google Scholar]
- Tarone G., Cirillo D., Giancotti F.G., Comoglio P.M., Marchisio P.C. Rous sarcoma virus-transformed fibroblasts adhere primarily at discrete protrusions of the ventral membrane called podosomes. Exp. Cell Res. 1985;159:141–157. doi: 10.1016/s0014-4827(85)80044-6. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J., Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol. Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- van Helden S.F., Oud M.M., Joosten B., Peterse N., Figdor C.G., van Leeuwen F.N. PGE2-mediated podosome loss in dendritic cells is dependent on actomyosin contraction downstream of the RhoA-Rho-kinase axis. J. Cell Sci. 2008;121:1096–1106. doi: 10.1242/jcs.020289. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Stephens L., Hawkins P. PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- Vogel V., Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Vogel V., Sheetz M.P. Cell fate regulation by coupling mechanical cycles to biochemical signaling pathways. Curr. Opin. Cell Biol. 2009;21:38–46. doi: 10.1016/j.ceb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrle-Haller B. Structure and function of focal adhesions. Curr. Opin. Cell Biol. 2012;24:116–124. doi: 10.1016/j.ceb.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Wu X., Gan B., Yoo Y., Guan J.L. FAK-mediated src phosphorylation of endophilin A2 inhibits endocytosis of MT1-MMP and promotes ECM degradation. Dev. Cell. 2005;9:185–196. doi: 10.1016/j.devcel.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Xu C., Watras J., Loew L.M. Kinetic analysis of receptor-activated phosphoinositide turnover. J. Cell Biol. 2003;161:779–791. doi: 10.1083/jcb.200301070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.H., Groves J.T. Engineering supported membranes for cell biology. Med. Biol. Eng. Comput. 2010;48:955–963. doi: 10.1007/s11517-010-0634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.H., Wu H.J., Kaizuka Y., Vale R.D., Groves J.T. Altered actin centripetal retrograde flow in physically restricted immunological synapses. PLoS ONE. 2010;5:e11878. doi: 10.1371/journal.pone.0011878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.H., Law J.B., Suryana M., Low H.Y., Sheetz M.P. Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation. Proc. Natl. Acad. Sci. USA. 2011;108:20585–20590. doi: 10.1073/pnas.1109485108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.H., Luo W., Sheetz M.P. Spatial-temporal reorganization of activated integrins. Cell Adhes. Migr. 2012;6:280–284. doi: 10.4161/cam.20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zech T., McDonald L., Gonzalez E.G., Li A., Macpherson I., Schwarz J.P., Spence H., Futó K., Timpson P. N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J. Cell Biol. 2012;199:527–544. doi: 10.1083/jcb.201203025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Actin at podosome core was visualized by mCherry-UtrCH. Both GFP-vinculin and RGD were recruited at podosome rings. The scale bar represents 10 μm.
Podosomes (red arrows) were identified by ring formations of both RGD and YFP-paxillin in the REF52 fibroblast. The scale bar represents 10 μm.
Actin at the podosome core was visualized by LifeAct-ruby in RPTPα+/+ mouse fibroblast cells. MLC-GFP was recruited around the dissociating actin core during the disassembly of podosomes. The scale bar represents 10 μm.
When a single cell adhered to both a continuous and a partitioned RGD membrane, podosomes formed only on the continuous region and did not form between the partitioning lines. RGD membrane with dense nanopartitions impeded podosome formation. Podosomes (red arrows) were identified by ring formations of both RGD and YFP-paxillin in the REF52 fibroblast. The scale bar includes 10 μm.
Class IA PI3K regulatory subunit p85beta was first recruited at prepodosomal RGD clusters, which was followed by F-actin polymerization in the podosome core. p85beta was subsequently reorganized into podosome rings. The scale bar represents 10 μm.
PIP3-bound RhoA-GAP ARAP3 was recruited at podosome cores during F-actin polymerization. The scale bar represents 10 μm.