Abstract
Background
Sepsis is one of the main causes of mortality and morbidity. The rapid detection of pathogens in blood of septic patients is essential for adequate antimicrobial therapy and better prognosis. This study aimed to accelerate the detection and discrimination of Gram-positive (GP) and Gram-negative (GN) bacteria and Candida species in blood culture samples by molecular methods.
Methods
The Real-GP®, -GN®, and -CAN® real-time PCR kit (M&D, Wonju, Republic of Korea) assays use the TaqMan probes for detecting pan-GP, pan-GN, and pan-Candida species, respectively. The diagnostic performances of the real-time PCR kits were evaluated with 115 clinical isolates, 256 positive and 200 negative blood culture bottle samples, and the data were compared to results obtained from conventional blood culture.
Results
Eighty-seven reference strains and 115 clinical isolates were correctly identified with specific probes corresponding to GP-bacteria, GN-bacteria and Candida, respectively. The overall sensitivity and specificity of the real-time PCR kit with blood culture samples were 99.6% and 89.5%, respectively.
Conclusions
The Real-GP®, -GN®, and -CAN® real-time PCR kits could be useful tools for the rapid and accurate screening of bloodstream infections (BSIs).
Keywords: Real-time polymerase chain reaction, Blood culture, Gram-positive bacteria, Gram-negative bacteria, Candida
Background
The incidence of sepsis in the United States is 240.4 cases per 100,000 people with an 8.7% annual increase during the last several years [1]. The mortality rate of sepsis ranges between 21% and 55%, and it has been unchanged during the last decade [2,3].
Gram-positive (GP) bacteria are the most frequent causative agents of bloodstream infections (BSIs) (30-50% of all cases), followed by Gram-negative (GN) bacteria in 25-30% and fungal infections representing 1-3% of all sepsis cases [4]. Although rapid and accurate diagnosis of sepsis plays an important role in the reduction of mortality caused by sepsis, at least in 30% of sepsis cases, the causative pathogen could not be detected [4]. Currently, blood culture is the standard method for the diagnosis of bacteremia. However, the final results from identification/antimicrobial susceptibility tests in the continuous monitoring blood culture system (CMBCS) require at least 48 to 72 hrs [5]. Moreover, the CMBCS may cause false-negative results when fastidious or slowly growing organisms are the causative pathogens or when blood specimens are collected after antimicrobial therapy has been started [6,7]. In order to reduce the turnaround time for blood culture, a number of molecular methods for rapid identification of pathogens in positive blood culture samples have been tried, including DNA microarrays [8], RNA-based fluorescence in situ hybridization probes [9], and PCR-based assays like real-time PCR [10,11]. Among these methods, PCR-based assays have been reported to provide an early and accurate diagnosis of bacteremia and candidemia [12]. Sequence analysis of the PCR product is time-consuming, but has improved the rate of microbial detection. The sequence of the 16S rRNA gene has been used to diagnose and identify bacterial infection in clinical practice [13,14]. Some PCR-based assays could identify specific bacterial pathogens [10,11], while broad range bacterial PCR can detect almost any bacterial species [15,16]. Broad-range bacterial PCR has a great advantage in that it is able to detect microorganisms that are found less frequently, or can even identify unknown causative agents of bacterial origin.
However, conventional PCR is inconvenient for use in routine rapid screening due to the time required for sample handling and the risk of contamination in post-PCR analysis. Thus, it is necessary to develop a reliable broad-range detection system for bacterial and fungal genomic DNA from clinical samples that is fast, easy to use and covers a wide range of clinically relevant microbes. Additionally, the simultaneous quantification and differentiation of a Gram stain in clinical blood samples with a broad-range real-time PCR assay is rarely described.
In this study, we used a GP-bacteria, GN-bacteria and Candida-specific TaqMan probe-based real-time PCR system, which targets the bacterial 16S rRNA gene and fungal 18S rRNA gene, allowing simultaneous detection and discrimination of clinically-relevant GP-bacteria, GN-bacteria and Candida species in a total of 87 reference strains, 115 clinical isolates and 456 blood culture bottle samples from BSI-suspected patients.
Materials and methods
Bacterial and fungal strains
In this study, to determine the specificity of the Real-GP®, -GN®, and -CAN® real-time PCR kit (M&D, Wonju, Republic of Korea), a total of 62 bacterial, 25 fungal reference strains (Table 1), and 115 clinical isolates from various specimen types were used (Table 2). To evaluate the performance of the Real-GP®, -GN®, and -CAN® real-time PCR assay with blood culture bottle samples, a total of 456 samples including 256 positive and 200 negative blood culture samples were collected. All bacterial strains and clinical specimens were collected from December of 2011 to January of 2013 at Yonsei University Wonju Severance Christian Hospital, Wonju, Republic of Korea. All bacterial strains except Mycobacterium spp. were grown on sheep blood agar and MacConkey agar (BD Diagnostic System, Spark, MD, USA) at 37°C overnight and identified by the microplate method [17], the MicroScan® system (Siemens Healthcare Diagnostics, Sacramento, CA, USA), and the Vitek 2 system (bioMérieux, Durham, NC, USA). All mycobacterial reference strains were grown on Lowenstein-Jensen media (Union Lab, Seoul, Republic of Korea) at 37°C under 5% CO2 for 7 days to 8 weeks at Department of Microbiology, College of Medicine, Yonsei University, Seoul, Republic of Korea. All fungal reference strains were grown on Saboraud dextrose agar (BD Diagnostic System, Spark, MD, USA) at 25°C for several days at Korea Culture Collection Medical Fungi (KCMF), Konyang University, Daejeon, Republic of Korea.
Table 1.
The specificity of Real-GP®, -GN®, and -CAN® real-time PCR assays for 62 bacterial and 25 fungal reference strains
| Genus | Species | Standard strains |
Real-time PCR TaqMan assay (C
T
value) |
||
|---|---|---|---|---|---|
| Real-GP® | Real-GN® | Real-CAN® | |||
|
Gram-positive bacteria |
|
|
|
|
|
|
Staphylococcus |
S. aureus |
29213 |
26.59 |
UD |
UD |
| |
S. aureus |
25923 |
28.42 |
UD |
UD |
| |
S. xylosus |
29971 |
20.81 |
UD |
UD |
|
Enterococcus |
E. hirae |
9790 |
26.31 |
UD |
UD |
| |
E. raffinosus |
49427 |
28.09 |
UD |
UD |
| |
E. sulfureus |
49903 |
28.40 |
UD |
UD |
| |
E. durans |
19432 |
23.48 |
UD |
UD |
| |
E. casseliflavus |
700327 |
25.98 |
UD |
UD |
| |
E. faecium |
19434 |
26.72 |
UD |
UD |
| |
E. faecalis |
29212 |
25.33 |
UD |
UD |
| |
E. mundtii |
43186 |
29.90 |
UD |
UD |
| |
E. cecorum |
43198 |
21.68 |
UD |
UD |
| |
E. flavescens |
49997 |
22.21 |
UD |
UD |
| |
E. gallinarum |
49573 |
23.88 |
UD |
UD |
| |
E. faecalis |
51299 |
24.10 |
UD |
UD |
| |
E. solitarius |
49428 |
30.44 |
UD |
UD |
| |
E. faecium |
35667 |
24.66 |
UD |
UD |
| |
E. malodoratus |
43197 |
27.02 |
UD |
UD |
| |
E. saccharolyticus |
43076 |
23.06 |
UD |
UD |
| |
E. casseliflavus |
25788 |
26.28 |
UD |
UD |
|
Streptococcus |
S. pneumoniae |
49619 |
21.11 |
UD |
UD |
| |
S. agalactiae |
13813 |
26.41 |
UD |
UD |
|
Micrococcus |
M. luteus |
49732 |
22.36 |
UD |
UD |
|
Mycobacterium |
M. avium |
25291 |
23.06 |
UD |
UD |
| |
M. chelonae |
35749 |
21.34 |
UD |
UD |
| |
M. gastri |
15754 |
23.90 |
UD |
UD |
| |
M. kansasii |
12478 |
22.17 |
UD |
UD |
| |
M. nonchromogenicum |
19530 |
18.31 |
UD |
UD |
| |
M. phlei |
11758 |
25.09 |
UD |
UD |
| |
M. smegmatis |
19420 |
24.40 |
UD |
UD |
| |
M. triviale |
23292 |
22.66 |
UD |
UD |
| |
M. aurum |
23366 |
24.83 |
UD |
UD |
| |
M. farcinogen |
35753 |
20.00 |
UD |
UD |
| |
M. gilvum |
43909 |
19.40 |
UD |
UD |
| |
M. neoaurum |
25795 |
17.83 |
UD |
UD |
| |
M. parafortuitum |
19686 |
19.06 |
UD |
UD |
| |
M. peregrinum |
14467 |
18.57 |
UD |
UD |
| |
M. septicum |
700731 |
23.47 |
UD |
UD |
| |
M. abscessus |
19977 |
21.73 |
UD |
UD |
|
Gram-negative bacteria |
|
|
|
|
|
|
Escherichia |
E. coli |
25922 |
UD |
20.46 |
UD |
| |
E. coli |
35218 |
UD |
17.90 |
UD |
|
Enterobacter |
E. aerogenes |
1304 |
UD |
19.09 |
UD |
|
Citrobacter |
C. freundii |
6750 |
UD |
14.19 |
UD |
|
Shigella |
S. boydii |
DML 399 |
UD |
25.59 |
UD |
| |
S. dysenteriae |
DML 400 |
UD |
18.38 |
UD |
| |
S. flexneri |
9199 |
UD |
21.82 |
UD |
|
Serratia |
S. liquifaciens |
27952 |
UD |
24.96 |
UD |
|
Salmonella |
S. typhi |
19430 |
UD |
16.54 |
UD |
| |
S. enteritidis |
13076 |
UD |
20.15 |
UD |
| |
S. paratyphi |
11511 |
UD |
19.61 |
UD |
| |
S. typhimurium |
13311 |
UD |
16.19 |
UD |
| |
S. newport |
6962 |
UD |
17.22 |
UD |
|
Klebsiella |
K. pneumoniae |
13883 |
UD |
20.60 |
UD |
| |
K. oxytoca |
700324 |
UD |
21.87 |
UD |
|
Proteus |
P. alcalifaciens |
51902 |
UD |
18.38 |
UD |
| |
P. vulgaris |
49132 |
UD |
17.13 |
UD |
| |
P. mirabilis |
49132 |
UD |
16.34 |
UD |
|
Pseudomonas |
P. cepacia |
25608 |
UD |
19.66 |
UD |
| |
P. aeruginosa |
27853 |
UD |
16.57 |
UD |
|
Haemophilus |
H. influenzae |
49247 |
UD |
18.61 |
UD |
|
Leclercia |
L. adecarboxylata |
23216 |
UD |
15.70 |
UD |
|
Bordetella |
B. bronchiseptica |
10580 |
UD |
19.44 |
UD |
|
Fungi |
|
|
|
|
|
|
Penicillium |
P. camemberti |
58608 |
UD |
UD |
UD |
| |
P. paneum |
KACC 44823 |
UD |
UD |
UD |
|
Aspergillus |
A. oryzae var oryzae |
KACC 44847 |
UD |
UD |
UD |
| |
A. oryzae var. effusus |
1010 |
UD |
UD |
UD |
| |
A. clavatus |
66443 |
UD |
UD |
UD |
| |
A. sydowii |
KACC 41869 |
UD |
UD |
UD |
| |
A. fumigatus |
KCMF 10773 |
UD |
UD |
UD |
| |
A. flavus |
KCMF 10777 |
UD |
UD |
UD |
| |
A. tamari |
20054 |
UD |
UD |
UD |
|
Fusarium |
F. acuminatum |
10466 |
UD |
UD |
UD |
|
Aureobasidium |
A. pullulans |
KACC 41291 |
UD |
UD |
UD |
|
Bipolaris |
B. sorokiniana |
KACC 44841 |
UD |
UD |
UD |
|
Cryptococcus |
C. neoformans |
KCMF 20047 |
UD |
UD |
UD |
|
Kodamea |
K. ohmeri |
KCMF 20430 |
UD |
UD |
UD |
|
Saccaromyces |
S. cerevisiae |
KCMF 50427 |
UD |
UD |
UD |
|
Trichophyton |
T. rubrum |
KCMF 10444 |
UD |
UD |
UD |
| |
T. mentagrophytes |
KCMF 10515 |
UD |
UD |
UD |
|
Microsporum |
M. canis |
KCMF 10531 |
UD |
UD |
UD |
|
Epidermophyton |
E. floccosum |
52063 |
UD |
UD |
UD |
|
Malassezia |
M. furfur |
KCMF 20409 |
UD |
UD |
UD |
|
Candida |
C. albicans |
36802 |
UD |
UD |
26.42 |
| |
C. tropicalis |
14506 |
UD |
UD |
25.98 |
| |
C. glabrata |
38326 |
UD |
UD |
17.09 |
| |
C. parapsilosis |
7330 |
UD |
UD |
24.27 |
| C. krusei | 20298 | UD | UD | 19.67 | |
Abbreviations:ATCC American type culture collection, DML Diagnostic Microbiology Laboratory, Biomedical laboratory science, Yonsei University; KACC Korean Agricultural Culture Collection; KCMF Korea Culture Collection Medical Fungi; UD Undetermined.
Table 2.
Real-GP®, -GN®, and -CAN® real-time PCR assay results for discriminating the Gram-positive and -negative bacteria and Candida species in 115 clinical isolates
| Culture identification | No. of samples (n) |
Real-time PCR TaqMan assay |
||
|---|---|---|---|---|
| GP/GN or Candida | Ranged C T value | Mean C T value | ||
|
Staphylococcus aureus |
12 |
GP |
22.44-26.65 |
24.47 |
|
Staphylococcus spp. (CoNS) |
8 |
GP |
19.46-28.71 |
21.5 |
|
Streptococcus spp. |
5 |
GP |
17.35-30.46 |
24.25 |
|
Enterococcus faecalis |
4 |
GP |
25.2-27.3 |
26.37 |
|
Enterococcus faecium |
10 |
GP |
21.3-31.6 |
26.58 |
|
Enterococcus mundtii |
1 |
GP |
27.85 |
27.85 |
|
Corynebacterium spp. |
1 |
GP |
24.51 |
24.51 |
|
Escherichia coli |
16 |
GN |
12.68-30.65 |
23.26 |
|
Klebsiella pneumoniae |
13 |
GN |
15.48-26.08 |
20.73 |
|
Pseudomonas aeruginosa |
13 |
GN |
15.23-19.96 |
18.11 |
|
Acinetobacter baumannii |
11 |
GN |
18.09-24.65 |
21.04 |
|
Enterobacter asburiae |
2 |
GN |
15.79 |
15.79 |
|
Enterocobacter cloacae |
1 |
GN |
15.41 |
15.41 |
|
Moraxella catarrhalis |
1 |
GN |
33.54 |
33.54 |
|
Serratia marcescens |
1 |
GN |
21.64 |
21.64 |
|
Providencia rettgeri |
1 |
GN |
24.3 |
24.30 |
|
Morganella morganii |
1 |
GN |
20.6 |
20.60 |
|
Proteus mirabilis |
1 |
GN |
24.88 |
24.88 |
|
Aeromonas spp. |
1 |
GN |
25.97 |
25.97 |
|
Citrobacter fruendii |
2 |
GN |
17.11-18.01 |
17.56 |
|
Candida albicans |
5 |
CAN |
17.61-29.56 |
23.9 |
|
Candida parapsilosis |
3 |
CAN |
24.73-30.95 |
27.58 |
|
Candida tropicalis |
1 |
CAN |
26.62 |
26.62 |
|
Candida glabrata |
1 |
CAN |
17.68 |
17.68 |
| Total | 115 | |||
Blood culture and collection of blood culture bottle samples
Three or two pairs of culture bottles for aerobes or anaerobes were incubated in the BacT/Alert 3D (bioMérieux) and BACTEC® 9240 system (Becton Dickinson Diagnostic System, Spark, MD, USA) or BACTEC FX (Becton Dickinson) blood culture systems for 5 days after inoculating blood drawn from the patient at the bedside. If no bacterial growth was detected within 5 days, the blood culture was considered negative. When bacterial growth was noted, the culture sample was inoculated into blood and MacConkey agar plates (BD Diagnostic Systems, Sparks, MD, USA), and then cultured overnight at 37°C in a 5% CO2 incubator. Isolates were identified based on the colony morphology, Gram stain, biochemical tests, and commercial identification kits. MicroScan® (Siemens Healthcare Diagnostics, Sacramento, CA, USA) overnight Pos BP Combo 28, MICroSTREP Plus, overnight Neg Combo 53, and Neg Combo 54 panels were used for the identification of GP, streptococci, and GN bacteria. For identification of Candida spp., a VITEK-2 (bioMérieux, Marcy l’Etoile, France) YST ID CARD was used.
DNA preparation
To prepare DNA templates for the real-time PCR TaqMan assay, one colony of each strain and clinical isolate was suspended in 100 μL of DNA extraction solution (M&D, Wonju, Republic of Korea). The suspended bacterial solution was boiled for 10 min. After centrifugation at 13,000 g for 10 min, the supernatant was used for DNA templates.
For preparation of DNA template from the blood culture bottle samples, 0.5 mL of blood suspension were taken directly from the blood culture bottle, and 1 mL of phosphate-buffered saline (pH 8.0) was added and centrifuged at 13,000 g for 1 min. The supernatant was removed, and the pellet was resuspended in 1 mL of ACK solution (0.15 M of NH4Cl, 1 mM of KHCO3, and 0.1 mM of Na2EDTA), and centrifuged at 13,000 g for 1 min. This washing step was repeated twice, and the pellet was resuspended in DNA extraction solution as described above for the clinical isolates.
Real-time PCR TaqMan assay
The real-time PCR TaqMan assay was carried out with the Real-GP®, -GN® and -CAN® real-time PCR assay kits (M&D), and a CFX-96 real-time PCR system (Bio-rad, Hercules, CA, USA) and an ABI 7500 FAST instrument (Applied Biosystem, Foster City, CA, USA) were used for the thermo-cycling and fluorescence detection. These real-time PCR assay kits are only able to determine GP bacteria, GN bacteria, and Candida, respectively for rapid screening of BSIs however they do not allow species or genus identification and antimicrobial susceptibility. The real-time PCR amplification was performed in a total volume of 25 μL that contained 12.5 μL of 2 × Thunderbird probe qPCR mix (Toyobo, Osaka, Japan), 5 μL of primer and TaqMan probe mixture, 5 μL of template DNA, and ddH2O was added to give a final volume of 25 μL for each sample.
Positive and negative controls were included throughout the procedure. No-template controls with ddH2O instead of template DNA were incorporated in each run under the following conditions: 95°C for 3 min and 40 cycles of 95°C for 20 s and 60°C for 40 s in single real-time PCR. The bacterial load was quantified by determining the cycle threshold (CT), the number of PCR cycles required for the fluorescence to exceed a value significantly higher than the background fluorescence.
Results
Sensitivity and specificity of the Real-GP®, -GN®, and -CAN® real-time PCR TaqMan assay with reference bacterial and fungal strains
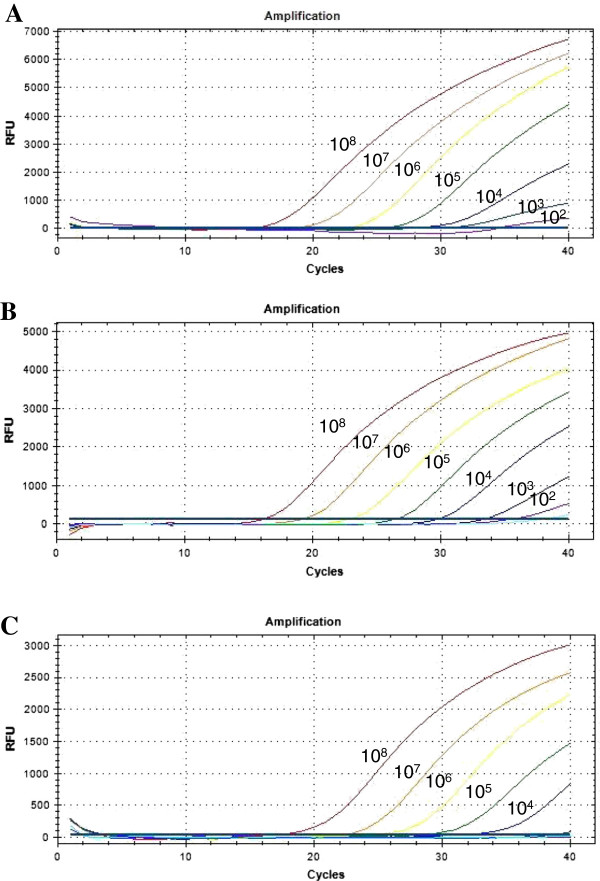
The detection limit of the real-time PCR TaqMan assay for GP-, GN-bacteria, and Candida was 103 CFU/mL, 103 CFU/mL, and 104 CFU/mL, respectively. The CT values for GP-, GN-bacteria, and Candida with each cell concentrate (108 - 102 CFU/mL) ranged from 16.17 to 32.46, 15.06 to 29.03, and 17.68 to 32.47, respectively (Figure 1).
Figure 1.
Detection limits of the three target probes from 10-fold serial diluted spiked samples. Serially diluted DNA amounts ranging from 108 to 10° CFU/mL were used to determine the detection limit of the multiplex real-time PCR assay. (A) amplification curve of GP-bacteria probe using Staphylococcus aureus, (B) amplification curve of GN-bacteria probe using Escherichia coli, (C) amplification curve of Candida probe using Candida glabrata. The overall detection limit of this assay for GP-, GN-bacteria, and Candida probe was approximately 103 to 104 CFU/mL.
All DNA extractions of bacterial and fungal reference strains showed the positive fluorescence signals with real-time PCR TaqMan assay. The CT values for GP-, GN-bacteria and Candida real-time PCR assays ranged from 17.83 to 30.44, 14.19 to 25.59, and 17.09 to 26.42, respectively (Table 1).
Results of Real-GP®, -GN®, and -CAN® real-time PCR TaqMan assay with clinical isolates
The results between subculture and real-time PCR assay were completely concordant (100%) in 115 clinical isolates. Forty-one GP clinical isolates, which included 12 Staphylococcus aureus, 10 Enterococcus faecium, 8 coagulase-negative staphylococci (CoNS), 5 Streptococcus spp., 4 E. faecalis, 1 Enterococcus mundtii, and 1 Corynebacterium spp., were positive by Real-GP® assay and sixty-four GN clinical isolates, which included 16 Escherichia. coli, 13 Klebsiella pneumoniae, 13 Pseudomonas aeruginosa, 11 Acinetobacter baumannii, two Citrobacter fruendii, two Enterobacter asburiae, one Enterobacter coloacae, one Aeromonas spp., one Moraxella catarrhalis, one Morganella morganii, one Proteus mirabilis, one Providencia rettgeri, and one Serratia marcescens, were positive by Real-GN® assay. Ten Candida spp. clinical isolates, which included five C. albicans, three C. parapsilosis, one C. tropicalis and one C. glabrata, were positive by Real-CAN® assay (Table 2). The CT values of clinical isolates of GP, GN, and Candida species ranged from 17.35 to 31.60, 12.68 to 33.54, and 17.61 to 30.95, respectively.
Results of Real-GP®, -GN®, and -CAN® real-time PCR TaqMan assay with positive and negative blood culture bottle samples
Of 256 positive blood culture bottles, GP-bacteria, GN-bacteria, and Candida were detected in 175, 70, and four bottles, respectively. Two kinds of microorganisms were detected from six bottles. Among 175 GP-bacteria-positive blood cultures, Staphylococcus epidermidis was the most prevalent at 26.7% (n = 47), followed by S. aureus (n = 24), Staphylococcus hominis (n = 17), GP rods (n = 16), and Staphylococcus capitis (n = 14). One S. hominis showed a negative result with real-time PCR. The CT values of 175 GP-bacteria ranged from 11.52 to 34.39. Seventy GN-bacteria from blood cultures included Escherichia coli (50%, n = 35), Klebsiella pneumoniae (n = 13), Acinetobacter baumannii (n = 5), and Pseudomonas aeruginosa (n = 3). The CT values of the 70 GN-bacteria ranged from 6.56 to 24.08. A total of four Candida-positive samples included two C. albicans, one C. parapsilosis and one C. tropicalis (Table 3). The CT values of the four Candida species ranged from 22.81 to 31.98.
Table 3.
Comparison of the results of Real-GP®, -GN®, and -CAN® real-time PCR assay and BACTEC 9240 for detection of bloodstream infection in positive and negative blood culture bottle samples
| Blood culture result |
Real-time PCR TaqMan assay (n) |
GP |
GN |
CAN |
|
|---|---|---|---|---|---|
| Positive | Negative | (C T range) | (C T range) | (C T range) | |
|
Blood culture positive (n = 256) |
255 |
1 |
|
|
|
|
Gram-positive bacteria (n = 176) |
175 |
1 |
|
|
|
|
Staphylococccus epidermidis (47) |
47 |
0 |
13.08-33.99 |
UD |
UD |
|
S. aureus (24) |
24 |
0 |
13.24-34.19 |
UD |
UD |
|
S. hominis (17) |
16 |
1 |
13.69-30.00 |
UD |
UD |
|
S. capitis (14) |
14 |
0 |
13.60-25.10 |
UD |
UD |
|
S. haemolyticus (8) |
8 |
0 |
14.68-33.30 |
UD |
UD |
|
S. warneri (1) |
1 |
0 |
18.50 |
UD |
UD |
|
S. saprophyticus (1) |
1 |
0 |
16.57 |
UD |
UD |
|
S. xylosus (1) |
1 |
0 |
21.23 |
UD |
UD |
|
S. chleiferi (1) |
1 |
0 |
20.44 |
UD |
UD |
|
Streptococcus salivarius (5) |
5 |
0 |
13.55-23.58 |
UD |
UD |
|
S. mitis (4) |
4 |
0 |
11.52-23.12 |
UD |
UD |
|
S. pneumoniae (4) |
4 |
0 |
16.37-17.77 |
UD |
UD |
|
S. agalactiae (2) |
2 |
0 |
21.52-25.95 |
UD |
UD |
|
S. pyogenes (1) |
1 |
0 |
22.10 |
UD |
UD |
|
S. dysgalactiae (1) |
1 |
0 |
15.81 |
UD |
UD |
|
S. parasangus (1) |
1 |
0 |
12.96 |
UD |
UD |
|
Streptococcus spp. (2) |
2 |
0 |
17.87-24.89 |
UD |
UD |
|
Enterococcus faecium (8) |
8 |
0 |
26.43-27.54 |
UD |
UD |
|
E. faecalis (1) |
1 |
0 |
14.50 |
UD |
UD |
|
Micrococcus spp. (5) |
5 |
0 |
20.96-31.33 |
UD |
UD |
|
Propionibacterium acnes (3) |
3 |
0 |
23.77-26.86 |
UD |
UD |
|
Peptostreptococcus asaccharolyticus (1) |
1 |
0 |
26.72 |
UD |
UD |
|
Peptostreptococcus micros (1) |
1 |
0 |
26.00 |
UD |
UD |
|
Corynebacterium spp. (6) |
6 |
0 |
26.47 |
UD |
UD |
| Gram positive rods (16) |
16 |
0 |
12.43-34.39 |
UD |
UD |
|
Gram-negative bacteria (n = 70) |
70 |
0 |
|
|
|
|
Escherichia coli (35) |
35 |
0 |
UD |
9.86-21.89 |
UD |
|
Klebsiella pneumoniae (13) |
13 |
0 |
UD |
12.84-13.20 |
UD |
|
Acinetobacter baumannii (5) |
5 |
0 |
UD |
10.53-11.89 |
UD |
|
A. woffii (1) |
1 |
0 |
UD |
12.22-15.11 |
UD |
|
Enterobacter spp. (2) |
2 |
0 |
UD |
6.56 |
UD |
|
Pseudomonas aeruginosa (3) |
3 |
0 |
UD |
12.08 |
UD |
|
Salmonella group D (1) |
1 |
0 |
UD |
13.62-21.48 |
UD |
|
Proteus mirabilis (1) |
1 |
0 |
UD |
14.38 |
UD |
|
Aeromonas spp. (2) |
2 |
0 |
UD |
20.10 |
UD |
|
Morganella morganii (1) |
1 |
0 |
UD |
10.39 |
UD |
|
Haemophillus influenzae (1) |
1 |
0 |
UD |
20.68 |
UD |
|
Chryseobacterium indologenes (1) |
1 |
0 |
UD |
14.40 |
UD |
|
Sphingomonas paucimobilis (1) |
1 |
0 |
UD |
24.08 |
UD |
|
Serratia marcescens (1) |
1 |
0 |
UD |
9.48 |
UD |
|
Citrobacter freundii (1) |
1 |
0 |
UD |
14.24 |
UD |
|
Candida
species (n = 4) |
4 |
0 |
|
|
|
|
Candida albicans (2) |
2 |
0 |
UD |
UD |
26.98-31.52 |
|
C. parapsilosis (1) |
1 |
0 |
UD |
UD |
31.98 |
|
C. tropicalis (1) |
1 |
0 |
UD |
UD |
22.81 |
|
*Polymicrobial infection (n = 6) |
6 |
0 |
|
|
|
|
Streptococcus agalactiae, Citrobacter koseri (1) |
1 |
- |
14.61 |
10.61 |
UD |
|
Enterococcus faecium, Candida albicans (1) |
1 |
- |
17.18 |
UD |
29.51 |
|
Enterococcus faecalis, Proteus mirabilis (1) |
1 |
- |
UD |
12.14 |
UD |
|
Escherichia coli, Enterococcus gallinarym (1) |
1 |
- |
UD |
13.25 |
UD |
|
Klebsiella pneumonia, Enterococcus casseliflavus (1) |
1 |
- |
UD |
13.31 |
UD |
|
Klebsiella pneumonia, Enterobacter cloacae (1) |
1 |
- |
UD |
14.42 |
UD |
|
Blood culture negative (n = 200) |
21 |
179 |
|
|
|
| Gram-positive bacteria |
7 |
193 |
17.54-27.43 |
- |
- |
| Gram-negative bacteria |
14 |
186 |
- |
20.48-31.26 |
- |
| Candida species | 0 | 200 | - | - | - |
*GP-, GN-bacteria or fungi mixed infection.
Among six cases of polymicrobial bacteremia, three were concordant between the standard identification method and real-time PCR assay, but the other three cases showed positive results for only a single organism by real-time PCR assay (Table 3).
Of 200 negative blood culture bottle samples, 21 samples (10.5%), including seven GP- and 14 GN-bacteria, had positive results with the real-time PCR assay (Table 3). The CT values of 7 GP-bacteria and 14 GN-bacteria ranged from 17.54 to 23.28, and 17.79 to 31.26, respectively.
The sensitivity of the real-time PCR kit for GP, GN, Candida and polymicrobial isolates was 99.4%, 100%, 100% and 100%, respectively, and the specificity of the real-time PCR kit for GP, GN and Candida was 96.5%, 93.0% and 100%, respectively. Therefore, the overall sensitivity and specificity of the real-time PCR kit compared with blood culture method were 99.6% and 89.5%, respectively.
Discussion
Blood culture is the current gold standard method for detecting BSI microbial pathogens; although it allows microbes to be identified and their susceptibility profiles to be tested, the method has several limitations. Lack of rapidity is a major problem because detection of bacterial and fungal growth requires approximately 12 to 48 hr, and it can take more time in the case of fastidious bacterial or invasive fungal infection [18,19].
Currently, novel diagnostic technique such as MALDI-TOF MS was developed and evaluated with positive blood culture bottle samples [20,21]. It could provide species or genus identification more rapidly for detecting BSIs than current molecular assays however it cannot replace to routine laboratory workflow in current clinical setting yet because of the cost, facility, and examiner. Current DNA-based Gram identification methods include Gram stain-specific PCR [22], nested PCR [23], and PCR probe hybridization [24-26], but all of these methods contain at least two sequential steps, and therefore they require a longer turnaround time to get a final result. For instance, the conventional PCR assays incorporate a pair of oligonucleotide primers to amplify a specific target gene that is then detected using agarose gel electrophoresis combined with an intercalating dye (e.g., ethidium bromide, EtBr) and UV light. The real-time PCR TaqMan assay is a promising tool for detecting bacterial genomic DNA from biological fluids (direct clinical specimens) such as blood, urine and sputum. Fluorescence hybridization probes result in fast detection of small amounts of bacterial genomic DNA and correct Gram classification [27]. It is not only applicable from one sample to a number of samples at once but also it can be commonly used for diagnostic purpose in current clinical laboratories cost effectively.
In this study, Real-GP®, -GN® and -CAN® real-time PCR TaqMan assays, which target the bacterial 16S rRNA and fungal 18S rRNA, were evaluated with reference bacterial and fungal strains, clinical isolates, and direct blood culture bottle samples. The results showed that the real-time PCR TaqMan assay was rapid; it usually took no more than 4 hr to complete the whole experiment, which included only 1 hr of sample preparation and 1.5 hr for DNA amplification, because thermal cycling is much faster and amplicon detection is performed in real time. It allowed for the rapid quantification and Gram classification of bacteria and fungi without the post-PCR process. Furthermore, it was very specific because the results were completely accurate when compared with the standard blood culture method.
In previous reports from other study groups, CoNS were reported to be the major causative microorganisms in sepsis [28,29]. In this study, S. epidermidis was identified as the most common in positive blood culture bottle samples, with a total of 47 cases, which is identical to the results from other studies. Therefore, future study of the role and the effect of S. epidermidis infection in the bloodstream might be essential in the Korean population. All GN-bacteria and Candida samples were positive and all GP-bacteria samples, except for one sample, were positive based on real-time PCR assay in clinical isolates and positive blood culture samples. Therefore, the sensitivity was sufficient for performance with positive blood culture bottle samples. Also, candidiasis is a yeast infection caused by different Candida species and can cause opportunistic infections of the skin and mucosa as well invasive infections. Candidiasis accounts for up to 10% of bloodstream infections and is associated with an exceptionally high mortality rate [30]. Invasive fungal infections are increasingly recognized as a primary cause of morbidity and mortality especially in immunocompromised patients. To reduce mortality in patients with invasive candidiasis, early diagnosis and rapid initiation of antifungal therapy is very important for the survival of patients. However, our research has shown that the test was just 4/256 (1.6%) in Candida species therefore it is necessary to be carried out more tests with larger number of samples. The positivity of the Real-GP®, -GN®, -CAN® real-time PCR assay (21/200; 10.5%) was significantly higher than that of blood culture (0/200; 0%). When blood culture was used as a standard control, the sensitivity of real-time PCR was 100% and the specificity was 89.5% (Table 4). The gold standard for diagnosing sepsis is now still blood culture, even though, in many cases, blood cultures are negative in the face of strong clinical indicators of sepsis [31]. Therefore, to effectively evaluate the real-time PCR kit for rapid screening of BSI, further evaluation is required with direct blood samples or a larger number of negative blood culture bottle samples from patients who are suspected to have sepsis.
Table 4.
The sensitivity and specificity of the Real-GP®, -GN®, and -CAN® real-time PCR assay with positive and negative blood culture bottle samples
| Blood culture |
Real-time PCR |
Sensitivity | Specificity | |
|---|---|---|---|---|
|
TaqMan assay (n) | ||||
| Positive | Negative | |||
|
Blood culture positive (n = 256) |
255 |
1 |
99.6% |
- |
| Gram-positive bacteria (176) |
175 |
1 |
99.4% |
- |
| Gram-negative bacteria (70) |
70 |
0 |
100% |
- |
|
Candida species (4) |
4 |
0 |
100% |
- |
| Multiple infection (6) |
6 |
0 |
100% |
- |
|
Blood culture negative (n = 200) |
21 |
179 |
- |
89.5% |
| Gram-positive bacteria |
7 |
193 |
- |
96.5% |
| Gram-negative bacteria |
14 |
186 |
- |
93.0% |
| Candida species | 0 | 200 | - | 100% |
Polymicrobial culture data show that six samples were infected with two microorganisms each. Among these six cases, the results of three were consistent between conventional culture and real-time PCR assay (showed double signal), however, three samples showed just a single positive signal with real-time PCR, even though two microorganisms were identified by the culture method. According to the real-time PCR results, GN signals were positive and GP signals were undetermined in those three samples. The reason might be due to the fact that the GP-bacteria, Enterococcus spp., grow more slowly, and thus, the number of GP-bacteria is much fewer than that of GN bacteria (E. coli and K. pneumoniae) in blood culture bottle samples, and the fluorescence signal for GN could not be detected.
Conclusions
The use of the molecular diagnostic assay, real-time PCR TaqMan assay, is more effective for rapid screening of BSI than microbiological diagnosis. In this study, this technique allowed the simultaneous detection, quantification, and Gram identification of bacterial and fungal organisms directly from blood culture bottle samples. Even culture, species or genus identification, and antimicrobial susceptibility testing could not be substituted by this real-time PCR TaqMan assay, it could not only differentiate bacterial and fungal from viral and other pathogens, but it could also classify Gram staining with a much shorter turnaround time than the gold standard culture method.
The Real-GP®, -GN®, and -CAN® real-time PCR assays may provide essential information to accelerate therapeutic decisions for earlier and adequate antibiotic treatment in the acute phase of sepsis.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HYW and SK carried out data organization and analysis and contributed to writing and to the interpretation of the results. HK, JK, YK, SDP, HJ, and YC carried out the evaluation of experiments and collected all clinical samples and data. HYL and YU contributed to the design of the study and assisted in the drafting of the manuscript. All authors have read and approved the manuscript.
Contributor Information
Hye-young Wang, Email: sapphire111@hanmail.net.
Sunghyun Kim, Email: shkim0423@yonsei.ac.kr.
Hyunjung Kim, Email: kimhj2786@gmail.com.
Jungho Kim, Email: jutosa70@naver.com.
Yeun Kim, Email: scaraby405@hotmail.com.
Soon-Deok Park, Email: mizpark66@naver.com.
Hyunwoo Jin, Email: jjinhw@cup.ac.kr.
Yeonim Choi, Email: yichoi@sonho.ac.kr.
Young Uh, Email: u931018@yonsei.ac.kr.
Hyeyoung Lee, Email: hyelee@yonsei.ac.kr.
Acknowledgements
This study was supported by a grant of the Korea Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A121030, H.L.). This work was supported (in part) by the Yonsei University Research Fund of 2013.
References
- Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;13:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- Engel C, Brunkhorst FM, Bone HG, Brunkhorst R, Gerlach H, Grond S, Gruendling M, Huhle G, Jaschinski U, John S, Mayer K, Oppert M, Olthoff D, Quintel M, Ragaller M, Rossaint R, Stuber F, Weiler N, Welte T, Bogatsch H, Hartog C, Loeffler M, Reinhart K. Epidemiology of sepsis in Germany: results from a national prospective multicenter study. Intensive Care Med. 2007;13:606–618. doi: 10.1007/s00134-006-0517-7. [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;13:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005;13:63–78. doi: 10.1016/S0140-6736(04)17667-8. [DOI] [PubMed] [Google Scholar]
- Schuurman T, de Boer RF, Kooistra-Smid AM, van Zwet AA. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for diagnosis of bacterial meningitis in a clinical setting. J Clin Microbiol. 2004;13:734–740. doi: 10.1128/JCM.42.2.734-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenollar F, Raoult D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int J Antimicrob Agents. 2007;13:S7–S15. doi: 10.1016/j.ijantimicag.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Horz HP, Vianna ME, Gomes BP, Conrads G. Evaluation of universal probes and primer sets for assessing total bacterial load in clinical samples: general implications and practical use in endodontic antimicrobial therapy. J Clin Microbiol. 2005;13:5332–5337. doi: 10.1128/JCM.43.10.5332-5337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleven BE, Palka-Santini M, Gielen J, Meembor S, Kronke M, Krut O. Identification and characterization of bacterial pathogens causing bloodstream infections by DNA microarray. J Clin Microbiol. 2006;13:2389–2397. doi: 10.1128/JCM.02291-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen GJ, Mooibroek M, Idema J, Harmsen HJ, Welling GW, Degener JE. Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J Clin Microbiol. 2000;13:814–817. doi: 10.1128/jcm.38.2.814-817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke D, Menard C, Picard FJ, Boissinot M, Ouellette M, Roy PH, Bergeron MG. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin Chem. 2000;13:324–331. [PubMed] [Google Scholar]
- van Haeften R, Palladino S, Kay I, Keil T, Heath C, Waterer GW. A quantitative LightCycler PCR to detect Streptococcus pneumoniae in blood and CSF. Diagn Microbiol Infect Dis. 2003;13:407–414. doi: 10.1016/S0732-8893(03)00129-9. [DOI] [PubMed] [Google Scholar]
- Clarridge JE 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;13:840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes U, Bohmeier B, Lippert H, König B, Meyer F. PCR-based rapid sepsis diagnosis effectively guides clinical treatment in patients with new onset of SIRS. Langenbecks Arch Surg. 2012;13:447–455. doi: 10.1007/s00423-011-0870-z. [DOI] [PubMed] [Google Scholar]
- Xu J, Millar BC, Moore JE, Murphy K, Webb H, Fox AJ, Cafferkey M, Crowe MJ. Employment of broad-range 16S rRNA PCR to detect aetiological agents of infection from clinical specimens in patients with acute meningitis-rapid separation of 16S rRNA PCR amplicons without the need for cloning. J Appl Microbiol. 2003;13:197–206. doi: 10.1046/j.1365-2672.2003.01839.x. [DOI] [PubMed] [Google Scholar]
- Klaschik S, Lehmann LE, Raadts A, Book M, Gebel J, Hoeft A, Stuber F. Detection and differentiation of in vitro-spiked bacteria by real-time PCR and melting-curve analysis. J Clin Microbiol. 2004;13:512–517. doi: 10.1128/JCM.42.2.512-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, Reesink HW, Vandenbroucke-Grauls CM, Savelkoul PH. Optimization of real-time PCR assay for rapid and sensitive detection of eubacterial 16S ribosomal DNA in platelet concentrates. J Clin Microbiol. 2003;13:4796–4798. doi: 10.1128/JCM.41.10.4796-4798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uh Y, Son JS, Hwang GY, Jang IH, Yoon KJ, Seo DM. Microplate identification system of Enterobacteriaceae. Korean J Clin Microbiol. 1999;13:135–143. [Google Scholar]
- Beekmann SE, Diekema DJ, Chapin KC, Doern GV. Effects of rapid detection of bloodstream infections on length of hospitalization and hospital charges. J Clin Microbiol. 2003;13:3119–3125. doi: 10.1128/JCM.41.7.3119-3125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RP, van Agtmael MA, Danner SA, Savelkoul PH, Vandenbroucke-Grauls CM. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis. 2004;13:751–760. doi: 10.1016/S1473-3099(04)01205-8. [DOI] [PubMed] [Google Scholar]
- Buchan BW, Riebe KM, Ledeboer NA. Comparison of the MALDI Bio-typer system using SeptityperTM specimen processing to routine microbiological methods for identification of bacteria from positive blood culture bottles. J Clin Microbiol. 2012;13:346–352. doi: 10.1128/JCM.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace’-Wiens PR, Adam HJ, Karlowsky JA, Nichol KA, Pang PF, Guenther J, Webb AA, Miller C, Alfa MJ. Identification of blood culture isolates directly from positive blood cultures by use of matrix-associated laser desorption ionization-time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J Clin Microbiol. 2012;13:3324–3328. doi: 10.1128/JCM.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausegger AM, Hell A, Berger K, Zinober S, Baier N, Jones N, Sperl W, Kofler B. Gram type-specific broad-range PCR amplification for rapid detection of 62 pathogenic bacteria. J Clin Microbiol. 1999;13:464–466. doi: 10.1128/jcm.37.2.464-466.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll NM, Jaeger EE, Choudhury S, Dunlop AA, Matheson MM, Adamson P, Okhravi N, Lightman S. Detection of and discrimination between gram-positive and gram-negative bacteria in intraocular samples by using nested PCR. J Clin Microbiol. 2000;13:1753–1757. doi: 10.1128/jcm.38.5.1753-1757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand AR, Madhavan HN, Therese KL. Use of polymerase chain reaction (PCR) and DNA probe hybridization to determine the Gram reaction of the infecting bacterium in the intraocular fluids of patients with endophthalmitis. J Infect. 2000;13:221–226. doi: 10.1053/jinf.2000.0731. [DOI] [PubMed] [Google Scholar]
- Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;13:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang S, Chen Z, Yu X. Detection of bacterial DNA by PCR and reverse hybridization in the 16S rRNA gene with particular reference to neonatal septicemia. Acta Paediatr. 2001;13:179–183. doi: 10.1111/j.1651-2227.2001.tb00281.x. [DOI] [PubMed] [Google Scholar]
- Klaschik S, Lehmann LE, Raadts A, Book M, Hoeft A, Stuber F. Real-time PCR for detection and differentiation of gram-positive and gram-negative bacteria. J Clin Microbiol. 2002;13:4304–4307. doi: 10.1128/JCM.40.11.4304-4307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucol F, Ammann RA, Berger C, Aebi C, Altwegg M, Niggli FK, Nadal D. Real-time quantitative broad-range PCR assay for detection of the 16S rRNA gene followed by sequencing for species identification. J Clin Microbiol. 2006;13:2750–2759. doi: 10.1128/JCM.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krediet TG, Mascini EM, van Rooij E, Vlooswijk J, Paauw A, Gerards LJ, Fleer A. Molecular epidemiology of coagulase-negative staphylococci causing sepsis in a neonatal intensive care unit over an 11-year period. J Clin Microbiol. 2004;13:992–995. doi: 10.1128/JCM.42.3.992-995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinghausen N, Siegel D, Winter J, Gebert S. Rapid diagnosis of candidaemia by real-time PCR detection of Candida DNA in blood samples. J Med Microbiol. 2009;13:1106–1111. doi: 10.1099/jmm.0.007906-0. [DOI] [PubMed] [Google Scholar]
- Kaufman D, Fairchild KD. Clinical microbiology of bacterial and fungal sepsis in very-low-birth-weight infants. Clin Microbiol Rev. 2004;13:638–680. doi: 10.1128/CMR.17.3.638-680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]